Abstract
Gap junctions are specialized membrane channels that enable coordination of cellular functions and whole-organ responses by facilitating both molecular and electrical communication between neighboring cells. Connexin43 (Cx43) is the most widely expressed and well-studied gap junction protein. In the heart, Cx43 is essential for normal cardiac development and function. Studies using a soluble version of the Cx43 carboxyl-terminal domain (Cx43CT; S255-I382) have established the central role it plays in channel regulation. However, in purifying and characterizing a more ‘native-like’ construct (Cx43CT attached to the fourth transmembrane domain (TM4-Cx43CT; D196-I382)), we have identified that the TM4-Cx43CT is a better model than the soluble Cx43CT to further investigate the mechanisms governing Cx43 channel regulation. Here, we report the backbone 1H, 15N, and 13C assignments and predicted secondary structure of the TM4-Cx43CT. Assignment of the TM4-Cx43CT is a key step towards a better understanding of the structural basis of Cx43 regulation, which will lead to improved strategies for modulation of junctional communication that has been altered due to disease or ischemic injury.
Keywords: Cx43, gap junction, LPPG detergent micelles, intrinsically disordered protein
Biological context
Gap junctions are integral membrane proteins that enable the direct cytoplasmic exchange of ions and low molecular mass metabolites between adjacent cells. They provide a pathway for propagating and/or amplifying the signal transduction cascades triggered by cytokines, growth factors, and other cell signaling molecules involved in growth regulation and development. Gap junctions are created by the apposition of two connexons from adjacent cells, where each connexon is formed by six connexin proteins. Connexins are a family of proteins that share a common topology; connexins are tetra-span, integral membrane proteins that contain two extracellular loops, one cytoplasmic loop (CL), and a cytoplasmic amino- and carboxyl-terminus. Though the 21 connexin isoforms (e.g. Cx43) share significant sequence homology, major sequence divergence occurs in the carboxyl-terminal (CT) domain, which is thought to be the main regulatory domain of most connexins. The CT domain plays a role in the trafficking, size, localization, and turnover of gap junctions, as well as the level of intercellular coupling mediated by the plaque via numerous post-translational modifications and protein-protein interactions.
Early structural studies of Cx43, which used cryo-electron microscopy, and the more recent X-ray structure of Cx26 have provided a significant amount of information about channel architecture as well as connexin topology (Unger et al. 1999; Maeda et al. 2009). However, neither technique was able to address the CT structure because of the dynamic nature of the domain. These same characteristics that interfere with crystallographic techniques make NMR an ideal tool for studying the CT. Structural studies by NMR determined that the soluble Cx43CT (residues S255-I382) was predominately disordered with two short α-helices (A315-T326; D340-A348) (Sorgen et al. 2004). The cytoplasmic domains of transmembrane proteins are often intrinsically disordered; analysis of eukaryotic genomes estimated that 41% of membrane proteins have intrinsically disordered regions with 30 or more consecutive residues that are preferentially localized at the cytoplasmic side (Uversky 2011). Disordered proteins fall into 4 groups: molecular recognition, molecular assembly, protein modifications, and entropic chains. The major characteristics of these, which include protein-protein binding, flexibility, and phosphorylation, all apply to the CT (Uversky 2011). A disordered CT would play an important role in regulation, signaling, and control pathways, where binding to multiple partners via high-specificity/low-affinity interactions is facilitated by disorder to order conformational transitions.
