Abstract
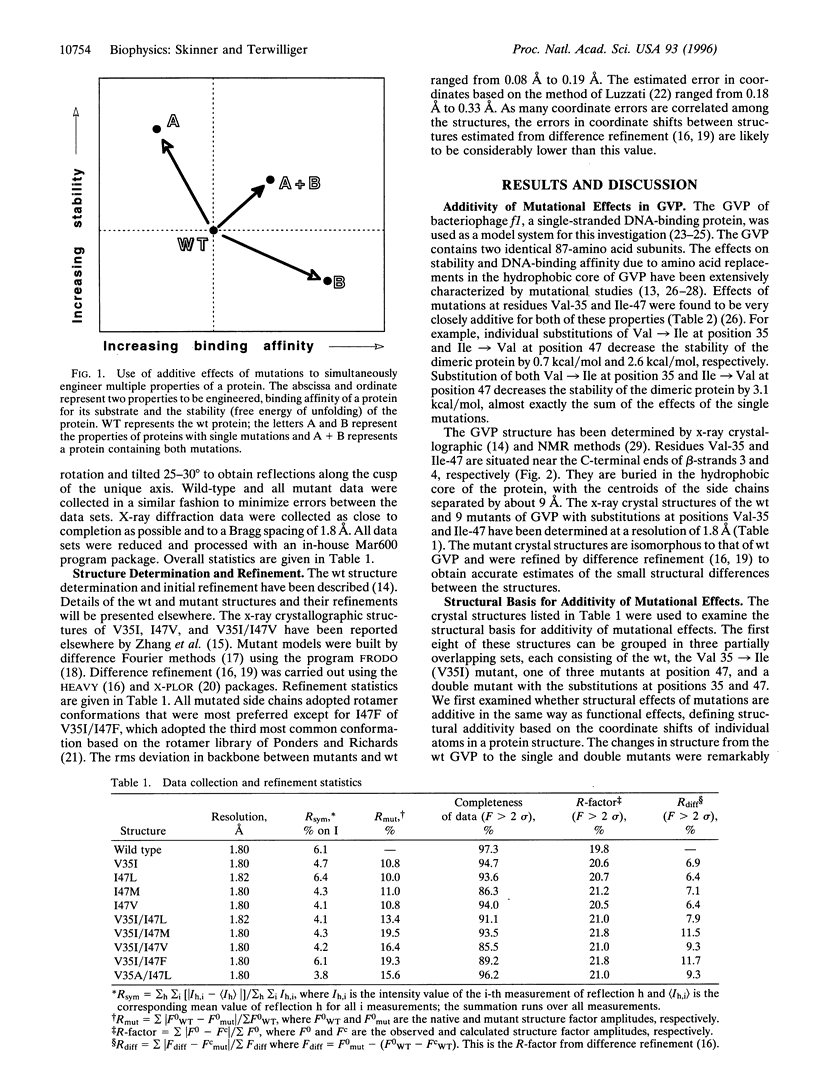
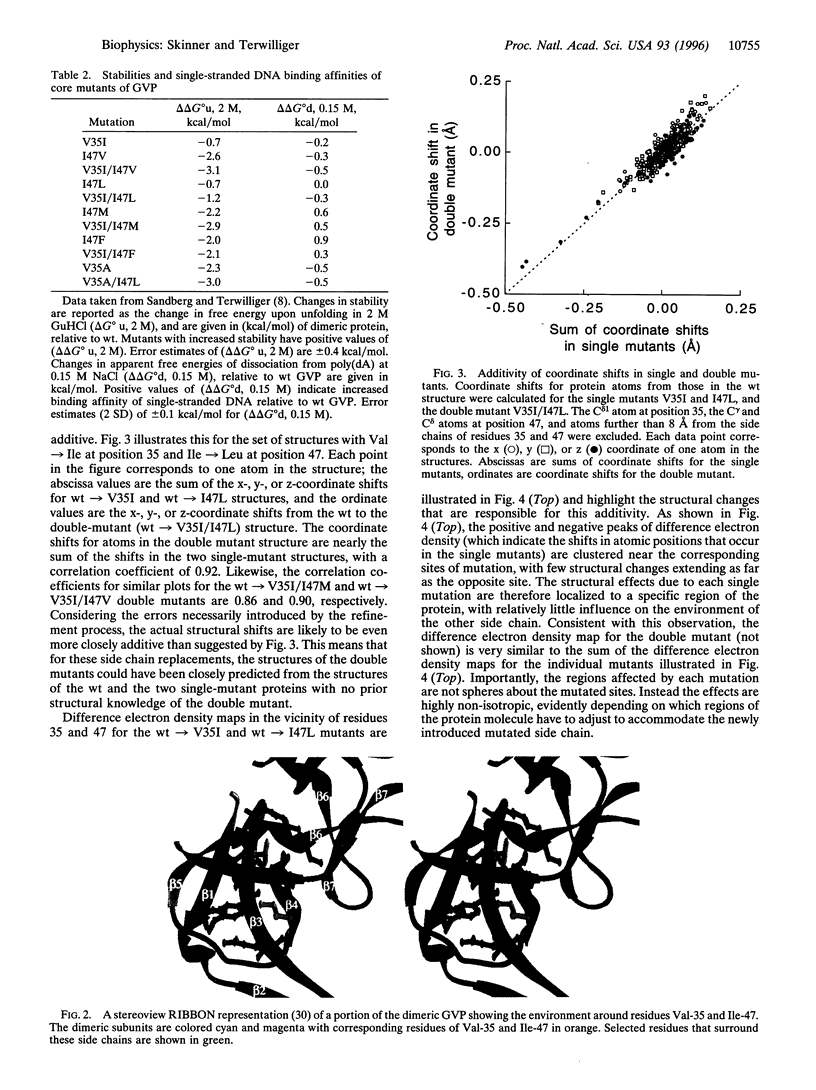
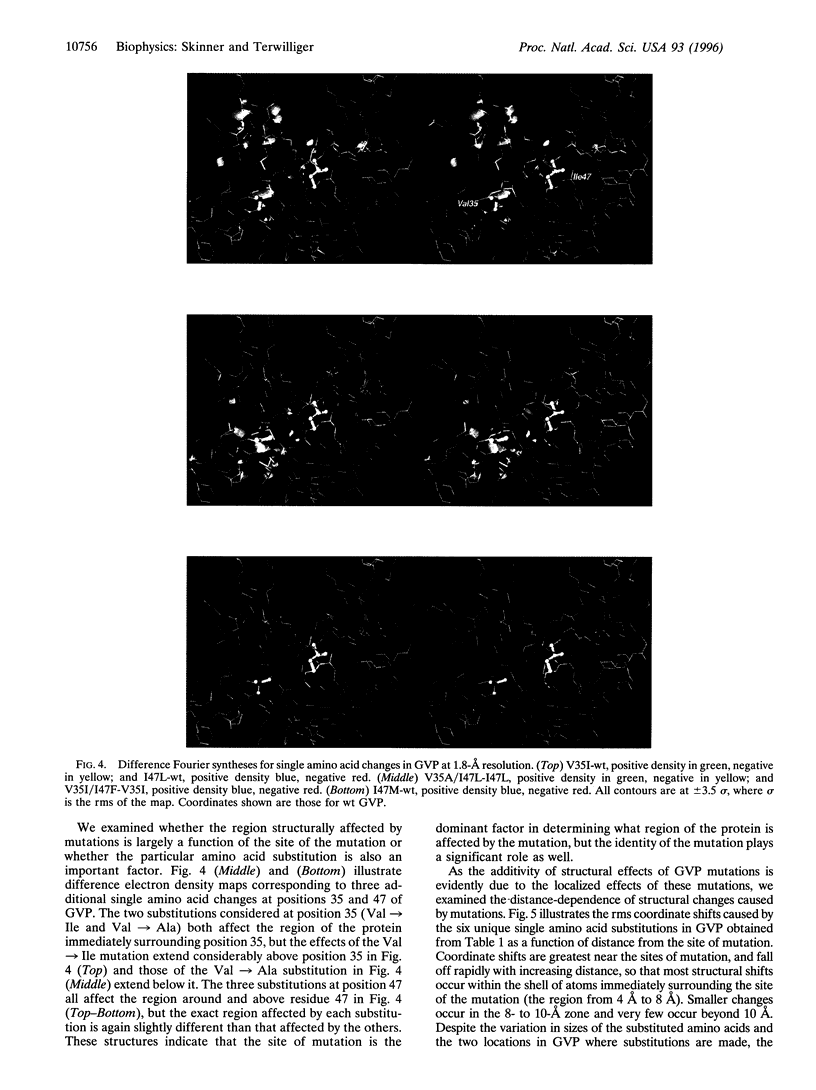
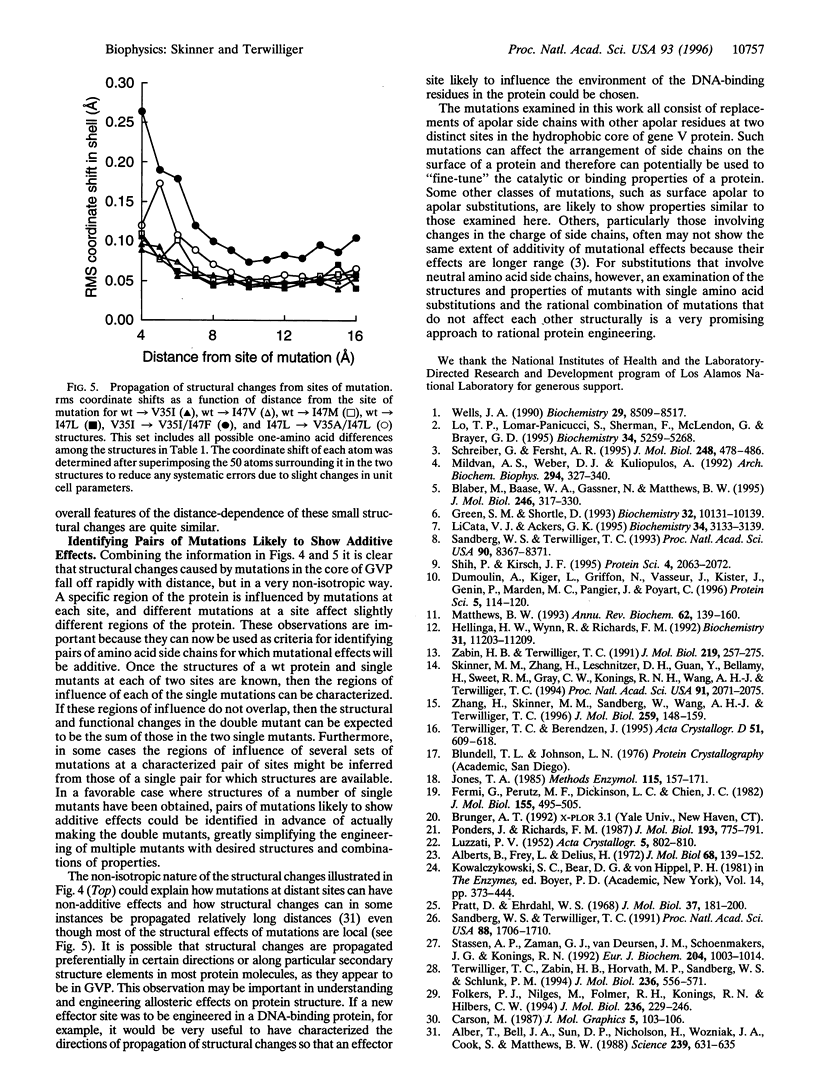
The problem of rationally engineering protein molecules can be simplified where effects of mutations on protein function are additive. Crystal structures of single and double mutants in the hydrophobic core of gene V protein indicate that structural and functional effects of core mutations are additive when the regions structurally influenced by the mutations do not substantially overlap. These regions of influence can provide a simple basis for identifying sets of mutations that will show additive effects.
