Abstract
Epithelial ovarian cancer (EOC) has the highest mortality rate of all gynaecological cancers. One of the greatest impediments to improving outcome is an incomplete understanding of the molecular underpinnings of EOC pathogenesis and progression. Recent studies suggest that microRNAs (miRNAs) are involved in ovarian tumorigenesis and cancer development. Several miRNA profiling studies have identified some consensus aberrantly expressed miRNAs in EOC tissues, and these EOC-related miRNAs may play critical roles in the pathogenesis and progression of EOC. Moreover, some of the miRNAs may have diagnostic or prognostic significance. In this review, recent progress to reveal the role of miRNAs in EOC will be addressed, and a model for miRNA functions in ovarian cancer initiation and progression will be proposed.
Keywords: microRNAs, ovarian cancer, tumorigenesis, metastasis
Introduction
Ovarian cancer is the fifth leading cause of cancer deaths among women in the Western world and has the highest mortality rate of all gynaecological cancers [1]. The poor survival from epithelial ovarian cancer (EOC) is due to the high percentage of patients diagnosed at an advanced stage who often develop resistance to combined chemotherapy and show substantially poor prognosis. Obviously, improved screening programs to detect cancers at an early-stage and more effective treatments for advanced disease are urgently required. Unfortunately, although efforts have been made in the last several decades towards discovering new serum biomarkers and therapeutic strategies such as gene therapy, analysis of trends in overall 5-year survival rates for women with ovarian cancer indicates only modest improvements for women diagnosed between 1996 and 2004, compared to the 1970s and 1980s [1]. Moreover, the complexity of diagnosis and treatment of ovarian cancer is further complicated by the significant heterogeneity within the EOC group. Histologically defined subgroups, such as serous, mucinous, endometrioid and clear cell carcinomas, all have variable clinical manifestations and underlying molecular genetic events [2]. However, it remains unclear what genetic elements direct the differentiation of ovarian epithelial cells. Thus, one of the greatest impediments to improving outcome is an incomplete understanding of the molecular underpinnings of ovarian cancer pathogenesis and progression. Although focusing on known genes has already yielded some valuable information, a recently discovered class of small RNAs, termed microRNAs (miRNAs), may lend insight into the biology of ovarian cancer.
miRNAs are single-stranded, evolutionarily conserved, small (17∼25 ribonucleotides) non-coding RNAs. They have been demonstrated to play important roles in essential processes, such as differentiation, cell growth, stress response and cell death, and are involved in several human diseases, including cancer. miRNAs have been shown to repress the expression of important cancer-related genes and can function as both tumour suppressors and oncogenes [3]. Increasing amounts of experimental evidence have shown that miRNAs are aberrantly expressed in ovarian cancer and can be associated with tumour chemo-resistance and clinical outcomes, indicating that miRNAs may be involved in ovarian tumorigenesis and cancer progression. Here we will review recent progress in understanding the functions of miRNAs in ovarian cancer. We will also discuss their possible use as markers of diagnosis and prognosis, and eventually as new targets or tools of a specific therapy.
Aberrant expression of miRNAs in ovarian cancer
Altogether eight groups [4–11] have investigated the expression profiles of miRNAs in relatively large (range n= 10–93) clinical ovarian cancer samples compared to normal ovarian tissues, ovarian epithelial cell lines, or fallopian tubes. The first detailed report was published by Zhang et al., who used an array comparative genomic hybridization approach to identify miRNA loci gained/lost in 93 primary ovarian cancer tissues and 16 cell lines. They determined that 37.1% (105/283) of the miRNA loci in the cancer tissues to be significantly altered in their copy number. Of these, 52% (55/105) had a gain in copy number, and 48% (50/105) had a loss in copy number [4]. Other studies have since addressed this issue at the transcriptional level by microarray profiling experiments or massive parallel pyrosequencing. Although all of these studies resulted in both up- and down-regulated miRNA patterns, miRNA down-regulation was more prominent during tumour progression, especially in late-stage and high-grade (HG) EOC [5]. Despite a great inconsistency between the de-regulated miRNA patterns of these studies, several miRNAs were identified as aberrantly expressed by more than one study (See Tables S1 and S2), suggesting that they are likely involved in ovarian tumorigenesis.
Among these consensus de-regulated miRNAs, several are well-known onco-miRNAs (miRNAs with oncogenic or tumour suppressive functions [3]), such as miR-20a, miR-106b, miR-143, miR-145, miR-125b and the let-7 family of miRNAs. Interestingly, epithelial-to-mesenchymal transition (EMT) regulating miRNAs, including the miR-200 family and miR-205 [12], show significant up-regulation in ovarian cancer tissues, suggesting that they may play an unique role in ovarian cancer initiation or progression (discussed later). Other miRNAs may have tumour inhibitory effects. For example, connective tissue growth factor, a verified target of miR-30 [13], was shown to be down-regulated in ovarian cancer [14, 15] and to inhibit cancer cell invasion [15]. Finally, several miRNAs such as miR-214, miR-199 and miR-16 have shown both down- and up-regulation in these studies. This may be due to the heterogeneity of ovarian cancer, as partly evidenced by Mor and colleagues [16] who showed that miR-199a was down-regulated in type I EOC cells which can constitutively secrete pro-tumour cytokines compared with type II EOC cells which do not. For other miRNAs, such as miR-99a, relatively little is known, and their potential roles in ovarian cancer warrant further investigation.
As EOC is a highly heterogeneous disease, it is of special interest to determine whether alterations in particular miRNAs are correlated with particular ovarian subtypes. In fact, although some miRNAs are conserved between EOC subtypes, there are pools of miRNAs that are specifically de-regulated in just one subtype. For example, miR-449a is serous specific, miR-499–5p/miR-375/ miR196a/miR-196b/miR-182 are endometrioid specific, and miR-486–5p/miR-144/miR-30a/miR-199a-5p are clear cell specific [9]. These subtype-specific miRNA patterns indicate their potential as new biomarkers for EOC subtype. Additionally, as miRNAs are key regulators of cell differentiation, we could hypothesize that the EOC subtype may be partly mediated by specific miRNA. For example, miR-196b, which can regulate multiple HOX genes and has a central role in posterior prevalence [17], is a consensus miRNA specifically de-regulated in endometrioid carcinoma [6, 9]. Because HOX genes play a central role in EOC subtype determination [18], we can speculate that miR-196b may also participate in EOC subtype differentiation, although further studies are needed to verify this hypothesis.
