Abstract
Telocytes (TCs) are interstitial cells with telopodes – very long prolongations that establish intercellular contacts with various types of cells. Telocytes have been found in many organs and various species and have been characterized ultrastructurally, immunophenotypically and electrophysiologically (http://www.telocytes.com). Telocytes are distributed through organ stroma forming a three-dimensional network in close contacts with blood vessels, nerve bundles and cells of the local immune system. Moreover, it has been shown that TCs express a broad range of microRNAs, such as pro-angiogenic and stromal-specific miRs. In this study, the gene expression profile of murine lung TCs is compared with other differentiated interstitial cells (fibroblasts) and with stromal stem/progenitor cells. More than 2000 and 4000 genes were found up- or down-regulated, respectively, in TCs as compared with either MSCs or fibroblasts. Several components or regulators of the vascular basement membrane are highly expressed in TCs, such as Nidogen, Collagen type IV and Tissue Inhibitor of Metalloproteinase 3 (TIMP3). Given that TCs locate in close vicinity of small vessels and capillaries, the data suggest the implication of TCs in vascular branching. Telocytes express also matrix metalloproteases Mmp3 and Mmp10, and thus could regulate extracellular matrix during vascular branching and de novo vessel formation. In conclusion, our data show that TCs are not fibroblasts, as the ultrastructure, immunocytochemistry and microRNA assay previously indicated. Gene expression profile demonstrates that TCs are functionally distinct interstitial cells with specific roles in cell signalling, tissue remodelling and angiogenesis.
Keywords: telocytes, mesenchymal stem cells, fibroblasts, gene expression profile, interstitial cells, stroma, connective tissue, lung
Introduction
Recent electron microscopic studies have identified telocytes (TCs), a distinct type of interstitial cells, in many cavitary and non-cavitary organs 1–20. Telocytes are defined by their very long prolongations – called telopodes (Tps; generally, 2–3/cell; length of up to hundreds of μm) – which emerge from a relatively small cellular body. It has been shown that TCs form a 3D network through the organ interstitium surrounding organ-specific structures, blood capillaries, immune cells and nerve endings. As a specific functional property, TCs are key players in intercellular signalling, at both short and long distance. Thus, the long Tps establish direct contacts (junctions) with neighbouring cells and contribute to the (directional) transport of long-range signals driven by TCs 21. Local (paracrine) signalling of TCs is achieved by shedding vesicles 8, 20, 22.
The ultrastructural portrait of TCs was recently complemented with the immunophenotypical and electrophysiological characterization and the specific microRNA expression signature 20, 22, 23. However, the gene expression profile for this type of cells has not been reported yet. Prompted by these studies, we sought to compare murine lung TCs with mesenchymal stem cells (MSCs) and fibroblasts to identify the genes which are specifically regulated in TCs. We choose lung TCs as these are well-characterized ultrastructurally and immunohistochemically in situ and in vitro 4, 5, 11, 16, 17.
Method and Materials
Cell lines and tissue sampling
Mouse colonies were maintained in Animal Research Center of Fudan University, Shanghai, China. Lung samples were obtained from 20 to 25 g male BABL/c mice, 4–6 weeks of age. The mice were killed with an overdose of anaesthetic and the lung tissues were harvested for the isolation of TCs. The animal study was approved by the Ethic Committee for Animal Care and Use, Fudan University. Mesenchymal stem cells and fibroblast cell lines were obtained from Sciencell Research Laboratories (Cat. no. M7500-57, Carlsbad, CA, USA) and from Chinese Academy of Science (Cat. no. GNM28, Shanghai, China) respectively.
Isolation and primary culture of telocytes from lung tissues
Lung tissues were cut into small pieces and harvested under sterile conditions and collected into sterile tubes containing Dulbecco's Modified Eagle's Medium (DMEM, Gibco, NY, USA), supplemented with 100 UI/ml penicillin and 0.1 mg/ml streptomycin (Sigma Chemical, St. Louis, MO, USA), and the samples were brought to the cell culture room immediately. Samples were further rinsed with sterile DMEM and minced into fragments about 1 mm3, which were then incubated at 37°C for 4 hrs on an orbital shaker, with 1 mg/ml type II collagenase (Sigma-Aldrich, St. Louis, MO, USA) in PBS without Ca2+ and Mg2+. Dispersed cells were separated from non-digested tissue by the filtration through a 40-μm-diameter cell strainer (BD Falcon, Franklin, NJ, USA), harvested by centrifugation, and resuspended in DMEM supplemented with 10% foetal calf serum (Gibco, NY, USA), 100 UI/ml penicillin and 0.1 mg/ml streptomycin. Cell density was counted in a haemocytometer and viability was assessed using the Trypan blue. Cells were distributed in 25 cm2 culture flasks at a density of 1 × 105 cells/cm2 and maintained at 37°C in a humidified atmosphere (5% CO2) until becoming semiconfluent (usually 4 days after plating). Culture medium was changed every 48 hrs. Cultured cells were examined by phase contrast microscope, under an inverted Olympus phase contrast microscope (1 × 51).
