Abstract
The purpose of this study was to investigate how human umbilical cord mesenchymal stem cells (HUMSCs) affect breast cancer tumourigenesis. To observe the influence of HUMSCs on tumourigenesis in vitro, we performed a co-culture of MDA MB-231 breast cancer cells with HUMSCs, and a result of HUMSCs on tumourigenesis in vivo was achieved by injection of HUMSCs into nonobese diabetic/severe combined immunodeficient mice following tumour establishment with MDA-MB231. During the co-culture, apoptosis of MDA-MB231 was noted, which was driven either by binding with HUMSC through direct cell–cell contact or by formation of a novel cell-in-cell phenomenon after internalization of HUMSC. Also, treatment with HUMSC injection was efficacious in both in situ and metastatic breast cancers in the animal models. Since HUMSCs were proved to efficaciously suppress breast cancer tumourigenesis both in vitro and in vivo, it is our expectation that treatment with HUMSCs can be a viable therapy for breast cancer in the near future. In addition, we share a new point of view on the role of HUMSCs in foetal development during pregnancy.
Keywords: human umbilical cord mesenchymal stem cell, breast cancer, cell-in-cell
Introduction
Mesenchymal stem cells (MSCs) have been harvested from bone marrow of the human foetus or from adults. Recently, MSCs have also been isolated from several compartments of the umbilical cord. Specifically, the MSCs have been isolated from umbilical cord blood, umbilical vein subendothelium and the Wharton’s jelly (the connective tissue surrounding the umbilical vessels that includes the perivascular zone, the intervascular zone and the subamnion zone). Like other MSCs, human umbilical cord mesenchymal stem cells (HUMSCs), derived from Wharton’s jelly, are characterized by their self-renewal and multipotency [1-3]. They are able to support the stem cell niche [4] and synthesize various cytokines [4], and they possess the properties of immunomodulation [5] and homing [6]. Studies regarding the immunophenotype of HUMSCs revealed that they are stained positively for markers of the mesenchymal cells and negatively for markers of the haematopoietic lineage. HUMSCs express CD73, CD90, CD105, CD10, CD13, CD29 and CD44 [1, 4, 7–9], and are negative for CD14, CD31 CD33, CD34, CD45 and CD56 [1, 8].
HUMSCs are earlier-stage cells than MSCs derived from adult fat or bone marrow [1, 10]. HUMSCs can differentiate into a variety of specialized cell types in vitro, including bone, cartilage, adipose, muscle, neural cells and pancreatic β-cells [1-3]. When compared with human bone marrow mesenchymal stem cells (HBMSCs), HUMSCs have greater expansion capability and faster growth in vitro. While HUMSCs have higher potency in forming chondrogenic and osteogenic cells, they are less successful in forming adipogenic cells [11]. In addition, it has been reported that HUMSCs produced cytokines similar to those of HBMSCs, and unlike HBMSCs, HUMSCs synthesized granulocyte macrophage colony stimulating factors and granulocyte colony stimulating factors [4].
The interactions between these MSCs and cells of other types have not been thoroughly studied yet. It has been documented that transfer of mitochondria from HBMSCs repaired A549 p° cells with non-functional mitochondria during co-culture [12]. In our previous study, HUMSCs were induced to differentiate into mature islet-like cell clusters and then transplanted into the liver of streptozotocin-induced diabetic rats. In addition to the existence of islet-like cell clusters in the liver, we noted some special liver cells characterized by human insulin and nuclei-positive staining that possessed secretory granules [3]. A cell-to-cell fusion was suspected, however, in need of further study.
Generally, HBMSCs are considered to enhance tumourigenesis since they are a source of carcinoma-associated fibroblasts [13], they help to establish a microenvironment contributory to tumour growth, and they increase the metastatic potency of breast cancer cells [14]. However, controversies over the role of HBMSCs in tumourigenesis still exist [15]. As for the role of HUMSCs in tumourigenesis, it is still an open question. In this study, we aimed to investigate how HUMSCs affect breast cancer tumourigenesis. To our surprise, we found that, during co-culture with HUMSCs, apoptosis of MDA-MB231 breast cancer cells occurred either after binding with HUMSC through direct cell–cell contact or after forming of a novel cell-in-cell phenomenon following internalization of HUMSC. Also, treatment with HUMSC injection was fairly efficacious in the animal models of both in situ and metastatic breast cancers. Since the ability of HUMSCs to suppress breast cancer tumourigenesis was proved, it is our expectation that HUMSCs can provide new treatment for breast cancer in the near future.
Materials and methods
Cell culture
HUMSCs and HBMSCs were purchased from PromoCell GmbH (Heidelberg, Germany). PromoCell HUMSCs were harvested from normal human umbilical cord matrix (Wharton’s jelly), while PromoCell HBMSCs were harvested from normal human bone marrow. The cells have been tested for their ability to differentiate in vitro into adipocytes, chondrocytes and osteoblasts, and have been tested: CD44/CD105 > 95% positive, CD31/CD45 > 95% negative. These HUMSCs and HBMSCs were cultured in mesenchymal stem cell growth medium (PromoCell) supplemented with penicillin-streptomycin. Cell passages 4–10 were used for the experiments. MDA-MB-231 breast cancer cells, obtained from ATCC, were cultured in Leibovitz’s L-15 Medium with 10% foetal bovine serum added and were incubated at 37°C without CO2. HS68 cells and SK-Hep1 cells were cultured in DMEM with 10% foetal bovine serum. WI38 cells were cultured in Eagle’s minimum essential medium with 10% foetal bovine serum.
Preparation of HUMSC-Luc-GFP (HUMSC-LG) and MDA-MB231-Luc-GFP (MDA-MB231-LG)
The vectors were constructed using standard cloning procedures. The pWPI was provided Addgene (http://www.addgene.org/pgvec1). To construct pWPI-Luc-GFP, the Luc cassette was excised from pGL3 basic (Promega, Madison, WI, USA) and cloned into pWPI, upstream of the IRES site. The 293T, HUMSC, MDA-MB231 cell lines were cultured in DMEM supplemented with 10% foetal bovine serum. Recombinant lentiviruses were produced by transient transfection of 293T cells according to standard protocols. Briefly, subconfluent 293T cells were cotransfected with 10 μg of a plasmid vector, 7.5 μg of psPAX2 and 2.5 μg of pMD2.G by calcium phosphate precipitation. After 16 hrs, the medium was changed, and recombinant lentiviral vector was harvested 24 hrs later. For transduction, HUMSCs or MDA-MB231 cells were placed on 6-well plate (1 × 105 cells/well), and after 16 hrs, medium containing lentivirus was added with 8 μg/ml of polybrene. The cells were harvested 5 days later and sorted by fluorescence-activated cell sorting.
Selection of ‘active’ HUMSCs
6-wells mothod: HUMSCs, labelled with CellTracker™ Red CMTPX 5 μM (Molecular Probes, Eugene, OR, USA), were cultured in each of 6 wells. Each 1.5 × 104 MDA-MB231-LG, labelled with CellTracker™ Green CMFDA 5 μM, was cultured in another 6-well plate for 12 hrs and then co-cultured with 3.0 × 104 HUMSCs from each of the 6 wells.
12-wells method: HUMSCs, labelled with CellTracker™ Red CMTPX 5 μM (Molecular Probes), were cultured in each of 12 wells. Each 1.0 × 104 MDA-MB231-LG, labelled with CellTracker™ Green CMFDA 5 μM, was cultured in another 12-well plate for 12 hrs and then co-cultured with 2.0 × 104 HUMSCs from each of the 12 wells.