As mentioned above, there is a great discrepancy between the results of these miRNA profiles in cancer. Several major reasons, in addition to the application of different microarray platforms, technique or analytical tools, contribute to the disagreement. First, the normal control tissues utilized were different. Several groups used normal ovarian tissues [6, 7], while others used human immortalized ovarian surface epithelial (HIOSE) cell lines [4, 5, 8, 9, 11]. Although it was thought that whole ovarian tissue would not be the most appropriate normal control, as the ovary is composed of several cell types, and that the epithelium only represents far less than 1% of the cellular content of the ovarian tissue, the use of an OSE cell line also has drawbacks. Since no commercial HIOSE cell is available, different approaches to establishing an HIOSE cell line may cause changes in miRNA expression. For example, two groups immortalized the OSE cells using HPV-derived viral oncoproteins E6 and E7 [8] or SV40 virus [11]; thus, it is not surprising that these two studies presented inconsistent and even conflict results. Additionally, primary cultured OSE cells [9] and, more recently, fallopian tube tissues [10] have also been used as normal controls. Second, as delineated before, the heterogeneity of EOC makes comparison of these results further difficult. Third, sources of EOC cells may be contaminated with adjacent benign tissue and/or stromal cells which could cause variations in the detected miRNAs. Therefore, accurate microarray analysis of miRNA expression remains a significant challenge. Several methods, including the use of canonical HIOSE normal control, the selection of subtype-specific ovarian cancer tissues and the application of laser-capture microdissection to enrich for the ovarian cancer cells from clinical samples should be helpful in overcoming these problems. Indeed, if identical EOC subtype tissues and normal controls are used, the de-regulated miRNA patterns would show highly consistent results [6, 7].
Regulation of microRNAs in ovarian cancer
Aberrant expression of miRNAs in human cancer can arise through several mechanisms, including genomic abnormalities, epigenetic factors, transcriptional regulation, mutations and polymorphisms (SNPs), and defects in the miRNA biogenesis machinery [19]. In EOC, the de-regulation of miRNAs can be caused partly by genomic abnormalities, since 37.1% of the genomic loci containing miRNA genes exhibit DNA number alterations by a array comparative genomic hybridization analysis, and miRNA copy number changes correlate with miRNA expression [4]. Epigenetic factors also directly impact miRNA expressions, and demethylating therapy has been shown to reverse hypermethylation and increase miRNA expression in EOC [5, 6]. In fact, genomic copy number loss and epigenetic silencing may account for the down-regulation of approximately 15% and at least 36% of miRNAs in advanced ovarian tumours, respectively [5]. In contrast, the overexpression of several miRNAs (miR-21, miR-203, miR-205) in vivo was thought to be caused by DNA hypomethylation [6].
Functional genetic mutations in miRNA genes are very rare in EOC, as described by Bearfoot et al. [20], who analysed 10 cancer-implicated miRNA genes for mutations in 90 EOC patients, and found no somatic mutations. Similarly, no miR-16–1 gene mutation were detected in 102 ovarian cancers [21]. However, a functional G to C single nucleotide polymorphism (SNP, rs2910164) within the pre-miR-146a sequence was identified in familial breast/ovarian cancers. The C containing allele increases mature miR-146a levels and creates a stronger match with the 3′-UTR of BRCA1, leading to an earlier age onset of familial breast and ovarian cancers [22].
miRNAs can also be regulated by transcriptional factors in EOC. A well-known example is p53, one of the most commonly mutated genes observed in EOC, which can robustly induce miR-34b and miR-34c in OSE cells [23]. Hormones such as oestrogen, progesterone, androgens and gonadotrophins have all been implicated in the aetiology of EOC by initiating and repressing transcription of key genes [24, 25]. It is therefore of much interest to elucidate the role of hormonal effects on miRNA expression. The expressions of miR-143, let-7a and miR-15b are under negative control by follicle stimulating hormone (FSH) in the ovary [26]. Additionally, it has been shown that miR-20a and miR-21 are oestrogen-responsive in endometrial stromal cells and leiomyoma smooth muscle cells [27, 28]. Given that both of these miRNAs are expressed in the OSE cells [6], these miRNAs may be similarly regulated in these cells.
Mature miRNA biogenesis is dependent on their processing enzymes, including two key RNase III enzymes, Dicer and Drosha [29]. Given that miRNA expressions are globally down-regulated in EOC, it is conceivable that the levels of Dicer and Drosha may also be decreased. However, Drosha or Dicer showed no significant differences [5] or even higher expression levels [30] between ovarian cancer cell lines and IOSE, indicating that the de-regulation of miRNAs in EOC cannot be attributed to the defects in the miRNA biogenesis machinery. In support of this contention, similar mRNA and protein levels of Drosha and Dicer were also found in EOC tumours at different stages, even though global miRNA down-regulation was found in early-stage EOC relative to late-stage EOC [5]. Similarly, Drosha and Dicer expression levels showed no significant difference between primary and recurrent ovarian cancers, although multiple miRNAs were de-regulated [31].
Biological function of miRNAs in epithelial ovarian cancer
The initiation and progression of EOC is a complex biological process involving cell de-differentiation and proliferation, angiogenesis, invasion and metastasis. Like other human cancer types, the inactivation of tumour suppressor genes and the activation of oncogenes may be major contributors to this disease. As miRNAs can function either as oncogenes or tumour suppressors, altered expression of miRNAs may contribute to ovarian malignant transformation and subsequent progression. Several EOC-related miRNAs were reported, and various studies have also begun to elucidate the biological significance of some of these miRNAs. Here, we summarize the available information on the function of specific miRNAs in EOC.