Full text
PDF
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alber T., Bell J. A., Sun D. P., Nicholson H., Wozniak J. A., Cook S., Matthews B. W. Replacements of Pro86 in phage T4 lysozyme extend an alpha-helix but do not alter protein stability. Science. 1988 Feb 5;239(4840):631–635. doi: 10.1126/science.3277275. [DOI] [PubMed] [Google Scholar]
- Alberts B., Frey L., Delius H. Isolation and characterization of gene 5 protein of filamentous bacterial viruses. J Mol Biol. 1972 Jul 14;68(1):139–152. doi: 10.1016/0022-2836(72)90269-0. [DOI] [PubMed] [Google Scholar]
- Blaber M., Baase W. A., Gassner N., Matthews B. W. Alanine scanning mutagenesis of the alpha-helix 115-123 of phage T4 lysozyme: effects on structure, stability and the binding of solvent. J Mol Biol. 1995 Feb 17;246(2):317–330. doi: 10.1006/jmbi.1994.0087. [DOI] [PubMed] [Google Scholar]
- Dumoulin A., Kiger L., Griffon N., Vasseur C., Kister I., Génin P., Marden M. C., Pagnier J., Poyart C. Two mutations in recombinant Hb beta F41(C7)Y, K82 (EF6)D show additive effects in decreasing oxygen affinity. Protein Sci. 1996 Jan;5(1):114–120. doi: 10.1002/pro.5560050114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fermi G., Perutz M. F., Dickinson L. C., Chien J. C. Structure of human deoxy cobalt haemoglobin. J Mol Biol. 1982 Mar 15;155(4):495–505. doi: 10.1016/0022-2836(82)90483-1. [DOI] [PubMed] [Google Scholar]
- Folkers P. J., Nilges M., Folmer R. H., Konings R. N., Hilbers C. W. The solution structure of the Tyr41-->His mutant of the single-stranded DNA binding protein encoded by gene V of the filamentous bacteriophage M13. J Mol Biol. 1994 Feb 11;236(1):229–246. doi: 10.1006/jmbi.1994.1132. [DOI] [PubMed] [Google Scholar]
- Green S. M., Shortle D. Patterns of nonadditivity between pairs of stability mutations in staphylococcal nuclease. Biochemistry. 1993 Sep 28;32(38):10131–10139. doi: 10.1021/bi00089a032. [DOI] [PubMed] [Google Scholar]
- Hellinga H. W., Wynn R., Richards F. M. The hydrophobic core of Escherichia coli thioredoxin shows a high tolerance to nonconservative single amino acid substitutions. Biochemistry. 1992 Nov 17;31(45):11203–11209. doi: 10.1021/bi00160a034. [DOI] [PubMed] [Google Scholar]
- Jones T. A. Diffraction methods for biological macromolecules. Interactive computer graphics: FRODO. Methods Enzymol. 1985;115:157–171. doi: 10.1016/0076-6879(85)15014-7. [DOI] [PubMed] [Google Scholar]
- LiCata V. J., Ackers G. K. Long-range, small magnitude nonadditivity of mutational effects in proteins. Biochemistry. 1995 Mar 14;34(10):3133–3139. doi: 10.1021/bi00010a001. [DOI] [PubMed] [Google Scholar]
- Lo T. P., Komar-Panicucci S., Sherman F., McLendon G., Brayer G. D. Structural and functional effects of multiple mutations at distal sites in cytochrome c. Biochemistry. 1995 Apr 18;34(15):5259–5268. doi: 10.1021/bi00015a041. [DOI] [PubMed] [Google Scholar]
- Matthews B. W. Structural and genetic analysis of protein stability. Annu Rev Biochem. 1993;62:139–160. doi: 10.1146/annurev.bi.62.070193.001035. [DOI] [PubMed] [Google Scholar]
- Mildvan A. S., Weber D. J., Kuliopulos A. Quantitative interpretations of double mutations of enzymes. Arch Biochem Biophys. 1992 May 1;294(2):327–340. doi: 10.1016/0003-9861(92)90692-p. [DOI] [PubMed] [Google Scholar]
