Abstract
Adeno-associated virus 2 (AAV2) is a helpervirus-dependent parvovirus with a bi-phasic life cycle comprising latency in absence and lytic replication in presence of a helpervirus, such as adenovirus (Ad) or herpes simplex virus type 1 (HSV-1). Helpervirus-supported AAV2 replication takes place in replication compartments (RCs) in the cell nucleus where virus DNA replication and transcription occur. RCs consist of a defined set of helper virus-, AAV2-, and cellular proteins. Here we compare the profile of cellular proteins recruited into AAV2 RCs or identified in Rep78-associated complexes when either Ad or HSV-1 is the helpervirus, and we discuss the potential roles of some of these proteins in AAV2 and helpervirus infection.
Keywords: AAV2, HSV-1, adenovirus, replication compartments.
ADENO-ASSOCIATED VIRUS - BIOLOGICAL PROPERTIES
AAV2 is one of the most promising vectors for human gene therapy [1]. The apparent lack of pathogenicity and low immunogenicity together with the capability to infect both dividing and non-dividing cells constitute perfect characteristics for a transgene delivery vector. AAV2 is a small, icosahedral and non-enveloped particle, which has a size of about 20-22 nm in diameter. Its genome consists of a linear single-stranded DNA with a size of 4.7kb and encodes two open reading frames (ORF), rep and cap. The rep ORF is regulated by two promoters (p5 and p19) which in cooperation with a common splicing site encode four Rep proteins termed Rep40, Rep52, Rep68 and Rep78, named according to their apparent molecular weight. The two large Rep proteins Rep68 and Rep78 are required for AAV2 DNA replication, self-regulation of transcription, site-specific integration as well as for inhibition of helpervirus replication, whereas the smaller Rep40 and Rep52 are believed to be required for packaging [2-4]. The cap ORF is controlled by the p40 promoter and encodes the three structural proteins VP1, VP2 and VP3 [5,6]. Moreover, a nested alternative ORF of the cap gene has been shown recently to encode a protein, designated assembly-activating protein (AAP), which is believed to be required for AAV2 capsid assembly in the nucleolus [7].
AAV2 belongs to the family of the Parvoviridae and the genus Dependovirus, as its replication depends on the simultaneous infection with a helper virus, such as Ad, HSV-1, or papillomavirus [8-12]. In absence of a helper virus however, AAV2 establishes a latent infection, either by maintaining its genome in an episomal state [13] or integrating it into a well-defined locus termed AAVS1 on human chromosome 19 (Ch19) at position 19q13.4 [14-18]. Upon infection with a helpervirus, AAV2 is rescued from latency and enters a lytic life cycle in which the viral DNA is replicated and progeny virus is produced.
The AAV2 DNA has a unique structure. Its two ends are characterized by 145 nt palindromic sequences termed inverted terminal repeats (ITR) which fold into hairpins. Just as unique as the AAV2 DNA structure is the rolling-hairpin mechanism of AAV2 DNA replication: the end of the 3’-ITR acts as the essential primer for second-strand synthesis, a process termed self-priming. The following asymmetric leading-strand DNA synthesis resembles a rolling-circle model of DNA replication, resulting in a closed-end intermediate at the 3’-ITR. This structure is resolved by the so called terminal resolution step, which involves a site- and strand-specific endonuclease cut performed by Rep68/78 at the so called terminal resolution site (TRS; [19-21]). The following unwinding of the remaining 3’-ITR allows to complete the replication process.
ADENOVIRUS AND HERPES SIMPLEX VIRUS TYPE 1 - BIOLOGICAL PROPERTIES
Adenovirus is the best characterized helper virus for AAV2. Adenoviruses are non-enveloped, double-stranded DNA viruses. Their capsid is characterized by a well-defined icosahedral structure consisting of three major proteins, hexon (II), penton base (III) and long knobbed projection termed fiber (IV), which is required to bind to the cell surface adenoviral receptor (CAR). The linear 26-45 kb dsDNA has inverted terminal repeats (ITRs) at each end, where a terminal protein (TP) is covalently attached to it [22]. The genome encodes four early transcription cassettes termed E1 (E1A and E1B), E2, E3 and E4, and five late gene products designated L1 to L5 which result from a series of splicing events of the major late (ML) transcript and constitute the structural proteins.
The adenovirus life cycle can be divided into two phases. The early phase is determined as the entry of the particle into the cell and the trafficking of the viral genome to the nucleus where expression of the early genes occurs. The second, or late, phase includes the virus DNA replication and the expression of late genes, followed by virus assembly and egress from the nucleus and the cell.
HSV-1 is another well characterized and efficient helper virus for AAV2 replication. It is a widespread human pathogen which has also both a lytic and a latent phase. It infects mainly mucosal tissue and neurons. The viral particle is composed of three structural compartments, (i) the viral envelope, (ii) the tegument consisting of a distinct set of viral proteins and (iii) the capsid which encloses the viral genome. The HSV-1 genome is a double-stranded DNA of 152 kbp which encodes at least 84 proteins.
HSV-1 can enter the cell both by fusion at the cell membrane or endocytosis and fusion with the endosomal membrane. The capsid is transported along microtubules to the nuclear pores, where the HSV-1 DNA, along with specific HSV-1 tegument proteins such as VP16 and the virion host shut-off protein, enters the nucleus. VP16 then transactivates the expression of the viral immediate early (IE) genes via the cellular RNA polymerase II [23]. The IE proteins have regulatory functions and activate the expression of the early (E) genes. Many of the E gene products are enzymes involved in DNA metabolism and their synthesis initiates the replication of the HSV-1 genome. The IE gene products and the replication of the HSV-1 DNA then activate expression of the late (L) genes, which encode structural components of the virion. HSV-1 can also establish latent infections, primarily in sensory neurons. During latency, HSV-1 gene expression is limited to the latency-associated transcripts (LATs) and no viral proteins are synthesized [24]. The mechanisms that control productive or latent infection of HSV-1 are not fully understood.
THE NUCLEAR REPLICATION COMPARTMENTS OF DNA VIRUSES
Viral RCs Develop Adjacent to PML Bodies
Similar to many other DNA viruses including Ad and HSV-1, AAV2 DNA replication takes place in so called viral replication compartments (RCs) or replication centers in the host cell nucleus [25-29]. The cell nucleus is a highly structured and compartmentalized space, containing not only the genome but also a multitude of proteins organized in subnuclear organelles [30]. Several of these nuclear bodies including clastosomes, nuclear speckles, nucleoli, or promyelocytic leukemia (PML) bodies contain proteins involved in central cellular processes such as proteolysis, transcription, apoptosis, or DNA-damage sensing and repair [31]. It seems to be a general feature of nuclear replicating DNA viruses that incoming viral genomes initially associate with PML bodies (also termed as ND10 nuclear bodies) and that the viral replication centers mature juxtaposed to these nuclear subcompartments [32-34]. PML bodies are functionally complex, containing proteins not only involved in DNA replication, transcription, or epigenetic silencing but also in the host defense mechanisms against viral infection [35]. It seems that viruses have evolved strategies to exploit PML bodies in order to initiate viral gene transcription and viral DNA replication [32,36-39]. For that reason, the often observed initial association of incoming viral genomes with PML bodies [32-34] might be a consequence of the cellular front line defense against viruses, however efficiently hijacked by the virus. During Ad supported AAV2 replication, AAV2 DNA has been found associated also with PML bodies [25], similar to cells infected with Ad alone [37,38], and AAV2 RCs mature juxtapose to PML bodies [25]. With the onset of infection, several replication and repair proteins, normally localized to PML bodies, have been found associated with Ad supported AAV2 RCs (discussed below). The HSV-1 ICP0 protein can efficiently disrupt PML bodies [40], and indeed, HSV-1-supported AAV2 RCs did not co-exist with PML bodies [25]. Nevertheless, integral cellular PML bodies might have an effect on initial AAV2 gene expression as well as the composition of cellular proteins in AAV2 RCs, as similar to Ad coinfection, several cellular replication and repair factors of PML bodies accumulate also in HSV-1 supported AAV2 RCs (discussed below).
Spatial and Temporal Organization of RCs
Viral RCs are composed of a multitude of cellular proteins. These factors may alter depending on the necessity of the different viruses but also depending on the different functions of viral RCs, which may change spatially and temporally over the course of replication. In HSV-1 infected cells, besides viral DNA and RNA metabolism, capsid assembly and genome packaging is also localized to viral RCs [41-43]. By contrast, Ad DNA replication and assembly is suggested to occur in distinct nuclear compartments [44]. AAV2 capsid assembly has been observed to occur also dislocated from viral RCs in nucleoli [45]. However, the site of genome packaging into pre-assembled capsids remains to be identified.
In the host cell, high transcriptional activity in G1 phase [46] is separated in a time dependent manner from genome replication in S-phase of the cell cycle. In contrast, viruses have to create an environment which enables high transcriptional activity of viral genes as well as the production of viral DNA for progeny virions in parallel. Therefore, viral RCs have to be well structured microenvironments not only in a temporal but also in a spatial manner. For example, Ad RCs are surrounded by ring-like structures called peripheral replicative zones, where Ad DNA replication, transcription, and pre-mRNA processing occur [47,48]. The defined pattern of VP5, ICP4, and ICP8 within HSV-1 RCs [41] also indicates a complex organization. In contrast, besides the well described vast growth dynamics of AAV2 RCs [25], not much is known about their spatial and temporal organization. The identification of factors associated with AAV2 RCs, as well as the definition of their localization may help to better understand the organization of helper supported AAV2 replication.
HSV-1 & AD HELPER VIRUS PROTEINS INVOLVED IN AAV2 REPLICATION
In addition to multiple cellular proteins, a defined set of helper virus proteins are involved in AAV2 replication. The best studied helper virus proteins for AAV2 replication are provided by Ad and HSV-1. In case of Ad, these helper factors include E1A, E1B55K, E2A, E4orf6, and the VA RNA [49-51]. The Ad E1A protein not only promotes transcription of the early Ad genes, but also transcription ofthe AAV2 Rep ORF [52]. E1B55K and E4orf6 form a heterodimer that supports AAV2 mRNA export from the nucleus [53], and the VA RNAs play a central role in AAV2 infection by maintaining protein translation [54]. The DNA-binding protein E2A supports multiple steps of the AAV2 life cycle including regulation of AAV2 gene expression, viral mRNA processing and export, and viral DNA replication [55,56].
The minimal set of HSV-1 helper proteins required for AAV2 replication includes the helicase-primase complex encoded by UL5, UL8, and UL52 and the major DNA binding protein ICP8 (UL29; [57-59]). All of these proteins are also mandatory for HSV-1 replication [60]. In addition, the HSV-1 origin binding protein (UL9), the viral DNA polymerase (UL30) and the ds DNA binding protein UL42 have been found to support AAV2 replication [57,58]. It has been shown that the UL5 helicase activity is necessary for efficient AAV2 DNA replication, whereas the primase-activity of UL52 is redundant [61]. Although not much is known about the detailed function of ICP8 in AAV2 replication, it seems that it is linked to the interaction with AAV2 Rep78, in an AAV2-ss DNA dependent manner [58,59,62].
The minimal set of HSV-1 helper proteins (UL5, UL8, UL29, UL52) together with two other HSV-1 proteins, the viral DNA polymerase (UL30) and the ds DNA binding protein UL42, along with AAV2 Rep68 have been shown to be also sufficient to initiate replication of duplex DNA, containing the AAV2 origins of DNA replication, in an in vitro replication system [59].
CELLULAR PROTEINS ASSOCIATED WITH HSV-1 OR AD SUPPORTED AAV2 RCs
While helper virus proteins involved in AAV2 replication are well studied [63], not much is known about the role of host proteins recruited to AAV2 RCs. Nevertheless, these proteins are likely implicated in several steps of the AAV2 life cycle including AAV2 DNA replication, gene expression and posttranscriptional modification. The recruitment of host cell proteins into AAV2 RCs may alter, depending on the type of helper virus. However, it is likely that AAV2 might also recruit a set of indispensable cellular proteins, independent of the type of helper virus.
In order to identify host proteins associated with AAV2 replication, co-immunoprecipitation (co-IP) assays of AAV2 Rep78 and immunofluorescence analysis in presence or absence of a helper virus have been performed (Tables 1 and 2; [64-73]). However, the majority of proteins were identified only by mass spectrometry of Rep78-associated complexes [64,68], and many of these interactions remain to be validated. Nevertheless, our aim was to analyze these cellular proteins concerning similarities and differences. We used the string database (http://string-db.org/) to reconstitute possible interaction networks between identified cellular proteins associated with AAV2 RCs in presence of Ad (Table 1, Fig. 1) or HSV-1 (Table 2, Fig. 2). The database lists confident known and predicted direct (physical) and indirect (functional) interactions between cellular proteins, based on experimental validations.
Table 1.
Proteins Identified in Rep78-Associated Complexes and/or AAV2 RCs During Ad Supported AAV2 Replication
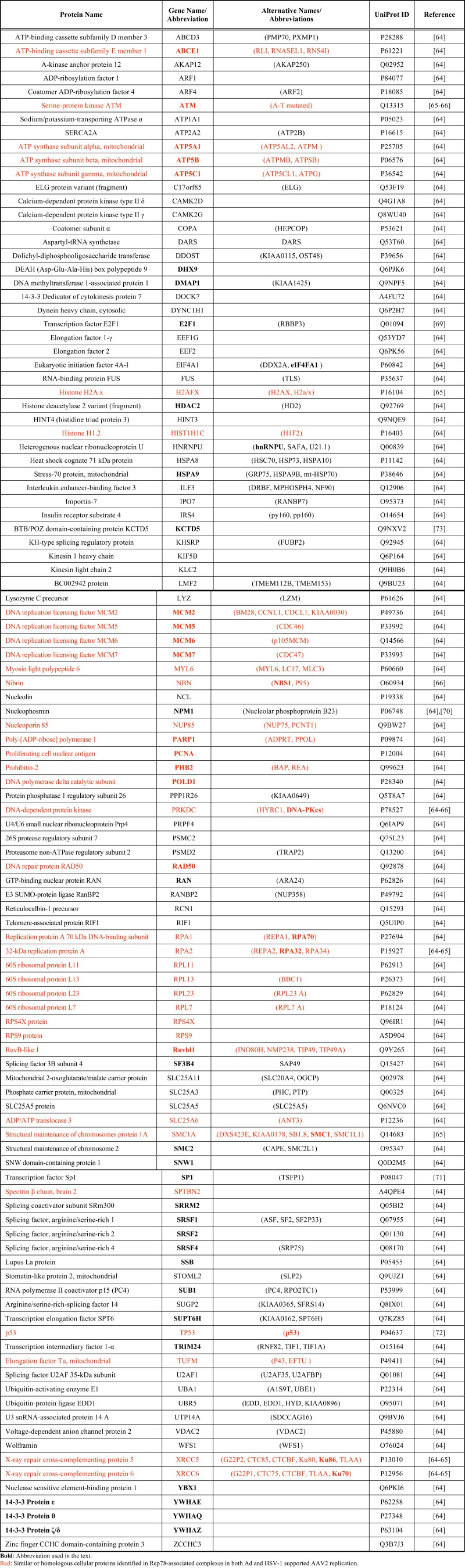 |
Table 2.
Proteins Identified in Rep78-Associated Complexes and/or AAV2 RCs during HSV-1 Supported AAV2 Replication
 |
Fig. (1).