Down-regulation of let-7: an early event during EOC initiation
Let-7 is the consensus miRNA down-regulated in almost all the expression profile studies in EOC [4–11]. The let-7 family is comprised 12 family members located on 8 different chromosomes with identical seed sequences [32]. As a ubiquitously expressed miRNA, let-7 has been shown to be a global regulator of cellular differentiation. Identified let-7 targets include a series of early embroyonic genes such as HMAG2, Mlin-41 and IMP-1 [33]. A high level of let-7 can permanently repress these genes and maintain the ‘differentiated’ status in the adult organism. As the early phase of carcinogenesis resembles embryonic development, often involving the re-expression of embryonic mesenchymal genes, it is reasonable that the de-differentiation of cancer cells caused by the reduction of let-7 is an early event in carcinogenesis [34]. This is further evidenced by the fact that the expression of the let-7 target gene, HMGA2, is higher in primary EOC tissues, but shows no difference between primary and metastastic sites [32].
A puzzle in EOC carcinogenesis is the apparent ‘differentiating up’ instead of ‘de-differentiation’, as EOCs morphologically resemble more committed epithelial phenotypes compared to uncommitted OSE cells [35]. This paradox has prompted the hypothesis which argues that EOCs may stem from the secondary mullerian system or the fallopian tube instead of OSE cells [36]. However, there is no direct experimental evidence to support this hypothesis. Recently, putative ovarian tumour initiating cells were found by several investigators [37–39] that lead to the hypothesis that the ‘differentiating up’ of EOCs is actually an example of aberrant recapitulation by tumours of organogenesis [35]. EOCs may arise from pluripotent ovarian tumour cells and differentiate along different lineages to form multiple cell subtypes resembling those of the specialized epithelia of the reproductive tract. Although it is not clear as to the origination of these so-called ovarian cancer stem cells, it is plausible that the loss of let-7 leads to a continuum of progressive de-differentiation, resulting in a cell at the end-point that has stem cell-like properties. In fact, it has been shown that let-7 can regulate stem cell-like properties of breast tumour-initiating cells through its target genes, HMGA2 and K-ras [40].
Multiple miRNAs as modulators of ovarian cancer cell proliferation
Multiple miRNAs may impact cancer cell proliferation and apoptosis during EOC initiation. The down-regulation of miR-143 [6, 7, 10] may promote cell proliferation via activation of the growth promoting proteins mitogen-activated protein kinase 7 (MAPK7) [41, 42] and KRAS [43], while the loss of miR-125a/b activity [6, 7, 11] may inhibit cell apoptosis by elevating ERBB2 levels [44], which are often seen in HG serous tumours. miR-21 [7, 10] and miR-214 [8, 9, 11], overexpressed in EOC, may suppress phosphatase and tensin homologue (PTEN) activity [11, 45] in endometroid cancer, while up-regulation of miR-106 [4, 9, 10] and miR-20a [7, 9, 10] may promote cell growth by targeting p21/CDKN1A [46, 47]. Additionally, miR-34s, which are induced by p53, had been shown to promote cell cycle arrest, apoptosis and senescence by repression of multiple target genes such as Bcl-2, Cdk4 and CCDN1 [23]. This is of particular importance as p53 mutation is one of the most common genetic alterations observed in EOC, especially in HG serous tumours [48]. However, reduction in miR-34s may also be caused by a p53-independent mechanism in low-grade (LG) serous tumours. For example, miR-34a is located in 1p36, a locus that is frequently deleted in LG serous tumours [49]. Moreover, miR-34a was shown to be up-regulated by ELK-1 during BRAF induced senescence [50], and mutation in BRAF has been described in 30∼50% of LG serous tumours [48]. Thus, the loss of miR-34s could be one of the mechanisms used by both LG and HG ovarian cancer cells to escape the control of a functioning p53 and BRAF and to survive oncogenic stimuli.
miR-199a and miR-9: link between tumour progression and chronic inflammation
miR-9 is expressed at a low level in ovarian cancer tissues [6, 51]. miR-9 can directly target NF-κB1, which is up-regulated in ovarian cancer [51]. In contrast, miR-199a can inhibit NF-κB activity through suppression of its upstream activator, IKKβ[16]. Suitably, miR-199a is also frequently de-regulated in EOC [6, 8].
Inflammation accompanying each ovulation event can stress OSE cells such that they are disposed to genetic damage [52]. Inflammatory processes can also promote cancer progression through the production of multiple cytokines and chemokines [53]. As NF-κB has a central role in the inflammatory response [53], it is possible that reduced expression of miR-199a and miR-9 work synergistically to promote an inflammatory environment by up-regulating NF-κB protein levels, leading to ovarian cancer initiation and progression. In fact, it has been demonstrated that ovarian cancer cells with low miR-199a expression have the capacity to constitutively secrete pro-inflammatory cytokines [16]. Moreover, another important pro-inflammatory factor, COX-2, is known to be suppressed by miR-199a*[54]. This further highlights the importance of miR-199a in the inflammatory response.
HIF-miR-210 promotes cell adaptation during hypoxia
Hypoxia is a common feature of pathological conditions such as inflammation and solid tumours. Multiple hypoxic responses impacting cell survival are mediated through hypoxia-inducible transcription factors (HIFs). HIFs, along with transcriptional cofactors, bind to hypoxia response elements, modulating the expression of multiple target genes, including miRNAs important for angiogenesis and cell survival [55]. Among the ‘hypoxia-responsive’ miRNAs, miR-210 has been reported to be the most prominent and consistent in ovarian cancer [56]. E2F transcription factor 3 (E2F3), a key protein in cell cycle, was directly targeted by miR-210 in EOC [56]. Further studies indicated there are up to 50 potential target genes for miR-210, a majority of which are implicated in adaptation and cell survival under hypoxic stress [57]. Thus, hypoxia-mediated induction of miR-210 modulates the expression of genes promoting cell survival during stress, followed by tumour progression under favourable conditions.
