Abstract
Telocytes (TCs) are a particular type of interstitial (stromal) cells defined by very long, moniliform telopodes. Their tissue location, between blood vessels and other cells such as cardiomyocytes (CMC) and neurons, suggests a role in intercellular signalling. In order to define a microRNA (miR) signature in cardiac TCs, we have found that miR-193 is differentially expressed between TCs and other interstitial cells. Because miR-193 regulates c-kit, our data support the previous finding that TCs express c-kit in certain circumstances. In addition, the miRs which are specific to CMC and other muscle cells (e.g. miR-133a, miR-208a) are absent in TCs. Overall the data reinforce the view that TCs are a particular type of interstitial (mesenchymal) cells.
Keywords: telocytes, telopodes, microRNA, laser capture microdissection
Introduction
Telocytes (TCs) are a recently described type of stromal cells defined by their ultrastructural and morphological features [1]. Telopodes (particularly long and thin prolongations) are the hallmark of this cell type as seen in several organs by electron microscopy [2–8], and have also offered a strong indication of TCs’ presence in cell cultures of different origins [9–11]. Although it has been shown that various proportions of TCs express c-kit [12], CD34, caveolin-1 or VEGF [5] in vivo and in vitro, the identification of additional molecular markers is still a challenge. Therefore, we took advantage of laser capture microdissection (LCM), which offers the possibility of isolating a single cell with particular morphology from tissues or cultures and enables their further accurate investigation.
MicroRNAs (miRs) are small, single-strand RNA molecules that regulate protein levels by repressing gene translation [reviewed in 13]. miRs can show tissue- and cell-type-specific restriction [14], have distinct expression patterns during differentiation and are involved in various pathological processes. miR-1, miR-133a and miR-208a are preferentially expressed in cardiomyocytes (CMC) and other muscle cells and are necessary for proper skeletal and cardiac muscle development and function. They also play a role in cardiac hypertrophy, preconditioning or ischemia-reperfusion injury [15]. miR-199a is up-regulated in hypertrophic heart, when it is predominantly expressed in CMC, but was also detected at low levels in cardiac fibroblasts [16].
Several reports indicate that miR-21, miR-22, miR-29 and miR-193 are expressed in embryonic and adult fibroblasts [17, 18]. miR-21 is also detected in cardiac fibroblasts, with levels selectively increased in interstitial fibrosis and cardiac hypertrophy [19]. It has been shown that miR-29 plays a role in cardiac fibrosis through down-regulation of miRs encoding collagens and extracellular matrix proteins [20]. Very recently, miR-193 was found to repress c-kit expression in acute myeloid leukaemia, functioning as a tumour suppressor gene [21].
In this study, we have investigated whether miRs known to be present and to play a functional role in myocardium are expressed in TCs isolated by LCM from cultured murine cardiac cells.
Material and methods
Cell cultures
Healthy adult C57 black mice were used in accordance with ethical guidelines to harvest heart tissue. After mechanical mincing, small fragments were rinsed in HBSS without calcium or magnesium and incubated in type II collagenase (Sigma-Aldrich, St. Louis, MO, USA) at a concentration of 250 U/ml for 15 min. at 37°C. Collagenase was inhibited using ice-cold HBSS and the resulting cell suspension was centrifuged, washed in culture medium and seeded at a density of 1 × 105 cells/cm2 in plastic culture dishes. Culture medium was DMEM-F12 (Invitrogen, Carlsbad, CA, USA) supplemented with 10% foetal calf serum. After 5 to 7 days, cells were detached using trypsin-ethylenediaminetetraacetic acid and replated at lower densities (1–2 × 105 cells/cm2) in DuplexDish 50 plates (Carl Zeiss MicroImaging GmbH, Jena, Germany), a PEN membrane-covered culture dish that allows further laser microdissection of target cells.
We also used controls consisting of both 3T3 fibroblasts in culture and cell suspensions enriched in murine CMC obtained through gradient centrifugation of collagenase-dissociated murine heart tissue.
Laser capture microdissection
Cell cultures were washed with phosphate-buffered saline and then fixed with absolute ethanol, Giemsa stained and air-dried. This allowed for a better visualization of morphology details as compared to phase-contrast examination. LCM was performed with the PALM MicroBeam system (Carl Zeiss MicroImaging GmbH), with 10× to 40× objectives and in accordance with the manufacturer protocols. The criteria used for TC identification were: mononucleated small cells with at least one process longer than 50 μm and thinner than 0.5 μm, with moniliform dilations. Control cells with fibroblast morphology, but no telopodes, were also microdissected. In two separate experiments we harvested a total of 500 TCs and 500 control cells. Cells were catapulted into 500 μm tube caps filled with 30 μm of lysing and extraction buffer.