After co-culture, the number of MDA-MB231-LG was counted every 3 hrs using the Cellomics ArrayScan VTI high-content screening system (Thermo Scientific, Pittsburg, PA, USA). Under examination by Cellomics ArrayScan, the HUMSCs in certain wells that significantly suppress MDA-MB231-LG cell growth were therefore selected for further experiments.
Co-culture of MDA-MB231 with selected HUMSC
Labelled with CellTracker™ Green CMFDA 5 μM, MDA-MB231-LG cells were cultured in 6-well plate for 12 hrs and then co-cultured with equivalent amount of selected HUMSCs, labelled with CellTracker™ Red CMTPX 5 μM. Images from time-lapse analysis under live cell microscopy (the Leica DMIRE2 inverted fluorescence microscope; Leica Microsystem GmbH, Wetzlar, Germany) were acquired every 3 hrs for 18 times after co-culture. Images from confocal microscopy were taken 3 days after co-culture.
TUNEL staining
TUNEL staining, represented by DAB, using a DeadEnd™ Colorimetric TUNEL System kit (Promega) was performed after co-culture of MDA-MB231-LG with selected HUMSC, HS68 and WI38, respectively, for 3 days. The number of TUNEL positive cell was counted in 10 pictures taken randomly under 20× microscopy in each group. The percentage of TUNEL positive cells was calculated by number of TUNEL positive cell/total cell count of co-culture.
Binding rate
MDA-MB231-LG cells were labelled with CellTracker™ Green CMFDA 5 μM, while the selected HUMSCs, HS68 and WI38 cells were labelled with CellTracker™ Red CMTPX 5 μM. Co-culture of MDA-MB231-LG with equivalent amount of selected HUMSC, HS68 cells and WI38, respectively, were performed in 4-wells chamber slide (Lab-Tek™ II Chamber Slide™ System, Nunc) for 3 days. The number of cells with single (red/green) or dual (red+green) fluorescence after co-culture was measured by flow cytometry in each of the four groups. The binding rate of each single cell population was calculated by number of cell with dual fluorescence/number of cell with one of each fluorescence.
Attenuation of MDA-MB231 cell growth by selected HUMSCs through transwell
Each 6-well plate was seeded with 2.0 × 104 MDA-MB231-LG, followed by adding four groups of transwell inserts (0.4 μm pore size, Corning) with culture medium alone, 3.0 × 104 selected HUMSC, 1.5 × 104 selected HUMSC plus 1.5 × 104 MDA-MB231-LG and 3.0 × 104 selected HUMSC plus 3.0 × 104 MDA-MB231-LG inside, respectively. MTT assay for MDA-MB231-LG in the wells was performed 3 days after the culture.
Inhibit the formation of cell-in-cell structure
MDA-MB231-LG cells were labelled with CellTracker™ Green CMFDA 5 μM, while the selected HUMSCs were labelled with CellTracker™ Red CMTPX 5 μM. Co-culture of MDA-MB231-LG was performed in four groups, with equivalent amount of selected HUMSC without pre-treatment, with selected HUMSC pre-treated by Y27632, blebbistatin and latrunculin B, respectively, for 2 hrs. Images on confocal microscopy were taken after co-culture for 6, 12 and 24 hrs. The number of cell-in-cell structure was counted in each of 10 pictures taken randomly under confocal microscopy in each group at different time points. The percentage of cell-in-cell structures was calculated by number of cell-in-cell structure/number of selected HUMSC.
Transmission electron microscopy (TEM) analysis
The cells, including HUMSC, selected HUMSC, MDA-MB231-LG and the selected HUMSC after co-culture with MDA-MB231-LG for 18 hrs, were fixed with 4% paraformaldehyde and 2.5% glutaraldehyde, and then the samples were embedded in epoxy resin (Spurr; EMS). Ultra-thin sections were cut in an ultramicrotome, stained with 2% uranyl acetate for 15 min and 3% lead citrate for 3 min. Then the sections were observed under a transmission electron microscope.
Genome-wide gene expression analysis by high-density oligonucleotide microarray
Total RNA corresponding to primary cultures of HUMSC and selected HUMSC was extracted using the Ribo pure kit (Ambion, Huntingdon, UK), following the manufacturers’ recommendations. Once isolated, RNA concentration and quality were determined by using Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Then, total cDNA was synthesized with a T7-polyT primer and a reverse transcriptase (Superscript II, Life Technologies, Inc., Carlsbad, CA, USA). In vitro transcription was carried out using biotinylated UTP and CTP (Enzo Diagnostics, Farmingdale, NY, USA). Afterward, labelled nucleic acid target was hybridized (37°C for 16 hrs) to Affymetrix Human Genome U133 plus 2.0 oligonucleotide arrays. Finally, after automated washing and staining, absolute values of expression were calculated and normalized from the scanned array by using Affymetrix software (Santa Clara, CA, USA). In this work, three samples corresponding to two different individuals were analysed for each cell type. The percentage of increase gene expression in selected HUMSCs compared with the other HUMSCs was determined by Gene Spring software version 11 (Silicon Genetics, Redwood City, CA, USA).
Animals
The nonobese diabetic/severe combined immunodeficient (NOD/SCID, NOD-CB17-Prkdcscid/J) mice, purchased from Jackson Laboratory (Bar Harbor, ME, USA), were raised in a pathogen-free environment in filtered cages. Filtered, sterilizing water and irradiated foods ad libitum were given to minimize infection. The female NOD/SCID mice at the age of 8 weeks were used for experiments. All animal experiments were evaluated and approved by the Institutional Animal Care and Use Committee of Academia Sinica.
Bioluminescence imaging
In vivo bioluminescence imaging was performed with an IVIS Imaging System (Xenogen, Alameda, CA, USA). Images and measurements of bioluminescence signals were acquired and analysed using Living Image software (Xenogen). Ten minutes prior to in vivo imaging, animals received the substrate D-luciferin (Biosynth, Staad, Switzerland) at 150 mg/kg in PBS by intraperitoneal injection and were anaesthetized using 2.5% isoflurane (Abbott Laboratories, North Chicago, IL, USA). Animals received continuous exposure to 2% isoflurane to sustain sedation during imaging. Imaging times ranged from 1 sec. to 5 min., depending on the bioluminescence of the tumours or metastatic lesions and two to five mice were imaged at a time. Regions of interest (ROI) from displayed images were drawn around the tumour sites and quantified as photons/second using the Living Image software (Xenogen).
Survival of HUMSCs in animal model
The NOD/SCID mice were injected with 5.0 × 105 HUMSC-LG (n = 5) and 5.0 × 105 selected HUMSC-LG (n = 5), respectively, into the fat pad on the right. In vivo bioluminescence images were taken with mice prone 7 days and 21 days post-HUMSC injection.
Homing of selected HUMSCs
In the model of tumour established for seven days, the NOD/SCID mice were injected with 2.5 × 105 MDA-MB231 to the fat pad on the right for tumour establishment. Both tumour-bearing mice (n = 5) and normal control mice (n = 5) were injected with 5.0 × 105 selected HUMSC-LG via tail vein seven days later. In vivo bioluminescence images were taken with the mice supine two days and four days post-HUMSC injection.
In the model of tumour established for 6 weeks, the NOD/SCID mice were injected with 2.5 × 105 MDA-MB231 to the fat pad on the right for tumour establishment. Both tumour-bearing mice (n = 5) and normal control mice (n = 5) were injected with 5.0 × 105 selected HUMSC-LG via tail vein 6 weeks later. In vivo bioluminescence images were taken with the mice prone 4 days and 6 days post-HUMSC injection.