In addition to miR-210, other miRNAs may participate in the hypoxia response by promoting tumour neo-angiogenesis. For example, the overexpression of miR-19a in ovarian cancer [9] can promote angiogenesis by directly repressing the anti-angiogenic factor TSP-1 [58].
miR-200 is a dynamic regulator during the transcoelomic metastasis of EOC
The miR-200 family genes, which include 5 members (miR-200a, miR-200b, miR-200c, miR-141 and miR-429) located within two clusters on separate chromosomes, was recently identified as both a marker and a powerful regulator of EMT by targeting two E box binding transcription factors, ZEB1 and ZEB2, both of which are key regulators of the expression of E-cadherin [59].
The most common route of metastasis from EOC occurs via the transcoelomic route. During the transcoelomic journey, tumour cells undergo EMT, detach from the primary tumour mass, and form spheroids in the ascetic fluid before they ultimately implant on the peritoneum [60]. miR-200 appears to be one of the critical molecular players during this process. The miR-200 family is a key regulator of EMT [12, 61], and miR-200c expression correlates with E-cadherin expression in ovarian cancer tissues [61], which mediate cell-to-cell adhesion. Metastasis-prone tumour cells also form highly polarized epithelial spheres in three dimensional cultures, while forcing expression of miR-200 abrogates the capacity of these cells to form spheroids and metastasis [62]. Although the expression of the miR-200 family miRNAs in ascites cells has not yet been examined, miR-200 expression was higher in stage I EOC compared to stage III EOC [63], indicating a down-regulation of miR-200 before tumour metastasis. Moreover, it has also been shown that the mir-200 target, ZEB2, was expressed in cells isolated from effusions [64, 65].
Although there is clear evidence showing the miR-200 family to be metastasis suppressive, their expression in EOC is complex. miR-200s have been reported to be up-regulated [6, 7, 9–11, 66], down-regulated [8] or even unchanged [5] in EOC. This discrepancy may partly be due to technical issues, as whole ovaries were used as normal controls in some studies, and the majority of the cells within ovaries are stromal cells which lack miR-200 expression [61]. However, it is more likely that miR-200 could be both up- and down-regulated during EOC progression. miR-200 may be down-regulated early when cancer cells acquire invasive behaviour, but may also be up-regulated again during the re-epithelialization of distal metastases when cells undergo mesenchymal-to-epithelial transition. Such stage specific regulation of miR-200 was partly demonstrated in a multistep tumorigenesis model, which showed that miR-200 began to be strongly down-regulated only when tumour progressed to metastasis [67]. Although direct evidence for miR-200 re-expression in metastastic sites is lacking, E-cadherin expression, which positively correlates with miR-200, is significantly increased in metastatic ovarian tumours compared with their primary tumours [68].
A proposed model for miRNA roles in ovarian cancer initiation and progression
miRNAs are implicated in different steps of ovarian tumorigenesis and progression. Ovarian cancer stem cells may be initially transformed from the normal stem cells, progenitor cells, or even mature, differentiated cells. This process is characterized by the reduction of let-7, which can repress multiple embryonic genes and maintain a ‘differentiated’ status [34], and the down-regulation of miR-200, which regulates EMT and represses stemness [69, 70]. The de-differentiated cancer stem cells may then proceed along different pathways: the HG pathway, which shows more aggressive phenotypes, and the LG pathway. Different miRNAs may act as important regulators during this process. For example, let-7 and miR-200 re-expression may occur in the LG pathway, resulting in cancer cells characteristic of the epithelial phenotype. However, let-7 and miR-200 may remain lowly expressed in mesenchymal HG tumours [71]. miR-143 and miR-21/miR-214, may be involved in the LG pathway by targeting K-ras [43] and PTEN [11, 45], which are frequently mutated in LG serous tumours [48]. Meanwhile, miR-125b and miR-146a may play a role in the HG grade pathway by repressing ERBB2 [41] and BRCA1 [22]. During ovarian cancer progression, miRNAs may facilitate tumour growth by promoting infiltration of inflammatory cells and inducing tumour angiogenesis or promoting cell adaptation during hypoxia. The dynamic expression of miR-200, which regulates EMT, may cause transcoelomic metastasis and ultimately macrometastasis formation of ovarian cancer. Therefore, multiple de-regulated miRNAs work synergistically during the initiation and multi-step progression of ovarian cancer. Figure 1 illustrates the proposed involvement of miRNAs in this complicated process.
Fig 1.
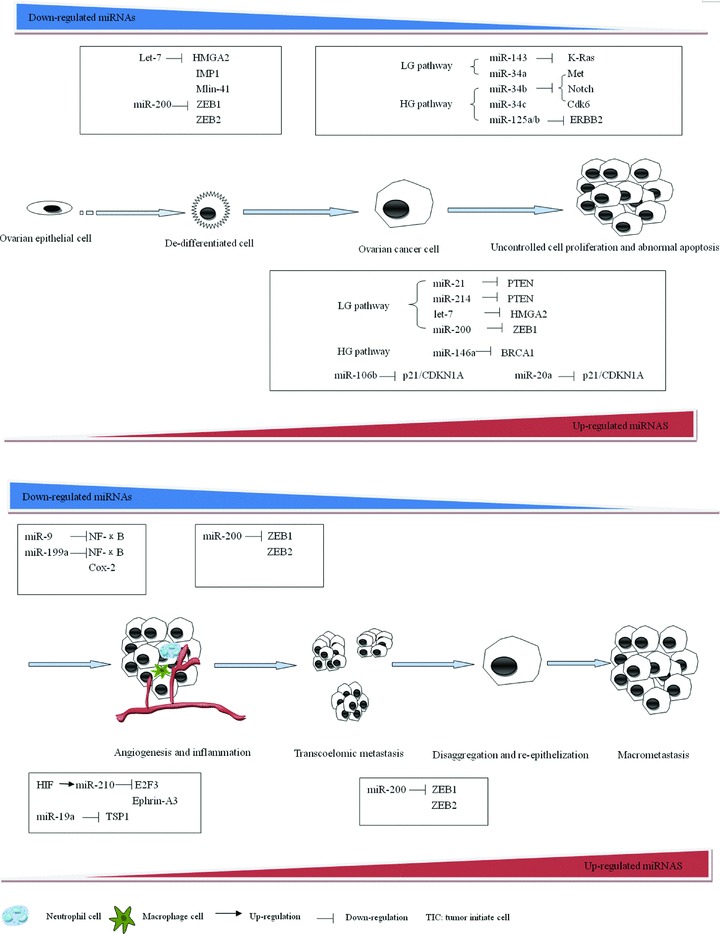
Biological function of miRNAs in EOC. In our proposed model for ovarian cancer development, EOC progresses through a process of de-differentiation, cell proliferation, angiogenesis and inflammation, transcoelomic metastasis, disaggregation and re-epithelization, and macromatasis formation. Let-7 may act as an important regulator of de-differentiation during ovarian tumour-initiating cells formation by targeting multiple oncofetal genes. Another key ‘stemness’-repressive miRNA, miR-200 may also down-regulated in this process. The de-differentiated cells may proceed along two pathways: the HG pathway, which showed more aggressive phenotypes, and the LG pathway. Different miRNAs may act as important regulators during this process. Let-7 and miR-200 re-expression may occur in the LG pathway, resulting in cancer cells characteristic of the epithelial phenotype. However, let-7 and miR-200 may remain lowly expressed in mesenchymal HG tumours. miR-21, miR-214, miR-143 may be involved in the LG pathway by targeting PTEN and K-ras, while miR-125b and miR-146a may play a role in the HG pathway by repressing ERBB2 and BRCA1. In addition, other miRNAs, such as miR-20a and miR-106b, may affect cell proliferation and apoptosis in both pathways. Subsequently, multiple miRNAs may play a role in chronic inflammation and angiogenesis, which are two main processes facilitating ovarian cancer progression. The dynamic expression of miR-200c, which regulates EMT, may cause transcoelomic metastasis and ultimately macrometastasis formation of ovarian cancer.