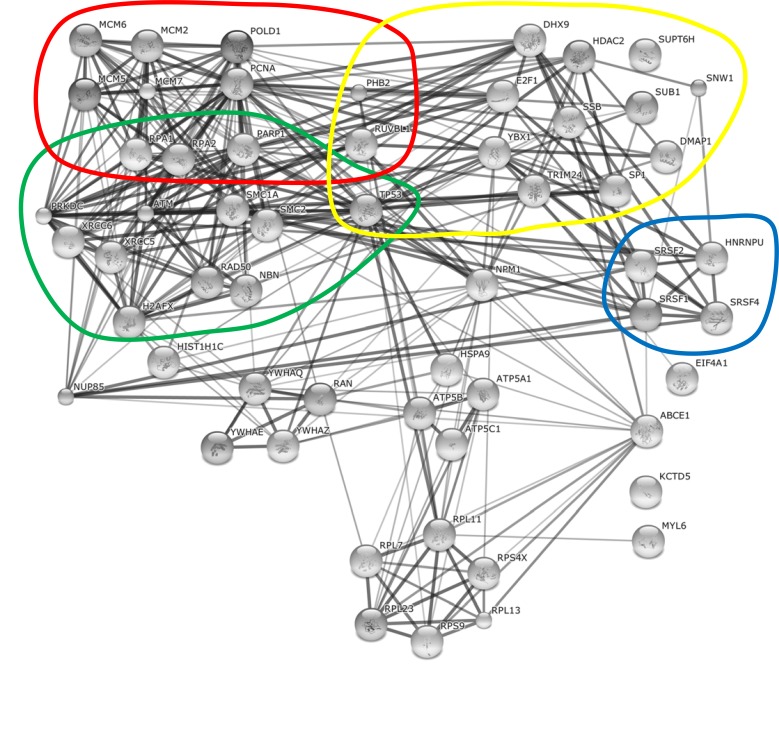
Cellular proteins associated with Ad supported AAV2 RCs. The String database (http://string-db.org/) was used to reconstitute the interaction networks between identified cellular proteins associated with Ad supported AAV2 RCs and/or the AAV2 Rep78 protein. The database lists confident known and predicted direct (physical) and indirect (functional) interactions between cellular proteins based on experimental validations. Four functional categories, mRNA processing (blue), DNA replication (red), transcription (yellow), and DNA damage sensing and repair (green) are highlighted.
Fig. (2).
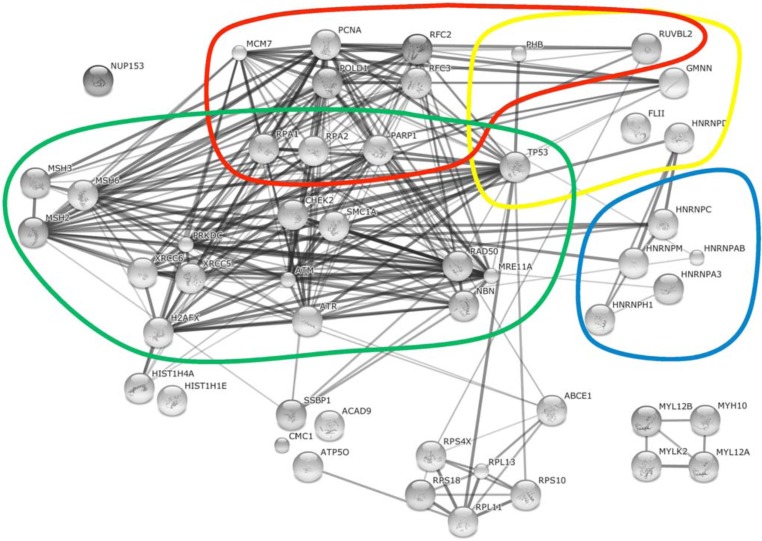
Cellular proteins associated with HSV-1 supported AAV2 RCs. The String database (http://string-db.org/) was used to reconstitute the interaction networks between identified cellular proteins (as described in Figure 1) associated with HSV-1 supported AAV2 RCs and/or the AAV2 Rep78 protein. Four functional categories, mRNA processing (blue), DNA replication (red), transcription (yellow), and DNA damage sensing and repair (green) are highlighted.
Based on the string database, the two main groups of cellular proteins of HSV-1 supported AAV2 RCs include DNA replication and repair proteins, and RNA splicing factors (Fig. 2). Similar to HSV-1 coinfection, a governing group of proteins associated with Ad supported AAV2 RCs belong to the cellular DNA replication and repair machinery (Fig. 1). In addition, a multitude of transcriptional regulators as well as RNA splicing factors and a noticeable network of mRNA and protein trafficking factors were identified in AAV2 and Ad coinfected cells (Fig. 1). Besides these main groups, also proteins involved in nuclear organization, translation, protein degradation, cytoplasmic signaling, and many more were identified in AAV2 Rep78 co-IP assays of cells coinfected with AAV2 and either HSV-1 or Ad as the helpervirus. In the following, we discuss a selection of these proteins in more detail including their potential functions in different steps of the AAV2 life cycle.
PROTEINS INVOLVED IN AAV2 DNA AND RNA METABOLISM
Cellular Proteins Promoting Initial AAV2 Second-Strand Synthesis and AAV2 DNA Replication
Due to the ss nature of the AAV2 genome, AAV2 replication initially requires DNA second-strand synthesis before viral genes can be expressed and viral DNA replication can occur. The formation of ds genomes is a critical step for wtAAV2 to initiate productive infection, but also for rAAV2 vectors, as this step is of importance for successful expression of transgene sequences [74]. Second-strand synthesis is believed to be mediated through the host replication machinery, but helper virus proteins might be involved as well [75].
Prior to polymerase-mediated displacement of the parental DNA strand, a pre-replication complex is formed, which loads the whole DNA replication machinery onto the DNA [76]. The MCM proteins 2-7 are essential components of this complex [76]. Several of these proteins were found in both Ad and HSV-1 supported AAV2 RCs (Tables 1 and 2, Fig. 3) and have been also found to be required to replicate AAV2 DNA from a ssDNA template in an in vitro replication assay [77,78]. Upon initial second-strand synthesis MCM might participate in the formation of a replication complex on the 3’ end of the incoming ss AAV2 DNA template. The complementary self-annealing sequence of the AAV2 ITR provides a base-paired 3’ hydroxyl group for initial unidirectional DNA synthesis [75]. Because AAV2 provides its own DNA primer, viral DNA replication exclusively uses leading-strand DNA synthesis and not lagging-strand synthesis. Although both helper viruses, Ad and HSV-1, express their own polymerase, AAV2 seems to use only the HSV-1 polymerase [57], while the Ad DNA polymerase has not been found to be involved in AAV2 DNA replication [49-51,79]. In contrast to AAV2 replication, Ad DNA synthesis uses a protein priming mechanism in which the viral polymerase (AdPol) recognizes and binds to the viral preterminal protein (pTP); the pTP-dCMP complex then serves as the primer for subsequent elongation catalyzed by AdPol via a strand displacement mechanism. Since the AdPol needs to form a stable heterodimer with pTP to initiate its DNA replication via a protein priming mechanism, and AAV2 replication uses a DNA primer to initiate viral DNA replication, it is not surprising that the Ad polymerase does not contribute to AAV2 DNA replication. HSV-1 replication is initiated by binding of the UL9 protein and unwinding of the HSV-1 ori [80]. Subsequently, the viral polymerase catalyzes DNA synthesis at the leading-strand and lagging-strand. At the eukaryotic replication fork, DNA polymerase delta (POLD) together with PCNA is suggested to build the lagging-strand replisome, and polymerase epsilon together with PCNA the leading-strand replisome [81]. However, PCNA together with POLD1 was found associated with AAV2-Rep78 during coinfection with Ad (Table 1, Fig. 1) and therefore might be the essential polymerase for leading-strand synthesis of the viral DNA. In addition, chromatin immunoprecipitation (ChIP) studies revealed that DNA POLD, but not polymerase alpha and epsilon, directly binds to UV inactivated AAV2 genomes, independent of the presence of a helper virus [82]. Collectively, these findings are in line with the observation that upon SV40 infection, POLD is also responsible for leading-strand synthesis of the viral DNA [83]. Interestingly, PCNA and POLD1 were also found in HSV-1 supported AAV2 RCs (Table 2, Fig. 2), although the HSV-1 polymerase complex has been shown to be sufficient to promote productive AAV2 infection [57,58]. Second-strand synthesis of the ssAAV2 genome leads to the formation of a duplex DNA molecule that is covalently closed at one end by the hairpin structure of the ITR initially used as a DNA primer. In order to complete replication, in a next step called terminal resolution, the remaining hairpin has to be resolved and replicated to yield a linear ds DNA [75]. For this, the AAV2 Rep protein binds to the RBS motif in order to promote cleavage of one strand at the unique TRS. The regenerated 3’hydroxyl group within the ITR then provides the basis for the replication through the viral ITR [75]. In a last step called reinitiation, a double-hairpinned intermediate is formed by denaturation and reannealing of the linear ITR [75]. This last step initiates a new round of strand displacement synthesis, generating a ss AAV2 genome that can be packaged into a capsid [75].
Fig. (3).
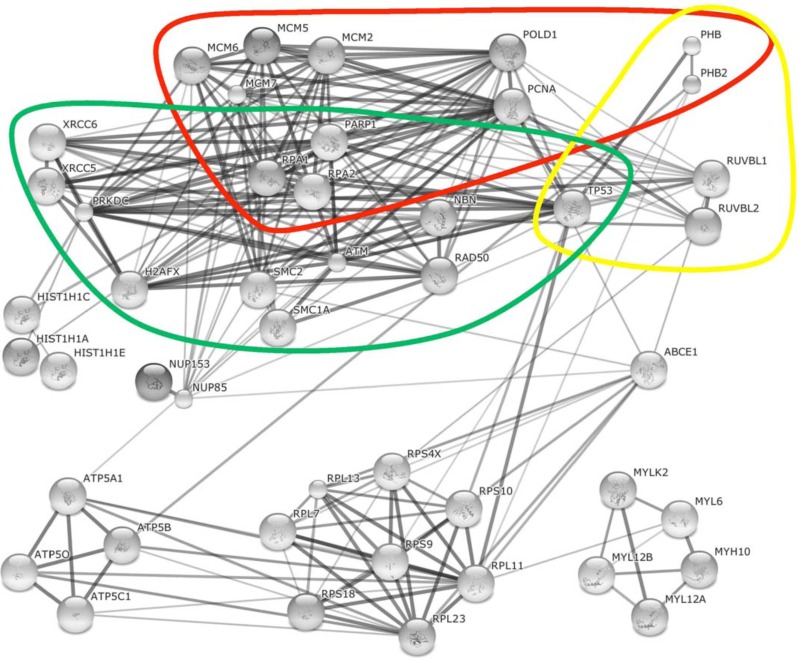
Cellular proteins and homologous proteins found in both Ad and HSV-1 supported AAV2 RCs. The String database (http://string-db.org/) was used to reconstitute the interaction networks between identified cellular proteins or homologous proteins (as described in Fig. 1) which were found associated with both Ad and HSV-1 supported AAV2 RCs and/or the AAV2 Rep78 protein. Three functional categories, DNA replication (red), transcription (yellow), and DNA damage sensing and repair (green) are highlighted.
Besides the MCM complex and the processive complex (containing PCNA and DNA polymerase), several other proteins are found in the cellular replisome complex including Claspin, And1, and RFC [84]. A combination of PCNA, POLD1, the helicase complex MCM, and RFC, together with AAV2 Rep have been shown to be required and sufficient to reconstitute efficient AAV2 DNA replication in an in vitro replication system [77,78] and all of these proteins were also found in both Ad and HSV-1 supported AAV2 RCs, except for RFC, which was found in AAV2 RCs only in cells coinfected with HSV-1. Rep78 and Rep 68 proteins have been shown to have site-specific DNA helicase and endonuclease activities required to carry out both terminal resolution and reinitiation in an in vitro assay [85-87]. Although AAV2 encodes its own helicase provided by Rep, the cellular helicase complex MCM seems to play a central role in AAV2 replication and may not only be involved in formation of the pre-initiation complex. Nash et al. suggested that a complex composed of Rep and MCM might bind to the 5′ hairpin after reinitiation of the ITR and unwind the displaced strand from the template, while POLD-PCNA extends the 3′ primer on the template strand [64]. Similar to eukaryotic DNA replication [84], the resulting ss DNA loop of the displaced strand may become coated with the host ss DNA binding protein RPA (composed of the subunits RPA70, RPA32, and RPA14), which is also found in both Ad and HSV-1 supported AAV2 RCs (Tables 1 and 2, Fig. 3). However, the observed phosphorylation in infected cells might change the function of RPA32 from DNA replication protein towards DNA damage signaling protein (discussed in more detail below). Premature activation of the DNA replication complex in G1 phase of the cell cycle has been shown to be regulated by binding of retinoblastoma protein (Rb) or prohibitin to MCM proteins [88,89]. Dissociation of prohibitin from MCM is suggested to be regulated by cell cycle regulated kinases and licenses DNA replication in S phase of the cell cycle [89]. Prohibitin (both, PHB and PHB2) was found associated with Rep in both Ad and HSV-1 supported AAV2 replication (Tables 1 and 2, Fig. 3; [64,68]). Prohibitin may have an inhibitory effect on regulation of AAV2 DNA replication; however, a direct impact has not yet been investigated. In addition to its role in preventing the activity of replisomes [89], prohibitin might also be involved in transcriptional regulation of viral genes (see below). Cyclin-dependent kinases (CDKs) and germinin are other MCM inhibitors [90]. Binding of both proteins results in the displacement of MCM from replicated DNA at fork termination in late S-phase and prevents re-replication of the cellular genome [90]. In cells coinfected with AAV2 and HSV-1, germinin was found recruited to AAV2 RCs (Table 2, Fig. 2; R. Vogel unpublished data). It is possible that a spatial and/or temporal regulation of the activity of the MCM complex by several regulators, including prohibitin and germinin, might also play a central role in the AAV2 life cycle. Spatial or temporal inhibition of AAV2 DNA replication within AAV2 RCs may be important to promote vast transcription of AAV2 genes from ds replication intermediates.
Immunofluorescence analysis revealed a more even distribution of MCM in the nucleus of cells coinfected with AAV2 and HSV-1 although it was also associated with Rep to a certain degree [68]. Similarly, besides significant overlap with Ad supported AAV2 RCs, MCM5 and MCM7 were also detected outside of AAV2 RCs [64]. It is suggested that during HSV-1-induced AAV2 DNA replication Ku proteins may substitute for MCM function in strand displacement activity [68], similar to their partial substitution of MCM in an in vitro AAV2 replication assay [64]. In addition, the essential HSV-1 helicase primase complex (UL5/8/52) is proposed to substitute for helicase activity of MCM upon strand displacement [68]. However, dislocation of MCM from AAV2 RCs might also be a consequence of a complex spatial and/or temporal regulation which might influence AAV2 DNA replication in coinfected cells.
The Role of Cellular DNA Damage Sensing and Repair Proteins in AAV2 DNA Replication
It is likely that the structure of incoming viral DNA as well as viral replication intermediates play a central role in the recruitment of cellular proteins into viral replication compartments. Both incoming Ad and HSV-1 DNA are linear ds molecules that seem to elicit an immediate DDR after entry into the nucleus [91-95]. The incoming AAV2 DNA is of ss nature, with a complex secondary structure. Therefore, it is not surprising that besides RPA several other ssDNA binding proteins including SSBP1, NPM1, RuvBL, and hnRNPU (Tables 1 and 2, Figs. 1, 2) were found associated with AAV2 RCs (discussed in more detail below). In addition, in absence of a helper virus, AAV2 DNA has been shown to elicit a DDR mediated by ATR [82], which is characteristic of the presence of ss DNA ends [96].
