Abstract
Connexin45 (Cx45) is a gap junction protein involved in cell-to-cell communication in the heart and other tissues. Here we report the 1H, 15N, and 13C resonance assignments for the monomer and dimer conformations of the Cx45 carboxyl terminal (Cx45CT) domain and provide evidence of dimerization using Diffusion Ordered Spectroscopy. The predicted secondary structure of the Cx45CT domain based on the chemical shifts identified one region of α-helical structure, which corresponds to the residues that broadened beyond detection in the dimer confirmation. Previous biophysical studies from our laboratory characterizing the CT domain from the other major cardiac connexins, Cx40 and Cx43, suggest that the amount of α-helical content may translate into the ability of a protein to dimerize. Even though the CT domain is thought to be the main regulatory domain of most connexins, the physiological role of CT dimerization is currently unknown. Therefore, these assignments will be useful for determining the intermolecular interactions that mediate Cx45CT dimerization, information that will be used to characterize dimerization in functional channels, as well as characterizing the binding sites for molecular partners involved in Cx45 regulation.
Keywords: Cx45, gap junction, dimerization, carboxyl terminus, intrinsically disordered protein
Biological context
Gap junctions are integral membrane proteins that enable the direct cytoplasmic exchange of ions and low molecular weight metabolites (<1 kDa) between adjacent cells. They provide a pathway for the propagation and/or amplification of signal transduction cascades triggered by cytokines, growth factors, and other cell signaling molecules involved in growth, regulation, and development. For example, gap junction intercellular communication is a principal determinant of myocardial conduction, and altered expression has been implicated in arrhythmogenesis (van Veen et al, 2006). Gap junctions are formed by the apposition of two connexons from adjacent cells, where each connexon is formed of six connexin proteins. Connexins are tetraspan transmembrane domain proteins with intracellular amino- and carboxyl-termini. There are 21 different connexin genes in the human genome with differential spatial-temporal expression throughout the body. In the heart, connexin45 (Cx45), connexin43 (Cx43), and connexin40 (Cx40) are found in distinctive combinations and relative quantities in different, functionally specialized subsets of cardiomyocytes. Though there is significant sequence homology among all connexins, the major divergence in primary structures occurs in the cytoplasmic loop and carboxyl terminal (CT) domains.
The CT domain is thought to be the main regulatory domain of most connexins. The CT domain plays a role in the trafficking, size, localization, and turnover of gap junctions, as well as the level of intercellular coupling mediated by the plaque via numerous post-translational modifications and protein-protein interactions. Early structural studies of Cx43, which used cryo-electron microscopy, and the more recent X-ray crystallographic studies of Cx26 have provided a significant amount of information about channel architecture as well as connexin topology (Unger et al, 1999; Maeda et al, 2009). However, neither technique was able to determine the CT structure because of the dynamic nature of the domain. These same characteristics that interfere with crystallographic techniques make nuclear magnetic resonance (NMR) an ideal tool for studying the CT domain.
Previously, we determined by NMR that the soluble Cx43CT structure was mostly random coiled, with two short α-helical domains (Sorgen et al, 2004). The soluble Cx40CT structure was entirely random coiled; however, one region (C267-G285) has a propensity to form α-helical structure (Bouvier et al., 2009). Sedimentation equilibrium analysis of the soluble Cx40CT and Cx43CT domains identified a correlation between the amount of α-helical content and the ability of the CT domain to dimerize (Bouvier et al., 2009). While the soluble CT domain retains biochemical and functional properties consistent with those found in a gap junction plaque, to date the physiological consequences of CT dimerization are unknown. In general, random coiled or intrinsically disordered domains have been identified as playing important roles in molecular recognition, molecular assembly, and protein modifications (Dunker et al, 2002). In the case of membrane proteins, like Cx40, Cx43, and Cx45, intrinsically disordered domains play an important role in cell signaling events by allowing many different binding partners with both high specificity and low affinity to rapidly switch between molecular partners, thus activating alternative signaling pathways (Dunker el al, 2002).