The soluble Cx43CT has proven to be a useful model for studying the structure-function mechanisms regulating channel gating. However, when studying the structure of a soluble domain from a membrane protein, an important question is whether it has the same structural characteristics as when attached to the membrane. Several results indicate that the soluble Cx43CT may not be the best model system. For example, although most of the CT domain was missing in the Cx43 cryo-electron microscopy studies, the α-helical conformation of the 4th transmembrane domain was projected to extend beyond the membrane several residues into the CT domain (Unger et al. 1999). However, the NMR cross-peaks of the soluble Cx43CT residues S255-K264, which overlap with the end of the CT-truncated Cx43 construct used in the cryo-electron microscopy study, are weak, suggesting these residues are in exchange between two conformations (i.e. unstructured and α-helical) (Sorgen et al. 2004). Additionally, not all of the expected NOEs were observed in the two α-helical regions of the soluble Cx43CT structure (Sorgen et al. 2002). Therefore, as a proof-of-concept test, we optimized the expression, purification, and solution conditions for NMR of a more native-like construct, the Cx43CT attached to the 4th transmembrane domain (TM4-Cx43CT), solubilized in detergent micelles (Kellezi et al. 2008; Grosely et al. 2010). We demonstrated that the TM4-Cx43CT construct is folded properly and retains its ability to bind the SH3 domain of Src, an established binding partner (Kellezi et al. 2008). Circular dichroism (CD) data indicated the TM4-Cx43CT was 46% α-helical at pH 5.8 and 33% α-helical at pH 7.5 (Grosely et al 2010). Given the TM4 portion only accounts for 15% of the total protein construct, the data suggest that tethering of the CT domain stabilizes α-helices extending out from the membrane and/or induces additional structure along portions of the CT. Furthermore, the data indicate that, unlike the soluble CT, the TM4-Cx43CT is structurally responsive to changes in pH (Grosely et al. 2010). Our hypothesis is that pH-dependent conformational changes in the CT alter the thermodynamic favorability of molecular binding partner interactions (e.g. Cx43CL) involved in Cx43 channel regulation. The structural responsiveness of the CT domain when tethered to the TM4 strongly suggests the TM4-Cx43CT is a better model system than the soluble Cx43CT for investigating the molecular mechanism of Cx43 regulation.
Here we report the backbone assignments for the TM4-Cx43CT in 1-palmitoyl-2-hydroxy-snglycero-3-[phospho-RAC-(1-glycerol)] (LPPG) detergent micelles at pH 5.8 with 10% 2,2,2-trifluoroethanol (TFE). TFE has been used in a multitude of studies ranging from investigating protein-folding pathways, to evaluating disease-related effects of amino acid mutations on structural propensities and protein partner interactions (e.g. (Libich and Harauz 2008)). In this study, TFE was used to stabilize the dynamic α-helices of the Cx43CT and improve the NMR spectra. Importantly, the inclusion of 10% TFE did not induce additional α-helical structure (Grosely et al. 2010). The rationale for the resonance assignments and eventual structure determination of the TM4-Cx43CT is that this construct will enable us to 1) define the CT residues that form α-helical structure in the presence of a membrane environment, 2) identify which CT residues have altered secondary structure due to changes in pH and/or phosphorylation state, 3) analyze if altering the CT secondary structure by pH and/or phosphorylation state is a mechanism that modulates the binding affinity for protein partners involved in Cx43 regulation, and 4) model the next generation of molecules that could potentially regulate Cx43 function.
Methods and experiments
Expression, purification, and optimization of solution NMR conditions for the TM4-Cx43CT have been previously described (Kellezi et al. 2008; Grosely et al. 2010). Quick Change Lightning (Agilent) was used to create several point mutants of the TM4-Cx43CT to aid in the assignments: TM4-Cx43CTC123,R124A; TM4-Cx43CTN125,Y126A; TM4-Cx43CTY247,Y265D; TM4-Cx43CTS255,S262D; TM4-Cx43CTS368,S372D; and TM4-Cx43CTS364,S365,S369,S373D. All mutants were purified using the purification procedure developed for the wild-type TM4-Cx43CT. NMR samples contained 850 µM of 15N-labeled or 13C, 15N-labeled protein in a buffer containing 20 mM 2-(N-morpholino) ethanesulfonic acid (MES), 50 mM sodium chloride, 1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM dithiothreitol (DTT), 8% (w/v) LPPG, 10% (v/v) TFE, and 7% (v/v) D2O.
NMR data were acquired at 42°C on either a Varian INOVA 600 MHz NMR spectrometer fitted with a cryo-probe at the University of Nebraska Medical Center (Omaha, NE) or a Bruker Avance 800 MHz NMR spectrometer fitted with a TCI cold probe at the University of Kansas (Lawrence, KS). Backbone resonance assignments were obtained using standard 2D and 3D experiments (15N-HSQC, HNCACB, CBCA(CO)NH, HNCO, HN(CA)CO, 15N-TOCSY-HSQC and 15N-NOESY-HSQC) using water as the reference. The pH and sample stability were monitored by comparison of 15N-HSQC spectra collected before and after each 3D experiment. All data were processed using NMRPipe/NMRDraw (Delaglio et al. 1995) and analyzed using NMRView (Johnson 2004). Secondary structure prediction for the TM4-Cx43CT was done with the chemical shift index (CSI) calculation function in NMRView using the Wutherich reference.