There is rising evidence that besides cellular replication factors, also proteins of the DNA sensing and repair machinery play central roles in replication of DNA viruses [91-95]. Three studies have intensively investigated DDR factors of Ad or HSV-1 supported AAV2 RCs [65-67]. The main kinase, required for activation of a robust DNA-damage response in AAV2 and Ad coinfected cells has been shown to be DNA-PK [65,66]. In cells coinfected with AAV2 and HSV-1, both ATM and DNA-PK seem to mediate signaling to downstream targets including p53 and Chk2, while signaling to RPA32 appears to be mediated by DNA-PK alone [67]. The DNA-PK complex colocalized with both HSV-1 and Ad supported AAV2 RCs [65,67], however, late upon coinfection with HSV-1, DNA-PKcs was degraded [67]. This might be the reason why DNA-PK was not found associated with AAV2 Rep protein in co-IP experiments [68]. Not only the AAV2 genome structure with its cis-acting replication element CARE, the ITRs, and the single-stranded nature, but also the AAV2 Rep proteins have been shown to evoke a cellular DDR and a cell cycle arrest in S phase [97,98]. The interference of AAV2 with the cellular DDR and repair machinery may not only allow control of cell cycle progression, but also promote second-strand synthesis of the AAV2 genome [74,99]. Cells exposed to a variety of genotoxic agents (e.g. HU or IR) have been shown to support significant helper-independent AAV2 rep expression and even DNA replication, although at very low levels [100-103].
Proteins of the DDR and repair machinery also influence rAAV2 vector fate [104-112]. It is suggested that the T-shaped hairpin structure within the AAV ITR and/or ss DNA-ds DNA junctions in the stem of the hairpin, may recruit DNA repair factors of the homologous recombination machinery [113]. While some proteins promote the formation of stable ds rAAV2 genomes, others decrease rAAV2 vector transduction efficiency [104-112]. In addition, genotoxic treatment has been shown to affect also integration rates or rAAV2 genomes; although these events occur at very low frequencies due to the absence of AAV2 rep expression [114,115].
Not only in absence of a helper virus, but also upon helper virus supported AAV2 replication, the accumulation of AAV2 replication intermediates harboring a covalently closed hairpin and a free DNA end is very likely to further activate and attract cellular components of the DDR signaling and repair machinery. Therefore, it is conceivably that the composition of cellular proteins in AAV2 RCs may change, depending on the prevalent structure of the AAV2 DNA during infection and replication.
The Role of the MRN Complex (and its Components) in AAV2 DNA Replication
Proteins of the MRN complex (composed of MRE11, NBS1, and RAD50) are among the first to appear at the site of DNA lesion [116]. MRN mediates downstream signaling to key proteins involved in sensing, signaling, and effector responses to DNA ds breaks [116]. In absence of a helper virus, the MRN complex has been shown to be recruited also to incoming AAV2 DNA [82,104,107]. The consequence of this recruitment on recombinant AAV2 transduction is still under debate.
An inhibitory effect of the MRN complex on transduction efficiency of recombinant AAV2 vectors due to binding of the MRN complex to AAV-ITRs has previously been reported [1,104,106,107,117]. However, upon infection with self-complementary recombinant AAV2 (scAAV) vectors, the MRE11 and NBS1 proteins of the MRN complex have been shown to be required for the formation of double-stranded circular episomes, the predominant form of AAV2 persisting in human tissue [107]. The MRN complex, although with opposite determination, is also a critical element for both helper viruses HSV-1 and Ad [118-125]. The interaction with MRN is one example how Ad and HSV-1 adopt very different ways to interfere with the host DDR. The disruption of the NHEJ machinery by E1b55K/E4orf6 mediated degradation of the MRN complex is absolutely required to prevent Ad genome concatemerization, which would prohibit viral genome packaging into virions [119,126,127]. In addition, Ad interference with these central elements of the DDR and repair system also prevents apoptosis during infection [118,119,128]. In contrast, the MRN complex is required for full activation of the ATM-mediated DDR pathway, which is suggested to support recombination dependent HSV-1 replication and the formation and stabilization of HSV-1 DNA concatemers and complex branched replication products [124,125]. Interestingly, in Ad supported AAV2 replication, the function of MRN for AAV2 replication seems to be linked to its effect on the helper virus. Ad mediated inhibition of the MRN complex has been shown to support AAV2 replication [117], and only the RAD50 and the NBS1 proteins of the MRN complex were detected in Ad supported AAV2 RCs (Table 1, Fig. 1). In contrast, all components of the MRN complex (NBS1, MRE11, and RAD50) were found recruited to HSV-1 supported AAV2 RCs (Table 2, Fig. 2; [67,68]) and so far, there is no indication of an inhibitory effect of the MRN complex on HSV-1 supported AAV2 replication. It is suggested that the utilization of the HSV-1 polymerase for AAV2 replication may bypass the inhibitory effects of MRN on second-strand synthesis and replication of AAV2 DNA under these helper virus conditions [117]. It remains to be investigated whether the MRN complex may even enhance HSV-1 supported AAV2 replication, as it enhances HSV-1 replication [124,125].
Independent of its function in the MRN complex, phosphorylated NBS1 is involved in the activation of the S-phase checkpoint [129]. In cells coinfected with AAV2 and HSV-1, abundant NBS1 was localized to AAV2 RCs [67], and it may be involved in the induction of a cell cycle arrest in these cells. Although NBS1 levels decrease with the onset of infection [66], a potential role in S-phase checkpoint activation remains to be determined in cells coinfected with AAV2 and Ad as the helpervirus.
In contrast to cells coinfected with AAV2 and Ad, but in line with the observed inhibitory effect of the MRN complex on rAAV2 vector transduction, cells deficient in the MRN target ATM, exhibit increased rAAV2 transduction efficiency [1]. It is suggested that ATM inhibits single-to double-strand conversion of rAAV2 vectors [1].
The Role of DDR Signaling via ATM and DNA-PK in AAV2 DNA Replication
Although the MRN complex is not functional in cells coinfected with AAV2 and Ad, ATM and DNA-PK, two main kinases in DDR [130] mediate downstream signaling to SMC1, Chk1, Chk2, H2AX, XRCC4, and RPA [65,66]. While H2AX, ATM and SMC1 were found in a pan-nuclear pattern, DNA-PK (DNA-PKcs, Ku70, and Ku86) and RPA32 accumulate within Ad supported AAV2 RCs (Table 1, Fig. 1; [65,66]). ATM, DNA-PKcs, Chk2, RPA32 and H2AX are phosphorylated and all but H2AX are recruited to HSV-1 supported AAV2 RCs (Table 2, Fig. 2; [67,68]). Since all proteins of the MRN complex are also associated with HSV-1 supported AAV2 RCs, it is not clear whether activation of ATM and DNA-PK signaling in these cells requires a functional MRN complex or not. Although the significance of DNA-PK in the AAV2 lytic cycle remains unclear, the activation of the ATM pathway appears to be beneficial for Ad supported AAV2 genome replication [66]. ATM signaling in cells coinfected with AAV2 and Ad may affect SMC1 and SMC2, two central components of condensin and cohesion complexes [131], which were found to be associated with Rep proteins in HSV-1 and Ad supported AAV2 replication (Tables 1 and 2, Fig. 3). Both proteins possess DNA ATPase activity and are required for the correct segregation of replicated chromosomes and many other events linked to chromatin dynamics and regulation of gene expression [131]. In addition, the cohesin complex works as a downstream effector in the ATM mediated DDR to control the S-phase checkpoint [132]. It is possible that these factors have also multiple functions in the AAV2 life cycle including checkpoint activation and viral gene expression.
Both ATM and DNA-PK are phosphatidylinositol 3-kinase-related kinases which possess serine/threonine kinase function [133]. 14-3-3 proteins (YWHA) bind to ATM and DNA-PK targets containing phospho-serine or phospho-threonine groups and regulate the function of these proteins implicated for example in cell cycle progression or apoptosis [134]. 14-3-3 proteins ε, θ, and ζ/δ were found associated with Rep proteins in cells coinfected with AAV2 and Ad (Table 1, Fig. 1). Although interaction of viral proteins with 14-3-3 proteins has been described for different viruses [135,136], it has not yet been described for HSV-1 or Ad. It is possible that the interaction of AAV2 Rep with 14-3-3 proteins in cells coinfected with Ad is part of the AAV2 mediated hijacking of cellular factors and might influence cell cycle regulation and/or apoptosis in coinfected cells.
The Multifunctional RPA Protein and AAV2 Replication
An important downstream target of the DDR is the cellular ss DNA binding protein RPA [137]. Besides its role in DNA replication, RPA is essential also for DNA recombination and repair processes [138]. In this case, the N-terminus of the 32-kDa subunit of human RPA becomes hyperphosphorylated by kinases of the cellular DDR (e.g. DNA-PK; [138]), which is suggested to cause a change in RPA conformation that down-regulates activity in DNA replication but does not affect DNA repair processes [138]. RPA was found in both, Ad and HSV-1 supported AAV2 RCs (Tables 1 and 2, Fig. 3) and DNA-PK-dependent phosphorylation of RPA32 has been observed also during both Ad and HSV-1 supported AAV2 replication [65-67]. It is not clear if non-phosphorylated and phosphorylated RPA coexist in AAV2 RCs. But it is possible that temporal phosphorylation of RPA may occur upon AAV2 replication which might influence its function. It is tempting to speculate that in contrast to non-phosphorylated RPA, phosphorylated RPA can participate in DDR signaling upon AAV2 replication and not directly promote the process of AAV2 strand displacement replication in vivo [77]. A similar phenomenon has been observed in cells infected with SV40 [139]. Although RPA maintained SV40 DNA replication in vitro, it has not been found to be involved in replication of SV40 DNA in vivo; more precisely, RPA is phosphorylated in vivo and therefore not able to localize to SV40 RCs [139].
Non-Homologous End Joining (NHEJ) and Homologous Recombination (HR)
While components of the non-homologous end joining (NHEJ) machinery have been shown to have both inhibitory (e.g. Ku70; [140]) and supportive (e.g. DNA ligaseIV/XRCC4; [141]) effects on HSV-1 replication, NHEJ proteins are not beneficial for Ad infection in general [118,119,126-128]. The impact of NHEJ proteins on AAV2 replication is less well understood. Besides the MRN complex and LigaseIV/XRCC4, the central element in NHEJ is the DNA-PK complex [130]. In both, Ad and HSV-1 supported AAV2 replication, DNA-PK was activated and recruited into AAV2 RCs (Tables 1 and 2, Fig. 3 [65,67]). In addition, signaling to XRCC4 was observed when Ad was the helpervirus [65]. It is suggested that DNA-PK induces the phosphorylation of histone 1 (H1) at DNA ds breaks in order to reduce its affinity for DNA and thereby support access of DNA ligaseIV/XRCC4 to broken DNA ends [142]. Histone 1 (H1F2 or H1E) was also detected associated with both Ad and HSV-1 supported AAV2 RCs (Tables 1 and 2, Fig. 3) and may therefore be involved in regulating the access of DNA repair factors to AAV2 DNA. However, another report showed that not the catalytic subunit (cs) of DNA-PK, but Ku70 and Ku80 are required to recruit ligaseIV/XRCC4 to sites of ds breaks and promote end-joining events [143]. It has been shown that Ku proteins directly interact with the AAV2 ITR hairpins [111]. Therefore, both Ku binding to AAV2 DNA [111] as well as the association of DNA-PKcs with Rep78 [64,144] may recruit the DNA-PK complex to AAV2 RCs. However, the role of DNA-PK and other factors of the NHEJ machinery in AAV2 replication is still under debate. In one report, rAAV2 replication in the presence of HSV-1 or Ad helper functions has been shown to be decreased in absence of DNA-PK [111] while another report showed enhanced wtAAV2 genome replication in absence of DNA-PK activity when Ad was used as the helper virus [66]. In the latter case, it has been suggested that loss of DNA-PK could lead to reduced circularization of the AAV2 genome which might promote AAV2 DNA replication events [66]. Indeed, DNA-PK has been shown to support the formation of stable ds rAAV2 vector genomes including circular monomers and concatemers [108,112]. Besides DNA-PKcs, the following repair proteins have been shown to also support rAAV2 recombination events: the MRN complex, ATM, Artemis, BLM, and WRN [105-110,112]. There is evidence that the hairpins at AAV-ITRs are targeted by the cellular DNA repair machinery, including DNA-PK [111] and Artemis [109] similar to DNA hairpin structures in mammalian cells emerging during the V(D)J recombination or NHEJ [145]; this may support viral genome recombination [109], self-circularization [108,112], concatemerization [146], and genomic integration [3] of the rAAV2 genomes. The impact of these proteins on helper virus supported AAV2 replication is not clear. Artemis and ATM are involved in both HR and NHEJ events, depending on the cell-cycle phase [147]. While ATM was found associated with both Ad and HSV-1 supported AAV2 replication (see above, Tables 1 and 2, Fig. 3), Artemis as well as BLM and WRN, two proteins of the RecQ helicases complex involved in HR events and stabilization of replicating DNA [148], were not found associated with AAV2 RCs. This is consistent with Ad infection, where BLM is degraded [149], but in contrast to HSV-1 infection, where BML is recruited into viral RCs [140].
Independent of its role in NHEJ events, there is preliminary evidence that DNA-PKcs may mediate also modification of the large Rep proteins upon AAV2 and Ad coinfection [66]. The group of J.P. Trempe showed that phosphorylation of the AAV2 Rep proteins alters their interactions with the AAV2 ITRs [150]; thus DNA-PK may play an essential role in regulating Rep mediated processes of viral DNA replication [65,66], including terminal resolution.
The Role of NHEJ in AAV2 Integration Events
Although the data is inconsistent, the NHEJ pathway seems to influence AAV2 integration events. In cell culture experiments, DNA-PKcs has been shown to enhance integration of both ss rAAV2 and ds rAAV2 vectors [106]. Another report showed that the NHEJ factor ligase IV in particular supports integration of incoming ss AAV2 vector genomes [151]. Another factor implicated in V(D)J recombination events is the high mobility group protein 1 (HMG1; [152]). Although not identified in presence of Ad and HSV-1, this protein has been shown to bind to Rep78 and enhance RBS binding and nicking activities of Rep as well as site-specific integration of the AAV2 genome in absence of a helper virus [153]. Although these data suggest a positive influence of NHEJ mechanisms on AAV2 integration in cell culture, rAAV2 genome integration has been shown to be strongly enhanced in DNA-PKcs-deficient SCID mice [154].
The ATR Pathway
The MRN complex and BLM as well as several factors of the ATR dependent DDR including ATR, TopBP1, Brca1, Rad17, RPA, Chk1, and Rad51 have been found recruited to AAV2 DNA in absence of a helper virus [82,104]. The group of P. Beard showed that ATR, Chk1, and BML are involved in an AAV2 DNA induced cell cycle arrest in G2 phase, likely by mimicking an aberrant cellular DNA replication fork [82]. Besides the MRN complex, RPA, and ATR, none of the other proteins (BLM, TopBP1, Brca1, Rad17, and Rad51) recruited to AAV2 DNA in absence of a helper virus were found associated with helper virus supported AAV2 RCs. Moreover, for both helper viruses, Ad5 and HSV-1, it has been shown that ATR signaling via Chk1 is inhibited in infected cells [67,118,155]. However, factors of the ATR pathway might still have an impact on viral replication, as it has been shown that ATR, RPA, TopBP1, clapsin, and CINP play a beneficial role in HSV-1 gene expression and virus production even in absence of ATR kinase activity [155,156]. The absence and/or obstruction of several proteins of the ATR pathway in presence of a helper virus implies the extensive influence of the helper virus on the cellular environment to promote AAV2 replication.