RNA extraction, reverse-transcription and microRNA qPCR
Total RNA was extracted with TRI Reagent (Sigma-Aldrich) and isolated using the protocol recommended by the manufacturer. The RNA samples were evaluated with NanoDrop ND-1000 Spectrophotometer (Thermo Scientific, Waltham, MA, USA).
For first-strand cDNA synthesis we used QuantiMir RT (System Biosciences, San Francisco, CA, USA) according to manufacturer instructions. Basically, small RNAs were poly-A tailed, and an oligo-dT adapter was annealed to serve as a reverse-transcription primer. cDNA templates were then quantified in a real-time SYBR Green qPCR reaction (iCycler system and software, Bio-Rad, Hercules, CA, USA) using universal reverse primers and miR-specific forward primers. Mouse U6 snRNA and RNU43 were used as endogenous control assays and for data normalization. Threshold cycles were calculated for analysed miRs and subtracted from those of endogenous controls. Numbers obtained were subtracted from an arbitrary value, and the resulting score was used to assess specific miR relative expression levels.
Statistics
The data are represented as mean + S.D. of three experiments and two-tailed P-values of less than 0.05 were considered as statistically significant.
Results and discussion
TCs are present in primary and first-passage cultures of murine cardiac tissue, as revealed by their typical morphology. Primary cultures had higher levels of debris and non-stromal cell types and reached 80% confluence 5 to 7 days after seeding. Two to 4 days after replating, variable numbers of TCs were visible, together with cells having ‘fibroblast-like’ morphology (e.g. spindle shaped, with a few short and thick end-processes).
Giemsa-stained TCs grown on membranes can be easily identified and accurately microdissected. Figure 1 shows the typical morphology of TCs that were harvested, as well as cells that were used as a negative control for miR expression relative quantification. Cells were isolated within 48 hrs of their staining and kept at 4°C between procedures.
fig 1.
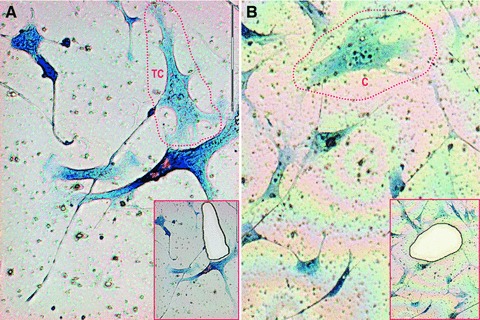
Laser microdissection of Giemsa-stained TCs in (A) and control cells (C) in (B) cultivated on membrane dishes. Insets show the corresponding areas after microdissection and correspond to areas enclosed in red discontinuous lines. 20× objective.
In order to validate our method in terms of specificity and sensitivity, we have tested the expression of several miRs known to be preferentially expressed in different types of cells within cardiac tissue (Fig. 2). Thus, the CMC-specific miRs, miR-133a and miR-208a, show high level of expression in the samples enriched in CMC (Fig. 2A), whereas miR-21, miR-29 and miR-199a-5p are preferentially expressed in cultured cardiac cells (CCC) and in fibroblasts (Fig. 2B). Noteworthy, miR-193 is expressed at lower level in CCC than in 3T3 fibroblasts, suggesting that, besides of cardiac fibroblasts, the cultures may contain cells where this miR is down-regulated (Fig. 2B).
fig 2.
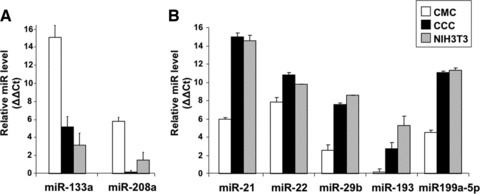
Expression levels of miRs specifically up-regulated in CMC (A) or in both cultured cardiac interstitial cells and NIH3T3 fibroblasts (B). The relative expression was determined by real-time PCR. RNU43 snoRNA and U6 snRNA were used as normalizing controls. The error bars are mean + S.D. (n = 3) and the data are representative for two replicate experiments. CMCs: cardiomyocytes; CCCs: cultured cardiac cells; NIH3T3: fibroblasts.