Structural information of the Cx43CT and Cx40CT domains have advanced our understanding of gap junction regulation via protein-protein interactions, phosphorylation induced structural changes, and channel regulation. However, to date, no structural information exists for the CT domain from the Cx45 isoform (Cx45CT). Here, we report the sequence-specific assignments for both the monomer and dimer conformations of the Cx45CT domain, confirm the dimerization using Diffusion Ordered Spectroscopy (DOSY), and identify the residues that broadened beyond detection in the dimer conformation are α-helical in the monomer. These assignments will be used in determining the intermolecular interactions that mediate Cx45CT dimerization, which will help investigate dimerization in functional channels, as well as characterizing the binding sites for molecular protein partners involved in the regulation of Cx45.
Methods and experiments
The protocols used to express and purify the recombinant rat Cx45CT (original Cx45 DNA was obtained as a generous gift from Dr. Steve Taffet) were based on those previously described for the Cx43CT and Cx40CT domains (Sorgen et al, 2004; Bouvier et al, 2009). Cx45CT K265-I296 was cloned into the pGEX-KT expression vector (ATCC) and overexpressed using Rosetta2 pLySs competent cells (Invitrogen). Transformed bacteria were grown at 37°C, 250 rpm in Luria Bertani (LB) media or enriched minimal media (15N or 15N13C) supplemented with 15N-ISOGRO or 15N13C-ISOGRO (1 g/1 L media; Isotec). Cultures were induced at an optical density of 0.6 at 600 nm with 1 mM isopropyl-1-thio-beta-D-galactopyranoside (final concentration). Growth was allowed to proceed for 4 hrs, then bacteria cultures were pelleted by centrifugation (3,500 rpm, 30 min, 4°C), washed in 1× phosphate-buffered saline (PBS), pelleted again by centrifugation (12,000 rpm, 12 min, 4°C), and stored at −20°C. Bacterial pellets were thawed and resuspended in 1× PBS (40 mL/6 L culture) containing 1% NP-40, PMSF, and Complete protease inhibitor (Roche Molecular Biochemicals), then lysed 4× using a French Press. The lysate was cleared by centrifugation (16,500 rpm, 1 hr, 4°C) and the presence of the fusion protein was confirmed using SDS-PAGE. The protein solution was incubated with glutathione-Sepharose 4B resin (20 mL/6 L culture; Amersham Biosciences) for 2-3 hrs with gentle rocking at 4°C. The resin was then washed with 2× PBS three times and 1× PBS three times to remove any non-specific-bound proteins. To cleave the Cx45CT from the GST-resin, the beads were incubated overnight with gentle rocking at 4°C with thrombin (120 units/6 L culture; Fisher Scientific). To remove thrombin, the eluate was incubated with 1 mL of benzamidine resin (GE Healthcare) for 1 hr at 4°C with gentle rocking. To remove residual contaminants, cation exchange chromatography was performed using a HiTrap SP HP column (GE Healthcare) and 0.02 M sodium phosphate buffer at pH 6.0 as the loading buffer. Cx45CT was eluted using a 50% gradient into 0.02 M sodium phosphate buffer containing 1 M sodium chloride. The fractions from the cation exchange chromatography were collected and screened using SDS-PAGE; the fractions containing the 15 kDa Cx45CT were pooled and concentrated using a 3 kDa Amicon Centriplus filter (Millipore). The concentrated protein solution was buffer exchanged into 1× PBS at pH 5.8 and protein concentration was measured using a Nanodrop. The recombinant protein product after cleavage from GST-resin contained the K265-I296 of Cx45CT preceded by two additional amino acids (GS) from the vector. The domain boundaries were chosen because they contain the Cx45CT residues that are expected to be outside the transmembrane domain and contain all of the predicted molecular partner binding and phosphorylation sites.