miRNA as diagnostic and prognostic tools in ovarian cancer
In EOC, early-stage diagnosis is very difficult due to its insidious asymptomatic nature in early onset and the lack of sensitive and specific biomarkers. Until now mRNA or protein profiling in order to find ideal biomarkers has not achieved satisfying results. However, miRNA expression profiles seem to be a promising method, since profiling a few hundred miRNAs would have stronger predictive power for cancer diagnosis than profiling several tens of thousands of mRNAs or proteins [72]. In fact, a list of 29 miRNAs is able to classify breast tumours and normal tissues with an accuracy of 100%[73]. As mentioned above, multiple studies showed distinctive miRNA signatures of EOC tissues compared with that of the OSE, and several consensus miRNAs characteristic of EOC were also found. These results suggest the possibility of discovering the combination of certain miRNAs that would represent a reliable biomarker for detection of ovarian cancer.
Serum biomarkers in EOC are important because they can be used as a screening strategy for ovarian cancer. Recent studies have shown that tumour-derived miRNAs are present in human serum in remarkable stable form and are protected from endogeneous ribonuclease activity [74]. The levels of plasma and serum miRNAs correlate strongly, suggesting that either plasma or serum can be used for investigation of these blood-based biomarkers [75]. Circulating miRNAs have been shown to be highly predictive of malignancy in colorectal cancer [76], diffuse large B cell lymphoma [77], and prostate cancer patients [74]. In ovarian cancer, it was demonstrated that eight miRNAs were differently expressed between normal and patient serum. Moreover, three miRNAs were overexpressed in three patients with normal pre-operative CA-125. This result highlighted the potential utility of these serum miRNAs as biomarkers, especially combined with Ca-125 [78]. Another study, focused on the circulating tumour exosomes of ovarian cancer patients, revealed a high correlation between the miRNA signature of exosomes and the primary tumour. More importantly, exosomal miRNA profiles were significantly distinct from that in benign disease and could not be detected in normal controls [79]. Altogether, these pilot results suggest that miRNA profiling, either of circulating tumour exosomes or patient sera, could potentially be used as diagnostic biomarkers.
miRNA profiles have been shown to be correlated with disease outcome in chronic lymphocytic leukaemia [80], lung cancer [81], breast cancer [82], colorectal cancer [83] and pancreatic cancer [84]. In ovarian cancer, the loss of let-7 and high expression of its target (HMGA2/let-7 ratio) was associated with unfavourable prognosis of ovarian cancer [32]. Similarly, reduced let-7i expression significantly increased the chemoresistance of ovarian cancer cells and was associated with the shorter progression-free survival [85]. More recently the prognostic value of miRNAs was studied by performing miRNA expression profiling analysis of 55 advanced ovarian tumours and correlating their expression level with cancer outcome [86]. Among the 96 cancer-related miRNAs, miR-200a was significantly associated with cancer survival. Elevated expression of miR-200 miRNAs predicted favourable outcomes [86]. However, inconsistent result was obtained by another group, who showed that tumours with higher miR-200a expression had poorer prognosis [7]. Moreover, miR-9 and miR-223 has been shown to be correlated with ovarian cancer recurrence [31], which greatly impacts patient outcomes. These results suggest that miRNAs can be potentially important as biomarkers of EOC prognosis and provide evidence for tailored therapy.
Therapeutic potential of miRNAs in ovarian cancer
The potential usefulness of a miRNA-based therapy in cancer has been exploited by different approaches to increase or decrease expression of different miRNAs. For oncogenic miRNAs, miRNAs can be silenced by antisense oligonucleotides and antagomirs (synthetic analogues of miRNAs) [87, 88]. For tumour suppressive miRNAs, overexpression of miRNAs can be induced by using synthetic miRNA mimics, chemically modified oligonucleotides [89], or adeno-associated virus based vector system [90]. The major advantage of miRNA based therapy is that a single miRNA can have roles in multiple aspects of cellular physiology; thus, the function of several pathways could be restored by a single hit. For example, ectopic expression of only miR-26a can potently suppress liver tumorigenesis in vivo[90].