RNA isolation and preparation
Mouse lung telocytes were isolated after 5 days of culture. Mouse MSCs and fibroblasts were cultured and collected on days 5 and 10 respectively. RNA preparation was performed using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) and the RNeasy kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions, including a DNase digestion treatment. The amount and quality of RNA were measured by NanoDrop-1000 spectrophotometer and with the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
RNA labelling, array hybridization and DNA microarray
The Mouse 4 × 44K Gene Expression Array (Agilent, Shanghai, China) with about 39,000+ mouse genes and transcripts represented with public domain annotations was applied for the analysis of gene profiles of mouse lung telocytes, MSCs and fibroblasts. Sample labelling and array hybridization were performed according to the protocol of One-Color Microarray-Based Gene Expression Analysis (Agilent Technology). Briefly, 1 μg of total RNA from each sample was linearly amplified and labelled with Cy3-dCTP. The labelled cRNAs were purified by RNAeasy Mini Kit (Qiagen). The concentration and specific activity of the labelled cRNAs (pmol Cy3/μg cRNA) were measured by NanoDrop ND-1000. One microgram of each labelled cRNA was fragmented by adding 11 μl 10 × Blocking Agent and 2.2 μl of 25 × Fragmentation Buffer, and heated at 60°C for 30 min. 55 μl 2 × GE Hybridization buffer was added to dilute the labelled cRNA. Hundred microlitre of hybridization solution was dispensed into the gasket slide and assembled to the gene expression microarray slide. The slides were incubated for 17 hrs at 65°C in an Agilent Hybridization Oven. The hybridized arrays were washed, fixed and scanned with the Agilent DNA Microarray Scanner (part number G2505B).
Data analysis
The acquired array images were analysed with Agilent Feature Extraction software (version 10.7.3.1). Quality normalization and subsequent data processing were performed with the GeneSpring GX v11.5.1 software package. The genes detected in all samples were chosen for further data analysis. Differentially expressed genes were identified through Fold Change filtering and hierarchically clustered by the Agilent GeneSpring GX software (version 11.5.1). Gene ontology and String Network analyses were performed with the standard enrichment computation method to study the relation among variant proteins expressed by variant genes. Fisher's exact test was used to find more overlaps between the descriptive list and the GO annotation list than would be expected by chance. The P-value denoted the significance of GO terms enrichment in the descriptive genes.
Results and discussions
The quality of gene data after filtering and the distribution of data sets were assessed and visualized by Box-Plot. There was no significant difference in distributions of log2 ratios among TCs, MSCs and fibroblasts (Figure S1).
Gene expression analysis
Gene expression array data show that more than 500 genes are at least 10 times higher expressed in TCs comparing with either MSCs or fibroblasts (Table 1). Several genes are found 100 times up-regulated in TCs versus fibroblasts (Cdh2, Cyba, Rnf128, Dpysl3, Fstl1, Rbp1, Gm12892, Cdh2, Aldh1a1, Gm5864) or MSCs (Rbp1 and Glipr1; Table 1A). Additional genes are significantly overexpressed in TCs comparing with MSCs or fibroblasts (Table 1B). Table 2 is a summary of genes found to be down-regulated in TCs. Although many genes are less expressed in TCs comparing with MSCs or fibroblasts, very few are found at least 100 times down-regulated in TCs. Table 2A and B show the genes with known functions that are found at least 30 times down-regulated specifically in TCs comparing with MSCs and fibroblasts.
