Abstract
Our previous works revealed that human ribosomal protein S13 (RPS13) was up-regulated in multidrug-resistant gastric cancer cells and overexpression of RPS13 could protect gastric cancer cells from drug-induced apoptosis. The present study was designed to explore the role of RPS13 in tumorigenesis and development of gastric cancer. The expression of RPS13 in gastric cancer tissues and normal gastric mucosa was evaluated by immunohistochemical staining and Western blot analysis. It was found RPS13 was expressed at a higher level in gastric cancer tissues than that in normal gastric mucosa. RPS13 was then genetically overexpressed in gastric cancer cells or knocked down by RNA interference. It was demonstrated that up-regulation of RPS13 accelerated the growth, enhanced in vitro colony forming and soft agar cologenic ability and promoted in vivo tumour formation potential of gastric cancer cells. Meanwhile, down-regulation of RPS13 in gastric cancer cells resulted in complete opposite effects. Moreover, overexpression of RPS13 could promote G1 to S phase transition whereas knocking down of RPS13 led to G1 arrest of gastric cancer cells. It was further demonstrated that RPS13 down-regulated p27kip1 expression and CDK2 kinase activity but did not change the expression of cyclin D, cyclin E, CDK2, CDK4 and p16INK4A. Taken together, these data indicate that RPS13 could promote the growth and cell cycle progression of gastric cancer cells at least through inhibiting p27kip1 expression.
Keywords: ribosomal protein S13 (RPS13), gastric cancer, tumorigenesis, RNA interference (RNAi), p27Kip1
Introduction
Ribosomal proteins (RPs) are the major components of ribosome, where they stabilize specific rRNA structures in mature ribosomal subunits, promote correct folding of rRNAs during ribosome assembly and coordinate the interaction between ribosome and mRNA as well as initiation and elongation factors. Interestingly, a variety of RPs have been found overexpressed in and associated with the development of malignant tumours. For example, the overexpression of RPS13, a 17 kD RP, was reported in colon cancer and small cell lung cancer cells [1, 2]. The corresponding mRNA of RPS13 was around 30-fold more abundant in the colon cancer cells than in normal colon [1] and about 50-fold more abundant in lung metastasis than in normal lung [2]. Moreover, the expression level of RPS13 correlated well with the growth rate of tumour cells [1]. RPP0 was identified as one differentially expressed gene between human hepatocellular carcinoma and matched non-tumourous liver tissues, which was further confirmed by pathological studies [3]. RPS27 level in hepatocytes increased in regenerating cirrhotic nodules compared to chronic hepatitis and reached the highest level in hepatocellular carcinoma [4]. Up-regulation of RPS2, RPL7a, RPL23a, RPS14 and RPL19 was identified in malignant prostate cancer cell lines and in prostate cancer tissue samples [5–7]. The genes encoding RPS2, RPS12, RPS27A, RPL5, RPL7a and RPL10a were enriched in a colonic adenoma minus normal mucosa suppression subtractive hybridization library [8]. In another study, it was found that RPS3, RPS6, RPS8, RPS12, RPL5 and RPP0 were expressed at higher levels in eight different colon adenocarcinomas and ade-nomatous polyps [9]. These results suggest that a select pool of RPs might be dysregulated in cancer cells and involved in tumorigenesis.
Previously, we analysed the mRNA profiles of a human gastric cancer cell line SGC7901 and its multidrug-resistant variant SGC7901/VCR by differential display PCR and suppression subtractive hybridization [10, 11]. RPS13 was identified as one of the genes overexpressed in SGC7901/VCR comparing to SGC7901, which was further confirmed by RT-PCR and Northern blot analysis [10]. Subsequent studies revealed that up-regulation of RPS13 could protect gastric cancer cells from vincristine-induced apop-tosis and enhance the resistance of gastric cancer cells to multiple chemotherapeutic drugs, including vincristine, adriamycin and 5-fludrouracil [12]. These data indicated that RPS13 could promote the malignant phenotype of gastric cancer cells, which led us to further explore the exact role of RPS13 in tumorigenesis and development of gastric cancer.