Several preliminary studies have demonstrated that EOC related miRNAs may be potential targets in ovarian cancer therapy. Overexpression of miR-34b and miR-34c can reduce proliferation and adhesion-independent growth of ovarian cancer cells in vitro[23]. Restoring miR-9 expression in the ovarian clear cancer cell line ES-2 can suppress cell growth [51]. Moreover, miRNAs involved in specific networks, such as the PTEN/Akt pathway, could likely influence the response to chemotherapy. For example, blocking miR-214 expression can up-regulate PTEN and sensitize the ovarian cancer cell A2780CP to cisplatin-induced apoptosis [11]. Forced expression of miR-200c can increase sensitivity to paclitaxel by targeting class III β-tubulin, whose expression is a common mechanism of resistance to microtubule-binding chemotherapeutic agents [91]. In fact, several reports indicate that ovarian cancer drug resistance is associated with a distinct miRNA fingerprint [63, 92, 93]. A seven-miRNA signature (up-regulated: miR-27a, miR-23a, miR-30c, let-7g, miR-199a-3p; down-regulated: miR-378 and miR-625) was representative of platinum-resistance [63], while down-regulation of three miRNAs (miR-30c, miR-130a, miR-335) suggested chemoresistance to platinum and paclitaxel [93]. Chemoresistance to platinum-based drugs coupled with paclitaxel is the main limitation to successful treatment of advanced ovarian cancer. Thus, miRNAs can be used as both a potential therapeutic tool for modulating the response to chemotherapy for ovarian cancer, and a prognostic tool to monitor the outcome.
Conclusions
The aberrantly expressed miRNAs in EOC have revealed novel mechanisms in ovarian tumorigenesis and progression. Moreover, the miRNA expression profiles in tissues and blood can potentially be used for the detection and surveillance of ovarian cancer. Additionally, miRNAs are implicated in aggressive tumour behaviour and chemotherapy resistance and may be a potentially useful tool for prognostic stratification and tailored therapy. Finally, miRNAs may be targeted by gene therapy to treat EOC. However, the study of miRNAs in EOC is a relatively new area. More efforts are needed to clarify the EOC-related miRNAs by using a uniform technical platform and appropriate normal controls, to elucidate key miRNAs as well as their signal transduction pathways. Such standard protocols and resulting findings will allow us to find ways to manipulate miRNAs for therapeutic benefit in a rational manner, and to validate experimental miRNAs therapy through studies involving different cohorts of patients before they can be introduced into clinical practice.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work is supported by the Shanghai Excellent Leading Scholars Program (08XD14032).
Supporting Information
Table S1 Consensus miRNAs up-regulated in EOC*
Table S2 Consensus miRNAs down-regulated in EOC*
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Köbel M, Kalloger SE, Boyd N, et al. Ovarian carcinoma subtypes are different diseases: implications for biomarker studies. PLoS Med. 2008;5:e232. doi: 10.1371/journal.pmed.0050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 4.Zhang L, Huang J, Yang N, et al. MicroRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA. 2006;103:9136–41. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang L, Volinia S, Bonome T, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci USA. 2008;105:7004–9. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iorio MV, Visone R, Di Leva G, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 7.Nam EJ, Yoon H, Kim SW, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14:2690–5. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 8.Dahiya N, Sherman-Baust CA, Wang TL, et al. MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PLoS One. 2008;3:e2436. doi: 10.1371/journal.pone.0002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wyman SK, Parkin RK, Mitchell PS, et al. Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA libraries. PLoS One. 2009;4:e5311. doi: 10.1371/journal.pone.0005311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee CH, Subramanian S, Beck AH, et al. MicroRNA profiling of BRCA1/2 mutation-carrying and non-mutation-carrying high-grade serous carcinomas of ovary. PLoS One. 2009;4:e7314. doi: 10.1371/journal.pone.0007314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang H, Kong W, He L, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–33. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 12.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat cell biol. 2008;10:593–60. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 13.Duisters RF, Tijsen AJ, Schroen B, et al. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res. 2009;104:170–8. doi: 10.1161/CIRCRESAHA.108.182535. [DOI] [PubMed] [Google Scholar]
- 14.Kikuchi R, Tsuda H, Kanai Y, et al. Promoter hypermethylation contributes to frequent inactivation of a putative conditional tumor suppressor gene connective tissue growth factor in ovarian cancer. Cancer Res. 2007;67:7095–105. doi: 10.1158/0008-5472.CAN-06-4567. [DOI] [PubMed] [Google Scholar]
- 15.Barbolina MV, Adley BP, Kelly DL, et al. Downregulation of connective tissue growth factor by three-dimensional matrix enhances ovarian carcinoma cell invasion. Int J Cancer. 2009;125:816–25. doi: 10.1002/ijc.24347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen R, Alvero AB, Silasi DA, et al. Regulation of IKKbeta by miR-199a affects NF-kappaB activity in ovarian cancer cells. Oncogene. 2008;27:4712–23. doi: 10.1038/onc.2008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yekta S, Tabin CJ, Bartel DP. MicroRNAs in the Hox network: an apparent link to posterior prevalence. Nat Rev Genet. 2008;9:789–96. doi: 10.1038/nrg2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng W, Liu J, Yoshida H, et al. Lineage infidelity of epithelial ovarian cancers is controlled by HOX genes that specify regional identity in the reproductive tract. Nat Med. 2005;11:531–7. doi: 10.1038/nm1230. [DOI] [PubMed] [Google Scholar]
- 19.Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bearfoot JL, Choong DY, et al. Genetic analysis of cancer-implicated MicroRNA in ovarian cancer. Clin Cancer Res. 2008;14:7246–50. doi: 10.1158/1078-0432.CCR-08-1348. [DOI] [PubMed] [Google Scholar]