Local HUMSC treatment for breast cancer in animal model
The NOD/SCID mice were injected with 2.5 × 105 MDA-MB231-LG to the fat pad on the right for tumour establishment and then divided randomly into HUMSC treatment group (n = 20), WI38 treatment group (n = 8) and breast cancer control group (n = 8). In HUMSC treatment group, 5.0 × 105 selected HUMSCs were injected locally into fat pad while 5.0 × 105 WI38 cells were injected locally into fat pad in WI38 treatment group 7 days post-tumour establishment. In vivo bioluminescence images were acquired with the mice prone 7 days and 14 days post-tumour establishment.
Prolonged observation was performed in another three groups, including HUMSC treatment group (n = 5), WI38 treatment group (n = 5) and breast cancer control group (n = 5). In HUMSC treatment group, 5.0 × 105 selected HUMSCs were injected locally into fat pad twice, seven days and 14 days post-tumour establishment, while, in WI38 treatment group, 5.0 × 105 WI38 cells were injected locally into fat pad twice, seven days and 14 days post-tumour establishment. In vivo bioluminescence images were acquired with the mice prone seven days, 14 days, 21 days, 28 days, 38 days and 58 days post-tumour establishment. The differences in ROI between three groups at different time points were analysed by Kruskal–Wallis test, and a P value less than 0.05 was considered to be statistically significant.
Systemic HUMSC treatment for breast cancer in animal model
The NOD/SCID mice of in situ model were injected with 2.5 × 105 MDA-MB231-LG to the fat pad on the right for tumour establishment and then divided randomly into HUMSC treatment group (n = 5) and breast cancer control group (n = 5). The NOD/SCID mice of metastasis model were intravenously injected with 2.5 × 105 MDA-MB231-LG via tail vein for tumour establishment and then divided randomly into HUMSC treatment group (n = 5) and breast cancer control group (n = 5). In HUMSC treatment group of both models, 5.0 × 105 selected HUMSCs were injected via tail vein seven days post-tumour establishment. In vivo bioluminescence images were acquired with the mice prone seven days and 14 days post-tumour establishment.
Suppression of breast cancer tumourigenesis by HBMSCs in animal model
The NOD/SCID mice were injected with 2.5 × 105 MDA-MB231-LG to the fat pad on the right for tumour establishment and then divided randomly into HBMSC treatment group (n = 5) and breast cancer control group (n = 5). In HBMSC treatment group, 5.0 × 105 HBMSCs were injected locally into fat pad seven days post-tumour establishment. In vivo bioluminescence images were acquired with the mice prone seven days and 14 days post-tumour establishment.
Suppression of breast cancer tumourigenesis by selected HUMSCs after freezing and thawing in animal model
The NOD/SCID mice were injected with 2.5 × 105 MDA-MB231-LG to the fat pad on the right for tumour establishment and then divided randomly into HUMSC treatment group (n = 6) and breast cancer control group (n = 6). The selected HUMSCs being preserved in the liquid nitrogen were thawed. In HUMSC treatment group, 5.0 × 105 thawed, selected HUMSCs were injected locally into fat pad seven days post-tumour establishment. In vivo bioluminescence images were acquired with the mice prone 7 days and 14 days post-tumour establishment.
Suppression of hepatocellular carcinoma (HCC) tumourigenesis by selected HUMSCs in animal model
The NOD/SCID mice were injected with 2.5 × 105 SK-Hep1 into the liver parenchyma for tumour establishment and then divided randomly into HUMSC treatment group (n = 4) and HCC control group (n = 4). In HUMSC treatment group, 5.0 × 105 selected HUMSCs were injected through tail vein 7 days post-tumour establishment. In vivo bioluminescence images were acquired with the mice supine 7 days and 14 days post-tumour establishment.
Histopathologic examination
Six weeks post-tumour establishment with local fat pad injection of 2.5 × 105 MDA-MB231-LG into NOD/SCID mice, these tumour-bearing mice were divided randomly into breast cancer control group (n = 7) and the HUMSC treatment group (n = 7) which underwent local fat pad injection of 5.0 × 105 selected HUMSCs. Both groups of mice were sacrificed 3 days later, the tumour nodules were removed for serial sections followed by haematoxylin and eosin staining, immunohistochemical staining for KI-67 and cleaved-caspase-3. The positive rate of KI-67 staining was calculated by number of positive-stained cells/total cell counts under high power field of the tumour nodule. The positive rate of cleaved-caspase-3 was calculated by number of positive-stained cells/total cell counts under high power field of the tumour nodule.
Results
Apoptosis of MDA-MB231 breast cancer cells after co-culture with HUMSCs
As a first step, we used Cellomics ArrayScan to observe how MDA-MB231 responds to HUMSC. In most of the co-cultured wells, the number of MDA-MB231 decreased slightly, whereas in some wells, the number of MDA-MB231 decreased dramatically. The frequency of the more ‘active’ HUMSCs versus the total number of sample tested was 1/6 (16.67%) (Fig. 1A). The HUMSCs were found to have the ability to suppress MDA-MB231 cell growth to different degrees, so the somehow more ‘active’ HUMSCs with better ability to suppress tumourigenesis were selected for further experiments.
Fig 1.
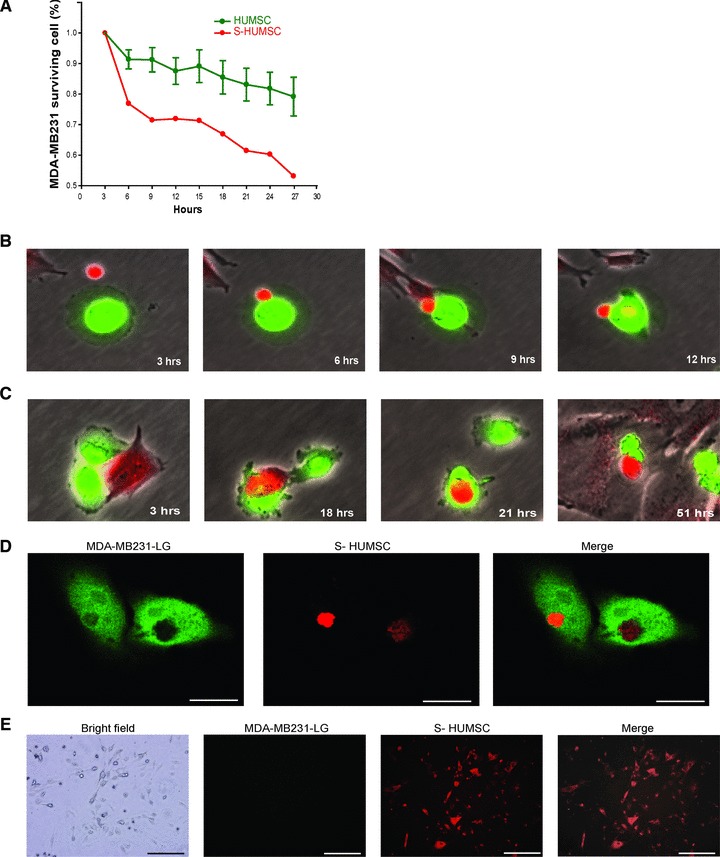
Effect and mechanism of selected HUMSC to suppress MDA-MB231. (A) Using Cellomics ArrayScan, the number of MDA-MB231 decreased slightly in five of the six co-cultured wells (HUMSC), whereas in one of the six wells (S-HUMSC), the number of MDA-MB231 dramatically decreased. The somehow more ‘active’ HUMSCs with better ability to suppress tumourigenesis were selected for further experiments. Data of HUMSC are means ± SEM (n = 5). (B) Fluoroscopic images from time-lapse analysis after co-culture showed the selected HUMSC (red) and MDA-MB231 (green) adhered on contact, and then the HUMSC infused some of its substance into MDA-MB231. (C) Fluoroscopic images from time-lapse analysis after co-culture showed the selected HUMSC (red) contacted with MDA-MB231 (green) and then internalized into MDA-MB231 to form a cell-in-cell structure. The MDA-MB231 shrank following their subsequent separation. (D) On confocal microscopy, simultaneous presentation of MDA-MB231 (green) and HUMSC (red) at the same site at a plane manifested the cell-in-cell structure of selected HUMSC internalized within MDA-MB231. Scale bars: 15 μm. (E) After co-culture of MDA-MB231 (green) with selected HUMSC (red), cells with dual fluorescence (red + green) were isolated and cultured for three more days. Only the red fluorescence was still visible on fluoroscopy. Scale bars: 100 μm.