In the present study, we examined the expression of RPS13 in gastric cancer tissues and normal gastric mucosa by immunohistochemical staining and Western blot. It was found RPS13 was expressed at a higher level in gastric cancer tissues than that in normal gastric mucosa. We then modulated RPS13 expression in gastric cancer cells by gene transfection or RNA interference (RNAi) techniques. It was demonstrated that RPS13 could promote gastric cancer cell growth in vitro as well as in vivo. Further studies revealed that RPS13 could promote cell cycle progression of gastric cancer cells at least through down-regulating p27kip1. For the first time, we have convincingly demonstrated that RPS13 is involved in the development of gastric cancer. It would be meaningful to further explore the possibility of RPS13 as a biomarker and therapeutic target of human gastric cancer.
Materials and methods
Tissue collection
For immunohistochemistry, serial sections of paraffin-embedded tissues from 61 patients with gastric cancer who underwent gastrectomy in our hospital from 2005 to 2006 were included. Twenty-four non-tumour gastric mucosa specimens were biopsies collected from routine upper gastrointestinal endoscopy. For Western blotting, fresh gastric cancer and adjacent non-tumourous tissues (>5 cm from the margin of the tumour) were obtained from 13 patients who underwent surgery at the department of general surgery in our hospital. All cases of gastric cancer and adjacent non-tumourous tissues were diagnosed clinically and pathologically. Patients contributed their fresh surgical tissues for the study signed informed consent. None of the patients had received preoperative radiotherapy or chemotherapy.
Cell culture
The human gastric cancer cell lines MKN28, SGC7901, AGS and MKN45, with various differentiation degrees, respectively, were preserved by our institute. The immortalized gastric epithelial mucosa cell line GES was established by the Beijing Cancer Institute. All cell lines were routinely maintained in RPMI1640 medium supplemented with 10% foetal calf serum (FCS) in a 37°C humidified incubator with a mixture of 95% air and 5% CO2.
Immunohistochemistry
The immunohistochemical study of RPS13 expression in gastric cancer and normal mucosa was performed by the avidin-biotin peroxidase staining technique as usual. The mouse anti-human-RPS13 polyclonal antibody was previously raised against purified His-tagged RPS13 protein by our group [13]. It has been demonstrated by Western blot analysis that this antibody could specifically recognize both recombinant RPS13 purified from recombinant E. coli BL21 cells and RPS13 protein extracted from human gastric cancer cells SGC7901/VCR [13]. Immunocytochemical staining using this antibody revealed that RPS13 was expressed at higher level in SGC7901/VCR cells than SGC7901 cells, which was consistent with previous findings [10]. These data supported us to employ this mouse anti-human-RPS13 antibody to further examine RPS13 expression in gastric cancer tissues. Tissue sections (4 μm) cut from the original paraffin wax-embedded blocks were dewaxed in xylene and rehydrated in graded alcohols and distilled water. The slides were washed in PBS three times for 5 min. Then the sections was treated in 0.3% (v/v) H2O2 for blocking endogenous peroxidase activity, 10% normal goat serum (Boster, Wuhan, Hubei, China) for 30 min. for blocking the non-specific binding, and incubated with homemade mouse anti-human-RPS13 antibody [13] (diluted 1:200) overnight at 4°C. After washing in PBS, the tissue sections were reacted with the biotinylated antimouse secondary antibody for 30 min. at room temperature, followed by incubation with peroxidase-conjugated avidin complexes and stained with 3,3-diaminobenzidine (Dako, Glostrup, Denmark). Finally, the sections were counter-stained with haematoxylin. For negative controls, the primary antibody was replaced by normal mouse serum. All sections were examined microscopically and scored by two independent pathologists in a blinded fashion in the dark of clinical and pathological information.
Depending on the percentage of positive cells and staining intensity, RPS13 staining was classified into three groups: negative, weak positive and strong positive. Specifically, the percentage of positive cells was divided into five grades (percentage scores): <10% (0), 10–25% (1), 25–50% (2), 50–75% (3) and >75% (4). The intensity of staining was divided into four grades (intensity scores): no staining (0), light brown (1), brown (2) and dark brown (3). RPS13 staining intensity was determined by the formula: overall scores = percentage score × intensity score. The overall score of ≤3 was defined as negative, of >3 positive.
DNA constructs and transfection
The cDNA fragment encoding human RPS13 was cloned into pcDNA3.1/V5-His (Invitrogen, Carlsbad, CA, USA) as previously described [12]. Two pairs of specific short interference RNA (siRNA) duplexes for RPS13 were designed homologous to the RPS13 mRNA consensus sequence (Table 1). A scramble DNA duplex was also designed as control siRNA (Table 1). The DNA duplexes were annealed, diluted and cloned into pSilencer3.1 (Ambion, Austin, TX, USA). Colonies were screened with ampicillin, identified by DNA sequencing.