Extent of assignments and data deposition
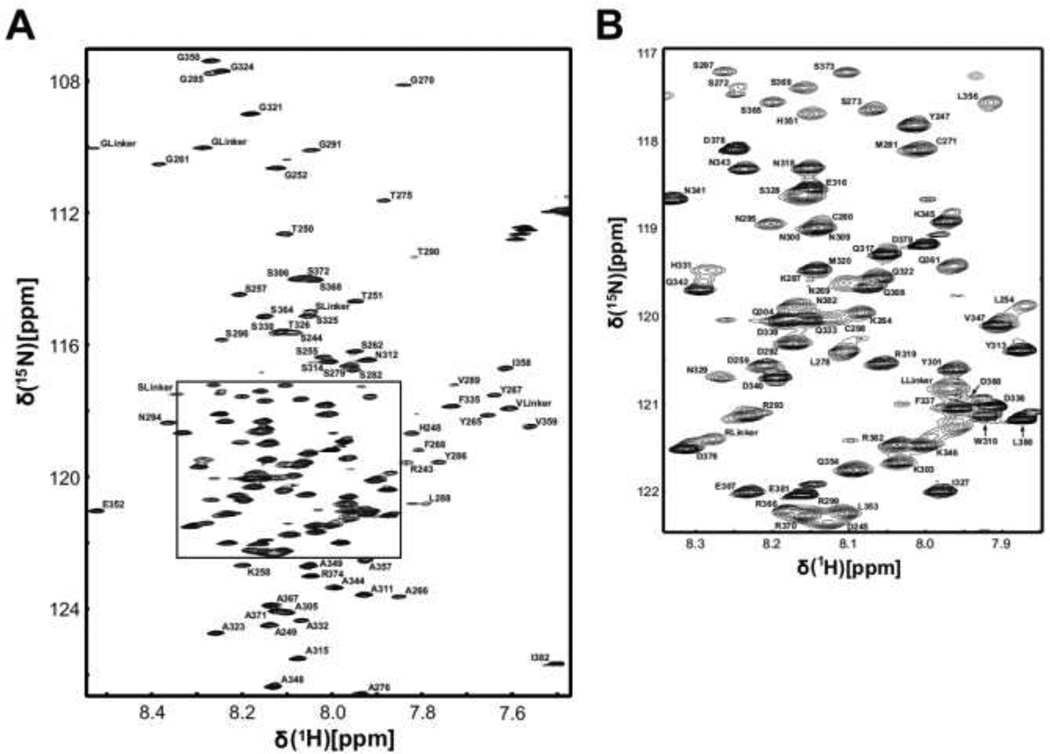
The TM4-Cx43CT construct contains 207 amino acids (17 prolines), the first 21 of which are a 6× His-tag followed by a linker sequence and part of the 2nd extracellular loop from Cx43. The amino acid sequence of the TM4-Cx43CT corresponds to Cx43 residues D197-I382 (186 residues, 15 prolines). Based on the TMpred prediction program results (http://www.ch.embnet.org/software/TMPRED_form.html), the TM4 is contained within residues I208-F233 and the CT portion comprises the remaining 149 residues (K234-I382). Figure 1 is the 15N-HSQC spectrum for the TM4-Cx43CT. With the exception of the first nine consecutive CT peaks (K234-G242), all remaining CT non-proline backbone 1H, 15N, and 13C resonances and proline backbone 13C resonances have been assigned (140 amino acids; V243-I382). Assignments for the TM4-Cx43CT were deposited in the BioMagResBank (http://www.bmrb.wisc.edu) under the accession number 18552.
Figure 1. 600 MHz 15N-HSQC spectra of the TM4-Cx43CT.
A) Peak assignments for the backbone amides are indicated with numbering corresponding to the full-length Cx43 protein. B) Magnification of the TM4-Cx43CT 15N-HSQC spectrum in panel A (boxed region).