To investigate further into the interaction between MDA-MB231 and selected HUMSC, we performed a time-lapse analysis under fluoroscopy. It was found during co-culture that the MDA-MB231 and HUMSC may adhere on contact at a variable ratio, from 1:1 to 9:1 (Fig. S1A), and all the MDA-MB231 cells shrank following contact with HUMSC. More specifically, there were two different interactions. Firstly, the MDA-MB231 may shrink immediately upon adhesion and infusion of some substance from HUMSC into MDA-MB231 (Fig. 1B). Secondly, the HUMSC may internalize into MDA-MB231 to form a cell-in-cell structure (Fig. 1C), where then later the two cells separated after a short period. The separated MDA-MB231 shrank, while the separated HUMSC may go on to contact with other MDA-MB231 cells and bring them to shrinkage (Fig. S1B). Since infusion of substance from HUMSC to MDA-MB231 was noted during the cell–cell contact, to distinguish from simple ‘adhesion’, the word ‘binding’ is used here to describe the unique interaction.
The cell-in-cell structure of selected HUMSC internalized within MDA MB-23l was manifested on confocal microscopy (Fig. 1D), with substance from HUMSC intermixed within MDA-MB231 noted (Fig. 2A). Although less dominant, it was also found that in a few cases, some substance from MDA-MB231 had been intermixed within HUMSC (Fig. S2A), implying that infusion of substance from MDA-MB231 into HUMSC also occurred during their interaction.
Fig 2.
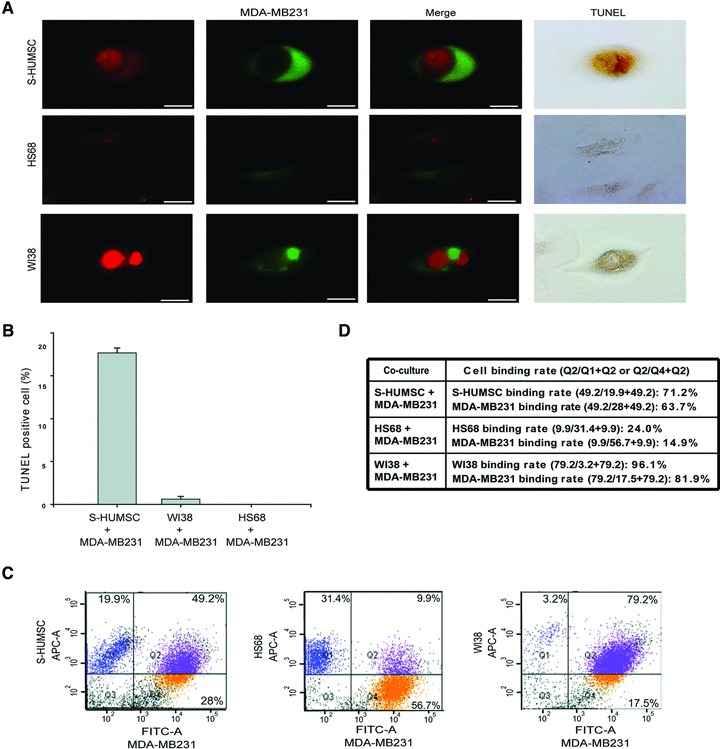
MDA-MB231 apoptosis following binding or formation of cell-in-cell structure with selected HUMSC. (A) Confocal microscopy demonstrated the cell-in-cell structure of selected HUMSC (red) internalized within MDA-MB231 (green) after co-culture. Simultaneous presentation of strong red fluorescence came from HUMSC within the region of green fluorescence delineated by MDA-MB231 suggested that some substance from HUMSC had been intermixed within MDA-MB231, and the MDA-MB231 was stained positively by TUNEL (top panels). Binding of HS68 (red) with MDA-MB231 (green) was rare, and no cells were TUNEL-positive (middle panels). The cell-in-cell structure of WI38 (red) internalized within MDA-MB231 (green) was stained negatively by TUNEL (bottom panels). Scale bars: 10 μm. (B) The percentage of TUNEL-positive cells after co-culture for 3 days was 17.66 ± 0.58% of co-cultured MDA-MB231 with selected HUMSC, but only 0.61 ± 0.31% of co-cultured MDA-MB231 with WI38 and 0 ± 0% of co-cultured MDA-MB231 with HS68 was TUNEL-positive. Data are means ± SEM. (C) Using flow cytometry, the number of cells with simply red (Q1), simply green (Q4) and dual (red + green) fluorescence (Q2) was measured. The percentage of cells with dual fluorescence was 49.2%, 9.9% and 79.2% after co-culture of MDA-MB231 with selected HUMSC, HS68 and WI38, respectively. (D) The binding rate of each cell population.
After co-culture of MDA-MB231 with selected HUMSC for 3 days, cells with dual fluorescence (of red and green) were isolated by flow cytometry sorting. These cells, considered to be mostly the cell-in-cell structures and a few from the cells after binding, were cultured for 3 more days for consequence inspection. On fluoroscopy, we found that only the red fluorescence, from the HUMSC, was still visible after the culture (Fig. 1E), which means, after binding or forming of the cell-in-cell structure with HUMSC, all the MDA-MB231 cells died while the selected HUMSC was the only cell population left alive.
To verify whether apoptosis of MDA-MB231 occurred, we performed TUNEL staining after co-culture for 3 days. It was noted that those MDA-MB231 which had been binding or had formed a cell-in-cell structure with HUMSC were all TUNEL-positive cells (17.66 ± 0.58%); and within the cell-in-cell structure, it was MDA-MB231 to be TUNEL-positive. Although almost all WI38 (a human embryonic lung fibroblast cell line) cells had been binding or had formed a cell-in-cell structure with MDA-MB231 after co-culture, very few TUNEL-positive cells were found (0.61 ± 0.31%). As for HS68 (a human foreskin fibroblast cell line), very few cells had been binding with MDA-MB231, and no TUNEL-positive cells were observed (0 ± 0%) after co-culture (Fig. 2A and B). To determine the binding rate (mainly the forming rate of cell-in-cell structure) of each cell population, measured by flow cytometry, we calculated the percentage of cells with single and dual fluorescence separately (Fig. 2C and D). These results indicate that, during co-culture of MDA-MB231 with selected HUMSC, the MDA-MB231 and HUMSC tended to bind with each other and form a cell-in-cell structure which then leads to MDA-MB231 apoptosis. In addition, we noted that it was the selected HUMSC to internalize into WI38 during co-culture of these two cells (Fig. S2C), but no cell death was observed following forming of the cell-in-cell structure (Fig. S2D).
Moreover, the results that Y27632 (inhibitor of ROCK protein), blebbistatin (myosin II inhibitor) and latrunculin B (inhibitor of actin polymerization), respectively, inhibited the formation of cell-in-cell structure during co-culture (Fig. S2A and S2B) implicate the role of myosin II, actin polymerization and Rho-ROCK-actin/myosin pathway in the internalization of selected HUMSC.