Table 1.
Summary of genes expressed preferentially in TCs, as compared with mesenchymal stem cells (MSCs) and fibroblasts (Fbs)
| Compared pairs/fold up-regulated | >2 | >10 | >30 | >100 |
|---|---|---|---|---|
| TCs vs. MSCs | 2921 | 500 | 174 | 44 |
| TCs vs. Fbs | 3173 | 661 | 295 | 85 |
| (A) Genes up-regulated more than 100-folds in telocytes (TCs) as compared with mesenchymal stem cells (MSCs) and fibroblasts (Fbs) | |||||
|---|---|---|---|---|---|
| TCs vs. Fbs | TCs vs. MSCs | ||||
| Gene | Folds | Gene | Folds | Gene | Folds |
| Ctgf | 6151 | Tm4sf1 | 217 | Sprr1a | 2971 |
| Sprr1a | 2593 | Sulf1 | 212 | Cck | 1242 |
| Myl9 | 1668 | Chi3l3 | 204 | Wfdc2 | 551 |
| Tagln | 1545 | Vopp1 | 198 | Serinc2 | 527 |
| Cck | 1206 | Mfhas1 | 198 | Chi3l3 | 369 |
| Nid1 | 1143 | Myh14 | 194 | Glipr1 | 355 |
| Sdpr | 1004 | Ogn | 185 | Eppk1 | 284 |
| Crlf1 | 942 | Dsp | 182 | Trf | 259 |
| Anxa8 | 799 | Mmp10 | 177 | Myh14 | 246 |
| Cd9 | 718 | Khdrbs3 | 175 | Gsta3 | 244 |
| Wfdc2 | 660 | Atp1b1 | 174 | Gpr56 | 222 |
| Sox4 | 501 | Papss2 | 171 | Cyb561 | 210 |
| Dhcr24 | 496 | Gprc5c | 168 | Gprc5c | 204 |
| Timp3 | 445 | Prl2c1 | 165 | Tjp2 | 202 |
| Trim44 | 410 | Gas6 | 165 | Atp1b1 | 194 |
| Serpine1 | 376 | Rbp1 | 161 | Lyz1 | 181 |
| Marcksl1 | 356 | Foxq1 | 156 | Aldh1a2 | 167 |
| Hs6st2 | 335 | Cblc | 149 | Gpx2 | 152 |
| Gpr56 | 331 | Aldh1a2 | 149 | Dsp | 150 |
| Nrg1 | 327 | Cdh2 | 136 | Khdrbs3 | 146 |
| Trf | 306 | Crct1 | 133 | Acp5 | 143 |
| Bmp4 | 298 | Mmp3 | 131 | Rbp1 | 141 |
| Cyba | 293 | Gpx2 | 126 | Gprc5c | 137 |
| Thy1 | 280 | Gprc5c | 125 | Clu | 131 |
| Lrrc32 | 278 | Fstl1 | 125 | Tmc4 | 128 |
| Rab34 | 269 | Lama2 | 120 | Acp5 | 114 |
| Dpysl3 | 263 | Tjp2 | 117 | Epb4.1l4b | 114 |
| Decr1 | 256 | Igsf9 | 116 | Mfsd6 | 109 |
| Gsta3 | 240 | Bcr | 110 | Cblc | 107 |
| Evl | 237 | Lce1i | 108 | Acta1 | 105 |
| Tmem45a | 233 | Rnf128 | 107 | F11r | 101 |
| Aldh1a1 | 225 | Klhl13 | 106 | ||
| Fzd1 | 223 | Echdc2 | 103 | ||
| Cryab | 219 | Trim16 | 101 | ||
| Lyz1 | 217 | ||||
| (B) Genes up-regulated between 30- and 100-folds in telocytes (TCs) as compared with mesenchymal stem cells (MSCs) and fibroblasts (Fbs) | |||||||
|---|---|---|---|---|---|---|---|
| TCs vs. Fbs | TCs vs. MSCs | ||||||
| Gene | Folds | Gene | Folds | Gene | Folds | Gene | Folds |
| Wnt11 | 100 | Letmd1 | 47 | Pdgfb | 97 | Pcgf5 | 36 |
| F3 | 98 | Rpgrip1 | 46 | Aldh1a1 | 93 | Fxyd3 | 36 |
| Pdgfb | 97 | Trp53i11 | 46 | Itpa | 90 | Ctsk | 35 |
| Fxyd6 | 94 | Hebp2 | 46 | Fxyd6 | 87 | Ctgf | 35 |
| Fhl2 | 94 | Dkk3 | 45 | Tns1 | 79 | Ckb | 35 |
| Nox4 | 93 | Cryab | 45 | St14 | 78 | Lama5 | 35 |
| Ptprf | 93 | Pvrl3 | 44 | Lce1i | 78 | Evpl | 34 |
| Tgfb1i1 | 93 | P2rx2 | 44 | Crip1 | 77 | Col4a6 | 34 |
| Ddah1 | 92 | A2bp1 | 43 | S100a16 | 76 | Chst4 | 34 |
| Cd99 | 92 | Cyba | 43 | Klhl13 | 74 | Apoe | 33 |
| Irx1 | 87 | Cyr61 | 42 | Tnk1 | 74 | Pik3r6 | 33 |
| Pdlim1 | 86 | Cobl | 42 | Mmrn2 | 74 | Panx1 | 33 |