NMR samples (1 mM) of purified isotopically labeled (15N or 15N13C) Cx45CT were prepared in 300 μL of 1× PBS at pH 5.8 with 10% D2O (dimer) or 300 μL 1× PBS at pH 5.8 with 30% deuterated acetonitrile and 10% D2O (monomer). The pH and sample stability were monitored by comparing 15N-HSQC spectra before and after each 3D NMR experiment. NMR data were acquired at 25°C (dimer) and 37°C (monomer) using an INOVA 600 MHz Varian spectrometer equipped with a cold probe, processed using NMRPipe/NMRDraw (Delaglio et al, 1995), and analyzed using NMRView (Johnson, 1994). Diffusion Ordered Spectroscopy (DOSY) data were collected using a 400 MHz Bruker spectrometer and processed using TopSpin (Bruker). Backbone assignments were obtained using the following 3D experiments: CBCANH, CBCA(CO)NH, HNCO, and HN(CA)CO. Side chain chemical shifts were obtained from 3D HCCH-TOCSY, 15N-TOCSY-HSQC, 15N-NOESY-HSQC, 13C-NOESY-HSQC, H(CCO)NH, and CC(CO)NH experiments.
Assignments and data deposition
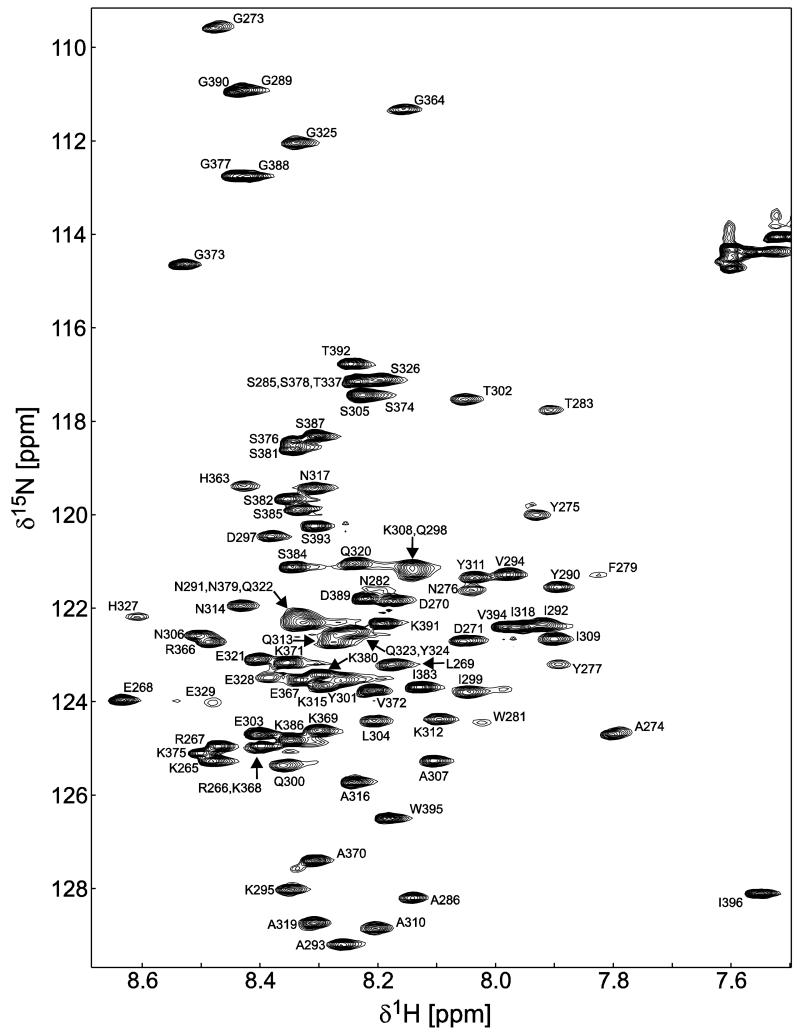
Data collection of the Cx45CT domain was initially performed in 1× PBS (pH 5.8), as previously collected for the Cx40CT and Cx43CT domains (Sorgen et al., 2004; Bouvier et al., 2009). The one difference was the Cx45CT domain showed the greatest amount of cross-peaks in the 15N-HSQC at 25°C, unlike that of the Cx43CT and Cx40CT domains (7°C). Initial investigation of the 15N-HSQC spectrum revealed approximately 100 of the possible 124 residues (82%) (Figure 1). Upon resonance assignment of the Cx45CT, we identified that residues A333-N361 had broadened beyond detection in all the collected experiments. Of the remaining 103 Cx45CT residues (plus a linker of two residues), assigned resonances included 96% of 1HN and 15N amides of the non-proline residues, 98% of all 13Cα, 97% of all 13Cβ, and 95% of all 13C’. All assignments, including most of the side chain resonances outside of residues A333-N361, were deposited in the BioMag ResBank database, accession number 18577. Based on the higher temperature requirement during NMR experiments and the inability of the Cx45CT domain to flow through a 30 kDa cut-off filter during its purification, we tested for Cx45CT oligomerization using DOSY at 25°C. Analysis of the DOSY data indicated dimerization of the Cx45CT domain (Table 1) and together with the assignments, suggests the dimerization domain is contained within residues A333-N361.