Attenuation of MDA-MB231 breast cancer cell growth by HUMSC through transwell
To clarify whether selected HUMSC secreted cytokines that caused MDA-MB231 death in addition to direct contact, we observed the influence of selected HUMSC on MDA-MB231 cell growth using a transwell co-culture system. It was found that MDA-MB231 cell growth was attenuated mildly by selected HUMSCs within the transwell insert and greatly by selected HUMSCs co-cultured with MDA-MB231 within the transwell insert (Fig. 3A). These results suggest that there should be some cytokines secreted by selected HUMSC and, predominantly, by selected HUMSC co-cultured with MDA-MB231 which worked to suppress MDA-MB231 cell growth through the microporous membrane without direct contact.
Fig 3.
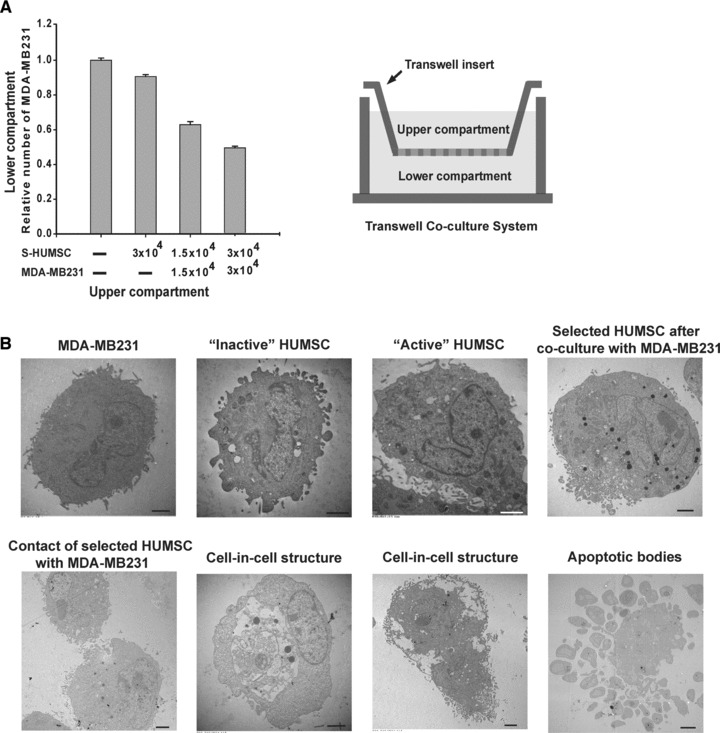
Attenuation of MDA-MB231 cell growth by selected HUMSCs through transwell; TEM analysis. (A) Compared with the group of only culture medium in the upper compartment, using MTT assay, the group of 3.0 × 104 selected HUMSCs showed little decline in the relative number of MDA-MB231, while the group of 1.5 × 104 selected HUMSCs plus 1.5 × 104 MDA-MB231 cells showed significant decline, and the group of 3.0 × 104 selected HUMSCs plus 3.0 × 104 MDA-MB231 cells showed the most decline in the relative number of MDA-MB231 in the lower compartment. Data are means ± SEM. (B) On TEM analysis, some dense bodies were noted in selected ‘active’ HUMSCs, but they were rare in ‘inactive’ HUMSCs. The dense bodies in selected HUMSCs became even more abundant after co-culture with MDA-MB231. On TEM images showing contact of selected HUMSC with MDA-MB231, there was excretion of some granules from HUMSC surface blebs in intercellular areas. Images of the cell-in-cell structure showed selected HUMSC, with prominent dense bodies, being internalized within MDA-MB231. Fragments of MDA-MB231 with some organelles inside and intact membranes manifested the typical apoptotic bodies. Scale bars: 3 μm.
Distinct features of selected HUMSCs
Under TEM, the selected ‘active’ HUMSCs showed distinct features as compared with ‘inactive’ HUMSCs and after co-culture with MDA-MB231. Also, typical apoptotic bodies of MDA-MB231 were observed (Fig. 3B). Moreover, within a total of 44,757 genes tested by DNA microarray [The raw data have been deposited in the GEO database under the accession number GSE27820 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=lzahxamisegckpq&acc=GSE27820)], 7454 genes in selected ‘active’ HUMSC were up-regulated more than 2-fold of ‘inactive’ HUMSC (data not shown), and 33 genes were up-regulated more than 3-fold of ‘inactive’ HUMSC (Table S1). Furthermore, in vivo bioluminescence imaging showed persistence of bioluminescence signals of selected ‘active’ HUMSCs (n = 5) 21 days post-injection, whereas no more bioluminescence signals of ‘inactive’ HUMSCs were visible (n = 5) on the same day (Fig. 4A).
Fig 4.
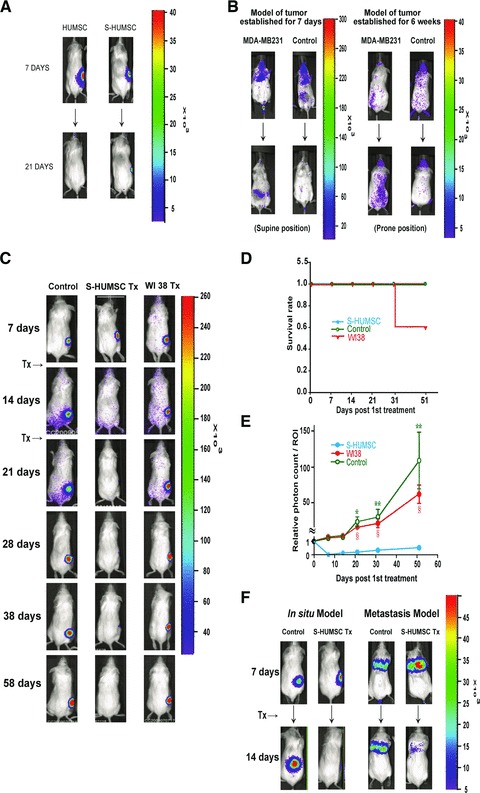
Suppression of breast cancer tumourigenesis by selected HUMSCs in animal models. (A) In vivo bioluminescence imaging seven days post-injection showed persistence of both ‘inactive’ HUMSCs (n = 5) and selected ‘active’ HUMSCs (n = 5) locally at the site of injection. However, no more bioluminescence signals of ‘inactive’ HUMSCs were visible (n = 5) 21 days post-injection whereas bioluminescence signals of selected ‘active’ HUMSCs were decreased but still visible (n = 5) on the same day. (B) In vivo bioluminescence images of tumour-bearing mice showed localization of selected HUMSCs in the fat pad where MDA-MB231 cells were injected in both the models of tumour established for seven days (n = 5) and for six weeks (n = 5), while the signals of HUMSCs almost disappeared in normal control mice (n = 5) of both models. (C) In local HUMSC treatment group (n = 5), bioluminescence signals of MDA-MB231 were completely invisible from seven days to 51 days post first HUMSC injection, but the signals kept increasing through the same period in breast cancer control group (n = 5) and WI38 treatment group (n = 5). (D) Survival rate of NOD/SCID mice. (E) Relative photon count level obtained at the end of the experiment was 0.27 in local HUMSC treatment group, 112.67 in breast cancer control group and 65.77 in WI38 treatment group. Data are means ± SEM. *P < 0.05, **P < 0.01 comparing HUMSC treatment with cancer control. §P < 0.05 comparing HUMSC treatment with WI38. (F) In systemic HUMSC treatment group, bioluminescence signals of MDA-MB231 were completely invisible in in situ model (n = 5) and nearly invisible in metastasis model (n = 5) seven days post-HUMSC injection, but the signals were increased in breast cancer control group (n = 5) of both models.