- 21.Yazici H, Zipprich J, Peng T, et al. Investigation of the miR16–1 (C > T) + 7 Substitution in seven different types of cancer from three ethnic groups. J Oncol. 2009;2009:827532. doi: 10.1155/2009/827532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen J, Ambrosone CB, DiCioccio RA, et al. A functional polymorphism in the miR-146a gene and age of familial breast/ ovarian cancer diagnosis. Carcinogenesis. 2008;29:1963–6. doi: 10.1093/carcin/bgn172. [DOI] [PubMed] [Google Scholar]
- 23.Corney DC, Flesken-Nikitin A, Godwin AK, et al. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 24.Lukanova A, Kaaks R. Endogenous hormones and ovarian cancer: epidemiology and current hypotheses. Cancer Epidemiol Biomarkers Prev. 2005;14:98–107. [PubMed] [Google Scholar]
- 25.Wong AS, Leung P. Role of endocrine and growth factors on the ovarian surface epithelium. J Obstet Gynaecol Res. 2007;33:3–16. doi: 10.1111/j.1447-0756.2007.00478.x. [DOI] [PubMed] [Google Scholar]
- 26.Yao N, Lu CL, Zhao JJ, et al. A network of miRNAs expressed in the ovary are regulated by FSH. Front Biosci. 2009;14:3239–45. doi: 10.2741/3447. [DOI] [PubMed] [Google Scholar]
- 27.Pan Q, Luo X, Chegini N. Differential expression of microRNAs in myometrium and leiomyomas and regulation by ovarian steroids. J Cell Mol Med. 2008;12:227–40. doi: 10.1111/j.1582-4934.2007.00207.x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Pan Q, Luo X, Toloubeydokhti T, et al. The expression profile of micro-RNA in endometrium and endometriosis and the influence of ovarian steroids on their expression. Mol Hum Reprod. 2007;13:797–806. doi: 10.1093/molehr/gam063. [DOI] [PubMed] [Google Scholar]
- 29.Lau PW, MacRae IJ. The molecular machines that mediate microRNA maturation. J Cell Mol Med. 2009;13:54–60. doi: 10.1111/j.1582-4934.2008.00520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flavin RJ, Smyth PC, Finn SP, et al. Altered eIF6 and Dicer expression is associated with clinicopathological features in ovarian serous carcinoma patients. Mod Pathol. 2008;21:676–84. doi: 10.1038/modpathol.2008.33. [DOI] [PubMed] [Google Scholar]
- 31.Laios A, O’Toole S, Flavin R, et al. Potential role of miR-9 and miR-223 in recurrent ovarian cancer. Mol Cancer. 2008;7:35. doi: 10.1186/1476-4598-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park SM, Shell S, Radjabi AR, et al. Let-7 prevents early cancer progression by suppressing expression of the embryonic gene HMGA2. Cell Cycle. 2007;6:2585–90. doi: 10.4161/cc.6.21.4845. [DOI] [PubMed] [Google Scholar]
- 33.Boyerinas B, Park SM, Shomron N, et al. Identification of let-7-regulated oncofetal genes. Cancer Res. 2008;68:2587–91. doi: 10.1158/0008-5472.CAN-08-0264. [DOI] [PubMed] [Google Scholar]
- 34.Peter M. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle. 2009;8:843–52. doi: 10.4161/cc.8.6.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naora H. The heterogeneity of epithelial ovarian cancers: reconciling old and new paradigms. Expert Rev Mol Med. 2007;9:1–12. doi: 10.1017/S1462399407000324. [DOI] [PubMed] [Google Scholar]
- 36.Crum CP, Drapkin R, Miron A, et al. The distal fallopian tube: a new model for pelvic serous carcinogenesis. Curr Opin Obstet Gynecol. 2007;19:3–9. doi: 10.1097/GCO.0b013e328011a21f. [DOI] [PubMed] [Google Scholar]
- 37.Zhang S, Balch C, Chan MW, et al. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008;68:4311–20. doi: 10.1158/0008-5472.CAN-08-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bapat SA, Mali AM, Koppikar CB, et al. Stem and progenitor-like cells contribute to the aggressive behavior of human epithelial ovarian cancer. Cancer Res. 2005;65:3025–9. doi: 10.1158/0008-5472.CAN-04-3931. [DOI] [PubMed] [Google Scholar]
- 39.Szotek PP, Pieretti-Vanmarcke R, Masiakos PT, et al. Ovarian cancer side population defines cells with stem cell-like characteristics and Mullerian Inhibiting Substance responsiveness. Proc Natl Acad Sci USA. 2006;103:11154–9. doi: 10.1073/pnas.0603672103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu F, Yao H, Zhu P, et al. Let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 41.Esau C, Kang X, Peralta E, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–5. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 42.Akao Y, Nakagawa Y, Kitade Y, et al. Downregulation of microRNAs-143 and -145 in B-cell malignancies. Cancer Sci. 2007;98:1914–20. doi: 10.1111/j.1349-7006.2007.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen X, Guo X, Zhang H, et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28:1385–92. doi: 10.1038/onc.2008.474. [DOI] [PubMed] [Google Scholar]
- 44.Scott GK, Goga A, Bhaumik D, et al. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem. 2007;282:1479–86. doi: 10.1074/jbc.M609383200. [DOI] [PubMed] [Google Scholar]
- 45.Meng F, Henson R, Wehbe-Janek H, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Inomata M, Tagawa H, Guo YM, et al. MicroRNA-17–92 down-regulates expression of distinct targets in different B-cell lymphoma subtypes. Blood. 2009;113:396–402. doi: 10.1182/blood-2008-07-163907. [DOI] [PubMed] [Google Scholar]
- 47.Kan T, Sato F, Ito T, et al. The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology. 2009;136:1689–700. doi: 10.1053/j.gastro.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Landen CN, Jr, Birrer MJ, Sood AK. Early events in the pathogenesis of epithelial ovarian cancer. J Clin Oncol. 2008;26:995–1005. doi: 10.1200/JCO.2006.07.9970. [DOI] [PubMed] [Google Scholar]
- 49.Kuo KT, Guan B, Feng Y, et al. Analysis of DNA copy number alterations in ovarian serous tumors identifies new molecular genetic changes in low-grade and high-grade carcinomas. Cancer Res. 2009;69:4036–42. doi: 10.1158/0008-5472.CAN-08-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Christoffersen NR, Shalgi R, Frankel LB, et al. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ. 2010;17:236–45. doi: 10.1038/cdd.2009.109. [DOI] [PubMed] [Google Scholar]
- 51.Guo LM, Pu Y, Han Z, et al. MicroRNA-9 inhibits ovarian cancer cell growth through regulation of NF-kappaB1. FEBS J. 2009;276:5537–46. doi: 10.1111/j.1742-4658.2009.07237.x. [DOI] [PubMed] [Google Scholar]
- 52.Ness RB, Cottreau C. Possible role of ovarian epithelial inflammation in ovarian cancer. J Natl Cancer Inst. 1999;91:1459–67. doi: 10.1093/jnci/91.17.1459. [DOI] [PubMed] [Google Scholar]
- 53.Chen R, Alvero AB, Silasi DA, et al. Inflammation, cancer and chemoresistance: taking advantage of the toll-like receptor signaling pathway. Am J Reprod Immunol. 2007;57:93–107. doi: 10.1111/j.1600-0897.2006.00441.x. [DOI] [PubMed] [Google Scholar]