Excluding the linker and tag, the first 46 residues (D197-G242) of the TM4-Cx43CT, which are the TM4 residues and residues immediately flanking the TM4, were not assigned. Assignment of these residues was most likely complicated by the increased linewidths associated with slower tumbling in the LPPG detergent micelles. Upon checking at the level of noise in the 15N-HSQC, we did observe very weak signals for ~20 peaks. In an effort to improve the signal for these weak peaks, we increased the protein concentration. However, attempts to increase the protein concentration beyond 850 µM actually caused a dramatic decrease in spectral quality, even when the concentration of LPPG micelles was increased concomitantly to maintain a 1:1 protein to micelle ratio. Assignment for the TM4 would have been ideal; nevertheless, the structure of the TM4 is well established as α-helical in the literature and our CD data indicated the TM4 portion of the TM4-Cx43CT is α-helical in the LPPG detergent micelles (Grosely et al. 2010). Conversely, very little is known about the structure of the CT domain when in its native environment (e.g. tethered to the membrane); assignments are a step towards this goal.
Assignment of the CT portion of the TM4-Cx43CT was not straightforward due to chemical shift overlap and sequence redundancy in the TM4-Cx43CT. Several point mutants were used to aid in the assignment of the TM4-Cx43CT. Point mutants TM4-Cx43CTC298,R299A and TM4-Cx43CTN300,Y301A were used to assign the specific mutated residues as well as confirm the assignment of residues S296, S297, R299, Y301 N302 and K303. Additionally, phospho-mimetic point mutations were constructed at sites of known Cx43CT phosphorylation (TM4-Cx43CTY247,Y265D, TM4-Cx43CTS255,S262D, TM4-Cx43CTS368,S372D, and TM4-Cx43CTS364,S365,S369,S373D). We have identified using CD and NMR that the α-helical content of the TM4-Cx43CT localized to the area of these phospho-mimetics is altered (data not shown). These changes in the 15N-HSQC were used to help assign the resonances in regions of sequence redundancy (e.g. S364SRASSRASSR374) and validate assignments throughout the CT portion of the TM4-Cx43CT.
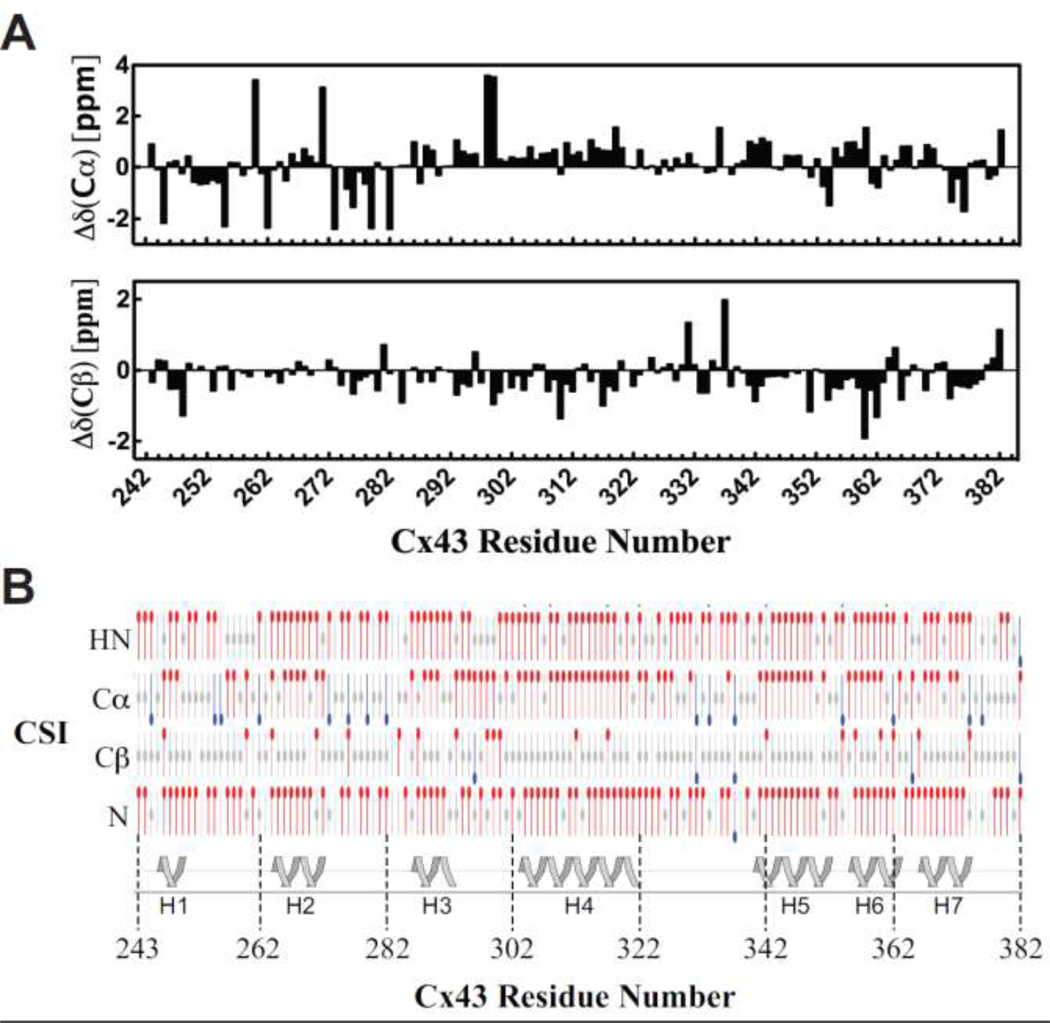