HUMSC treatment for breast cancer in animal models
To investigate the homing ability of HUMSCs, we intravenously injected selected HUMSCs into NOD/SCID mice. In vivo bioluminescence images showed eventual localization of HUMSCs in the fat pad where MDA-MB231 cells were injected in both the model of tumour established for 7 days (n = 5), with the tumours not palpable yet, and the model of tumour established for 6 weeks (n = 5), with the tumours about 0.5 cm in size (Fig. 4B). Since the homing ability of HUMSCS was confirmed, we then performed local and systemic treatment with HUMSCs in NOD/SCID mice following tumour establishment.
In WI38 treatment group (n = 8), in vivo bioluminescence signals of MDA-MB231 were all increased (n = 8) 7 days post-WI38 injection (Fig. S3A). In contrast, in local HUMSC treatment group (n = 20), bioluminescence signals of MDA-MB231 were either completely invisible (n = 15) or nearly invisible (n = 4) 7 days post-HUMSC injection, with no significant change of bioluminescence signals in a single case (n = 1) (data not shown). Here, the in vivo breast cancer tumourigenesis was proved to be efficaciously suppressed by local treatment with selected HUMSCs, and the amelioration rate was 95% 7 days post-HUMSC injection. In addition, the selected HUMSCs after freezing and thawing were as well proved to suppress in vivo breast cancer tumourigenesis (Fig. S3B), while HBMSCs, comparatively, did not suppress breast cancer tumourigenesis at all (Fig. S3A).
To investigate the cancer-free stage after initial successful treatment with local HUMSC injection, we performed another prolonged observation. In local HUMSC treatment group (n = 5), in vivo bioluminescence signals of MDA-MB231 were all completely invisible (n = 5) from 7 days to 51 days post first injection, and the tumour masses were not palpable anymore from that time on (Fig. 4C). In contrast, in WI38 treatment group (n = 5), bioluminescence signals of MDA-MB231 were all gradually increasing (n = 5) through the same period, and death of two cases occurred 33 days post first injection (Fig. 4D), which lowered the photon count level of the ROI (Fig. 4E). This prolonged observation under bioluminescence imaging revealed a cancer-free stage of at least 50 days after local HUMSC treatment, and the relative photon count level of ROI obtained at the end of the experiment was only 0.27 as compared with that right before HUMSC treatment started (Fig. 4E).
In systemic HUMSC treatment group, in vivo bioluminescence signals of MDA-MB231 were all completely invisible in in situ model (with local injection of MDA-MB231-LG into the fat pad) (n = 5) and all nearly invisible in metastasis model (with intravenous injection of MDA-MB231-LG via tail vein) (n = 5) 7 days post-HUMSC injection (Fig. 4F). Thus, the in vivo breast cancer tumourigenesis of both in situ and metastasis models was proved to be efficaciously suppressed by systemic therapy with selected HUMSCs. Furthermore, a cancer-free stage of at least 35 days after systemic HUMSC treatment was revealed in the prolonged observation of in situ model (Fig. S3C).
Histopathologic evidence
Compared with breast cancer control group (n = 7), tumour cells in HUMSC treatment group (n = 7) were sparse and loose, with condensation of nuclei noted on haematoxylin and eosin staining. Very few cells with positive KI-67 (a cellular marker for proliferation) staining but lots of cells with positive cleaved-caspase-3 staining were found within the tumour nodule in HUMSC treatment group (Fig. 5A). In breast cancer control group, the KI-67 positive staining rate was 51.21 ± 1.72%, and the cleaved-caspase-3 positive staining rate was 0 ± 0% under high power field of the tumour nodule. However, in HUMSC treatment group, the KI-67 positive staining rate was 10.47 ± 1.61%, while the cleaved-caspase-3 positive staining rate was 75.04 ± 4.65% under high power field of the tumour nodule (Fig. 5B). These findings demonstrate that the tumour cells were no longer largely proliferating and growing, and most of them went into apoptosis following HUMSC treatment.
Fig 5.
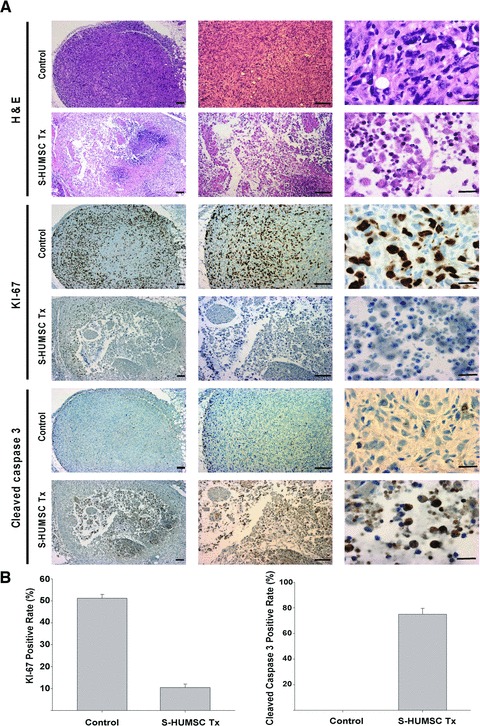
Histopathologic evidence of the suppression of breast cancer tumourigenesis by selected HUMSCs. (A) On haematoxylin and eosin staining, the tumour cells in breast cancer control group (n = 7) were dense and tight, but the tumour cells in HUMSC treatment group (n = 7) were sparse and loose, and condensation of nuclei was observed. On immunohistochemical staining of KI-67, lots of positive cells were noted within the tumour nodule in breast cancer control group (n = 7), but very few positive cells were found in HUMSC treatment group (n = 7). On immunohistochemical staining of cleaved-caspase-3, very few positive cells were found in breast cancer control group (n = 7), but lots of positive cells were noted within the tumour nodule in HUMSC treatment group (n = 7). Scale bars: 20 μm (right) or 100 μm (all others). (B) The KI-67 positive staining rate under high power field of the tumour nodule was 51.21 ± 1.72% in breast cancer control group but was only 10.47 ± 1.61% in HUMSC treatment group. The cleaved-caspase-3 positive staining rate under high power field of the tumour nodule was 0 ± 0% in breast cancer control group but was 75.04 ± 4.65% in HUMSC treatment group.
Discussion
The interactions we have observed between selected HUMSC and MDA-MB231, which caused MDA-MB231 breast cancer cell death, include (1) binding mechanism: breast cancer cell apoptosis from direct cell–cell contact with HUMSC, at a various adhesion ratio from 1:1 to 9:1, and infusion of some substance into cancer cell by HUMSC; (2) cell-in-cell mechanism: breast cancer cell apoptosis from internalization of HUMSC; (3) indirect (cytokine) mechanism: attenuation of breast cancer cell growth from one or more cytokines secreted, predominantly, by co-cultured HUMSC and MDA-MB231 or by HUMSC alone, without direct contact with cancer cells. Though infusion of some substance from MDA-MB231 into HUMSC was also noticed in a few cases, substance transfer from HUMSC to MDA-MB231 was strikingly more dominant and may be the key factor to initiate MDA-MB231 apoptosis. The phenomenon we discovered here of a cell-in-cell structure of HUMSC internalized within MDA-MB231 resulting in MDA-MB231 apoptosis is different from phagocytosis, cell cannibalism and a recently depicted non-apoptotic cell death mechanism named entosis [16], and it has not yet been discussed in the literature. It is known that the substance or the cell being engulfed would be the one to degrade during phagocytosis, cell cannibalism and entosis. In the phenomenon we discovered here, after internalization of HUMSC, it was MDA-MB231 that died. Similar to that during entosis, formation of this previously unrecognized cell-in-cell structure involved Rho-ROCK-actin/myosin pathway. However, the host cell took an apoptosis path to death in this cell-in-cell structure, whereas death of the internalized cell was suggested to have a lysosomal involvement during entosis. Additionally, we found that selected HUMSCs inclined to internalize during co-culture with WI38, yet forming of cell-in-cell structure with WI38 did not cause any cell death, suggesting that the utility of HUMSC to initiate apoptosis was specifically aimed at breast cancer cells, but not at normal cells.