PARP
Another multifunctional cellular repair protein found associated with AAV2 RCs in presence of both Ad and HSV-1 is poly(ADP-ribose) polymerase-1 (PARP-1). This nuclear enzyme mediates poly-ADP-ribosylation (PARylation) of cellular proteins involved in several different processes including replication, recombination, repair, and cell death [157]. The addition of poly-ADP-ribosyl polymers is implicated in the recruitment of DNA damage repair factors to sites of single- and double-strand breaks [157]. In addition, PARP supports DNA stability by protecting DNA ends from nucleases [157]. A recent report showed that upon HSV-1 infection, PARP1/2 is involved in increasing total protein PARylation levels [158], which may occur in the context of DDR activation by HSV-1 [158]. There is one report, showing that the HSV-1 ICP4 protein is PARylated upon infection [159]; however, which effect this modification as well as the overall upregulation of protein PARylation has on HSV-1 infection remains to be investigated. Due to its presence in both Ad and HSV-1 supported AAV2 RCs (Tables 1 and 2, Fig. 3), it is very likely that PARP might also affect the function of viral proteins and/or cellular repair and replication factors in AAV2 RCs. In addition, PARP-1 may also be involved in the integration of the AAV2 genome into the host chromosome, similar to its role in HIV integration [160].
RUVBL1 and RUVBL2
The RuvB-like proteins (RUVBL1 and RUVBL2), also termed pontin and reptin, are members of the family of ATPases and are associated with diverse cellular activities including regulation of cell proliferation, apoptosis, transcription and DNA repair [161]. Both, pontin and reptin were found associated with Ad and HSV-1 supported AAV2 RCs (Tables 1 and 2, Fig. 3). In AAV2 replication, these proteins may be exploited to regulate cell cycle progression or apoptosis. In addition, similar to the AAV2 Rep proteins [75], pontin and reptin both possess ATPase and helicase activity [161] which might also support AAV2 DNA replication.
NUP85 and NUP153
The roles of nucleoporins in DNA repair are less well described. Two nucleoporins were found associated with AAV2 RCs, NUP85 and NUP153 (Tables 1 and 2, Fig. 3). In yeast, nucleoporins have been shown to be involved in sequestration of active sites of DNA repair to the nuclear periphery [162,163]. In human cells, there is evidence that NUP153 is essential for proper activation of the DNA damage checkpoints and that it promotes NHEJ over HR events [164]. NUP153 may also be involved in supporting NHEJ events in AAV2 infection. In addition, it is tempting to speculate that similar to their role in spatial organization of DNA repair centers in yeast [162,163], nucleoporins may play a role in spatial organization of AAV2 replication compartments.
Mismatch Repair Proteins
Another group of DNA repair proteins found in HSV-1 supported AAV2 RCs belong to the family of mismatch repair (MMR) factors. In general, MMR factors are required to maintain DNA integrity [165]. These proteins are highly conserved from prokaryotes to humans and are involved in the recognition of DNA loop or base-base mismatches, resulting from insertion/deletion or DNA polymerase proofreading errors, respectively [165]. Two mismatch repair complexes composed of MSH2 and 3 (MutSβ), and MSH2 and 6 (MutSα) are involved in the recognition of DNA mismatches. MMR factors are also involved in DDR signaling and the control of HR [165]. Similar to HSV-1 infected cells, where MSH2 and 6 have been found associated with the HSV-1 ICP8 protein [140], AAV2 Rep was found associated with MSH2, MSH3, and MSH6 (Table 2, Fig. 2; [68]) and the localization of these proteins to HSV-1 supported AAV2 RCs was confirmed by immunofluorescence experiments [68]. It is possible that MSH proteins localize only to HSV-1 but not to Ad supported AAV2 RCs because they bind to the HSV-1 ICP8 protein, which has been shown to both interact with MSH proteins [140] and localize to AAV2 RCs in coinfected cells [68]. Upon HSV-1 infection, MMR proteins have been shown to be not only recruited to viral RCs, but also to be required for efficient HSV-1 replication [156]. It is suggested that besides their proofreading role in viral DNA replication, MMR proteins may play a role in IE gene expression prior to HSV-1 DNA replication [156]. The expression of HSV-1 IE genes is necessary to promote efficient AAV2 replication. In contrast, proteins of the cellular mismatch repair machinery have not been found associated with Ad RCs and do not seem to be involved in efficient virus replication, which may also explain their absence in AAV2 and Ad coinfection. The precise function of MMR proteins for HSV-1 supported AAV2 replication remains to be elucidated.
B23/Nucleophosmin
In addition to cellular proteins directly involved in DNA- RNA- or protein-metabolism, some unexpected cellular factors were found associated with AAV2 Rep proteins. For instance B23/nucleophosmin (NPM; Table 1, Fig. 1; [64,70]), which is a multifunctional protein involved in duplication of centrosomes [166,167] and protein shuttling [168]. Although it is unclear which of the several functions of NPM may be involved in AAV2 amplification, it has been shown that nucleophosmin participates in Rep-mediated nicking at the AAV2 TRS [70]. Moreover, it is suggested that nucleophosmin has a role also in virion assembly via formation of Rep-Cap-NPM complexes [70].
Transcriptional Regulation
The AAV2 transcripts are generated from three different promoters (p5, p19, and p40) and all pre-mRNAs are generated by the cellular RNA polymerase II complex. Although none of the subunits of the TATA-box dependent transcription initiation complex (formed by TFII, TBP, and Pol II) were identified associated with AAV2 RCs in presence of a helper virus, at least TAF1 (a subunit of TFII; [144]) and TBP [169] were found associated with AAV2 Rep in absence of a helper virus. It is possible that these cellular initiation complex factors are also involved in the initiation of AAV2 transcription, similar to their role in HSV-1 [170] and Ad [171] infection. In addition, it has been shown that the TATA box in cis and the TBP in trans are involved in Rep-dependent replication from the minimal replication origin present within the AAV2 p5 promoter region [172].
Transcriptional activity is not only dependent on the formation of the initiation complex, but is tightly regulated by transcription factors and coactivators [173]. Several transcriptional regulator proteins including Sp1, E2F, p53, and PHB2 as well as transcriptional coactivators including SUB1 and FLII were found in Ad and HSV-1 supported AAV2 RCs (Tables 1 and 2, Figs. 1, 2; [64,72,100,169, 174]). It is worth mentioning that many more proteins involved in transcription (string data base) were found in Ad supported AAV2 RCs (E2F, PHB2, RuvBL1, SSB, SNW1, Sp1, SuB1, SUPT6H, p53, TRIM24, HDAC2, DMAP1, DHX9, YBX1; Table 1, Fig. 1) compared to HSV-1 supported AAV2 RCs (PHB, RuvBL2, FLII, hnRNPD, germinin, p53; Table 2, Fig. 2). The proteins found in Ad coinfection cover diverse steps of cellular transcription including regulation (E2F or PHB2, string data base), elongation (SUPT6H, string data base), and termination (SSB, string data base), while transcription proteins found in presence of HSV-1 coinfection are mainly regulators of initiation (string data base). Interestingly, in absence of a helper virus, several more cellular transcriptional regulators were found associated with AAV2 Rep including JUN, HMG1, and topors [153,175,176]. It is possible that certain helper virus proteins can substitute for cellular proteins involved in regulation, elongation, and termination of AAV2 transcripts in coinfected cells. For example the HSV-1 ICP4 protein, which is involved in several steps of HSV-1 transcription, including elongation [177], might also be involved in elongation of AAV2 transcripts. For other helper virus proteins, namely the Ad E1A and DBP [52,56,178] as well as the HSV-1 ICP0 protein [57], a positive impact on rep gene expression has been shown previously.
Histones and Histone-Modulating Proteins
As mentioned above, prohibitin is another cellular protein that is recruited into both Ad and HSV-1 supported AAV2 RCs (Tables 1 and 2, Fig. 3). Prohibitin is a potential tumor suppressor protein [179]. Similar to Rb, prohibitin mediates suppression of the E2F transcription factor [179]. Prohibitin is recruited into SV40 RCs and represses transcription from SV40 promoters together with histone-deacetylase and N-CoR [180]. Deacetylation of histones leads to a tight binding of histones to the DNA [181] and therefore transcriptional repression.
Upon HSV-1 infection, HDAC2 in a complex with CoREST, LSD1 and REST is involved in the temporal regulation of HSV-1 gene expression by repression of premature transcription of E and L genes [182]. Also upon Ad infection, histone-deacetylase has been shown to interact with E1A in order to regulate temporal expression of the Ad genes [183]. Although not found during AAV2 and HSV-1 coinfection, HDAC2 was found associated with Ad supported AAV2 RCs (Table 1; Fig. 1). As already mentioned, E1A has been shown to be required directly for the activation of the AAV2 p5 promoter [56]. Therefore, it is possible that Ad E1A together with prohibitin and histone-deacetylase regulates temporal expression of AAV2 genes via modification of histones associated with viral DNA.
Indeed several histone proteins were found in both Ad and HSV-1 supported AAV2 RCs (Tables 1 and 2, Fig. 3). This is in line with a report showing that within hours of infection ds AAV2 DNA is associated with nucleosome-like structures, independent of the presence or absence of a helper virus [184]. ds AAV2 DNA associated with nucleosomes may be created either by reassociation of incoming parental ss DNA or by integration into the cellular genome [184]. Similar results have been observed also for autonomous parvoviruses, such as minute virus of mice (MVM; [185]). Histones have also been found to associate with rAAV genomes and induce gene silencing, but expression can be rescued by treatment of the cells with HDAC inhibitor [186].
POSTTRANSCRIPTIONAL MODIFICATION
Polyadenylation Capping, and Cap Recognition
In cells pre-mRNA cap formation and polyadenylation are coupled to transcription [187]. Similar to the host RNAs, viral RNAs need to be equipped with 5’ Methyl-Gppp caps and 3’ poly-adenylated tails or proteins that substitute for these functions, in order to allow nuclear export, translation, and stability of mRNA. All AAV2 transcripts contain a polyadenylation site. Similar to its helper viruses Ad and HSV-1, AAV2 polyadenylation and capping may be performed by host enzymes. Nevertheless, none of the 6 cellular proteins necessary for mRNA polyadenylation cleavage and polyadenylation specificity factor (CPSF), cleavage stimulation factor (CstF), cleavage factors Im and IIm, poly(A) polymerase, and poly(A)-binding protein II (PABP [188]), have been identified as associated with AAV2 RCs. However, upon HSV-1 infection it has been shown that hnRNPL, a component of the hnRNP complex[189], functions as an adaptor protein to recruit proteins necessary for viral mRNA polyadenylation and nucleocytoplasmic export [190]. Several proteins of the hnRNP complex were indeed found associated with AAV2 RCs when HSV-1 (hnRNPA3, AB, C, D, H1 and M) or Ad (hnRNP U) was the helper virus (Tables 1 and 2, Figs. 1, 2). These components of the hnRNP complex are involved in several functions including polyadenylation, splicing (see below), and localization of mRNAs [189]. Similar to their role in HSV-1 infection, hnRNPs proteins may be involved in polyadenylation and nucleocytoplasmic export of AAV2 mRNAs [190]. In particular hnRNPH, which has been shown to be directly involved in the polyadenylation of cellular mRNAs [189], may support polyadenylation of AAV2 mRNAs in presence of HSV-1.
The eukaryotic initiation factor eIF4F recognizes 5′ caps of mRNAs and mediates initiation of translation via recruitment of ribosomes to mRNA [191]. The different functions that can be contributed to its subunits include recognition of the mRNA 5′ cap structure (eIF4E), delivery of an RNA helicase to the 5′ region (eIF4A), bridging of the mRNA and the ribosome (eIF4G), and circularization of the mRNA via interaction with PABP [191]. The helicase component eIF4A1 was found associated with Ad supported AAV2 RCs (Table 1, Fig. 1).
Even if not found in cells coinfected with AAV2 and HSV-1, eIF4A is also involved in HSV-1 infection, as the HSV-1 endoribonuclease vhs binds to eIF4A to promote degradation of cellular mRNAs [192,193], while HSV-1 RNAs are largely spared from vhs-eIF4A mediated degradation [194]. Which role aIF4A plays for AAV2 replication remains to be investigated and might be dependent on the type of helper virus.
Although not much is known about polyadenylation of AAV2 mRNAs, the presence of AAV2 polyadenylation sites has been shown to be necessary for Rep supported AAV2 pre-mRNA splicing in cells coinfected with Ad [195,196].
Pre-mRNA Splicing
All AAV2 pre-mRNAs contain introns allowing alternative RNA processing of the overlapping transcription products [195]. Transcription from the p5 and p19 promoters generates mRNAs which encode Rep 78 and Rep 52, respectively; while spliced mRNAs from these promoters encode Rep 68 and Rep 40 [195]. Not only splicing but also alternative splicing plays an important role in the generation of the two different mRNAs generated from the p40 promoter, for translation of the AAV2 capsid proteins VP1 as well as VP2 and VP3 [195]. In addition, the use of two different start codons on the 2.3-kb mRNA, gives rise to VP2 and VP3 [197,198]. As splicing of the nascent AAV2 transcripts is an essential step in AAV2 replication, it is not surprising that one of the main groups of proteins found in Ad and HSV-1 supported AAV2 RCs belong to the cellular splicing machinery including serine/arginine (SR)-rich proteins (SRSFs) and heterogeneous nuclear ribonucleoproteins (hnRNPs; [199]). SRSFs are involved in recruiting the splicing machinery to pre-mRNAs (also called heterogeneous nuclear RNA (hnRNA)), thereby supporting splicing together with hnRNA-binding proteins (hnRNPs; [199]). However, hnRNPs are also implicated in the repression of splicing events by blocking spliceosome assembly [199]. Both SRSF and hnRNP binding sites are located at exon/intron junctions of pre-RNAs [199]. The interplay between SRSF-mediated support and hnRNP-mediated repression of splicing has been found to influence constitutive and alternative splicing events of cellular pre-mRNAs [199]. It is tempting to speculate that similar to their function on cellular pre-mRNAs, SRSFs and/or hnRNPs found in AAV2 RCs (Tables 1 and 2, Fig. 1 and 2) regulate constitutive splicing of the two Rep pre-mRNAs from the p5 and p19 promoter [195] as well as alternative splicing of the capsid pre-mRNA from the p40 promoter [195]. Besides the cellular splicing machinery, the AAV2 Rep78/68 proteins as well as helper virus factors are involved in efficient processing of nascent AAV2 transcripts [200]. The Ad factors E2a, E4, and VA in combination with Rep as well as the HSV-1 gene products UL5, UL8, UL52 and UL29 have been shown to stimulate splicing of AAV2 RNA [195]. The AAV2 Rep proteins and helper virus factors might influence the composition and activity of RNA processing factors associated with the RNA polymerase II complex upon AAV2 gene expression; however, the detailed mechanism of Rep and helper virus proteins supported AAV2 mRNA splicing is not yet determined.
Besides their role in mRNA processing, hnRNPs might also be involved in transcriptional regulation, recombination, and telomere maintenance [201]. The reported hnRNP function in protecting single-stranded telomeric DNA repeats from nuclease attacks [201] might also be important for maintaining the integrity of ss AAV2 genomes.
mRNA EXPORT
mRNA export is intimately coupled to splicing, via exon junction complex mediated escort by the export receptor TAP/NXF 1 [202]. However, this factor has not been found associated with AAV2 RCs. Besides TAP/NXF 1, nucleoporins are other central proteins involved in mRNA nuclear export, which were found associated with both Ad (Nup85) and HSV-1 (Nup153) supported AAV2 RCs (Tables 1 and 2, Fig. 3). Nucleoporins may contribute to the export of AAV2 mRNAs in coinfected cells. In addition to their function in mRNA export, there is evidence that nucleoporins may also play a role in DNA repair (see above). Two other proteins, hnRNPA3 and RAN, which were found associated with HSV-1 and Ad supported AAV2 RCs, respectively, may also be implicated in viral mRNA trafficking [203].