Table 1.
The DNA sequence of siRNA duplexes used in this study
| Direction | Sequence | Targeting site in RPS13 (NM_001017.2) | |||
|---|---|---|---|---|---|
| First siRNA duplexes for RPS13 | Forward | 5′- GAT CCA TTC CGT CTG ATT CTA ATA TTC AAG AGA TAT TAG AAT CAG ACG GAA TTT TTT TGG AAA-3′ | nt368–386 | ||
| Reverse | 5′- AGC TTT TCC AAA AAA ATT CCG TCT GAT TCT AAT ATC TCT TGA ATA TTA GAA TCA GAC GGA ATG-3′ | ||||
| Second siRNA duplexes for RPS13 | Forward | 5′- GAT CCG AAT TCT TAA GTC TAA GGG TTC AAG AGA CCC TTA GAC TTA AGA ATT CTT TTT TGG AAA-3′ | nt250–268 | ||
| Reverse | 5′- AGC TTT TCC AAA AAA GAA TTC TTA AGT CTA AGG GTC TCT TGA ACC CTT AGA CTT AAG AAT TCG-3′ | ||||
| Scramble control siRNA duplexes | Forward | 5′- GAT CCG ACA TGT AGA GTC TGC CAG TTC AAG AGA CTG GCA GAC TCT ACA TGT CTT TTT TGG AAA-3′ | NO | ||
| Reverse | 5′- AGC TTT TCC AAA AAA GAC ATG TAG AGT CTG CCA GTC TCT TGA ACT GGC AGA CTC TAC ATG TCG-3′ | ||||
The plasmids were transduced into SGC7901 cells using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instruction. Twenty-four hours after transfection, G418 (400 μg/ml) was added into the culture medium for establishing stable clones. The mixed clones were expanded for an additional 2 months. The gastric cancer cells stably expressing exogenic RPS13 were named as SGC7901/RPS13. The G418-resistant cells harbouring RPS13-specific siRNA were designated as SGC7901/siRPS13–1 (transfected with the first pair of siRNA duplex) or SGC7901/siRPS13–2 (transfected with the second pair of siRNA duplex). The empty vector pcDNA3.1/V5-His and scramble control siRNA vector were also transduced into SGC7901 and the G418-resistant cell variants were named as SGC7901/pcDNA3.1 and SGC7901/siControl, respectively.
Western blot analysis
For preparation of protein extracts from tissues, approximately 500 mg of frozen tumour and adjacent non-tumour tissues from 13 patients were lysed by sonication. For preparation of whole-cell extracts, approximately 1 × 107 cells at log phase were collected and homogenized in 300 μl of lysis buffer [50 mM Tris-HCl, pH 7.5, 1% Triton X-100, 10% Glycerol, 5 mM ethylenedi-aminetetraacetic acid, 150 mM NaCl, 5 mM CaCl2, 10 mM MgCl2 with 10 μg/ml leupeptin, 10 μg/ml aprotinin, 5 μg/ml pepstatin and 1 mM phenylmethanesulfonyl fluoride (PMSF)]. The protein concentrations of samples were measured based on the Bradford's method. For Western blotting, cell and tissue protein extracts (50–100 μg/lane) were resolved over 12% SDS-PAGE and transferred to nitrocellulose membranes (0.22 μm, Millipore, Bedford, MA, USA). After blocking with 5% non-fat milk in TBST (Tris buffered saline containing 0.1% Tween-20), the blots were stained with first antibodies overnight at 4°C, followed by incubation with peroxidase-conjugated secondary antibodies for 60 min. at room temperature. Immunoreactive bands were detected and developed in enhanced chemilu-minescence system (Amersham Pharmacia Biotechnology, Uppsala, Sweden) with X-ray film. Antibodies against p27Kip1, cyclin E, CDK2, p16INK4A, cyclin D1, CDK4 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-β-actin was from Sigma (St. Louis, MO, USA). Peroxidase-conjugated goat antimouse and goat anti-rabbit IgG were purchased from Boster (Wuhan, Hubei, China).
MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide) assay
The cell growth was evaluated by MTT assay. SGC7901 and its derivates at log phase were seeded in 96-well plates at a density of 2 × 103 cells/well in RPMI1640 containing 10% FCS. The cells were cultured 8 days. Each day, viable cells were incubated with MTT for 4 hrs. Following MTT addition, the supernatant were carefully removed and 150 μl dimethyl sulfoxide (DMSO) was used to lyse the cells for 10 min. and the absorbance at 490 nm was obtained using a microplate reader.
Cell cycle analysis
For analysis of cell cycle phase distribution, SGC7901 and its variants at log phase were harvested. For analysis of cell cycle progression, SGC7901 and its variants were synchronized to the G1/S boundary by double thymidine block as previously described [14] and then harvested at 4 hr intervals for 16 hrs after releasing. Cells were fixed with 70% ethanol and then incubated in propidium iodide/RNase staining buffer (BD Biosciences, Franklin Lakes, NJ, USA) at room temperature for 15 min. The DNA content of at least 1 × 105 cells was analysed by FACScan (BD Biosciences) using the CellQuest flow cytometric analysis software. The experiment was repeated four times.
Colony formation assay
Colony formation assay was performed to determine the effects of RPS13 on colony-forming ability of gastric cancer cells. Briefly, a total of 200 cells were plated in 60 mm dish and then cultured for approximately 2 weeks. The colonies were fixed with 10% (v/v) methanol for 15 min. and stained with 5% (g/v) Giemsa solution (Sigma) for 20 min. Colonies that consisted of more than 50 cells were scored. The ratio of colony formation was calculated as: (colonies formed/cells seeded) × 100%. Data were expressed as the mean and standard deviation of triplicates in one experiment.
Soft agar clonogenic assay
Anchorage-independent growth as a characteristic of in vitro tumorigenic-ity was assessed by soft agar clonogenic assay. Briefly, cells were detached and plated in 0.3% agarose with a 0.5% agarose underlay (1 × 104/well in 6-well plates). The number of foci >100 μm was counted after 14 days.
Tumour formation in nude mice
Tumour formation was carried out to assess the effects of RPS13 on tumorigenicity in vivo. BALB/c nude mice of 4 to 6 weeks were provided by Shanghai Cancer Institute and housed in micro-isolator cages under positive air pressure, and maintained at a constant temperature (22°C) and humidity for the tumorigenicity study. Approximately 3 × 106 cells at log phase were collected and injected subcutaneously into the right upper back of BALB/c nude mice. Tumour formation was monitored daily and recorded. At least three nude mice were used for each group. The mice were killed 4 weeks later and tumour weight of each mouse was evaluated. All procedures for animal experimentation were performed in accordance with the Institutional Animal Care and Use Committee guidelines.
in vitro CDK2 kinase assay
Whole cell extracts (500 μg) from gastric cancer cells were pre-cleared with protein A agarose conjugates, incubated with anti-CDK2 antibody overnight at 4°C and precipitated with protein A agarose beads at 4°C for 1 hr. The immunoprecipitates were washed and incubated at 30°C for 30 min. with histone H1 (100 μg/ml) and [γ-32P]ATP (10 μCi) in kinase buffer [50 mM N-2-hydroxyethylpiperazine-N-ethane-sulphonic acid (HEPES) pH 7.0, 5 mM MgCl2, 10 mM DTT]. The reactions were terminated by boiling in SDS sample buffer, resolved over SDS-PAGE and transferred to nitrocellulose. Phosphorylated proteins were visualized by autoradiogra-phy and the membranes were then probed with anti-CDK2 antibody to confirm equal loading of CDK2 protein.
Real-time RT-PCR
Real-time RT-PCR was employed for the quantification of p27kip1 mRNA expression. Total RNA was extracted from SGC7901 and its variants using RNeasy kit (QIAGEN, Beijing, China). One microgram of total RNA was reverse transcribed with Superscript II RNase H reverse transcriptase using oligo (dT) according to the manufacturer's instructions (Invitrogen). Realtime RT-PCR was performed using the following primers: p27 primers, forward 5′-CTGCCCTCCCCAGTCTCTCT-3′ and reverse 5′-CAAGCACCTCG-GATTTT-3′; β-actin primers, forward 5′-GATGAGATTGGCATGGCTTT-3′ and reverse 5′-CACCTTCACCGTTCCAGTTT-3′. All samples were analysed in triplicate. The mRNA expression of p27 was normalized to the level of β-actin.