In our experiment, some HUMSCs were found to be more ‘active’ as they had better ability to suppress breast cancer cell growth. The features of ‘active’ and ‘inactive’ HUMSCs were different under TEM, and in vivo survival of these ‘active’ HUMSCs appeared longer. Furthermore, the markedly different gene expressions during microarray analysis manifested distinct phenotypes of the ‘active’ and ‘inactive’ HUMSCs. For example, among the 33 genes in the ‘active’ HUMSC being up-regulated more than 3-fold of the ‘inactive’ HUMSC, some have been documented to be associated with migration (e.g. CTNNA1, MALAT1, SEMA3C, WASF2, CCDC88A, IQGAP1), filopodia formation (e.g. MyO10, SH3PXD2A), apoptosis (e.g. IL6ST), tumour cell death (e.g. TCF25), immune response (e.g. NFAT5) and vesicle secretion (e.g. SCAMP1, MYO6), which seemed quite consistent with the unique characteristics that we have observed on the ‘active’ HUMSC. In addition, these 33 up-regulated genes differ from those which are highly expressed in HBMSC, adipose and umbilical cord blood MSCs during differentiation, and from those in HUMSC during transdifferentiation to endothelial cell [17], indicating that the ‘active’ HUMSC should be an active form of HUMSC rather than a product from HUMSC differentiation.
In the literature, the function of HUMSC to attenuate breast cancer cell growth has been reported once [18]. Different from their report, in our study, the ‘active’ HUMSCs were selected, the mechanisms of HUMSCs suppressing MDA-MB231 were disclosed, and apoptosis of MDA-MB231 following HUMSC treatment was corroborated both in vitro and in vivo. In addition, our study revealed that the medium of co-cultured HUMSC with MDA-MB231 delivered a much better result in attenuating breast cancer cell growth than the medium of HUMSC alone.
In this present study, the selected HUMSCs showed their homing ability to breast cancer, either 6 weeks or only 7 days post-tumour establishment. The ability of selected HUMSC to suppress breast cancer tumourigenesis was proved both in vitro and in vivo. Treatment with selected HUMSCs was fairly efficacious in both in situ and metastatic breast cancers without considerations of HLA mismatch, and the cancer-free stage could last for 50 days. Also, the result of selected HUMSC after freezing and thawing in suppressing breast cancer tumourigenesis is uplifting. In addition to breast cancer, treatment with selected HUMSCs was proved to be efficacious in HCC (Fig. S3D). It is our expectation that this newly established HUMSCs treatment model can serve as a viable therapy for breast cancer or other tumours in the near future.
Acknowledgments
This work was supported by Academia Sinica Peak Project (grant number 2371, 4012, http://www.sinica.edu.tw). K.C.C. designed the study and performed the research; K.C.C. and H.T.Y. analysed the data and wrote the paper; M.W.C. provided technical advice. We thank Professor Wen-Hwa Lee and Jin-Yuh Shew for their support and advice on the study.
Conflict of interest
The authors confirm that there are no conflicts of interest.
Supporting Information
Mechanism of selected HUMSCs to suppress MDA-MB231 cell growth. (A) After co-culture, adhesion of nine MDA-MB231 cells (green) to one selected HUMSC (red) was noted. (B) Fluoroscopic images from time-lapse analysis were acquired every 3 hrs after co-culture of MDA-MB231 (green) with selected HUMSC (red). The HUMSC contacted with MDA-MB231 and then internalized into MDA-MB231 to form a cell-in-cell structure. The MDA-MB231 shrank following their subsequent separation, while the separated HUMSC went on to contact with the other MDA-MB231.
Fig. S2. Formation of cell-in-cell structure. (A) On confocal microscopy, moderate amount of cell-in-cell structure was found during co-culture of MDA-MB231 with selected HUMSC without pre-treatment (control). Simultaneous presentation of some weak green fluorescence came from MDA-MB231 within the region of red fluorescence delineated by HUMSC (arrow) suggested that some substance from MDA-MB231 had been intermixed within HUMSC. In contrast, internalization of selected HUMSC pre-treated by Y27632, blebbistatin and latrunculin B, respectively, was rare during co-culture with MDA-MB231. (B) Formation of cell-in-cell structure reached a peak at about 12 hrs after co-culture of MDA-MB231 with selected HUMSC without pre-treatment (control), with a percentage (number of cell-in-cell structures/number of selected HUMSCs) about 33.51%. In contrast, the percentage of cell-in-cell structures was no more than 4.46% at the same time after co-culture of MDA-MB231 with selected HUMSC pre-treated by Y27632, blebbistatin and latrunculin B, respectively. Data are presented as means ± SEM. (C) Confocal microscopy demonstrated the cell-in-cell structures of selected HUMSC (red) internalized within WI38 (green) after co-culture. (D) Confocal microscopy demonstrated the cell-in-cell structure of selected HUMSC (red) internalized within WI38 (green) after co-culture. The cell-in-cell structure was stained negatively by TUNEL. Scale bars: 10 μm.
Fig. S3. Suppression of breast cancer and hepatocellular carcinoma (HCC) tumourigenesis in animal models. (A) In HBMSC treatment group, in vivo bioluminescence signals of MDA-MB231 were increased (n = 5) 7 days post-HBMSC injection. In WI38 treatment group, bioluminescence signals of MDA-MB231 were increased (n = 8) 7 days post-WI38 injection. (B) To explore the tumour suppression effect of selected HUMSCs after preservation, treatment was performed with selected HUMSCs after freezing and thawing. In the group of treatment with thawed HUMSC, in vivo bioluminescence signals of MDA-MB231 were nearly invisible in 2/3 (n = 4) and were completely invisible in 1/3 (n = 2) 7 days post-HUMSC injection. In contrast, in breast cancer control group, bioluminescence signals of MDA-MB231 were increased (n = 6) on the same day. (C) In systemic HUMSC treatment group, in vivo bioluminescence signals of MDA-MB231 were completely invisible from 7 days to 42 days post first HUMSC injection. In contrast, bioluminescence signals of MDA-MB231 kept gradually increasing in breast cancer control group through the same period. (D) In HUMSC treatment group, in vivo bioluminescence signals of SK-Hep1 were nearly invisible (n = 4) 7 days post-HUMSC injection. In contrast, bioluminescence signals of SK-Hep1 were increased in HCC control group (n = 4) on the same day. Selected HUMSCs were proved to suppress in vivo HCC tumourigenesis as well.
Table S1 Genes in ‘active’ HUMSC up-regulated more than 3-fold of ‘inactive’ HUMSC.