CMC-specific miRs are not expressed by TCs, as expected. Microdissected TCs and control cells did not express any of the CMC-specific miRs (miR-1, 133a or 208a), but, in accordance with their mesenchymal origin, showed various levels of miR-21, 22, 29 and 199a-5p (Fig. 3A).
fig 3.
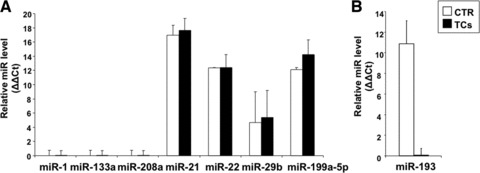
miR detection in laser microdissected cells. The results represent real-time PCR data were U6 snRNA is the internal control. (A) miR-1, miR-133 and miR-208a are not detected; the interstitial cell specific miRs miR-21, miR-22, miR-29b and miR-199a-5p are present in both fibroblast-like cells and TCs. In case of miR-29b, because the expression level is reduced the data variation from one experiment to another is high. (B) miR-193 is not expressed in TCs, whereas it is present in control cells. The error bars are mean + S.D. (A, n = 2; B, n = 3) and the data are representative for two replicate experiments. CTR: control fibroblast-like cells; TC: telocyte.
TCs do not express miR-193. In all experiments, miR-193 expression was below the detection level in microdissected TCs, whereas neighbouring fibroblasts-like cells were positive for it (Fig. 3B). The result correlates with the lower detection of miR-193 in CCC than in fibroblasts (Fig. 2B) because the cardiac culture contains both types of cells, positive and negative for miR-193. The data support the view that TCs are a distinct cell type also in terms of gene expression and regulation pathways. Noteworthy, c-kit is predicted by TargetScan (http://www.targetscan.org) to be a target of miR-193, an interaction that has been validated experimentally [21]. Thus, our results concur with previous observations that cardiac TCs can be differentiated in situ from surrounding stromal cells by their c-kit expression [12]. Other predicted targets of miR-193 include matrix metallopeptidase 19-an enzyme that facilitates cell motility by breaking down extracellular matrix in normal processes such as tissue remodelling and in pathological circumstances [22]; ρ guanine nucleotide exchange factor 12-a ρ GTPase involved in repulsive guidance of cell migration [23] and the chromobox protein CBX7 that positively regulates E-cadherin expression by interacting with the histone deacetylase protein 2 [24].
In conclusion, a functional picture of TCs is emerging, where this particular type of stromal cells is responsible, as opposed to canonical fibroblasts, for establishing and maintaining tissue scaffold that is involved in cell guidance and cell-to-cell signalling. Moreover, we think that TCs act in tandem with cardiac (progenitor) stem cells [25, 26].
Acknowledgments
This paper is partially supported by the Sectorial Operational Programme Human Resources Development (SOPHRD), financed from the European Social Fund and by the Romanian Government under the contract number POSDRU/89/1.5/S/64153 (to V.B.C.) and under the contract number POSDRU/89/1.5/S/64109 (to E.R.). We thank Dr. Laura C. Suciu for constant help, Teodor Regalia for technical assistance and Dr. Catalin G. Manole for editorial assistance.
Conflicts of interest
The authors confirm that there are no conflicts of interest.