Statistical analysis
For immunohistochemistry study, the two-tailed χ2 test was performed to determine the significance of the difference. Numeral data were expressed as mean ± S.D. (standard division). Statistical analyses were performed with SPSS statistical software (SPSS, Inc., Chicago, IL, USA). Student's t-test and one-way ANOVA followed by Dunnett's multiple comparison tests were adopted. Significance was defined as P < 0.05.
Results
RPS13 was overexpressed in human gastric cancer specimens
Our previous works indicated that RPS13 was up-regulated in multidrug-resistant gastric cancer cells and could protect gastric cancer cells from vincristine-induced apoptosis [10, 12]. To further explore the role of RPS13 in gastric cancer, we examined the expression of RPS13 protein in human gastric cancer tissue samples in the present studies. The expression of RPS13 was firstly evaluated in 13 paired samples of human gastric cancer and adjacent non-tumourous gastric mucosa tissues by Western blot. The results of four representative pairs of tissues were presented in Fig. 1A. In 11 out of 13 cases, RPS13 was expressed at much higher level in cancerous tissues than adjacent non-tumourous tissues. To further confirm the overex-pression of RPS13 in gastric cancer, we carried out immunohis-tochemical staining of RPS13 in 61 gastric cancer and 24 non-tumour gastric mucosa specimens. As shown in Fig. 1B, RPS13 was predominantly expressed in the cytoplasm of non-tumour gastric mucosa cells and gastric cancer cells. While the fundic actively growing gastric epithelia cells in normal tissues displayed weak staining of RPS13, gastric cancer cells exhibited strong positive expression of RPS13 (Fig. 1B). The overall positive percentage of gastric cancer cases was 74%, whereas only 42% of the normal tissues were positive of RPS13 (Fig. 1C). These data indicated that RPS13 was overexpressed in human gastric cancer tissues.
Fig 1.

RPS13 expression in gastric cancer tissues. (A) Western blot analysis of RPS13 protein in gastric tissues. Proteins extracted from gastric cancer (T) and adjacent normal (N) tissues taken from 13 patients were separated by 12% SDS-PAGE and subjected to Western blot analysis using anti-RPS13 and anti-β-actin specific antibodies. Four representative pairs of tissues are presented. (B) Immunohistochemical staining of RPS13 in 24 non-tumour gastric mucosa tissues and 61 gastric cancer tissues. Expression of RPS13 was determined by immunohisto-chemical staining with anti-RPS13 antibody. Shown are representative staining of RPS13 in non-tumour gastric mucosa tissues (a, fundic actively growing gastric epithelia cells display positive staining), well-differentiated gastric carcinoma (b), moderately differentiated gastric carcinoma (c) and poorly differentiated gastric carcinoma (d). Original magnifications: 100×. (C) Percentage of cases showing RPS13 immunoreactivity in 24 normal gastric mucosa tissues and 61 gastric cancer tissues. The immunostaining pattern was scored according to a four-tiered system based on the percentage of immunoreactive tumour cells. *P < 0.01 versus normal tissues.
The clinicalpathological characteristics of the 61 gastric cancer specimens were further analysed. There was no correlation of RPS13 with patient's gender, age, tumour differentiation, tumour stage and lymph node metastasis status (Table 2).
Table 2.
Clinicopathological associations of RPS13 expression in patients with gastric cancer
| Characteristics | Case number (n = 61) | Staining intensity | P–value | |||
|---|---|---|---|---|---|---|
| Negative (n = 16) | Positive (n = 45) | |||||
| Gender | ||||||
| Male | 48 | 12 (25%) | 36 (75%) | 0.728 | ||
| Female | 13 | 4 (31%) | 9 (69%) | |||
| Age | ||||||
| ≥65 | 23 | 7 (30%) | 16 (70%) | 0.565 | ||
| <65 | 38 | 9 (24%) | 29 (76%) | |||
| Differentiation | ||||||
| (1) Well | 12 | 4 (33%) | 8 (67%) | 0.232 (1 versus 2) | ||
| (2) Moderately | 26 | 4 (15%) | 22 (85%) | 0.183 (2 versus 3) | ||
| (3) Poorly | 23 | 8 (35%) | 15 (65%) | 1.000 (1 versus 3) | ||
| Stage (TNM) | ||||||
| I | 27 | 7 (26%) | 20 (74%) | 0.716 (I versus II) | ||
| II | 17 | 3 (18%) | 14 (82%) | 0.438 (II versus III) | ||
| III | 17 | 6 (35%) | 11 (65%) | 0.521 (I versus III) | ||
| Lymph node metastasis | ||||||
| With | 17 | 6 (35%) | 11 (65%) | 0.344 | ||
| Without | 44 | 10 (23%) | 34 (77%) | |||
RPS13 was up-regulated in gastric cancer cell lines
The expression of RPS13 protein in human gastric cancer cell lines, including MKN28, MKN45, AGS, SGC7901 and immortalized gastric mucosa epithelial cell line GES were analysed by Western blot. It was found RPS13 was ubiquitously expressed in those cell lines (Fig. 2A). However, all the cancer cell lines showed a higher level of RPS13 than GES cells, which suggested up-regulation of RPS13 in human gastric cancer cells and coincided with above data from gastric cancer specimens.