CYTOPLASMIC PROTEINS
Several cytoplasmic and mitochondrial proteins were found associated with AAV2 Rep in co-IP experiments in both Ad and HSV-1 supported AAV2 replication [64,68]. The significance of Rep associated with ribosomal factors (RPS and RPL proteins; Tables 1 and 2, Fig. 3) is not clear. It is assumed that because of their abundance, ribosomal proteins are often found in proteomic screens [64], and therefore could likely by a contamination of the purified extracts [68]. But associations of Rep with cytoplasmic proteins in general is not entirely unexpected, considering that Rep68 is detected not only in the nuclear [27,45,85], but also in the cytoplasmic fraction of cells coinfected with AAV2 and Ad [85]. In addition, Rep 78/68 proteins have been previously shown to interact with another cytoplasmic protein, KCTD5, in cells coinfected with AAV2 and Ad [73]. KCTD5 has been shown to act as a substrate-specific adaptor in multimeric cullin E3 ligase reactions, by recruiting proteins for ubiquitination and subsequent proteasome-dependent degradation [204]. It is likely that although only present in small amounts, AAV2 Rep78/68 proteins may have also important functions in the regulation of cytoplasmic proteins such as KCTD5.
Another group of proteins found associated with AAV2 Rep belong to the cellular myosin network (Tables 1 and 2, Fig. 3). For HSV-1, interference with cellular myosin has been shown to be implicated in HSV-1 entry processes [205] as well as virion egress [206,207]. In addition, alterations in the distribution of myosin (and actin) filaments in cells infected with HSV-1 have been observed [206-208]. Rep association with these factors might influence the interference of HSV-1 with the cytoskeleton in coinfected cells.
Another cytoplasmic protein associated with Rep78 during both, Ad and HSV-1 supported AAV2 replication is ABCE1 (Tables 1 and 2, Fig. 3). In the ATP-binding cassette (ABC) multigene family, the ABCE subfamily is involved in regulation of protein synthesis [209]. Binding of ABCE1 to the eukaryotic initiation factor 2 initiates translation [210]. In addition, ABCE1 interferes with the interferon mediated cellular response against viruses, via ribonuclease L (RNase L) of the 2'-5' oligoadenylate/RNase L (2-5A/RNase L) pathway [211]. Several viruses appear to have developed strategies to counteract the antiviral activity of the 2-5A/RNase L pathway including HSV-1 and vaccinia virus [212,213]. It is possible that interaction of Rep 78 with ABCE1 is implicated in counteracting the 2-5A/RNase L activity. On the other hand ABCE1 might support AAV2 capsid assembly, similar to its role in HIV infection, where ABCE1 binds to the HIV-1 Gag and functions as a chaperone by promoting ATP-dependent conformational changes important for HIV capsid assembly and RNA packaging [214].
MITOCHONDRIAL PROTEINS
As mentioned above, mitochondrial proteins were found associated with AAV2 Rep in co-IP experiments including ATP5B, ATP5C1, HSPA9, ACAD9, ATP5O, SSBP1, and CMC1 (Tables 1 and 2; Figs. 1, 2; [64,68]). It is suggested that the interaction of Rep with several mitochondrial proteins may be a consequence of AAV2 interference with the cells apoptotic pathway [68]. Besides their central role in apoptosis, mitochondria are also central elements of the host defense against viral infections [215]. Therefore, AAV2 interference with mitochondrial proteins may also support productive viral infection by preventing excessive antiviral responses of the host cell.
CONCLUDING REMARKS
Co-IP experiments as well as immunofluorescence analysis of cells coinfected with AAV2 and helper virus, either Ad or HSV-1, identified numerous cellular proteins that interact with Rep78 and/or are recruited into AAV2 RCs. Clearly additional experiments will be needed to confirm these interactions and to investigate the role of most of these cellular proteins identified in AAV2 RCs in AAV2 replication. In this article, we nevertheless suggest potential functions in fundamental steps of AAV2 replication.
It is conspicuous that irrespective of the type of helper virus (Ad or HSV-1), the largest functional categories of cellular proteins of AAV2 RCs concern DNA replication and repair. In addition several hnRNPs involved in RNA metabolism as well as a minor group of cytoplasmic and mitochondrial proteins were identified as Rep78 interacting partners in presence of either helper virus. By taking a closer look at the composition of the functional categories, it is apparent that the participation of cellular proteins in AAV2 DNA and RNA metabolism depends not only on the specific replication strategy of AAV2, but also on the available helper virus in coinfected cells. Ad and HSV-1 follow very different strategies to replicate their genomes. Ad DNA replication occurs via a strand displacement mechanism in which both strands are synthesized in a continuous fashion after protein-primed initiation. In contrast, the basic model for HSV-1 replication is a rolling-circle mechanism including leading- and lagging-stand synthesis. Due to the different modes of replication, it is not surprising that proteins involved in AAV2 replication alter depending on the type of helper virus. Since AAV2 replication occurs by a mechanism distinct from that of both helper viruses, only some of the helper virus replication proteins participate in AAV2 replication, while others have to be substituted by cellular proteins. For example, while Ad supported AAV2 replication uses central cellular components of the DNA replication machinery, such as the MCM complex and polymerase delta, HSV-1 supported AAV2 replication uses helperviral DNA replication proteins including the HSV-1 helicase-primase complex (UL5/UL8/UL52) and the HSV-1 polymerase (UL30; [57-59]). The presence of helper virus substitutes of cellular proteins, and vice versa, may allow AAV2 to expand the host range. Besides cellular and helper virus DNA replication proteins, several factors involved in DNA damage sensing and repair seem to be important for AAV2 replication. It was shown that cells exposed to genotoxic agents (e.g. HU or IR) support efficient helper-independent AAV2 rep expression and low levels of viral DNA replication [100-103]. In addition, for the autonomous parvovirus minute virus of mice, the induction of a DNA damage response facilitates viral replication [216]. Upon coinfection with a helper virus, a defined set of DNA damage sensing and repair proteins were found associated with AAV2 RCs. Some of these proteins, e.g. MMR proteins, were found exclusively in HSV-1 supported AAV2 RCs. MMR proteins may be implicated in proofreading of the viral polymerase and efficient expression of helper virus IE genes [155], which in turn would enhance AAV2 replication. Also, the role of the MRN complex in AAV2 replication seems to depend on its differential effect on the helper virus. Although the effect of many cellular proteins on AAV2 replication is indirect and linked to their effect on the helper virus, other proteins seem to have a more direct role in AAV2 replication. A large number of DNA damage sensing and repair proteins were identified associated with both Ad and HSV-1 supported AAV2 RCs (e.g. RPA, ATM, H2AX, XRCC5/6, and p53). In this context, AAV2 was shown to modulate the interaction of the helper virus with the cellular DDR [65-67]. Interestingly, while Ad and HSV-1 induce and inactivate different pathways of the DDR to promote their replication [92,94,95], cellular DDR is more similar when coinfected with AAV2. For example, numerous proteins involved in NHEJ are recruited into HSV-1 as well as Ad supported AAV2 RCs [64-68], while in cells infected with Ad alone NHEJ is virtually inactivated. A potential role of NHEJ proteins in AAV2 RCs is to maintain integrity of the viral genome, by protecting the AAV2 DNA from further processing by the AAV2 Rep78 endonuclease activity after nicking at the TRS. In addition, NHEJ components seem to be involved in circularization of incoming AAV2 genomes [108,112], a similar role has been described for retrovirus infection [217]. But not only AAV2 modulates the helper virus induced DDR. The DDR induced by AAV2 alone is completely different from that induced upon coinfection with AAV2 and a helper virus. In that sense, the helper viruses strongly manipulate the AAV2 induced DDR. For example, several proteins of the ATR mediated DDR activated and recruited to AAV2 DNA in absence of a helper virus [82,104] were not found associated with helper virus supported AAV2 RCs. Moreover, in cells coinfected with AAV2 and HSV-1, ATR signaling via Chk1 seems to be abrogated [67].
When comparing the minimal set of Ad and HSV-1 helper factors for AAV2 replication, it is conspicuous that several of the helper proteins from Ad but none of the helper proteins provided by HSV-1 are involved in AAV2 RNA metabolism [53,55-59]. Also more cellular proteins involved in RNA metabolism were found associated with Ad supported AAV2 RCs than with HSV-1 supported AAV2 RCs. It is possible that other HSV-1 proteins that do not belong to the minimal set of helper factors can substitute for the cellular RNA metabolism proteins involved AAV2 replication.
As all AAV2 transcripts are spliced [195], it is not surprising that several hnRNPs were found associated with HSV-1 supported AAV2 RCs. Cellular mRNA splicing proteins, including hnRNPs and SRSFs, are even more abundant in Ad supported AAV2 RCs. It would be interesting to define the precise localization of these proteins during AAV2 replication in order to spatially monitor transcriptional activity within AAV2 RCs. For some proteins of the DDR, such a monitoring within/around AAV2 RCs has been performed [65-67] and the data support the idea that AAV2 RCs are well-structured and sub-compartmentalized. In this context, it is suggested that the nuclear matrix acts as a scaffold to which different proteins are recruited and retained [218]. Further studies are needed to determine the importance of such subcompartments for AAV2 replication.
Besides cellular proteins involved in DNA and RNA metabolism, several cytoplasmic and mitochondrial proteins were found associated with AAV2 Rep in co-IP experiments in both Ad and HSV-1 supported AAV2 replication [64,68]. Although the significance of these factors is not known, it suggests that additional cellular pathways not directly involved in DNA and RNA metabolism are targeted by Rep. For example, interaction with cytoplasmic proteins may influence AAV2 virion trafficking while interaction with mitochondrial proteins may interfere with intrinsic responses of the host cell against viral infection [215].
The identification of cellular proteins within helper virus supported AAV2 RCs forms the basis for the investigation of the functional roles of these proteins in AAV2 infection. The identification of cellular inhibitors and enhancers of helper virus supported AAV2 replication will contribute to a better understanding of the complex mechanisms of interaction between AAV2, its helper viruses, and the coinfected cell. Moreover, as AAV2 is a widely used vector in biomedical applications, the detailed knowledge of the functions of cellular proteins in AAV2 gene expression and DNA replication may also help to improve vector production and transduction efficiency.
SUMMARY
Adeno-associated virus type 2 (AAV2) DNA replication takes place in the host cell nucleus in viral replication compartments (RCs). These compartments have a defined composition of viral and cellular proteins. Co-immunoprecipitation (co-IP) experiments as well as immunofluorescence analysis of cells coinfected with AAV2 and one of its helper viruses, adenovirus (Ad) or herpes simplex virus type 1 (HSV-1), revealed numerous cellular proteins that are recruited into AAV2 RCs or interact with Rep78. The largest functional categories of cellular proteins associated with AAV2 RCs correspond to factors of the DNA replication and repair machinery, independent of the type of helper virus. In addition, proteins involved in RNA metabolism as well as a minor group of cytoplasmic and mitochondrial proteins were identified as Rep78 interacting partners. Although the majority of cellular proteins associated with Ad supported AAV2 RCs was also present in HSV-1 supported AAV2 RCs, some proteins were found associated with AAV2 RCs only in presence of either Ad or HSV-1. The importance of most of the cellular proteins identified in AAV2 RCs for AAV2 replication is not known. Nevertheless, based on the roles of these proteins in cellular processes and in the replication of other viruses, we can make some predictions concerning their functions in different steps of the AAV2 life cycle.