| Epb4.1l3 | 86 | Pdlim3 | 41 | Rpgrip1 | 72 | Rnu1b6 | 33 |
| Tuft1 | 86 | Map3k9 | 41 | Gsta3 | 71 | Nppb | 33 |
| Msln | 83 | Tlr13 | 41 | Endod1 | 71 | Sema6a | 33 |
| Panx1 | 83 | Tjp3 | 41 | Scnn1a | 69 | Serpinb6b | 33 |
| Clic5 | 83 | Grhl2 | 41 | Tacstd2 | 69 | Apoc2 | 32 |
| Ggh | 83 | Sdcbp2 | 41 | Mboat1 | 68 | Vill | 32 |
| Bst1 | 79 | Cd14 | 41 | Gas6 | 67 | Irx1 | 31 |
| Mansc1 | 79 | Krt17 | 41 | Dapk2 | 66 | Isyna1 | 30 |
| Slco3a1 | 78 | Loxl2 | 40 | Cpsf3l | 65 | Map3k9 | 30 |
| Tnfsf15 | 78 | Cald1 | 40 | Plac9 | 64 | ||
| Il6 | 78 | Brsk1 | 40 | Krtcap3 | 63 | ||
| Saa3 | 77 | Ppp1r9a | 40 | Mapkapk3 | 62 | ||
| Fgd3 | 77 | Stxbp2 | 39 | Tbc1d2 | 62 | ||
| Echdc2 | 77 | Rab25 | 39 | Tbc1d2 | 61 | ||
| Mapk13 | 75 | Stfa3 | 39 | Cytip | 60 | ||
| Tnfrsf11b | 75 | Cald1 | 39 | Spint1 | 60 | ||
| Basp1 | 70 | Brsk1 | 39 | Lcp1 | 60 | ||
| Slc4a11 | 70 | Lmo7 | 38 | Grhl2 | 59 | ||
| Bst1 | 69 | Timp1 | 38 | Wnt11 | 59 | ||
| F3 | 69 | Slc35f5 | 38 | Rarb | 57 | ||
| Ubqln2 | 69 | Id1 | 38 | Ctsh | 57 | ||
| Adam8 | 68 | Rnf130 | 37 | Mansc1 | 56 | ||
| Parp8 | 67 | Serping1 | 37 | Mmp10 | 56 | ||
| Sox4 | 67 | Csf2rb | 37 | Ephx1 | 55 | ||
| Egfl7 | 66 | Olfr1383 | 37 | Coro1a | 55 | ||
| Gsta3 | 64 | Sulf2 | 37 | Rpgrip1 | 53 | ||
| Tnk1 | 64 | Nhsl1 | 37 | Cd36 | 53 | ||
| Fzd2 | 64 | Itm2a | 37 | Klf6 | 52 | ||
| Gpm6b | 63 | Slamf9 | 37 | Heph | 52 | ||
| Cgn | 62 | Cacnb3 | 36 | Nipsnap1 | 50 | ||
| Unc13b | 61 | Spint1 | 36 | Arhgef16 | 50 | ||
| Celsr1 | 61 | Tuba1a | 36 | Atp9a | 50 | ||
| Mmrn2 | 61 | Rgs17 | 36 | Bst1 | 49 | ||
| Dok2 | 61 | Col4a6 | 36 | Adm | 49 | ||
| Tpm2 | 60 | Tpm1 | 36 | Elovl7 | 49 | ||
| Ppfibp2 | 60 | Scnn1a | 35 | Fcgr2b | 49 | ||
| Npr3 | 60 | Sirpb1a | 35 | Tjp3 | 48 | ||
| Cpsf3l | 59 | Clic3 | 35 | Hic1 | 48 | ||
| Peg13 | 59 | Klf13 | 35 | Rab25 | 47 | ||
| Arhgef16 | 59 | Lrrc33 | 35 | Serpine1 | 47 | ||
| Lass3 | 58 | Gprc5a | 35 | Abcc3 | 47 | ||
| Dapk2 | 58 | Sgk1 | 35 | Psmg2 | 47 | ||
| Plac9 | 58 | Ankrd1 | 34 | Col4a4 | 46 | ||
| Msrb2 | 58 | Mid1ip1 | 34 | Csf2rb2 | 45 | ||
| Ckb | 57 | Coro1a | 34 | Tmem88 | 45 | ||
| Fam83h | 57 | Cd248 | 34 | Cd97 | 45 | ||
| Vcan | 56 | Acta1 | 34 | Ppl | 45 | ||
| Acp5 | 56 | Inadl | 33 | P2rx2 | 44 | ||
| Csf1r | 56 | Sesn3 | 33 | A2bp1 | 43 | ||
| Ap1s3 | 56 | Evpl | 33 | Akr1c13 | 43 | ||
| Pbx3 | 56 | C3 | 33 | St6gal1 | 42 | ||
| Tmc4 | 56 | Tpm2 | 33 | Efnb1 | 41 | ||
| Rpgrip1 | 55 | Pilra | 33 | Dok2 | 41 | ||
| Ctsw | 55 | H19 | 33 | Adam8 | 41 | ||
| Wwc1 | 54 | Pfkfb3 | 32 | Clic5 | 41 | ||
| Glipr1 | 54 | Zfhx3 | 32 | Sh3bgr | 40 | ||
| Hes6 | 54 | Fcer1g | 32 | Fgd3 | 39 | ||
| Tacstd2 | 54 | Stab 1 | 32 | Csf2rb | 39 | ||
| Nsd1 | 54 | Col1a2 | 32 | Olfr1383 | 39 | ||
| Cyb561 | 53 | Igfbp2 | 31 | H19 | 39 | ||
| Fcgr2b | 53 | Vcam1 | 31 | Sirpb1a | 39 | ||
| Cdc42ep5 | 53 | Chpf2 | 31 | Fcer1g | 38 | ||
| Mdfi | 52 | Nppb | 31 | Slc39a4 | 38 | ||
| Galntl4 | 52 | Ccl27a | 31 | Fcgr4 | 38 | ||
| Anxa8 | 52 | Ccl2 | 31 | Sh3bgr | 38 | ||
| Plcg2 | 52 | Tnfaip3 | 31 | Slc22a18 | 38 | ||
| Col4a4 | 51 | Fnbp1l | 31 | Alcam | 38 | ||
| Acp5 | 50 | Marveld3 | 31 | Stfa3 | 38 | ||
| Btg3 | 49 | Spint2 | 30 | Ppfibp2 | 37 | ||
| Ltbp2 | 48 | Sh3bgr | 30 | Clic3 | 37 | ||
| Cd93 | 47 | Adamts9 | 30 | Csf1r | 37 | ||
| Gadd45b | 47 | Abcc3 | 30 | Spint2 | 36 | ||
| Afap1l2 | 47 | Lcp1 | 30 | Lamc2 | 36 | ||
Table 2.