Fig 2.

RPS13 expression in gastric cancer cell lines. RPS13 expression in human gastric cancer cell lines was determined by Western blot analysis with anti-RPS13 antibody. β-actin was probed as loading control. (A) Detection of RPS13 in human gastric cancer cell lines MKN28, MKN45, SGC7901, AGS and immortalized gastric mucosa epithelial cell line GES. (B) Expression of RPS13 in SGC7901 and its derivates. SGC7901 cells were transfected with RPS13 construct (in pcDNA3.1), RPS13-specific siRNA constructs (designated as siRPS13, in pSilencer) or the corresponding control vectors. The transfectants were selected with G418 for 2 months. The mixed cell pools of G418-resistant clones were used for analysing the expression of RPS13 by Western blot.
The cell line SGC7901 was chosen for subsequent experiments because it had a moderate expression level of RPS13 (Fig. 2A). SGC7901 cells were transfected with vectors expressing exogenic RPS13 or RPS13-specific siRNA. The G418-resistant mixed clones were isolated. The gastric cancer cells stably expressing exogenic RPS13 were named as SGC7901/RPS13. The G418-resistant cells harbouring RPS13-specific siRNA were designated as SGC7901/siRPS13–1 (transfected with the first pair of siRNA duplex) or SGC7901/siRPS13–2 (transfected with the second pair of siRNA duplex). The cell transfected with empty vectors pcDNA3.1/V5-His and scramble control siRNA vectors were named as SGC7901/pcDNA3.1 and SGC7901/siControl, respectively. As illustrated by Fig. 2B, RPS13 was successfully up-regulated in SGC7901/RPS13 cells but down-regulated in SGC7901/siRPS13–1 and SGC7901/siRPS13–2 cells. Transfection with the empty vectors pcDNA3.1 and scramble control siRNA vector did not change RPS13 expression in SGC7901 cells. These SGC7901-derived cell variants displayed different expression levels of RPS13 and provided us good cell models to further explore the exact role of RPS13 in gastric cancer.
RPS13 promoted the growth of gastric cancer cells
It was demonstrated by immunohistochemical staining and Western blot analysis, as described above, that RPS13 was up-regulated in human gastric cancer tissues and cell lines. The over-expression of RPS13 might be involved in the tumorigenesis or might be a result of tumorigenesis of gastric cancer. To clarify if RPS13 played a role in the development of gastric cancer, we evaluated the effects of RPS13 on growth of gastric cancer cells. The growth of SGC7901 cell variants was determined by MTT assay. As indicated by cell growth curves, SGC7901/RPS13 cell grew faster than SGC7901 and SGC7901/pcDNA3.1 cells (Fig. 3A), whereas SGC7901/siRPS13 cells grew slower than SGC7901 and SGC7901/siControl cells (Fig. 3B). It was clear that RPS13 expression level correlated very well with the growth rate of gastric cancer cells. These suggested that RPS13 could promote the growth of gastric cancer cells.
Fig 3.

The growth curves of SGC7901 cell derivates. Cell growth was determined by MTT assays as described in ‘Materials and methods’. Values represent the mean ± S.D. of triplicates in one experiment. Shown are representative of at least three separate experiments. *P < 0.05 versus empty vector controls and SGC7901 cells. #P < 0.01 versus scramble control and SGC7901 cells.