ACKNOWLEDGEMENTS
This work was supported by the Swiss National Science Foundation grants # 31003A_124938 and 31003A_144097.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
REFERENCES
- 1.Sanlioglu S, Monick MM, Luleci G, Hunninghake GW, Engelhardt JF. Rate limiting steps of AAV transduction and implications for human gene therapy. Curr Gene Ther. 2001;1:137–47. doi: 10.2174/1566523013348788. [DOI] [PubMed] [Google Scholar]
- 2.Chejanovsky N, Carter BJ. Mutagenesis of an AUG codon in the adeno-associated virus rep gene: effects on viral DNA replication. Virology. 1989;173:120–8. doi: 10.1016/0042-6822(89)90227-4. [DOI] [PubMed] [Google Scholar]
- 3.McCarty DM, Young SM, Samulski RJ. Integration of Adeno-Associated Virus (AAV) and Recombinant AAV Vectors. Annu. Rev. Genet. 2004;38:819–45. doi: 10.1146/annurev.genet.37.110801.143717. [DOI] [PubMed] [Google Scholar]
- 4.King JA, Dubielzig R, Grimm D, Kleinschmidt JA. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J. 2001;20:3282–91. doi: 10.1093/emboj/20.12.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cassinotti P, Weitz M, Tratschin JD. Organization of the adeno-associated virus (AAV) capsid gene: mapping of a minor spliced mRNA coding for virus capsid protein 1. Virology. 1988;167:176–84. [PubMed] [Google Scholar]
- 6.Becerra SP, Koczot F, Fabisch P, Rose JA. Synthesis of adeno-associated virus structural proteins requires both alternative mRNA splicing and alternative initiations from a single transcript. J Virol. 1988;62:2745–54. doi: 10.1128/jvi.62.8.2745-2754.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sonntag F, Schmidt K, Kleinschmidt JA. A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proc Natl Acad Sci USA. 2010;107:10220–5. doi: 10.1073/pnas.1001673107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atchison RW, Casto BC, Hammon WM. Adenovirus-Associated Defective Virus Particles Adenovirus-Associated Defective Virus Particles. Science. 1965;149:754–6. doi: 10.1126/science.149.3685.754. [DOI] [PubMed] [Google Scholar]
- 9.Hoggan MD, Blacklow NR, Rowe WP. Studies of small DNA viruses found in various adenovirus preparations: physical, biological, and immunological characteristics. Proc Natl Acad Sci USA. 1966;55:1467–74. doi: 10.1073/pnas.55.6.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomson BJ, Weindler FW, Gray D, Schwaab V, Heilbronn R. Human herpesvirus 6 (HHV-6) is a helper virus for adeno-associated virus type 2 (AAV-2) and the AAV-2 rep gene homologue in HHV-6 can mediate AAV-2 DNA replication and regulate gene expression. Virology. 1994;204:304–11. doi: 10.1006/viro.1994.1535. [DOI] [PubMed] [Google Scholar]
- 11.Buller RM, Janik JE, Sebring ED, Rose JA. Herpes simplex virus types 1 and 2 completely help adenovirus-associated virus replication. J Virol. 1981;40:241–7. doi: 10.1128/jvi.40.1.241-247.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walz C, Deprez A, Dupressoir T, Durst M, Rabreau M, Schlehofer JR. Interaction of human papillomavirus type 16 and adeno-associated virus type 2 co-infecting human cervical epithelium. J Gen Virol. 1997;78(Pt 6):1441–52. doi: 10.1099/0022-1317-78-6-1441. [DOI] [PubMed] [Google Scholar]
- 13.Schnepp BC, Jensen RL, Chen C, Johnson PR, Clark KR. Characterization of Adeno-Associated Virus Genomes Isolated from Human Tissues. J Virol. 2005;79:14793–803. doi: 10.1128/JVI.79.23.14793-14803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kotin RM, Linden RM, Berns KI. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 1992;11:5071–8. doi: 10.1002/j.1460-2075.1992.tb05614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kotin RM, Menninger JC, Ward DC, Berns KI. Mapping and direct visualization of a region-specific viral DNA integration site on chromosome 19q13-qter. Genomics. 1991;10:831–4. doi: 10.1016/0888-7543(91)90470-y. [DOI] [PubMed] [Google Scholar]
- 16.Kotin RM, Siniscalco M, Samulski RJ, et al. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci USA. 1990;87:2211–5. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samulski RJ, Zhu X, Xiao X, et al. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991;10:3941–50. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheung A, Hoggan M, Hauswirth W, Berns K. Integration of the adeno-associated virus genome into cellular DNA in latently infected human Detroit 6 cells. J Virol. 1980;33:739–48. doi: 10.1128/jvi.33.2.739-748.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Im DS, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–57. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 20.Snyder RO, Im DS, Muzyczka N. Evidence for covalent attachment of the adeno-associated virus (AAV) rep protein to the ends of the AAV genome. J Virol. 1990;64:6204–13. doi: 10.1128/jvi.64.12.6204-6213.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong G, Ward P, Berns KI. In vitro replication of adeno-associated virus DNA. Proc Natl Acad Sci USA. 1992;89:4673–7. doi: 10.1073/pnas.89.10.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rekosh DMK, Russell WC, Bellet AJD, Robinson AJ. Identification of a protein linked to the ends of adenovirus DNA. Cell. 1977;11:83–95. doi: 10.1016/0092-8674(77)90045-9. [DOI] [PubMed] [Google Scholar]
- 23.Roizman B. The checkpoints of viral gene expression in productive and latent infection: the role of the HDAC/CoREST/LSD1/REST repressor complex. J Virol. 2011;85:7474–82. doi: 10.1128/JVI.00180-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mador N, Panet A, Steiner I. The latency-associated gene of herpes simplex virus type 1 (HSV-1) interferes with superinfection by HSV-1. J Neurovirol. 2002;8:97–102. doi: 10.1080/13550280290167920. [DOI] [PubMed] [Google Scholar]
- 25.Fraefel C, Bittermann AG, Bueler H, Heid I, Bachi T, Ackermann M. Spatial and temporal organization of adeno-associated virus DNA replication in live cells. J Virol. 2004;78:389–98. doi: 10.1128/JVI.78.1.389-398.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heilbronn R, Engstler M, Weger S, Krahn A, Schetter C, Boshart M. ssDNA-dependent colocalization of adeno-associated virus Rep and herpes simplex virus ICP8 in nuclear replication domains. Nucleic Acids Res. 2003;31 :6206–13. doi: 10.1093/nar/gkg827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hunter LA, Samulski RJ. Colocalization of adeno-associated virus Rep and capsid proteins in the nuclei of infected cells. J Virol. 1992;66:317–24. doi: 10.1128/jvi.66.1.317-324.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stracker TH, Cassell GD, Ward P, et al. The rep protein of adeno-associated virus type 2 interacts with single-stranded DNA-binding proteins that enhance viral replication. J Virol. 2004;78:441–53. doi: 10.1128/JVI.78.1.441-453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weitzman MD, Fisher KJ, Wilson JM. Recruitment of wild-type and recombinant adeno-associated virus into adenovirus replication centers. J Virol. 1996;70:1845–54. doi: 10.1128/jvi.70.3.1845-1854.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Bortle K, Corces VG. Nuclear Organization and Genome Function. Annu. Rev. Cell Dev. Biol. 2012;28:163–87. doi: 10.1146/annurev-cellbio-101011-155824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends in Genetics. 2011;27:295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Everett RD, Sourvinos G, Leiper C, Clements JB, Orr A. Formation of Nuclear Foci of the Herpes Simplex Virus Type 1 Regulatory Protein ICP4 at Early Times of Infection: Localization, Dynamics, Recruitment of ICP27, and Evidence for the De Novo Induction of ND10-Like Complexes. J Virol. 2004;78:1903–17. doi: 10.1128/JVI.78.4.1903-1917.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Everett RD, Sourvinos G, Orr A. Recruitment of herpes simplex virus type 1 transcriptional regulatory protein ICP4 into foci juxtaposed to ND10 in live, infected cells. J Virol. 2003;77:3680–9. doi: 10.1128/JVI.77.6.3680-3689.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maul GG, Ishov AM, Everett RD. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217:67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- 35.Lallemand-Breitenbach V. The H de. PML Nuclear Bodies. Cold Spring Harbor Perspectives in Biology. 2010;2:a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahn JH, Hayward GS. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol. 1997;71:4599–613. doi: 10.1128/jvi.71.6.4599-4613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doucas V, Ishov AM, Romo A, et al. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 38.Leppard KN, Everett RD. The adenovirus type 5 E1b 55K and E4 Orf3 proteins associate in infected cells and affect ND10 components. J Gen Virol. 1999;80(Pt 4):997–1008. doi: 10.1099/0022-1317-80-4-997. [DOI] [PubMed] [Google Scholar]
- 39.Wilkinson GW, Kelly C, Sinclair JH, Rickards C. Disruption of PML-associated nuclear bodies mediated by the human cytomegalovirus major immediate early gene product. J Gen Virol. 1998;79(Pt 5):1233–45. doi: 10.1099/0022-1317-79-5-1233. [DOI] [PubMed] [Google Scholar]
- 40.Maul GG, Everett RD. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol. 1994;75(Pt 6):1223–33. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 41.Bruyn Kops A de, Knipe DM. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell. 1988;55:857–68. doi: 10.1016/0092-8674(88)90141-9. [DOI] [PubMed] [Google Scholar]
- 42.Lamberti C, Weller SK. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J Virol. 1998;72:2463–73. doi: 10.1128/jvi.72.3.2463-2473.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ward PL, Ogle WO, Roizman B. Assemblons: nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus 1. J Virol. 1996;70:4623–31. doi: 10.1128/jvi.70.7.4623-4631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hasson TB, Ornelles DA, Shenk T. Adenovirus L1 52- and 55-kilodalton proteins are present within assembling virions and colocalize with nuclear structures distinct from replication centers. J Virol. 1992;66:6133–42. doi: 10.1128/jvi.66.10.6133-6142.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wistuba A, Kern A, Weger S, Grimm D, Kleinschmidt JA. Subcellular compartmentalization of adeno-associated virus type 2 assembly. J Virol. 1997;71:1341–52. doi: 10.1128/jvi.71.2.1341-1352.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yonaha M, Chibazakura T, Kitajima S, Yasukochi Y. Cell cycle-dependent regulation of RNA polymerase II basal transcription activity. Nucleic Acids Res. 1995;23:4050–4. doi: 10.1093/nar/23.20.4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bridge E, Carmo-Fonseca M, Lamond A, Pettersson U. Nuclear organization of splicing small nuclear ribonucleoproteins in adenovirus-infected cells. J Virol. 1993;67:5792–802. doi: 10.1128/jvi.67.10.5792-5802.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pombo A, Ferreira J, Bridge E, Carmo-Fonseca M. Adenovirus replication and transcription sites are spatially separated in the nucleus of infected cells. EMBO J. 1994;13:5075–5085. doi: 10.1002/j.1460-2075.1994.tb06837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Janik JE, Huston MM, Rose JA. Locations of adenovirus genes required for the replication of adenovirus-associated virus. Proc Natl Acad Sci USA. 1981;78:1925–9. doi: 10.1073/pnas.78.3.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richardson WD, Westphal H. A cascade of adenovirus early functions is required for expression of adeno-associated virus. Cell. 1981;27:133–41. doi: 10.1016/0092-8674(81)90367-6. [DOI] [PubMed] [Google Scholar]
- 51.Samulski RJ, Shenk T. Adenovirus E1B 55-Mr polypeptide facilitates timely cytoplasmic accumulation of adeno-associated virus mRNAs. J Virol. 1988;62:206–10. doi: 10.1128/jvi.62.1.206-210.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tratschin JD, Miller IL, Carter BJ. Genetic analysis of adeno-associated virus: properties of deletion mutants constructed in vitro and evidence for an adeno-associated virus replication function. J Virol. 1984;51:611–9. doi: 10.1128/jvi.51.3.611-619.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pilder S, Moore M, Logan J, Shenk T. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol Cell Biol. 1986;6:470–6. doi: 10.1128/mcb.6.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.West MH, Trempe JP, Tratschin JD, Carter BJ. Gene expression in adeno-associated virus vectors: the effects of chimeric mRNA structure, helper virus, and adenovirus VA1 RNA. Virology. 1987;160:38–47. doi: 10.1016/0042-6822(87)90041-9. [DOI] [PubMed] [Google Scholar]
- 55.Carter BJ, Antoni BA, Klessig DF. Adenovirus containing a deletion of the early region 2A gene allows growth of adeno-associated virus with decreased efficiency. Virology. 1992;191:473–6. doi: 10.1016/0042-6822(92)90213-9. [DOI] [PubMed] [Google Scholar]
- 56.Chang L, Shenk T. The adenovirus DNA-binding protein stimulates the rate of transcription directed by adenovirus and adeno-associated virus promoters. J Virol. 1990;64:2103–9. doi: 10.1128/jvi.64.5.2103-2109.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alazard-Dany N, Nicolas A, Ploquin A, et al. Definition of herpes simplex virus type 1 helper activities for adeno-associated virus early replication events. PLOS Pathogens. 2009;5:e1000340. doi: 10.1371/journal.ppat.1000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weindler FW, Heilbronn R. A subset of herpes simplex virus replication genes provides helper functions for productive adeno-associated virus replication. J Virol. 1991;65:2476–83. doi: 10.1128/jvi.65.5.2476-2483.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ward P, Falkenberg M, Elias P, Weitzman M, Linden RM. Rep-dependent initiation of adeno-associated virus type 2 DNA replication by a herpes simplex virus type 1 replication complex in a reconstituted system. J Virol. 2001;75:10250–8. doi: 10.1128/JVI.75.21.10250-10258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Challberg MD. A method for identifying the viral genes required for herpesvirus DNA replication. Proc Natl Acad Sci USA. 1986;83:9094–8. doi: 10.1073/pnas.83.23.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Slanina H, Weger S, Stow ND, Kuhrs A, Heilbronn R. Role of the herpes simplex virus helicase-primase complex during adeno-associated virus DNA replication. J Virol. 2006;80:5241–50. doi: 10.1128/JVI.02718-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alex M, Weger S, Mietzsch M, Slanina H, Cathomen T, Heilbronn R. DNA-binding activity of adeno-associated virus rep is required for inverted terminal repeat-dependent complex formation with herpes simplex virus ICP8. J Virol. 2012;86:2859–63. doi: 10.1128/JVI.06364-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Geoffroy M, Salvetti A. Helper functions required for wild type and recombinant adeno-associated virus growth. Curr Gene Ther. 2005;5:265–71. doi: 10.2174/1566523054064977. [DOI] [PubMed] [Google Scholar]
- 64.Nash K, Chen W, Salganik M, Muzyczka N. Identification of Cellular Proteins That Interact with the Adeno-Associated Virus Rep Protein. J Virol. 2008;83:454–69. doi: 10.1128/JVI.01939-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schwartz RA, Carson CT, Schuberth C, Weitzman MD. Adeno-associated virus replication induces a DNA damage response coordinated by DNA-dependent protein kinase. J Virol. 2009;83:6269–78. doi: 10.1128/JVI.00318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Collaco RF, Bevington JM, Bhrigu V, Kalman-Maltese V, Trempe JP. Adeno-associated virus and adenovirus coinfection induces a cellular DNA damage and repair response via redundant phosphatidylinositol 3-like kinase pathways. Virology. 2009;392:24–33. doi: 10.1016/j.virol.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vogel R, Seyffert M, Strasser R, et al. Adeno-Associated Virus Type 2 Modulates the Host DNA Damage Response Induced by Herpes Simplex Virus 1 during Coinfection. J Virol. 2011;86:143–55. doi: 10.1128/JVI.05694-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nicolas A, Alazard-Dany N, Biollay C, et al. Identification of Rep-Associated Factors in HSV-1-Induced AAV-2 Replication Compartments. J Virol. 2012 doi: 10.1128/JVI.00725-10. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Batchu RB. Dual Level Inhibition of E2F-1 Activity by Adeno-associated Virus Rep78. J Biol Chem. 2001;276:24315–22. doi: 10.1074/jbc.M008154200. [DOI] [PubMed] [Google Scholar]
- 70.Bevington JM, Needham PG, Verrill KC, Collaco RF, Basrur V, Trempe JP. Adeno-associated virus interactions with B23/Nucleophosmin: Identification of sub-nucleolar virion regions. Virology. 2007;357:102–13. doi: 10.1016/j.virol.2006.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pereira DJ, Muzyczka N. The cellular transcription factor SP1 and an unknown cellular protein are required to mediate Rep protein activation of the adeno-associated virus p19 promoter. J Virol. 1997;71:1747–56. doi: 10.1128/jvi.71.3.1747-1756.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Batchu RB, Shammas MA, Wang JY, Munshi NC. Interaction of adeno-associated virus Rep78 with p53: implications in growth inhibition. Cancer Res. 1999;59:3592–5. [PubMed] [Google Scholar]