Previously, CD data showed that the TM4-Cx43CT has more α-helical content than can be attributed to just adding the TM4 to the soluble Cx43CT suggesting the TM4 provides structural stability to the CT (Grosely et al. 2008). To identify which region(s) of the CT are α-helical, the chemical shift index (CSI) values for the 13C and 13C were plotted as a function of residue number (Figure 2A) and used, along with the CSI values for the 1HN and 15N to predict the secondary structure (Figure 2B). The results from the CSI analysis identified seven intermittent regions along the CT portion of the TM4Cx43CT to be α-helical (H, helix; H1, Y247-T251; H2, K264-272; H3, K287-D292; H4, K303-A322; H5, D340-L353; H6, L356-R362; H7, R366-R374). H1, H2, and H3 are consistent with previous cryo-electron microscopy studies that projected the α-helical conformation of the TM4 to extend several residues beyond the membrane into the Cx43CT domain. Additionally, a 26mer peptide of the Cx43 tubulin binding domain (K234-D259), which has been shown by NMR to adopt an α-helical conformation upon binding to tubulin, overlaps with H1 and H2 (Saidi Brikci-Nigassa et al. 2012). The two α-helical regions along the CT identified by the NMR solution structure of the soluble Cx43CT are contained within H4 and H5 of the TM4-Cx43CT (Sorgen et al. 2004). Interestingly, the H4 and H5 span a greater area further suggesting that the structural stability provided by TM4 propagates along the CT and is not just limited to regions of the CT directly adjacent to the TM4. Finally, the seven α-helical regions of the CT (~30% of the TM4-Cx43CT) together with the α-helical TM4 portion (~15% of the TM4-Cx43CT) are consistent with the total amount of α-helical content observed from CD data of the whole TM4-Cx43CT construct (Grosely et al. 2008).
Figure 2. Chemical shift index (CSI) of the carboxyl terminus (CT) portion of the TM4-Cx43CT.
A) Deviations from random coil 13Cα (upper panel) and 13Cβ (lower panel) chemical shifts plotted as a function of residue number. B) Graphical representation of results from CSI calculations using the Wutherich reference for 1HN, 13Cα, 13Cβ, and 15N atoms: red circles at the top are chemical shifts consistent with α-helical structure; blue circles at the bottom are chemical shifts consistent with sheet structure; and gray circles at the center are for intermediate shifts. The cartoon illustrates regions of the CT portions of the TM4-Cx43CT determined to α-helical based on the CSI values. The seven α-helical (H) regions are numbered H1–H7.