RPS13 enhanced tumorigenesis of gastric cancer cells
To determine whether elevated RPS13 affected tumorigenesis of gastric cancer, we carried out in vitro colony-forming assay of SGC7901 and its variants. As indicated in Fig. 4A, SGC7901/RPS13 cells showed much more cell colonies than control cells, while SGC7901/siRPS13 cells exhibited much less cell colonies than their control cells. In addition, the anchorage-independent growth of SGC7901 and its variants was also evaluated by soft agar clonogenic assay. It was clear that overexpression of RPS13 induced a significant increase in the colony numbers whereas knocking down of RPS13 resulted in a substantial reduction in colony formation (Fig. 4B).
Fig 4.

Effects of RPS13 on in vitro tumorigenesis of SGC7901 cells. (A) in vitro colony formation assay. Cells were placed into 60 mm dish and cultured for approximately 2 weeks. The cell colonies were fixed with methanol and visualized with Giemsa staining. The ratio of colony formation was calculated as: (colonies formed/cells seeded) × 100%. (B) Soft agar clono-genic assay. Cells were plated in 0.3% agarose with a 0.5% agarose underlay. Two weeks later, the formed colonies were counted. Values represent the mean ± S.D. of triplicates in one experiment. Shown are representative of at least three separate experiments. *P < 0.01 versus SGC7901 and SGC7901/pcDNA3.1 cells. #P < 0.01 versus SGC7901 and SGC7901/siControl cells.
To further confirm the effects of RPS13 on tumorigenesis of gastric cancer cells, tumour formation assay was performed in nude mice. SGC7901 cell derivates were subcutaneously inoculated into the right upper back of athymic nude mice. Four weeks later, mice were killed and the transplanted tumours were excised (Fig. 5A) and tumour weight was evaluated (Fig. 5B). There was a significant increase of size and weight of xenografts in the presence of RPS13 overexpression. Meanwhile, down-regulation of RPS13 resulted in smaller tumours in nude mice. These data indicated that up-regulation of RPS13 could promote gastric cancer cell growth in vivo and interference of RPS13 expression led to growth suppression.
Fig 5.

Effects of RPS13 on in vivo tumori-genesis of SGC7901 cells. Nude mice were injected subcutaneously with 3 × 106 cells into the right upper back. Four weeks later, mice were killed and the transplanted tumours were excised (A) and tumour weight was evaluated (B). *P < 0.05 versus SGC7901/pcDNA3.1 cells. #P < 0.01 versus SGC7901/si Control cells.
RPS13 promoted G1/S transition of gastric cancer cells
To address whether RPS13 promoted gastric cancer cell growth through regulating cell cycle progression, cell cycle analysis was performed on SGC7901 and its derivates. Gastric cancer cells at log phase were harvested and analysed for cell cycle phase distribution (Fig. 6A). SGC7901/RPS13 cells displayed a significantly increased percentage of S phase comparing to their parental cells SGC7901 and vector control cells SGC7901/pcDNA3.1 (Fig. 6B). SGC7901/siRPS13–1 and SGC7901/siRPS13–2 cells that had a lower level of RPS13 exhibited a significantly increased percentage of G0/G1 phase (Fig. 6B) comparing to their control cells. These suggested RPS13 might be involved in the regulation of G1/S transition of gastric cancer cells.
Fig 6.
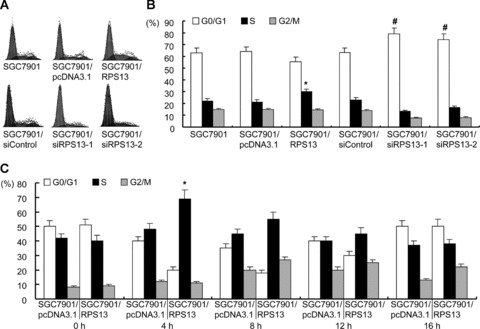
Cell cycle analysis of SGC7901 variants. (A) SGC7901 and its derivates in log phase were harvested and subjected to cell cycle analysis. The cell cycle phase distribution was evaluated by FACScan. Shown are representative of four separate experiments. (B) The cell cycle distribution was calculated and expressed as mean ± S.D. of four separate experiments described in (A). *P < 0.01 versus SGC7901 and SGC7901/pcDNA3.1 cells. #P < 0.01 versus SGC7901 and SGC7901/siControl cells. (C) SGC7901/pcDNA3.1 and SGC7901/RPS13 cells were synchronized to the G1/S boundary by double thymidine block and then harvested at 4 hr intervals for 16 hrs after releasing. The cell cycle was evaluated by FACScan and expressed as mean ± S.D. of four separate experiments. *P < 0.01 versus SGC7901/pcDNA3.1 cells.