- 73.Weger S, Hammer E, Gotz A, Heilbronn R. Identification of a cytoplasmic interaction partner of the large regulatory proteins Rep78/Rep68 of adeno-associated virus type 2 (AAV-2) Virology. 2007;362:192–206. doi: 10.1016/j.virol.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 74.Ferrari FK, Samulski T, Shenk T, Samulski RJ. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J Virol. 1996;70:3227–34. doi: 10.1128/jvi.70.5.3227-3234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weitzman MD, Linden RM. In: Adeno-Associated Virus. Snyder RO, Moullier P, editors. Totowa, NJ: Humana Press; 2011. pp. 1–23. [Google Scholar]
- 76.Diffley JFX. Regulation of early events in chromosome replication. Curr Biol. 2004;14:R778–86. doi: 10.1016/j.cub.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 77.Nash K, Chen W, Muzyczka N. Complete in vitro reconstitution of Adeno-associated virus DNA replication requires the minichromosome maintenance complex proteins. J Virol. 2008;82:1159–64. doi: 10.1128/JVI.01968-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ni TH, McDonald WF, Zolotukhin I, et al. Cellular proteins required for adeno-associated virus DNA replication in the absence of adenovirus coinfection. J Virol. 1998;72:2777–87. doi: 10.1128/jvi.72.4.2777-2787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Myers MW, Laughlin CA, Jay FT, Carter BJ. Adenovirus helper function for growth of adeno-associated virus: effect of temperature-sensitive mutations in adenovirus early gene region 2. J Virol. 1980;35:65–75. doi: 10.1128/jvi.35.1.65-75.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee SS, Lehman IR. Unwinding of the box I element of a herpes simplex virus type 1 origin by a complex of the viral origin binding protein, single-strand DNA binding protein, and single-stranded DNA. Proc Natl Acad Sci USA. 1997;94:2838–42. doi: 10.1073/pnas.94.7.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Prindle MJ, Loeb LA. DNA polymerase delta in dna replication and genome maintenance. Environ Mol Mutagen. 2012;53:666–82. doi: 10.1002/em.21745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jurvansuu J, Raj K, Stasiak A, Beard P. Viral Transport of DNA Damage That Mimics a Stalled Replication Fork. J Virol. 2004;79:569–80. doi: 10.1128/JVI.79.1.569-580.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Waga S, Bauer G, Stillman B. Reconstitution of complete SV40 DNA replication with purified replication factors. J Biol Chem. 1994;269:10923–34. [PubMed] [Google Scholar]
- 84.Leman A, Noguchi E. The Replication Fork: Understanding the Eukaryotic Replication Machinery and the Challenges to Genome Duplication. Genes. 2013;4:1–32. doi: 10.3390/genes4010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Im DS, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–57. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 86.Snyder RO, Im DS, Muzyczka N. Evidence for covalent attachment of the adeno-associated virus (AAV) rep protein to the ends of the AAV genome. J Virol. 1990;64:6204–13. doi: 10.1128/jvi.64.12.6204-6213.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou X, Zolotukhin I, Im DS, Muzyczka N. Biochemical characterization of adeno-associated virus rep68 DNA helicase and ATPase activities. J Virol. 1999;73:1580–90. doi: 10.1128/jvi.73.2.1580-1590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sterner JM, Dew-Knight S, Musahl C, Kornbluth S, Horowitz JM. Negative regulation of DNA replication by the retinoblastoma protein is mediated by its association with MCM7. Mol Cell Biol. 1998;18:2748–57. doi: 10.1128/mcb.18.5.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rizwani W, Alexandrow M, Chellappan S. Prohibitin physically interacts with MCM proteins and inhibits mammalian DNA replication. Cell Cycle. 2009;8:1621–9. doi: 10.4161/cc.8.10.8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol. 2005;6:476–86. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jiang M, Imperiale MJ. Design stars: how small DNA viruses remodel the host nucleus. Future Virology. 2012;7:445–59. doi: 10.2217/FVL.12.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Turnell AS, Grand RJ. DNA viruses and the cellular DNA-damage response. J Gen Virol. 2012;93:2076–97. doi: 10.1099/vir.0.044412-0. [DOI] [PubMed] [Google Scholar]
- 93.Nikitin PA, Luftig MA. At a crossroads: human DNA tumor viruses and the host DNA damage response. Future Virology. 2011;6:813–30. doi: 10.2217/fvl.11.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lilley CE, Schwartz RA, Weitzman MD. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 2007;15:119–26. doi: 10.1016/j.tim.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 95.Weitzman MD, Carson CT, Schwartz RA, Lilley CE. Interactions of viruses with the cellular DNA repair machinery. DNA Repair (Amst) 2004;3:1165–73. doi: 10.1016/j.dnarep.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 96.Shiotani B, Zou L, editors. A Human Cell Extract-Based Assay for the Activation of ATM and ATR Checkpoint Kinases. New York, NY: Humana Press, c/o Springer Science+Business Media; 2011. [DOI] [PubMed] [Google Scholar]
- 97.Berthet C, Raj K, Saudan P, Beard P. How adeno-associated virus Rep78 protein arrests cells completely in S phase. Proc Natl Acad Sci USA. 2005;102:13634–9. doi: 10.1073/pnas.0504583102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Saudan P, Vlach J, Beard P. Inhibition of S-phase progression by adeno-associated virus Rep78 protein is mediated by hypophosphorylated pRb. EMBO J. 2000;19:4351–61. doi: 10.1093/emboj/19.16.4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fisher KJ, Gao GP, Weitzman MD, DeMatteo R, Burda JF, Wilson JM. Transduction with recombinant adeno-associated virus for gene therapy is limited by leading-strand synthesis. J Virol. 1996;70:520–32. doi: 10.1128/jvi.70.1.520-532.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nicolas A, Jolinon N, Alazard-Dany N, et al. Factors influencing helper-independent adeno-associated virus replication. Virology. 2012;432:1–9. doi: 10.1016/j.virol.2012.05.027. [DOI] [PubMed] [Google Scholar]
- 101.Yalkinoglu AO, Heilbronn R, Burkle A, Schlehofer JR, Zur Hausen H. DNA amplification of adeno-associated virus as a response to cellular genotoxic stress. Cancer Res. 1988;48:3123–9. [PubMed] [Google Scholar]
- 102.Yakobson B, Hrynko TA, Peak MJ, Winocour E. Replication of adeno-associated virus in cells irradiated with UV light at 254 nm. J Virol. 1989;63:1023–30. doi: 10.1128/jvi.63.3.1023-1030.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yakobson B, Koch T, Winocour E. Replication of adeno-associated virus in synchronized cells without the addition of a helper virus. J Virol. 1987;61:972–81. doi: 10.1128/jvi.61.4.972-981.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cervelli T, Palacios JA, Zentilin L, et al. Processing of recombinant AAV genomes occurs in specific nuclear structures that overlap with foci of DNA-damage-response proteins. J Cell Sci. 2008;121:349–57. doi: 10.1242/jcs.003632. [DOI] [PubMed] [Google Scholar]
- 105.Sanlioglu S, Benson P, Engelhardt JF. Loss of ATM function enhances recombinant adeno-associated virus transduction and integration through pathways similar to UV irradiation. Virology. 2000;268:68–78. doi: 10.1006/viro.1999.0137. [DOI] [PubMed] [Google Scholar]
- 106.Cataldi MP, McCarty DM. Differential effects of DNA double-strand break repair pathways on single-strand and self-complementary adeno-associated virus vector genomes. J Virol. 2010;84:8673–82. doi: 10.1128/JVI.00641-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Choi VW, McCarty DM, Samulski RJ. Host cell DNA repair pathways in adeno-associated viral genome processing. J Virol. 2006;80:10346–56. doi: 10.1128/JVI.00841-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Duan D, Yue Y, Engelhardt JF. Consequences of DNA-dependent protein kinase catalytic subunit deficiency on recombinant adeno-associated virus genome circularization and heterodimerization in muscle tissue. J Virol. 2003;77:4751–9. doi: 10.1128/JVI.77.8.4751-4759.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Inagaki K, Ma C, Storm TA, Kay MA, Nakai H. The role of DNA-PKcs and artemis in opening viral DNA hairpin termini in various tissues in mice. J Virol. 2007;81:11304–21. doi: 10.1128/JVI.01225-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nakai H, Storm TA, Fuess S, Kay MA. Pathways of removal of free DNA vector ends in normal and DNA-PKcs-deficient SCID mouse hepatocytes transduced with rAAV vectors. Hum Gene Ther. 2003;14:871–81. doi: 10.1089/104303403765701169. [DOI] [PubMed] [Google Scholar]
- 111.Choi YK, Nash K, Byrne BJ, Muzyczka N, Song S. The effect of DNA-dependent protein kinase on adeno-associated virus replication. PloS ONE. 2010;5:e15073. doi: 10.1371/journal.pone.0015073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Song S, Laipis PJ, Berns KI, Flotte TR. Effect of DNA-dependent protein kinase on the molecular fate of the rAAV2 genome in skeletal muscle. Proc Natl Acad Sci USA. 2001;98:4084–8. doi: 10.1073/pnas.061014598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Adachi K, Nakai H, editors. The Role of DNA Repair Pathways in Adeno-Associated Virus Infection and Viral Genome Replication / Recombination / Integration. New York, NY: InTech; 2011. [Google Scholar]
- 114.Russell DW, Alexander IE, Miller AD. DNA synthesis and topoisomerase inhibitors increase transduction by adeno-associated virus vectors. Proc Natl Acad Sci USA. 1995;92:5719–23. doi: 10.1073/pnas.92.12.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fragkos M, Breuleux M, Clement N, Beard P. Recombinant Adeno-Associated Viral Vectors Are Deficient in Provoking a DNA Damage Response. J Virol. 2008;82:7379–87. doi: 10.1128/JVI.00358-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Williams GJ, Lees-Miller SP, Tainer JA. Mre11-Rad50-Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double-strand breaks. DNA Repair (Amst) 2010;9:1299–306. doi: 10.1016/j.dnarep.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schwartz RA, Palacios JA, Cassell GD, Adam S, Giacca M, Weitzman MD. The Mre11/Rad50/Nbs1 complex limits adeno-associated virus transduction and replication. J Virol. 2007;81:12936–45. doi: 10.1128/JVI.01523-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Carson C, Schwartz R, Stracker T, Lilley C, Lee D, Weitzman M. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 2003;22:6610–20. doi: 10.1093/emboj/cdg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stracker TH, Carson CT, Weitzman MD. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature. 2002;418:348–52. doi: 10.1038/nature00863. [DOI] [PubMed] [Google Scholar]
- 120.Karen KA, Hearing P. Adenovirus core protein VII protects the viral genome from a DNA damage response at early times after infection. J Virol. 2011;85:4135–42. doi: 10.1128/JVI.02540-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sohn S, Hearing P. Adenovirus regulates sumoylation of Mre11-Rad50-Nbs1 components through a paralog-specific mechanism. J Virol. 2012;86:9656–65. doi: 10.1128/JVI.01273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Araujo FD, Stracker TH, Carson CT, Lee DV, Weitzman MD. Adenovirus type 5 E4orf3 protein targets the Mre11 complex to cytoplasmic aggresomes. J Virol. 2005;79:11382–91. doi: 10.1128/JVI.79.17.11382-11391.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu Y, Shevchenko A, Shevchenko A, Berk AJ. Adenovirus exploits the cellular aggresome response to accelerate inactivation of the MRN complex. J Virol. 2005;79:14004–16. doi: 10.1128/JVI.79.22.14004-14016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lilley CE, Carson CT, Muotri AR, Gage FH, Weitzman MD. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc Natl Acad Sci USA. 2005;102:5844–9. doi: 10.1073/pnas.0501916102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wilkinson DE, Weller SK. Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J Virol. 2004;78:4783–96. doi: 10.1128/JVI.78.9.4783-4796.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Baker A, Rohleder KJ, Hanakahi LA, Ketner G. Adenovirus E4 34k and E1b 55k Oncoproteins Target Host DNA Ligase IV for Proteasomal Degradation. J Virol. 2007;81:7034–40. doi: 10.1128/JVI.00029-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Boyer J, Rohleder K, Ketner G. Adenovirus E4 34k and E4 11k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology. 1999;263:307–12. doi: 10.1006/viro.1999.9866. [DOI] [PubMed] [Google Scholar]
- 128.Querido E, Blanchette P, Yan Q, et al. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 2001;15:3104–17. doi: 10.1101/gad.926401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lavin MF, Kozlov S. ATM activation and DNA damage response. Cell Cycle. 2007;6:931–42. doi: 10.4161/cc.6.8.4180. [DOI] [PubMed] [Google Scholar]
- 130.Giglia-Mari G, Zotter A, Vermeulen W. DNA damage response. Cold Spring Harbor perspectives in biology. Genes Dev. 2011;3:a000745. doi: 10.1101/cshperspect.a000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Legagneux V, Cubizolles F, Watrin E. Multiple roles of Condensins: a complex story. Biol Cell. 2004;96:201–13. doi: 10.1016/j.biolcel.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 132.Yazdi PT, Wang Y, Zhao S, Patel N, Lee EYP, Qin J. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev. 2002;16:571–82. doi: 10.1101/gad.970702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Abraham RT. PI 3-kinase related kinases: 'big' players in stress-induced signaling pathways. DNA Repair. 2004;3:883–7. doi: 10.1016/j.dnarep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 134.Gardino AK, Yaffe MB. 14-3-3 proteins as signaling integration points for cell cycle control and apoptosis. Semin Cell Dev Biol. 2011;22:688–95. doi: 10.1016/j.semcdb.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pei Z, Harrison MS, Schmitt AP. Parainfluenza virus 5 m protein interaction with host protein 14-3-3 negatively affects virus particle formation. J Virol. 2011;85:2050–9. doi: 10.1128/JVI.02111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Aoki H, Hayashi J, Moriyama M, Arakawa Y, Hino O. Hepatitis C virus core protein interacts with 14-3-3 protein and activates the kinase Raf-1. J Virol. 2000;74:1736–41. doi: 10.1128/jvi.74.4.1736-1741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang H, Guan J, Perrault AR, Wang Y, Iliakis G. Replication protein A2 phosphorylation after DNA damage by the coordinated action of ataxia telangiectasia-mutated and DNA-dependent protein kinase. Cancer Res. 2001;61:8554–63. [PubMed] [Google Scholar]
- 138.Binz SK, Sheehan AM, Wold MS. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair. 2004;3:1015–24. doi: 10.1016/j.dnarep.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 139.Vassin VM, Wold MS, Borowiec JA. Replication protein A (RPA) phosphorylation prevents RPA association with replication centers. Mol Cell Biol. 2004;24:1930–43. doi: 10.1128/MCB.24.5.1930-1943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Taylor TJ, Knipe DM. Proteomics of herpes simplex virus replication compartments. J Virol. 2004;78:5856–66. doi: 10.1128/JVI.78.11.5856-5866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Muylaert I, Elias P. Knockdown of DNA Ligase IV/XRCC4 by RNA Interference Inhibits Herpes Simplex Virus Type I DNA Replication. J Biol Chem. 2007;282:10865–72. doi: 10.1074/jbc.M611834200. [DOI] [PubMed] [Google Scholar]
- 142.Kysela B, Chovanec M, Jeggo PA. Phosphorylation of linker histones by DNA-dependent protein kinase is required for DNA ligase IV-dependent ligation in the presence of histone H1. Proc Natl Acad Sci USA. 2005;102:1877–82. doi: 10.1073/pnas.0401179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mari P, Florea BI, Persengiev SP, et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc Natl Acad Sci USA. 2006;103:18597–602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pegoraro G, Marcello A, Myers MP, Giacca M. Regulation of adeno-associated virus DNA replication by the cellular TAF-I/set complex. J Virol. 2006;80:6855–64. doi: 10.1128/JVI.00383-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Le Deist F, Poinsignon C, Moshous D, Fischer A, Villartay J de. Artemis sheds new light on V(D)J recombination. Immunol Rev. 2004;200:142–55. doi: 10.1111/j.0105-2896.2004.00169.x. [DOI] [PubMed] [Google Scholar]