Summary of genes less expressed in TCs, as compared with mesenchymal stem cells (MSCs) and fibroblasts (Fbs)
| Compared pairs/fold down-regulated | >2 | >10 | >30 | >100 |
|---|---|---|---|---|
| TCs vs. MSCs | 4365 | 175 | 32 | 5 |
| TCs vs. Fbs | 5451 | 326 | 63 | 16 |
| (A) Genes down-regulated more than 100-folds in telocytes (TCs) as compared with mesenchymal stem cells (MSCs) and fibroblasts (Fbs) | ||||
|---|---|---|---|---|
| TCs vs. Fbs | TCs vs. MSCs | |||
| Gene | Folds | Gene | Folds | |
| Car6 | 323 | Ccl5 | 282 | |
| Odz4 | 275 | Hoxc6 | 146 | |
| Tenm4 | 269 | Cdsn | 159 | |
| Pla2g2e | 253 | Ifi203 | 63 | |
| Cdsn | 229 | Gdpd2 | 85 | |
| Glod5 | 209 | |||
| Rarres2 | 180 | |||
| Hoxc6 | 152 | |||
| Ndufa4l2 | 150 | |||
| Hoxc10 | 133 | |||
| Rhd | 122 | |||
| Plin4 | 113 | |||
| Gm2022 | 105 | |||
| Car9 | 102 | |||
| (B) Genes down-regulated between 30- and 100-folds in telocytes (TCs) as compared with mesenchymal stem cells (MSCs) and fibroblasts (Fbs) | |||||
|---|---|---|---|---|---|
| TCs vs. Fbs | TCs vs. MSCs | ||||
| Gene | Folds | Gene | Folds | Gene | Folds |
| Serpinb9f | 95 | Tbx15 | 44 | Tbx15 | 93 |
| Foxg1 | 94 | Dmrtc1c2 | 42 | Hoxc10 | 92 |
| Mst1 | 88 | Igf2bp3 | 41 | Nkx2-5 | 84 |
| Ifi203 | 82 | Itk | 41 | Gbp3 | 72 |
| Avil | 75 | Paip1 | 38 | Lpar4 | 67 |
| Hsd17b14 | 69 | Rps3a | 38 | Hoxb9 | 66 |
| Acacb | 68 | Slx | 37 | Odz4 | 58 |
| Angpt1 | 67 | Gchfr | 35 | Eif2s1 | 58 |
| Csprs | 67 | Hc | 35 | Pde8b | 54 |
| Gm4951 | 67 | Ptgir | 33 | Ebf3 | 46 |
| Mtap1b | 65 | Accn2 | 32 | Angpt1 | 46 |
| Serpinb9e | 59 | Masp2 | 32 | Rsad2 | 45 |
| Cox6a2 | 59 | Cbr2 | 31 | Ifi202b | 45 |
| Matn2 | 57 | Col5a3 | 30 | Fbln1 | 37 |
| Pla2g2e | 54 | Ifi204 | 35 | ||
| Nrxn3 | 49 | Thbs2 | 35 | ||
| Cbr2 | 49 | Mx2 | 34 | ||
| Ebf3 | 48 | Ndufa4l2 | 34 | ||
| Cldn15 | 47 | Tgfbr3 | 31 | ||
| Ppargc1a | 45 | Car6 | 31 | ||
Hierarchical cluster and gene ontology analyses
The hierarchical cluster of the genes with more than twofold changes among telocytes, MSCs and fibroblasts is shown in Figure 1. Remarkably, the MCSs and fibroblast gene expression profiles relate each other to higher extent than to TCs supporting the view that TCs have a distinct gene expression pattern. In fact this is an important additional proof that TCs and fibroblasts are different cells. The GO analysis indicates that the genes differentially expressed in TCs are mainly involved in development, in tissue and organ morphogenesis and in transport and maintenance of a biological compound to a specific location (Fig. 2A). In addition, many of the differentially expressed genes likely function in extracellular compartments (Fig. 2B) and may play roles in cell survival, growth and differentiation through autocrine and paracrine activity (Fig. 2C). The relationships, including direct (physical) and indirect (functional) associations, of those genes were analysed by String Network analysis (http://www.string-db.org). Among the 156 co-expressed genes, 46 genes were found to have certain interactions (Fig. 3).
Fig. 1.
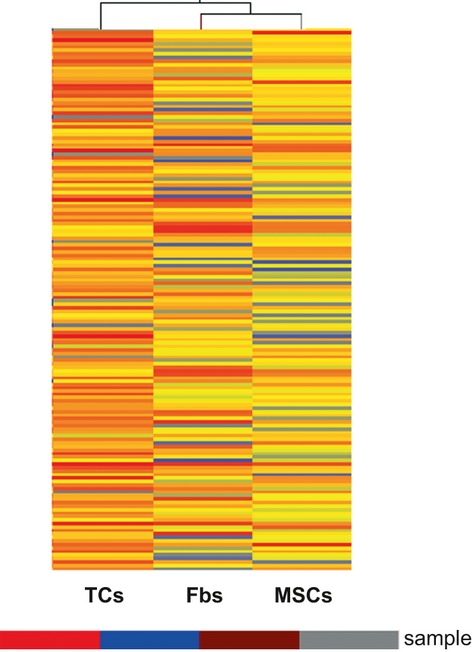
Hierarchical cluster analysis of the differentially expressed genes among telocytes (TCs), mesenchymal stem cells (MSCs) and fibroblasts (Fbs).
Fig. 2.