Figure 1. 15N-HSQC spectrum of the Cx45CT dimer in phosphate-buffered saline (pH 5.8) at 25°C.
Peak assignments for the backbone amides are indicated with numbering corresponding to the full-length Cx45 protein. The amide peaks that represent the dimerization domain (A333-N361) are not present in the spectrum due to the cross-peaks broadening beyond detection.
Table 1. Diffusion Ordered Spectroscopy analysis of the Cx45CT in solution conditions that stabilize the dimer and monomer conformations.
| Solution Conditions |
Diffusion Constant (m2/s) |
Hydrodynamic Radius (Å) |
Calculated Number of Amino Acids |
Average Molecular Weight (kDa) |
Conformation |
|---|---|---|---|---|---|
| 1× PBS | 1.291E-10 | 25.1 | 313 | 34.41 | Dimer |
| 1× PBS with 30% acetonitrile |
1.275E-09 | 19.4 | 127 | 14.00 | Monomer |
Using the Stokes-Einstein equation, the hydrodynamic radius for each solution condition was calculated with the experimentally collected diffusion constant; D=KBT/(6ηπrH). The number of amino acids was then calculated based on the hydrodynamic radius (rH=4.75N^0.29), which was converted into an average molecular weight.
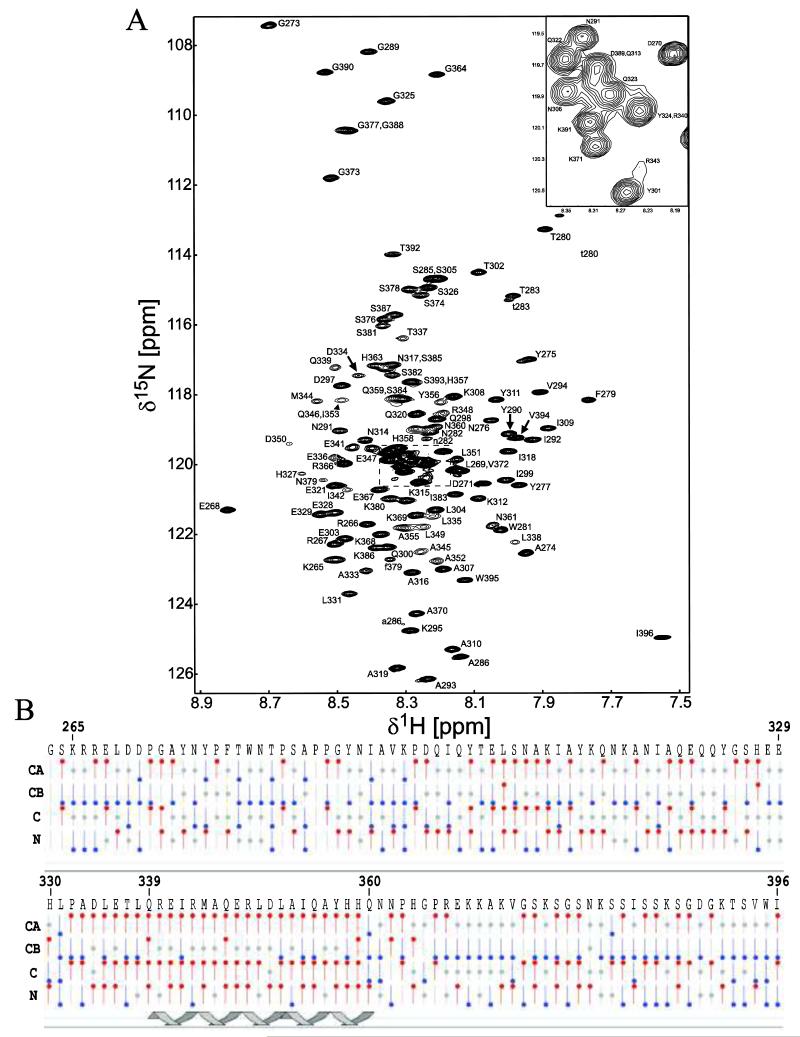
Although the physiological role of CT dimerization is currently unknown, dimerization has been shown in vitro using isolated protein fragments and then corroborated in functional channels (Leach et al., 2003). In order to address the importance of Cx45CT dimerization in functional channels, we need to structurally characterize the dimerization interface. Therefore, we optimized solution conditions that would disrupt the Cx45CT dimer and, depending on the conditions needed to break the dimer, model structures of the dimer based on the type of interface that was disrupted by the buffer. Many different buffer conditions were tested and evaluated using DOSY for dimerization (data not shown). Solution conditions that disrupted dimerization were