Results of the CSI calculations were confirmed by evaluation of NOE data (data not shown). Numerous, medium range, backbone proton NOEs expected for these α-helical regions are present; however, not all expected NOEs were observed. The lack of NOEs supports the hypothesis that these CT α-helical regions are dynamic; a key attribute for the function of an intrinsically disordered protein. The additional α-helical regions identified in the TM4-Cx43CT not present in the soluble Cx43CT indicate that the CT domain has different structural characteristics when attached to the TM4 further supporting the TM4-Cx43CT as a better, more native-like model system compared to the soluble CT for studying the structure/function mechanism of Cx43 gap junction channel regulation.
Acknowledgements
This work is funded by the United States Public Health Service Grant, GM072631. Rosslyn Grosely is funded by the Graduate Assistance in Areas of National Need (GAANN) Fellowship, McDonald Fellowship, Regents Tuitions Fellowship, and the Fred W. Upson Grant. We would like to acknowledge Dr. Asokan Anbanandam for his assistance in collecting data at the University of Kansas NIH Center of Biomedical Research Excellence in Protein Structure and Function, which is supported by NIH grant RR-017708; Ed Ezell, who manages the NMR facility at the University of Nebraska Medical Center; and Jeff Lovelace for his heroic efforts in our computer lab.
Footnotes
This work was supported by United States Public Health Service Grant GM072631.
References
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. Journal of biomolecular NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Grosely R, Kieken F, Sorgen PL. Optimizing the solution conditions to solve the structure of the Connexin43 carboxyl terminus attached to the 4(th) transmembrane domain in detergent micelles. Cell communication & adhesion. 2010;17(2):23–33. doi: 10.3109/15419061.2010.487956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods in molecular biology (Clifton, NJ. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- Kellezi A, Grosely R, Kieken F, Borgstahl GE, Sorgen PL. Purification and reconstitution of the connexin43 carboxyl terminus attached to the 4th transmembrane domain in detergent micelles. Protein expression and purification. 2008;59(2):215–222. doi: 10.1016/j.pep.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libich DS, Harauz G. Solution NMR and CD spectroscopy of an intrinsically disordered, peripheral membrane protein: evaluation of aqueous and membrane-mimetic solvent conditions for studying the conformational adaptability of the 18.5 kDa isoform of myelin basic protein (MBP) Eur Biophys J. 2008;37(6):1015–1029. doi: 10.1007/s00249-008-0334-8. [DOI] [PubMed] [Google Scholar]
- Maeda S, Nakagawa S, Suga M, Yamashita E, Oshima A, Fujiyoshi Y, Tsukihara T. Structure of the connexin 26 gap junction channel at 3.5 A resolution. Nature. 2009;458(7238):597–602. doi: 10.1038/nature07869. [DOI] [PubMed] [Google Scholar]
- Saidi Brikci-Nigassa A, Clement MJ, Ha-Duong T, Adjadj E, Ziani L, Pastre D, Curmi PA, Savarin P. Phosphorylation controls the interaction of the connexin43 C-terminal domain with tubulin and microtubules. Biochemistry. 2012;51(21):4331–4342. doi: 10.1021/bi201806j. [DOI] [PubMed] [Google Scholar]
- Sorgen PL, Duffy HS, Cahill SM, Coombs W, Spray DC, Delmar M, Girvin ME. Sequence-specific resonance assignment of the carboxyl terminal domain of Connexin43. Journal of biomolecular NMR. 2002;23(3):245–246. doi: 10.1023/a:1019892719979. [DOI] [PubMed] [Google Scholar]
- Sorgen PL, Duffy HS, Sahoo P, Coombs W, Delmar M, Spray DC. Structural changes in the carboxyl terminus of the gap junction protein connexin43 indicates signaling between binding domains for c-Src and zonula occludens-1. J Biol Chem. 2004;279(52):54695–54701. doi: 10.1074/jbc.M409552200. [DOI] [PubMed] [Google Scholar]
- Unger VM, Kumar NM, Gilula NB, Yeager M. Three-dimensional structure of a recombinant gap junction membrane channel. Science (New York, NY. 1999;283(5405):1176–1180. doi: 10.1126/science.283.5405.1176. [DOI] [PubMed] [Google Scholar]
- Uversky VN. Intrinsically disordered proteins from A to Z. The international journal of biochemistry & cell biology. 2011;43(8):1090–1103. doi: 10.1016/j.biocel.2011.04.001. [DOI] [PubMed] [Google Scholar]