- 54.Chakrabarty A, Tranguch S, Daikoku T, et al. MicroRNA regulation of cyclooxygenase-2 during embryo implantation. Proc Natl Acad Sci USA. 2007;104:15144–9. doi: 10.1073/pnas.0705917104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kulshreshtha R, Davuluri RV, Calin GA, et al. A microRNA component of the hypoxic response. Cell Death Differ. 2008;15:667–71. doi: 10.1038/sj.cdd.4402310. [DOI] [PubMed] [Google Scholar]
- 56.Giannakakis A, Sandaltzopoulos R, Greshock J, et al. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol Ther. 2008;7:255–64. doi: 10.4161/cbt.7.2.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang X, Ding L, Bennewith KL, et al. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell. 2009;35:856–67. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dews M, Homayouni A, Yu D, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–5. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gregory PA, Bracken CP, Bert AG, et al. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle. 2008;7:3112–8. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- 60.Tan DS, Agarwal R, Kaye SB. Mechanisms of transcoelomic metastasis in ovarian cancer. Lancet Oncol. 2006;7:925–34. doi: 10.1016/S1470-2045(06)70939-1. [DOI] [PubMed] [Google Scholar]
- 61.Park SM, Gaur AB, Lengyel E, et al. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gibbons DL, Lin W, Creighton CJ, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23:2140–51. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eitan R, Kushnir M, Lithwick-Yanai G, et al. Tumor microRNA expression patterns associated with resistance to platinum based chemotherapy and survival in ovarian cancer patients. Gynecol Oncol. 2009;114:253–9. doi: 10.1016/j.ygyno.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 64.Elloul S, Elstrand MB, Nesland JM, et al. Snail, Slug, and Smad-interacting protein 1 as novel parameters of disease aggressiveness in metastatic ovarian and breast carcinoma. Cancer. 2005;103:1631–43. doi: 10.1002/cncr.20946. [DOI] [PubMed] [Google Scholar]
- 65.Elloul S SI, Trope CG, Benshushan A, et al. Expression of E-cadherin transcriptional regulators in ovarian carcinoma. Virchows Arch. 2006;449:520–8. doi: 10.1007/s00428-006-0274-6. [DOI] [PubMed] [Google Scholar]
- 66.Bendoraite A, Knouf EC, Garg KS, et al. Regulation of miR-200 family microRNAs and ZEB transcription factors in ovarian cancer: evidence supporting a mesothelial-to-epithelial transition. Gynecol Oncol. 2010;116:117–25. doi: 10.1016/j.ygyno.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Olson P LJ, Zhang H, Shai A, et al. MicroRNA dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Genes Dev. 2009;23:2152–65. doi: 10.1101/gad.1820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Imai T, Horiuchi A, Shiozawa T, et al. Elevated expression of E-cadherin and alpha-, beta-, and gamma-catenins in metastatic lesions compared with primary epithelial ovarian carcinomas. Hum Pathol. 2004;35:1469–76. doi: 10.1016/j.humpath.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 69.Wellner U, Schubert J, Burk UC, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–95. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 70.Shimono Y, Zabala M, Cho RW, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shell S, Park SM, Radjabi AR, et al. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci USA. 2007;104:11400–5. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Calin GA, Corce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 73.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 74.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gilad S, Meiri E, Yogev Y, et al. Serum microRNAs are promising novel biomarkers. Plos one. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang Z, Huang D, Ni S, et al. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2009 doi: 10.1002/ijc.25007. Oct 28 DOI: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 77.Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–5. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 78.Resnick KE, Alder H, Hagan JP, et al. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. 2009;112:55–9. doi: 10.1016/j.ygyno.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 79.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 80.Calin GA, Ferracin M, Cimmino A, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 81.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 82.Yan LX, Huang XF, Shao Q, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–60. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schepeler T, Reinert JT, Ostenfeld MS, et al. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68:6416–24. doi: 10.1158/0008-5472.CAN-07-6110. [DOI] [PubMed] [Google Scholar]
- 84.Bloomston M, Frankel WL, Petrocca F, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–8. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 85.Yang N, Kaur S, Volinia S, et al. MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer Res. 2008;68:10307–14. doi: 10.1158/0008-5472.CAN-08-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hu X, Macdonald DM, Huettner PC, et al. A miR-200 microRNA cluster as prognostic marker in advanced ovarian cancer. Gynecol Oncol. 2009;114:457–64. doi: 10.1016/j.ygyno.2009.05.022. [DOI] [PubMed] [Google Scholar]
- 87.Krützfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 88.Elmén J, Lindow M, Silahtaroglu A, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–62. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–60. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 90.Kota J, Chivukula R, O’Donnell KA, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–17. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cochrane DR, Spoelstra NS, Howe EN, et al. MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Mol Cancer Ther. 2009;8:1055–66. doi: 10.1158/1535-7163.MCT-08-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Boren T, Xiong Y, Hakam A, et al. MicroRNAs and their target messenger RNAs associated with ovarian cancer response to chemotherapy. Gynecol Oncol. 2009;113:249–55. doi: 10.1016/j.ygyno.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 93.Sorrentino A, Liu CG, Addario A, et al. Role of microRNAs in drug-resistant ovarian cancer cells. Gynecol Oncol. 2008;111:478–86. doi: 10.1016/j.ygyno.2008.08.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Consensus miRNAs up-regulated in EOC*
Table S2 Consensus miRNAs down-regulated in EOC*