all hydrophobic (e.g. 15% glycerol, 3 M urea, and 30% acetonitrile). Using resolution as a guide in the 15N-HSQC experiment, we determined that the optimal solution condition was 1× PBS (pH 5.8) containing 30% acetonitrile at 37°C. Resonances assigned for the Cx45CT plus linker (in total 134 residues) included 98% of 1HN and 15N amides of the non-proline residues, 99% of all 13Cα and 13Cβ, and 95% of all 13C’. Additionally, most side chain resonances of the Cx45CT monomer were assigned and all data were deposited in the BioMagResBank database, accession number 18578. Figure 2a shows the 15N-HSQC spectrum and assignments for the Cx45CT monomer. Interestingly, in the presence of the acetonitrile, most of the serines and asparagines did not have carbonyl peaks in the HNCO experiment. Additionally, seven peaks (F279, T280, W281, N282, T283, S285, A286) displayed a second weaker peak in the 15N-HSQC spectrum. These residues are flanked by prolines (P278, P284, P287, and P288), suggesting proline cis-trans isomerization. The presence of a prolyl bond undergoing cis-trans isomerization can provide a binding site for potential molecular partners (Sarkar et al., 2007) and thus presents a novel mechanism for the regulation of gap junctions.
Figure 2. 15N-HSQC spectrum and chemical shift index of the Cx45CT monomer in phosphate-buffered saline with 30% deuterated-acetonitrile (pH 5.8) at 37°C.
A) Peak assignments for the backbone amides are indicated with numbering corresponding to the full-length Cx45 protein. The seven residues (F279, T280, W281, N282, T283, S285, A286) affected by proline cis-trans isomerization have a second weaker peak in the 15N-HSQC spectrum and are labeled with a lowercase letter. B) Graphical representation of results from CSI calculations using the Wutherich reference for Cα, Cβ, C, and N atoms: red circles at the top are chemical shifts consistent with helical structure; blue circles at the bottom are chemical shifts consistent with sheet structure; and gray circles at the center are for intermediate shifts. The cartoon illustrates one region of the Cx45CT (Q339-Q360) determined to be α-helical based on the CSI values.