- 146.Nakai H, Thomas CE, Storm TA, et al. A limited number of transducible hepatocytes restricts a wide-range linear vector dose response in recombinant adeno-associated virus-mediated liver transduction. J Virol. 2002;76:11343–9. doi: 10.1128/JVI.76.22.11343-11349.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Goodarzi AA, Jeggo P, Lobrich M. The influence of heterochromatin on DNA double strand break repair: Getting the strong, silent type to relax. DNA Repair. 2010;9:1273–82. doi: 10.1016/j.dnarep.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 148.Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer. 2009;9:644–54. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 149.Orazio NI, Naeger CM, Karlseder J, Weitzman MD. The adenovirus E1b55K/E4orf6 complex induces degradation of the Bloom helicase during infection. J Virol. 2011;85:1887–92. doi: 10.1128/JVI.02134-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Narasimhan D, Collaco R, Kalman-Maltese V, Trempe JP. Hyper-phosphorylation of the adeno-associated virus Rep78 protein inhibits terminal repeat binding and helicase activity. Biochim Biophys Acta. 2002;1576:298–305. doi: 10.1016/s0167-4781(02)00394-9. [DOI] [PubMed] [Google Scholar]
- 151.Daya S, Cortez N, Berns KI. Adeno-associated virus site-specific integration is mediated by proteins of the nonhomologous end-joining pathway. J Virol. 2009;83:11655–64. doi: 10.1128/JVI.01040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lieber MR, Wilson TE. SnapShot: Nonhomologous DNA end joining (NHEJ) Cell. 2010;142:496–496.e1. doi: 10.1016/j.cell.2010.07.035. [DOI] [PubMed] [Google Scholar]
- 153.Costello E, Saudan P, Winocour E, Pizer L, Beard P. High mobility group chromosomal protein 1 binds to the adeno-associated virus replication protein (Rep) and promotes Rep-mediated site-specific cleavage of DNA, ATPase activity and transcriptional repression. EMBO J. 1997;16:5943–54. doi: 10.1093/emboj/16.19.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Song S, Lu Y, Choi YK, et al. DNA-dependent PK inhibits adeno-associated virus DNA integration. Proc Natl Acad Sci USA. 2004;101:2112–6. doi: 10.1073/pnas.0307833100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mohni KN, Dee AR, Smith S, Schumacher AJ, Weller SK. Efficient Herpes Simplex Virus 1 Replication Requires Cellular ATR Pathway Proteins. J Virol. 2012;87:531–42. doi: 10.1128/JVI.02504-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mohni KN, Mastrocola AS, Bai P, Weller SK, Heinen CD. DNA Mismatch Repair Proteins Are Required for Efficient Herpes Simplex Virus 1 Replication. J Virol. 2011;85:12241–53. doi: 10.1128/JVI.05487-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Thomas C, Tulin AV. Poly-ADP-ribose polymerase: Machinery for nuclear processes. Mol Aspects Med. 2013 doi: 10.1016/j.mam.2013.04.001. doi: pii: S0098-2997 (13)00025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Grady SL, Hwang J, Vastag L, Rabinowitz JD, Shenk T. Herpes simplex virus 1 infection activates poly(ADP-ribose) polymerase and triggers the degradation of poly(ADP-ribose) glycohydrolase. J Virol. 2012;86:8259–68. doi: 10.1128/JVI.00495-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Blaho JA, Michael N, Kang V, et al. Differences in the poly(ADP-ribosyl) ation patterns of ICP4, the herpes simplex virus major regulatory protein, in infected cells and in isolated nuclei. J Virol. 1992;66:6398–407. doi: 10.1128/jvi.66.11.6398-6407.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ha HC. Poly(ADP-ribose) polymerase-1 is required for efficient HIV-1 integration. Proc Natl Acad Sci USA. 2001;98:3364–8. doi: 10.1073/pnas.051633498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Huber O, Menard L, Haurie V, Nicou A, Taras D, Rosenbaum J. Pontin and reptin, two related ATPases with multiple roles in cancer. Cancer Res. 2008;68:6873–6. doi: 10.1158/0008-5472.CAN-08-0547. [DOI] [PubMed] [Google Scholar]
- 162.Nagai S, Dubrana K, Tsai-Pflugfelder M, et al. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 2009;23:912–27. doi: 10.1101/gad.1782209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lemaitre C, Fischer B, Kalousi A, et al. The nucleoporin 153, a novel factor in double-strand break repair and DNA damage response. Oncogene. 2012;31:4803–9. doi: 10.1038/onc.2011.638. [DOI] [PubMed] [Google Scholar]
- 165.Jiricny J. MutLalpha: at the cutting edge of mismatch repair. Cell. 2006;126:239–41. doi: 10.1016/j.cell.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 166.Okuda M. The role of nucleophosmin in centrosome duplication. Oncogene. 2002;21:6170–4. doi: 10.1038/sj.onc.1205708. [DOI] [PubMed] [Google Scholar]
- 167.Okuda M, Horn HF, Tarapore P, et al. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell. 2000;103:127–40. doi: 10.1016/s0092-8674(00)00093-3. [DOI] [PubMed] [Google Scholar]
- 168.Szebeni A, Herrera JE, Olson MO. Interaction of nucleolar protein B23 with peptides related to nuclear localization signals. Biochemistry. 1995;34:8037–42. doi: 10.1021/bi00025a009. [DOI] [PubMed] [Google Scholar]
- 169.Hermonat PLea. The adeno-associated virus Rep78 major regulatory protein binds the cellular TATA-binding protein in vitro and in vivo. Virology. 1998;245:120–7. doi: 10.1006/viro.1998.9144. [DOI] [PubMed] [Google Scholar]
- 170.Grondin B, DeLuca N. Herpes simplex virus type 1 ICP4 promotes transcription preinitiation complex formation by enhancing the binding of TFIID to DNA. J Virol. 2000;74:11504–10. doi: 10.1128/jvi.74.24.11504-11510.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kawase H, Okuwaki M, Miyaji M, et al. NAP-I is a functional homologue of TAF-I that is required for replication and transcription of the adenovirus genome in a chromatin-like structure. Genes Cells. 1996;1:1045–56. doi: 10.1046/j.1365-2443.1996.d01-223.x. [DOI] [PubMed] [Google Scholar]
- 172.Francois A, Guilbaud M, Awedikian R, Chadeuf G, Moullier P, Salvetti A. The cellular TATA binding protein is required for rep-dependent replication of a minimal adeno-associated virus type 2 p5 element. J Virol. 2005;79:11082–94. doi: 10.1128/JVI.79.17.11082-11094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Maston GA, Evans SK, Green MR. Transcriptional regulatory elements in the human genome. Annu Rev Genomics Hum Genet. 2006;7:29–59. doi: 10.1146/annurev.genom.7.080505.115623. [DOI] [PubMed] [Google Scholar]
- 174.Pereira DJ, McCarty DM, Muzyczka N. The adeno-associated virus (AAV) Rep protein acts as both a repressor and an activator to regulate AAV transcription during a productive infection. J Virol. 1997;71:1079–88. doi: 10.1128/jvi.71.2.1079-1088.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Prasad C, Meyers C, Zhan D, et al. The adeno-associated virus major regulatory protein Rep78-c-Jun-DNA motif complex modulates AP-1 activity. Virology. 2003;314:423–31. doi: 10.1016/s0042-6822(03)00439-2. [DOI] [PubMed] [Google Scholar]
- 176.Weger S, Hammer E, Heilbronn R. Topors, a p53 and topoisomerase I binding protein, interacts with the adeno-associated virus (AAV-2) Rep78/68 proteins and enhances AAV-2 gene expression. J Gen Virol. 2002;83:511–6. doi: 10.1099/0022-1317-83-3-511. [DOI] [PubMed] [Google Scholar]
- 177.Wagner LM, Lester JT, Sivrich FL, DeLuca NA. The N terminus and C terminus of herpes simplex virus 1 ICP4 cooperate to activate viral gene expression. J Virol. 2012;86:6862–74. doi: 10.1128/JVI.00651-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Laughlin CA, Jones N, Carter BJ. Effect of deletions in adenovirus early region 1 genes upon replication of adeno-associated virus. J Virol. 1982;41:868–76. doi: 10.1128/jvi.41.3.868-876.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wang S, Nath N, Adlam M, Chellappan S. Prohibitin, a potential tumor suppressor, interacts with RB and regulates E2F function. Oncogene. 1999;18:3501–10. doi: 10.1038/sj.onc.1202684. [DOI] [PubMed] [Google Scholar]
- 180.Wang S, Fusaro G, Padmanabhan J, Chellappan SP. Prohibitin co-localizes with Rb in the nucleus and recruits N-CoR and HDAC1 for transcriptional repression. Oncogene. 2002;21:8388–96. doi: 10.1038/sj.onc.1205944. [DOI] [PubMed] [Google Scholar]
- 181.Hayakawa T, Nakayama J. Physiological roles of class I HDAC complex and histone demethylase. J Biomed Biotechnol. 2011;2011:129383. doi: 10.1155/2011/129383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Roizman B, Zhou G, Du T. Checkpoints in productive and latent infections with herpes simplex virus 1: conceptualization of the issues. J Neurovirol. 2011;17:512–7. doi: 10.1007/s13365-011-0058-x. [DOI] [PubMed] [Google Scholar]
- 183.Gallimore PH, Turnell AS. Adenovirus E1A: remodelling the host cell, a life or death experience. Oncogene. 2001;20:7824–35. doi: 10.1038/sj.onc.1204913. [DOI] [PubMed] [Google Scholar]
- 184.Marcus-Sekura CJ, Carter BJ. Chromatin-like structure of adeno-associated virus DNA in infected cells. J Virol. 1983;48:79–87. doi: 10.1128/jvi.48.1.79-87.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ben-Asher E, Bratosin S, Aloni Y. Intracellular DNA of the parvovirus minute virus of mice is organized in a minichromosome structure. J Virol. 1982;41:1044–54. doi: 10.1128/jvi.41.3.1044-1054.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Okada T, Uchibori R, Iwata-Okada M, et al. A histone deacetylase inhibitor enhances recombinant adeno-associated virus-mediated gene expression in tumor cells. Mol Ther. 2006;13:738–46. doi: 10.1016/j.ymthe.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 187.Yonaha M, Proudfoot NJ. Transcriptional termination and coupled polyadenylation in vitro. EMBO J. 2000;19:3770–7. doi: 10.1093/emboj/19.14.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhao J, Hyman L, Moore C. Formation of mRNA 3' ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Boil. 1999;63:405–45. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Chaudhury A, Chander P, Howe PH. Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: Focus on hnRNP E1's multifunctional regulatory roles. RNA. 2010;16:1449–62. doi: 10.1261/rna.2254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Guang S, Felthauser AM, Mertz JE. Binding of hnRNP L to the pre-mRNA processing enhancer of the herpes simplex virus thymidine kinase gene enhances both polyadenylation and nucleocytoplasmic export of intronless mRNAs. Mol Cell Biol. 2005;25:6303–13. doi: 10.1128/MCB.25.15.6303-6313.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–63. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 192.Feng P, Everly DN, Read GS. mRNA Decay during Herpesvirus Infections: Interaction between a Putative Viral Nuclease and a Cellular Translation Factor. J Virol. 2001;75:10272–80. doi: 10.1128/JVI.75.21.10272-10280.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Feng P, Everly DN, Read GS. mRNA Decay during Herpes Simplex Virus (HSV) Infections: Protein-Protein Interactions Involving the HSV Virion Host Shutoff Protein and Translation Factors eIF4H and eIF4A. J Virol. 2005;79:9651–64. doi: 10.1128/JVI.79.15.9651-9664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Taddeo B, Zhang W, Roizman B. The herpes simplex virus host shutoff RNase degrades cellular and viral mRNAs made before infection but not viral mRNA made after infection. J Virol. 2013;87:4516–22. doi: 10.1128/JVI.00005-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Qiu J, Pintel D. Processing of adeno-associated virus RNA. Front Biosci. 2008;1:3101–15. doi: 10.2741/2912. [DOI] [PubMed] [Google Scholar]
- 196.Mouw MB, Pintel DJ. Adeno-associated virus RNAs appear in a temporal order and their splicing is stimulated during coinfection with adenovirus. J Virol. 2000;74:9878–88. doi: 10.1128/jvi.74.21.9878-9888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Becerra SP, Rose JA, Hardy M, Baroudy BM, Anderson CW. Direct mapping of adeno-associated virus capsid proteins B and C: a possible ACG initiation codon. Proc Natl Acad Sci USA. 1985;82:7919–23. doi: 10.1073/pnas.82.23.7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Muralidhar S, Becerra SP, Rose JA. Site-directed mutagenesis of adeno-associated virus type 2 structural protein initiation codons: effects on regulation of synthesis and biological activity. J Virol. 1994;68:170–6. doi: 10.1128/jvi.68.1.170-176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Busch A, Hertel KJ. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev RNA. 2012;3:1–12. doi: 10.1002/wrna.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Qiu J, Pintel DJ. The adeno-associated virus type 2 Rep protein regulates RNA processing via interaction with the transcription template. Mol Cell Biol. 2002;22:3639–52. doi: 10.1128/MCB.22.11.3639-3652.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Krecic AM, Swanson MS. hnRNP complexes: composition, structure, and function. Curr Opin Cell Biol. 1999;11:363–71. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- 202.Reed R, Magni K. A new view of mRNA export: separating the wheat from the chaff. Nat Cell Biol. 2001;3:E201–4. doi: 10.1038/ncb0901-e201. [DOI] [PubMed] [Google Scholar]
- 203.Ma ASW, Moran-Jones K, Shan J, et al. Heterogeneous nuclear ribonucleoprotein A3, a novel RNA trafficking response element-binding protein. J Biol Chem. 2002;277:18010–20. doi: 10.1074/jbc.M200050200. [DOI] [PubMed] [Google Scholar]
- 204.Bayon Y, Trinidad AG, La Puerta ML de, et al. KCTD5, a putative substrate adaptor for cullin3 ubiquitin ligases. FEBS J. 2008;275:3900–10. doi: 10.1111/j.1742-4658.2008.06537.x. [DOI] [PubMed] [Google Scholar]
- 205.Arii J, Goto H, Suenaga T, et al. Non-muscle myosin IIA is a functional entry receptor for herpes simplex virus-1. Nature. 2010;467:859–62. doi: 10.1038/nature09420. [DOI] [PubMed] [Google Scholar]
- 206.van Leeuwen H, Elliott G, O'Hare P. Evidence of a role for nonmuscle myosin II in herpes simplex virus type 1 egress. J Virol. 2002;76:3471–81. doi: 10.1128/JVI.76.7.3471-3481.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Roberts KL, Baines JD. Myosin Va enhances secretion of herpes simplex virus 1 virions and cell surface expression of viral glycoproteins. J Virol. 2010;84:9889–96. doi: 10.1128/JVI.00732-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Winkler M, Dawson GJ, Elizan TS, Berl S. Distribution of actin and myosin in a rat neuronal cell line infected with herpes simplex virus. Arch Virol. 1982;72:95–103. doi: 10.1007/BF01314454. [DOI] [PubMed] [Google Scholar]
- 209.Tyzack JK, Wang X, Belsham GJ, Proud CG. ABC50 interacts with eukaryotic initiation factor 2 and associates with the ribosome in an ATP-dependent manner. J Biol Chem. 2000;275:34131–9. doi: 10.1074/jbc.M002868200. [DOI] [PubMed] [Google Scholar]
- 210.Chen Z, Dong J, Ishimura A, Daar I, Hinnebusch AG, Dean M. The essential vertebrate ABCE1 protein interacts with eukaryotic initiation factors. J Biol Chem. 2006;281:7452–7. doi: 10.1074/jbc.M510603200. [DOI] [PubMed] [Google Scholar]
- 211.Bisbal C, Silhol M, Laubenthal H, et al. The 2'-5' oligoadenylate/RNase L/RNase L inhibitor pathway regulates both MyoD mRNA stability and muscle cell differentiation. Mol Cell Biol. 2000;20:4959–69. doi: 10.1128/mcb.20.14.4959-4969.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Cayley PJ, Davies JA, McCullagh KG, Kerr IM. Activation of the ppp(A2'p)nA system in interferon-treated, herpes simplex virus-infected cells and evidence for novel inhibitors of the ppp(A2'p)nA-dependent RNase. Eur J Biochem. 1984;143:165–74. doi: 10.1111/j.1432-1033.1984.tb08355.x. [DOI] [PubMed] [Google Scholar]
- 213.Paez E, Esteban M. Resistance of vaccinia virus to interferon is related to an interference phenomenon between the virus and the interferon system. Virology. 1984;134:12–28. doi: 10.1016/0042-6822(84)90268-x. [DOI] [PubMed] [Google Scholar]
- 214.Lingappa JR, Dooher JE, Newman MA, Kiser PK, Klein KC. Basic residues in the nucleocapsid domain of Gag are required for interaction of HIV-1 gag with ABCE1 (HP68), a cellular protein important for HIV-1 capsid assembly. J Biol Chem. 2006;281:3773–84. doi: 10.1074/jbc.M507255200. [DOI] [PubMed] [Google Scholar]
- 215.Ohta A, Nishiyama Y. Mitochondria and viruses. Mitochondrion. 2011;11:1–12. doi: 10.1016/j.mito.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Adeyemi RO, Landry S, Davis ME, Weitzman MD, Pintel DJ. Parvovirus minute virus of mice induces a DNA damage response that facilitates viral replication. PLOS Pathogens. 2010;6:e1001141. doi: 10.1371/journal.ppat.1001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Li L, Olvera JM, Yoder KE, et al. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 2001;20:3272–81. doi: 10.1093/emboj/20.12.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Jong RN de, van der Vliet PC, Brenkman AB. Adenovirus DNA replication: protein priming, jumping back and the role of the DNA binding protein DBP. Curr Top Microbiol Immunol. 2003;272:187–211. doi: 10.1007/978-3-662-05597-7_7. [DOI] [PubMed] [Google Scholar]