Gene ontology of the genes with at least twofolds difference among telocytes (TCs), mesenchymal stem cells (MSCs) and fibroblast (Fbs), analysed under following categories: Biological Processes (A), Cellular Components (B) and Molecular Function (C). (P ≤ 0.01).
Fig. 3.
String Network of the proteins that are differentially expressed among telocytes (TCs), mesenchymal stem cells (MSCs) and fibroblast (Fbs). A group of 46 genes are found connected functionally. Strong associations are represented by thick lines.
TCs are potentially involved in tissue remodelling and basement membrane homeostasis
A set of genes are specifically up- or down-regulated in TCs comparing with both fibroblasts and MSC (Table 3). As last two cell types are developmentally and functionally quite different, one being progenitors and the other differentiated, specialized cells, this set of genes should connect to the specific biological activities of TCs among the other stromal cells. Thus, we have found that several genes with roles in tissue remodelling and repair are significantly up-regulated in TCs (Tables 1A and 3): connective tissue growth factor (CTGF) 24, 25, Transgelin (Tagln) 26, Nidogen 1 (Nid1) 27, 28, tissue inhibitor of metalloproteinase 3 (TIMP3) 29, collagen type IV, alpha (Col4a4, Col4a6, Col4a5) 28, 30, Matrix Metallopeptidase 10 (Mmp10) 31–33, Matrix Metallopeptidase 3 (Mmp3) 31–33 and Retinol-binding protein 1 (RBP1). RBP1 (also known as CRABP-I, CRBP, CRBP1, CRBPI, RBPC) is required in tissue remodelling 34. Regarding the molecular mechanisms, RBP1 delivers vitamin A to other cells through the plasma membrane protein STRA6 involved in JAK/STAT signalling and the intracellular metabolism of the vitamin 35. Remarkably, two main components of basement membrane, Collagen type IV and Nidogen 1 are up-regulated in the cultured TCs comparing with both MSCs and fibroblasts. Moreover, TIMP3 is an extracellular matrix-anchored metalloproteinase inhibitor that acts specifically to increase vascular (endothelial) basement membrane stability 36, 37. As TCs express Matrix Metalloproteases Mmp3 and Mmp10 also, it is likely that TCs are involved in both basement membrane assembly (stability) and surrounding extracellular matrix remodelling.
Table 3.
Genes up- or down-regulated in telocytes (TCs) relative to both mesenchymal stem cells (MSCs) and fibroblasts (Fbs)
| TCs vs. Fbs | TCs vs. MSCs | |||
|---|---|---|---|---|
| Gene name | Fold change | Reg | Fold change | Reg |
| Ctgf | 6150 | Up | 35 | Up |
| Mmp10 | 177 | Up | 56 | Up |
| Mmp3 | 131 | Up | 25 | Up |
| Col4a4 | 46 | Up | 51 | Up |
| Col4a6 | 34 | Up | 36 | Up |
| Col4a5 | 8 | Up | 32 | Up |
| Unc13b | 61 | Up | 7 | Up |
| Mapk13 | 75 | Up | 13 | Up |
| Igsf9 | 115 | Up | 3 | Up |
| Glipr1 | 54 | Up | 355 | Up |
| Clic5 | 83 | Up | 41 | Up |
| Myh14 | 194 | Up | 245 | Up |
| Aldh1a1 | 225 | Up | 92 | Up |
| Aldh1a2 | 148 | Up | 167 | Up |
| Rbp1 | 161 | Up | 141 | Up |
| Gprc5c | 125 | Up | 136 | Up |
| Gsta3 | 64 | Up | 70 | Up |
| Plac9 | 57 | Up | 63 | Up |
| Fgd3 | 77 | Up | 39 | Up |
| Dok2 | 60 | Up | 41 | Up |
| Scnn1a | 35 | Up | 68 | Up |
| Car6 | 323 | Down | 31 | Down |
| Odz4 | 275 | Down | 59 | Down |
| Oz/ten-m | 269 | Down | 56 | Down |
| Cdsn | 229 | Down | 153 | Down |
| Hoxc6 | 152 | Down | 207 | Down |
| Ifi203 | 82 | Down | 150 | Down |
Concluding remarks
Overall, the data indicate that TCs are clearly distinct from both MSCs and fibroblasts, and the gene signature of TCs suggests specific biological functions in (a) development and tissue morphogenesis, (b) biological compound transport and (c) extracellular matrix remodelling. It has been proposed that TCs play essential roles in angiogenesis given that TCs are frequently found in close vicinity of small vessels and express angiogenesis-related factors (VEGF, NO) and pro-angiogenic microRNAs 22. The data presented here bring additional support to this view suggesting that TCs may also regulate vascular basement membrane remodelling as key step in vascular branching and de novo vessel formation.