Based on the bias of the connexin CT domains to form α-helical structure, a helical wheel analysis of residues A333-N361 was performed and identified a hydrophobic face (L335, L338, I342, L349, I353, and Y356), which is consistent with the ability of hydrophobic compounds to disrupt the dimer. To identify if any of the residues are forming α-helical structure, chemical shift index (CSI) analysis of the Cx45CT monomeric conformation was performed using NMRView. The results indicate residues Q339-Q360, which are found within the hydrophobic face, are α-helical (Figure 2b). Subsequent circular dichroism studies using a peptide corresponding to residues A333-N361 and the Cx45CT domain in both dimeric and monomeric solution conditions demonstrated that all contained a similar amount of α-helical content (data not shown). This indicates that the 30% acetonitrile is not altering the secondary structure and the α-helical structure is contained within residues A333-N361.
In summary, we have assigned the resonances for both the monomeric and dimeric conformations of the Cx45CT domain, determined the domain involved in the Cx45CT dimerization, and optimized a solution condition to disrupt the Cx45CT dimer. Assignments of the monomer will be used to solve the Cx45CT structure and model the dimer conformation, while the assignments of the dimer will be useful for characterizing the effects of phosphorylation and protein-protein interactions on the Cx45CT structure.
Acknowledgements
This work is funded by the United States Public Health Sevice Grant, GM072631. Jennifer Kopanic is funded by the Graduate Assistance in Areas of National Need (GAANN) Fellowship. We would like to thank and acknowledge Ed Ezell manager of the Nuclear Magnetic Resonance Laboratory Manager at the University of Nebraska Medical Center for his assistance with DOSY analysis and Dr. Fabien Kieken for his invaluable NMR expertise.
Footnotes
This work was supported by United States Public Health Service Grant GM072631.
References
- Bouvier D, Spagnol G, Chenavas S, Kieken F, Vitrac H, Brownell S, Kellezi A, Forge V, Sorgen PL. Characterization of the structure and intermolecular interactions between the connexin40 and connexin43 carboxyl-terminal and cytoplasmic loop domains. J Biol Chem. 2009;284:34257–34271. doi: 10.1074/jbc.M109.039594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- Johnson BA. NMRView: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Leach RN, Boyett MR, Findlay JB. Expression, purification and spectroscopic studies of full-length Kir3.1 channel C-terminus. Biochim Biophys Acta. 2003;1652:83–90. doi: 10.1016/j.bbapap.2003.07.001. [DOI] [PubMed] [Google Scholar]
- Maeda S, Nakagawa S, Suga M, Yamashita E, Oshima A, Fujiyoshi Y, Tsukihara T. Structure of the connexin 26 gap junction channel at 3.5 A resolution. Nature. 2009;458:597–602. doi: 10.1038/nature07869. [DOI] [PubMed] [Google Scholar]
- Sarkar P, Reichman C, Saleh T, Birge RB, Kalodimos CG. Proline cis-trans isomerization controls autoinhibition of a signaling protein. Mol Cell. 2007;25:413–426. doi: 10.1016/j.molcel.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorgen PL, Duffy HS, Spray DC, Delmar M. pH-Dependent Dimerization of the Carboxyl Terminal Domain of Cx43. Biophys J. 2004;87:574–581. doi: 10.1529/biophysj.103.039230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger VM, Kumar NM, Gilula NB, Yeager M. Three-dimensional structure of a recombinant gap junction membrane channel. Science. 1999;283:1176–1180. doi: 10.1126/science.283.5405.1176. [DOI] [PubMed] [Google Scholar]
- van Veen TA, van Rijen HV, Jongsma HJ. Physiology of cardiovascular gap junctions. Adv Cardiol. 2006;42:18–40. doi: 10.1159/000092560. [DOI] [PubMed] [Google Scholar]