References
- 1.Wang HS, Hung SC, Peng ST, et al. Mesenchymal stem cells in the Wharton’s jelly of the human umbilical cord. Stem Cells. 2004;22:1330–7. doi: 10.1634/stemcells.2004-0013. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell KE, Weiss ML, Mitchell BM, et al. Matrix cells from Wharton’s jelly form neurons and glia. Stem Cells. 2003;21:50–60. doi: 10.1634/stemcells.21-1-50. [DOI] [PubMed] [Google Scholar]
- 3.Chao KC, Chao KF, Fu YS, et al. Islet-like clusters derived from mesenchymal stem cells in Wharton’s Jelly of the human umbilical cord for transplantation to control type 1 diabetes. PLoS One. 2008 doi: 10.1371/journal.pone.0001451. . doi:10.1371/journal.pone.0001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu LL, Liu YJ, Yang SG, et al. Isolation and characterization of human umbilical cord mesenchymal stem cells with hematopoiesis-supportive function and other potentials. Haematologica. 2006;91:1017–26. [PubMed] [Google Scholar]
- 5.Cho PS, Messina DJ, Hirsh EL, et al. Immunogenicity of umbilical cord tissue derived cells. Blood. 2008;111:430–8. doi: 10.1182/blood-2007-03-078774. [DOI] [PubMed] [Google Scholar]
- 6.Rachakatla RS, Marini F, Weiss ML, et al. Development of human umbilical cord matrix stem cell-based gene therapy for experimental lung tumours. Cancer Gene Ther. 2007;14:828–35. doi: 10.1038/sj.cgt.7701077. [DOI] [PubMed] [Google Scholar]
- 7.Karahuseyinoglu S, Cinar O, Kilic E, et al. Biology of stem cells in human umbilical cord stroma: in situ and in vitro surveys. Stem Cells. 2007;25:319–31. doi: 10.1634/stemcells.2006-0286. [DOI] [PubMed] [Google Scholar]
- 8.Weiss ML, Medicetty S, Bledsoe AR, et al. Human umbilical cord matrix stem cells: preliminary characterization and effect of transplantation in a rodent model of Parkinson’s disease. Stem Cells. 2006;24:781–92. doi: 10.1634/stemcells.2005-0330. [DOI] [PubMed] [Google Scholar]
- 9.Fu YS, Shih YT, Cheng YC, et al. Transformation of human umbilical mesenchymal cells into neurons in vitro. J Biomed Sci. 2004;11:652–60. doi: 10.1007/BF02256131. [DOI] [PubMed] [Google Scholar]
- 10.Wu KH, Zhou B, Lu SH, et al. In vitro and in vivo differentiation of human umbilical cord derived stem cells into endothelial cells. J Cell Biochem. 2007;100:608–16. doi: 10.1002/jcb.21078. [DOI] [PubMed] [Google Scholar]
- 11.Troyer DL, Weiss ML. Wharton’s jelly-derived cells are a primitive stromal cell population. Stem Cells. 2008;26:591–9. doi: 10.1634/stemcells.2007-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spees JL, Olson SD, Whitney MJ, et al. Mitochondrial transfer between cells can rescue aerobic respiration. Proc Natl Acad Sci USA. 2006;103:1283–8. doi: 10.1073/pnas.0510511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Direkze NC, Hodivala-Dilke K, Jeffery R, et al. Bone marrow contribution to tumour-associated myofibroblasts and fibroblasts. Cancer Res. 2004;64:8492–5. doi: 10.1158/0008-5472.CAN-04-1708. [DOI] [PubMed] [Google Scholar]
- 14.Karnoub AE, Dash AB, Vo AP, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–63. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 15.Mishra PJ, Glod JW, Banerjee D. Mesenchymal stem cells: flip side of the coin. Cancer Res. 2009;69:1255–8. doi: 10.1158/0008-5472.CAN-08-3562. [DOI] [PubMed] [Google Scholar]
- 16.Overholtzer M, Mailleux AA, Mouneimne G, et al. A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell. 2007;131:966–79. doi: 10.1016/j.cell.2007.10.040. [DOI] [PubMed] [Google Scholar]
- 17.Menicanin D, Bartold PM, Zannettino AC, et al. Genomic profiling of mesenchymal stem cells. Stem Cell Rev. 2009;5:36–50. doi: 10.1007/s12015-009-9056-2. [DOI] [PubMed] [Google Scholar]
- 18.Ayuzawa R, Doi C, Rachakatla RS, et al. Naive human umbilical cord matrix derived stem cells significantly attenuate growth of human breast cancer cells in vitro and in vivo. Cancer Lett. 2009;280:31–7. doi: 10.1016/j.canlet.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mechanism of selected HUMSCs to suppress MDA-MB231 cell growth. (A) After co-culture, adhesion of nine MDA-MB231 cells (green) to one selected HUMSC (red) was noted. (B) Fluoroscopic images from time-lapse analysis were acquired every 3 hrs after co-culture of MDA-MB231 (green) with selected HUMSC (red). The HUMSC contacted with MDA-MB231 and then internalized into MDA-MB231 to form a cell-in-cell structure. The MDA-MB231 shrank following their subsequent separation, while the separated HUMSC went on to contact with the other MDA-MB231.
Fig. S2. Formation of cell-in-cell structure. (A) On confocal microscopy, moderate amount of cell-in-cell structure was found during co-culture of MDA-MB231 with selected HUMSC without pre-treatment (control). Simultaneous presentation of some weak green fluorescence came from MDA-MB231 within the region of red fluorescence delineated by HUMSC (arrow) suggested that some substance from MDA-MB231 had been intermixed within HUMSC. In contrast, internalization of selected HUMSC pre-treated by Y27632, blebbistatin and latrunculin B, respectively, was rare during co-culture with MDA-MB231. (B) Formation of cell-in-cell structure reached a peak at about 12 hrs after co-culture of MDA-MB231 with selected HUMSC without pre-treatment (control), with a percentage (number of cell-in-cell structures/number of selected HUMSCs) about 33.51%. In contrast, the percentage of cell-in-cell structures was no more than 4.46% at the same time after co-culture of MDA-MB231 with selected HUMSC pre-treated by Y27632, blebbistatin and latrunculin B, respectively. Data are presented as means ± SEM. (C) Confocal microscopy demonstrated the cell-in-cell structures of selected HUMSC (red) internalized within WI38 (green) after co-culture. (D) Confocal microscopy demonstrated the cell-in-cell structure of selected HUMSC (red) internalized within WI38 (green) after co-culture. The cell-in-cell structure was stained negatively by TUNEL. Scale bars: 10 μm.
Fig. S3. Suppression of breast cancer and hepatocellular carcinoma (HCC) tumourigenesis in animal models. (A) In HBMSC treatment group, in vivo bioluminescence signals of MDA-MB231 were increased (n = 5) 7 days post-HBMSC injection. In WI38 treatment group, bioluminescence signals of MDA-MB231 were increased (n = 8) 7 days post-WI38 injection. (B) To explore the tumour suppression effect of selected HUMSCs after preservation, treatment was performed with selected HUMSCs after freezing and thawing. In the group of treatment with thawed HUMSC, in vivo bioluminescence signals of MDA-MB231 were nearly invisible in 2/3 (n = 4) and were completely invisible in 1/3 (n = 2) 7 days post-HUMSC injection. In contrast, in breast cancer control group, bioluminescence signals of MDA-MB231 were increased (n = 6) on the same day. (C) In systemic HUMSC treatment group, in vivo bioluminescence signals of MDA-MB231 were completely invisible from 7 days to 42 days post first HUMSC injection. In contrast, bioluminescence signals of MDA-MB231 kept gradually increasing in breast cancer control group through the same period. (D) In HUMSC treatment group, in vivo bioluminescence signals of SK-Hep1 were nearly invisible (n = 4) 7 days post-HUMSC injection. In contrast, bioluminescence signals of SK-Hep1 were increased in HCC control group (n = 4) on the same day. Selected HUMSCs were proved to suppress in vivo HCC tumourigenesis as well.
Table S1 Genes in ‘active’ HUMSC up-regulated more than 3-fold of ‘inactive’ HUMSC.