References
- 1.Popescu LM, Faussone-Pellegrini MS. Telocytes – a case of serendipity: the winding way from interstitial cells of Cajal (ICC), via Interstitial Cajal-Like Cells (ICLC) to Telocytes. J Cell Mol Med. 2010;14:729–40. doi: 10.1111/j.1582-4934.2010.01059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gherghiceanu M, Popescu LM. Cardio-myocyte precursors and telocytes in epicardial stem cell niche: electron microscope images. J Cell Mol Med. 2010;14:871–7. doi: 10.1111/j.1582-4934.2010.01060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bani D, Formigli L, Gherghiceanu M, et al. Telocytes as supporting cells for myocardial tissue organization in developing and adult heart. J Cell Mol Med. 2010;14:2531–8. doi: 10.1111/j.1582-4934.2010.01119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Popescu LM, Manole CG, Gherghiceanu M, et al. Telocytes in human epicardium. J Cell Mol Med. 2010;14:2085–93. doi: 10.1111/j.1582-4934.2010.01129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suciu L, Popescu LM, Gherghiceanu M, et al. Telocytes in human term placenta: morphology and phenotype. Cells Tissues Organs. 2010;192:325–39. doi: 10.1159/000319467. [DOI] [PubMed] [Google Scholar]
- 6.Carmona IC, Bartolome MJ, Escribano CJ. Identification of telocytes in the lamina propria of rat duodenum: transmission electron microscopy. J Cell Mol Med. 2011;15:26–30. doi: 10.1111/j.1582-4934.2010.01207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hinescu ME, Gherghiceanu M, Suciu L, et al. Telocytes in pleura: two- and three-dimensional imaging by transmission electron microscopy. Cell Tissue Res. 2011;343:389–97. doi: 10.1007/s00441-010-1095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cantarero I, Luesma M, Junquera C. The primary cilium of telocytes in the vasculature: electron microscope imaging. J Cell Mol Med. 2011 doi: 10.1111/j.1582-4934.2011.01312.x. 10.1111/j.1582-4934.2011.01312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suciu L, Popescu LM, Regalia T, et al. Epicardium: interstitial Cajal-like cells (ICLC) highlighted by immunofluorescence. J Cell Mol Med. 2009;13:771–7. doi: 10.1111/j.1582-4934.2009.00756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suciu L, Nicolescu MI, Popescu LM. Cardiac telocytes: serial dynamic images in cell culture. J Cell Mol Med. 2010;14:2687–92. doi: 10.1111/j.1582-4934.2010.01185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou J, Zhang Y, Wen X, et al. Telocytes accompanying cardiomyocyte in primary culture: two- and three-dimensional culture environment. J Cell Mol Med. 2010;14:2641–5. doi: 10.1111/j.1582-4934.2010.01186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kostin S. Myocardial telocytes: a specific new cellular entity. J Cell Mol Med. 2010;14:1917–21. doi: 10.1111/j.1582-4934.2010.01111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight. Nat Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 14.Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Townley-Tilson WH, Callis TE, Wang D. MicroRNAs 1, 133, and 206: critical factors of skeletal and cardiac muscle development, function, and disease. Int J Biochem Cell Biol. 2010;42:1252–5. doi: 10.1016/j.biocel.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song XW, Li Q, Lin L, et al. MicroRNAs are dynamically regulated in hypertrophic hearts, and miR-199a is essential for the maintenance of cell size in cardiomyocytes. J Cell Physiol. 2010;225:437–43. doi: 10.1002/jcp.22217. [DOI] [PubMed] [Google Scholar]
- 17.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–8. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 18.Koh W, Sheng CT, Tan B, et al. Analysis of deep sequencing microRNA expression profile from human embryonic stem cells derived mesenchymal stem cells reveals possible role of let-7 microRNA family in downstream targeting of hepatic nuclear factor 4 alpha. BMC Genomics. 2010;11:S6. doi: 10.1186/1471-2164-11-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thum T, Gross C, Fiedler J, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signaling in fibroblasts. Nature. 2008;456:980–4. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 20.van Rooij E, Sutherland LB, Thatcher JE, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA. 2008;105:13027–32. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao XN, Lin J, Li YH, et al. MicroRNA-193a represses c-kit expression and functions as a methylation-silenced tumor suppressor in acute myeloid leukemia. Oncogene. 2011 doi: 10.1038/onc.2011.62. ; doi: 10.1038/onc.2011.62. [DOI] [PubMed] [Google Scholar]
- 22.Konttinen YT, Ainola M, Valleala H, et al. Analysis of 16 different matrix metalloproteinases (MMP-1 to MMP-20) in the synovial membrane: different profiles in trauma and rheumatoid arthritis. Ann Rheum Dis. 1999;58:691–7. doi: 10.1136/ard.58.11.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hata K, Kaibuchi K, Inagaki S, et al. Unc5B associates with LARG to mediate the action of repulsive guidance molecule. J Cell Biol. 2009;184:737–50. doi: 10.1083/jcb.200807029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Federico A, Pallante P, Bianco M, et al. Chromobox protein homologue 7 protein, with decreased expression in human carcinomas, positively regulates E-cadherin expression by interacting with the histone deacetylase 2 protein. Cancer Res. 2009;69:7079–87. doi: 10.1158/0008-5472.CAN-09-1542. [DOI] [PubMed] [Google Scholar]
- 25.Popescu LM. Telocytes and stem cells: a tandem in cardiac regenerative medicine. Curr Med Chem. 2011;18:94. [Google Scholar]
- 26.Popescu LM. The tandem: telocytes – stem cells. J Biol Biomed Eng. 2011 ; [in press. [Google Scholar]