- Ponder J. W., Richards F. M. Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. J Mol Biol. 1987 Feb 20;193(4):775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- Pratt D., Erdahl W. S. Genetic control of bacteriophage M13 DNA synthesis. J Mol Biol. 1968 Oct 14;37(1):181–200. doi: 10.1016/0022-2836(68)90082-x. [DOI] [PubMed] [Google Scholar]
- Sandberg W. S., Terwilliger T. C. Energetics of repacking a protein interior. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1706–1710. doi: 10.1073/pnas.88.5.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg W. S., Terwilliger T. C. Engineering multiple properties of a protein by combinatorial mutagenesis. Proc Natl Acad Sci U S A. 1993 Sep 15;90(18):8367–8371. doi: 10.1073/pnas.90.18.8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber G., Fersht A. R. Energetics of protein-protein interactions: analysis of the barnase-barstar interface by single mutations and double mutant cycles. J Mol Biol. 1995 Apr 28;248(2):478–486. doi: 10.1016/s0022-2836(95)80064-6. [DOI] [PubMed] [Google Scholar]
- Shih P., Kirsch J. F. Design and structural analysis of an engineered thermostable chicken lysozyme. Protein Sci. 1995 Oct;4(10):2063–2072. doi: 10.1002/pro.5560041011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner M. M., Zhang H., Leschnitzer D. H., Guan Y., Bellamy H., Sweet R. M., Gray C. W., Konings R. N., Wang A. H., Terwilliger T. C. Structure of the gene V protein of bacteriophage f1 determined by multiwavelength x-ray diffraction on the selenomethionyl protein. Proc Natl Acad Sci U S A. 1994 Mar 15;91(6):2071–2075. doi: 10.1073/pnas.91.6.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stassen A. P., Zaman G. J., van Deursen J. M., Schoenmakers J. G., Konings R. N. Selection and characterization of randomly produced mutants of gene V protein of bacteriophage M13. Eur J Biochem. 1992 Mar 15;204(3):1003–1004. doi: 10.1111/j.1432-1033.1992.tb16722.x. [DOI] [PubMed] [Google Scholar]
- Terwilliger T. C., Berendzen J. Difference refinement: obtaining differences between two related structures. Acta Crystallogr D Biol Crystallogr. 1995 Sep 1;51(Pt 5):609–618. doi: 10.1107/S0907444994013247. [DOI] [PubMed] [Google Scholar]
- Terwilliger T. C., Zabin H. B., Horvath M. P., Sandberg W. S., Schlunk P. M. In vivo characterization of mutants of the bacteriophage f1 gene V protein isolated by saturation mutagenesis. J Mol Biol. 1994 Feb 18;236(2):556–571. doi: 10.1006/jmbi.1994.1165. [DOI] [PubMed] [Google Scholar]
- Wells J. A. Additivity of mutational effects in proteins. Biochemistry. 1990 Sep 18;29(37):8509–8517. doi: 10.1021/bi00489a001. [DOI] [PubMed] [Google Scholar]
- Zabin H. B., Terwilliger T. C. Isolation and in vitro characterization of temperature-sensitive mutants of the bacteriophage f1 gene V protein. J Mol Biol. 1991 May 20;219(2):257–275. doi: 10.1016/0022-2836(91)90566-o. [DOI] [PubMed] [Google Scholar]
- Zhang H., Skinner M. M., Sandberg W. S., Wang A. H., Terwilliger T. C. Context dependence of mutational effects in a protein: the crystal structures of the V35I, I47V and V35I/I47V gene V protein core mutants. J Mol Biol. 1996 May 31;259(1):148–159. doi: 10.1006/jmbi.1996.0309. [DOI] [PubMed] [Google Scholar]