Acknowledgments
The authors would like to thank Hongjian Gao, Department of Electronic Microscopy, Shanghai Medical College, Fudan University, for the technical assistance in TEM; Biomedical Research Center of Fudan University Zhongshan Hospital for technical supports and facility supplies. The work was supported by Shanghai Leading Academic Discipline Project (Project Number: B115), Fudan University (Distinguished Professor Grant), Shanghai Science & Technology Committee Grants for International Collaboration (11410708600), Project of Science and Technology Innovation Plan in Biomedicine, National Natural Science foundation of China (H0108) and National Natural Key Science foundation of China: ‘Lung injury of ischemic reperfusion’ (30930090). This study is partially supported by the Sectorial Operational Programme Human Resources Development (SOPHRD), financed from the European Social Fund and by the Romanian Government under the contract number POSDRU/89/1.5/S/64153 (to V.B.C) and by grant 350/2012 PN-II-ID-PCE-2011-3-0134 of the Romanian National Authority for Scientific Research, CNCS – UEFISCDI (to L.M.P).
Conflict of interest
The authors confirm that there are no conflicts of interest.
Supporting information
Additional Supporting Information may be found in the online version of this article:
Figure S1 Box-Plot of Quality assessment of gene data after filtering. After normalization, the distributions of log2 ratios among all samples are nearly the same.
References
- 1.Popescu LM, Faussone-Pellegrini MS. TELOCYTES - a case of serendipity: the winding way from Interstitial Cells of Cajal (ICC), via Interstitial Cajal-Like Cells (ICLC) to TELOCYTES. J Cell Mol Med. 2010;14:729–40. doi: 10.1111/j.1582-4934.2010.01059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gherghiceanu M, Popescu LM. Cardiomyocyte precursors and telocytes in epicardial stem cell niche: electron microscope images. J Cell Mol Med. 2010;14:871–7. doi: 10.1111/j.1582-4934.2010.01060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suciu L, Popescu LM, Gherghiceanu M, et al. Telocytes in human term placenta: morphology and phenotype. Cells Tissues Organs. 2010;192:325–339. doi: 10.1159/000319467. [DOI] [PubMed] [Google Scholar]
- 4.Popescu LM, Gherghiceanu M, Suciu LC, et al. Telocytes and putative stem cells in the lungs: electron microscopy, electron tomography and laser scanning microscopy. Cell Tissue Res. 2011;345:391–403. doi: 10.1007/s00441-011-1229-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinescu ME, Gherghiceanu M, Suciu L, et al. Telocytes in pleura: two- and three-dimensional imaging by transmission electron microscopy. Cell Tissue Res. 2011;343:389–397. doi: 10.1007/s00441-010-1095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Popescu LM, Manole E, Serboiu CS, et al. Identification of telocytes in skeletal muscle interstitium: implication for muscle regeneration. J Cell Mol Med. 2011;15:1379–1392. doi: 10.1111/j.1582-4934.2011.01330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gevaert T, De Vos R, Van Der Aa F, et al. Identification of telocytes in the upper lamina propria of the human urinary tract. J Cell Mol Med. 2011;16:2085–93. doi: 10.1111/j.1582-4934.2011.01504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicolescu MI, Popescu LM. Telocytes in the interstitium of human exocrine pancreas: ultrastructural evidence. Pancreas. 2012;41:949–56. doi: 10.1097/MPA.0b013e31823fbded. [DOI] [PubMed] [Google Scholar]
- 9.Cretoiu D, Cretoiu SM, Simionescu AA, et al. Telocytes, a distinct type of cell among the stromal cells present in the lamina propria of jejunum. Histol Histopathol. 2012;27:1067–1078. doi: 10.14670/HH-27.1067. [DOI] [PubMed] [Google Scholar]
- 10.Cantarero I, Luesma MJ, Junquera C. The primary cilium of telocytes in the vasculature: electron microscope imaging. J Cell Mol Med. 2011;15:2594–2600. doi: 10.1111/j.1582-4934.2011.01312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng Y, Manole CG, Bai C, et al. Telocytes in trachea and lungs. J Cell Mol Med. 2011;15:2262–2268. doi: 10.1111/j.1582-4934.2011.01404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rusu MC, Nicolescu MI, Jianu AM, et al. Esophageal telocytes and hybrid morphologies. Cell Biol Int. 2012;36:1079–88. doi: 10.1042/CBI20120007. [DOI] [PubMed] [Google Scholar]
- 13.Popescu BO, Gherghiceanu M, Kostin S, et al. Telocytes in meninges and choroid plexus. Neurosci Lett. 2012;516:265–269. doi: 10.1016/j.neulet.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 14.Nicolescu MI, Bucur A, Dinca O, et al. Telocytes in parotid glands. Anat Rec. 2012;295:378–385. doi: 10.1002/ar.21540. [DOI] [PubMed] [Google Scholar]
- 15.Ceafalan L, Gherghiceanu M, Popescu LM, et al. Telocytes in human skin; are they involved in skin regeneration. J Cell Mol Med. 2012;16:1405–20. doi: 10.1111/j.1582-4934.2012.01580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng Y, Bai C, Wang X. Potential significance of telocytes in the pathogenesis of lung diseases. Expert Rev Respir Med. 2012;6:45–9. doi: 10.1586/ers.11.91. [DOI] [PubMed] [Google Scholar]
- 17.Zheng Y, Bai C, Wang X. Telocyte morphologies and potential roles in diseases. J Cell Physiol. 2012;227:2311–7. doi: 10.1002/jcp.23022. [DOI] [PubMed] [Google Scholar]
- 18.Cretoiu SM, Cretoiu D, Popescu LM. Human myometrium - the ultrastructural 3D network of telocytes. J Cell Mol Med. 2012;16:2844–9. doi: 10.1111/j.1582-4934.2012.01651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cretoiu SM, Cretoiu D, Marin A, et al. Telocytes: ultrastructural, immunohistochemical and electrophysiological characteristics in human myometrium. Reproduction. 2013 doi: 10.1530/REP-12-0369. doi: 10.1530/REP-12-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gherghiceanu M, Popescu LM. Cardiac telocytes - their junctions and functional implications. Cell Tissue Res. 2012;348:265–79. doi: 10.1007/s00441-012-1333-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Popescu LM, Nicolescu MI. Telocytes and stem cells. In: Goldenberg RCdS, Campos de Carvalho AC., editors. Resident stem cells and regenerative therapy. Oxford: Academic Press/Elsevier; 2012. pp. 205–31. [Google Scholar]
- 22.Manole CG, Cismasiu V, Gherghiceanu M, et al. Experimental acute myocardial infarction: telocytes involvement in neo-angiogenesis. J Cell Mol Med. 2012;15:2284–96. doi: 10.1111/j.1582-4934.2011.01449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cismasiu VB, Radu E, Popescu LM. MiR-193 expression differentiates telocytes from other stromal cells. J Cell Mol Med. 2011;15:1071–4. doi: 10.1111/j.1582-4934.2011.01325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sonnylal S, Shi-Wen X, Leoni P, et al. Selective expression of connective tissue growth factor in fibroblasts in vivo promotes systemic tissue fibrosis. Arthritis Rheum. 2010;62:1523–32. doi: 10.1002/art.27382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lipson KE, Wong C, Teng Y, et al. CTGF is a central mediator of tissue remodeling and fibrosis and its inhibition can reverse the process of fibrosis. Fibrogenesis Tissue Repair. 2012;5:S24. doi: 10.1186/1755-1536-5-S1-S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nair RR, Solway J, Boyd DD. Expression cloning identifies transgelin (SM22) as a novel repressor of 92-kD type IV collagenase (MMP-9) expression. J Biol Chem. 2006;281:26424–36. doi: 10.1074/jbc.M602703200. [DOI] [PubMed] [Google Scholar]
- 27.Marionnet C, Pierrard C, Vioux- Chagnoleau C, et al. Interactions between fibroblasts and keratinocytes in morphogenesis of dermal epidermal junction in a model of reconstructed skin. J Invest Dermatol. 2006;126:971–9. doi: 10.1038/sj.jid.5700230. [DOI] [PubMed] [Google Scholar]
- 28.Stratman AN, Malotte KM, Mahan RD, et al. Pericyte recruitment during vasculogenic tube assembly stimulates endothelial basement membrane matrix formation. Blood. 2009;114:5091–101. doi: 10.1182/blood-2009-05-222364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Limana F, Esposito G, D'Arcangelo D, et al. HMGB1 attenuates cardiac remodeling in the failing heart via enhanced cardiac regeneration and miR-206-mediated inhibition of TIMP-3. PLoS ONE. 2011;6:e19845. doi: 10.1371/journal.pone.0019845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abreu-Velez AM, Howard MS. Collagen IV in normal skin and in pathological processes. N Am J Med Sci. 2012;4:1–8. doi: 10.4103/1947-2714.92892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Girard MT, Matsubara M, Kublin C, et al. Stromal fibroblasts synthesize collagenase and stromelysin during long-term tissue remodeling. J Cell Sci. 1993;104:1001–11. doi: 10.1242/jcs.104.4.1001. [DOI] [PubMed] [Google Scholar]
- 32.Rodgers WH, Matrisian LM, Giudice LC, et al. Patterns of matrix metalloproteinase expression in cycling endometrium imply differential functions and regulation by steroid hormones. J Clin Invest. 1994;94:946–53. doi: 10.1172/JCI117461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner NA, Warburton P, O'Regan DJ, et al. Modulatory effect of interleukin-1α on expression of structural matrix proteins, MMPs and TIMPs in human cardiac myofibroblasts: role of p38 MAP kinase. Matrix Biol. 2010;29:613–20. doi: 10.1016/j.matbio.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu M, Ishibashi-Ueda H, Ohta-Ogo K, et al. Transient expression of cellular retinol-binding protein-1 during cardiac repair after myocardial infarction. Pathol Int. 2012;62:246–53. doi: 10.1111/j.1440-1827.2012.02802.x. [DOI] [PubMed] [Google Scholar]
- 35.Berry DC, O'Byrne SM, Vreeland AC, et al. Cross talk between signaling and vitamin A transport by the retinol-binding protein receptor STRA6. Mol Cell Biol. 2012;32:3164–75. doi: 10.1128/MCB.00505-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baker AH, Edwards DR, Murphy G. Metalloproteinase inhibitors: biological actions and therapeutic opportunities. J Cell Sci. 2002;115:3719–27. doi: 10.1242/jcs.00063. [DOI] [PubMed] [Google Scholar]
- 37.Ma DH, Chen JI, Zhang F, et al. Inhibition of fibroblast-induced angiogenic phenotype of cultured endothelial cells by the overexpression of tissue inhibitor of metalloproteinase (TIMP)-3. J Biomed Sci. 2003;10:526–34. doi: 10.1007/BF02256114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