The role of RPS13 in cell cycle regulation was further analysed by synchronization. SGC7901/pcDNA3.1 and SGC7901/RPS13 cells were synchronized to the G1/S boundary by double thymi-dine block. It was found that after cell cycle blocking by addition of thymidine, cells were mostly arrested at G1 phase (Fig. 6C). Four hours after releasing, the cells began entering into S phase (Fig. 6C). The average releasing rate of cells from G1 to S phase in SGC7901/RPS13 was 51.3% (Fig. 6C), which was significantly higher than those of SGC7901/pcDNA3.1 cells (33.2%). Twelve hours after releasing, most cells had gone into G2 phase. By 16 hrs, cells entered a new cell cycle (Fig. 6C). No significant difference was found in cell cycle profile between SGC7901/pcDNA3.1 and SGC7901/RPS13 cells at 8 hrs and later times after releasing from thymidine block. Taken together, these data strongly indicated that RPS13 played an important role in promoting the growth of gastric cancer cells through accelerating G1 to S phase transition in the cell cycle.
RPS13 down-regulated p27Kip1 in gastric cancer cells
It is well known that G1-S progression is controlled by cyclinD/CDK4 and cyclinE/CDK2 complexes, which are regulated by INK4 family members and p27Kip1, respectively. The expression of these regulators in gastric cancer cells was determined by Western blot analysis. As shown in Fig. 7A, overexpression of RPS13 induced a significant reduction of p27Kip1 whereas suppression of RPS13 caused an up-regulation of p27Kip1 in gastric cancer cells. Meanwhile, RPS13 displayed no effect on the expression of cyclin D1, cyclin E, CDK2, CDK4 and p16INK4A (Fig. 7A). p27Kip1 is an important negative regulator of cyclinE/CDK2. It inhibits the phosphorylation and activation of CDK2 and thus induces G1 arrest [15]. To confirm the effects of RPS13 on p27Kip1 expression, we investigated CDK2 activity in gastric cancer cells by in vitro kinase assay. It was demonstrated that CDK2 activity was increased in SGC7901/RPS13 cells but decreased in SGC7901/siRPS13 cells (Fig. 7B), which coincided with the down-regulation of p27Kip1 in SGC7901/RPS13 cells and up-regulation of p27Kip1 in SGC7901/siRPS13 cells (Fig. 7A).
Fig 7.

Effects of RPS13 on p27Kip1 expression in SGC7901 cells. (A) Detection of cell cycle associated proteins by Western blot. Proteins extracted from SGC7901 and its variants were resolved over 12% SDS-PAGE, transferred to nitrocellulose membrane and sequentially blotted with antibodies specific to p27Kip1, cyclin E, CDK2, p16INK4A, cyclin D1, CDK4 and β-actin. (B) in vitro CDK2 kinase assay. CDK2 was immunopre-cipitated from total cell lysates of SGC7901 and its derivates. Immunoprecipitates were employed for in vitro kinase assay using histone H1 as substrate, then resolved over SDS-PAGE and transferred to nitro-cellulose membrane. Phospho-labelled histone H1 was visualized by radioautography (upper panel). Afterward, the membrane was probed for CDK2 to confirm equal loading of kinase assay (lower panel). (C) To assess if RPS13 influenced degradation of p27Kip1, the proteasome inhibitor MG132 was added to the cells 2 hrs before harvest. p27Kip1 was detected by Western blot. (D) Detection of p27Kip1 by real-time RT-PCR. Total RNA extracted from SGC7901 and its variants was subjected to real-time PCR to determine p27Kip1 mRNA level. β-actin was used as normalizer. The histograms indicate the expression of p27Kip1 mRNA relative to the level of β-actin mRNA. Data shown represent the mean ± S.D. of three independent experiments. *P < 0.01 versus SGC7901 and SGC7901/pcDNA3.1 cells. #P < 0.01 versus SGC7901 and SGC7901/siControl cells.
The mechanisms underlying the inhibitory effects of RPS13 on p27Kip1 expression were further investigated. The immuno-precipitates of p27Kip1 were analysed by Western blot analysis but no co-immunoprecipitation of RPS13 was found (data no shown), which suggested there was no interaction between p27Kip1 and RPS13. p27Kip1 expression is mainly regulated by proteosome-mediated degradation at the G1 to S phase transition [15]. To assess if RPS13 influenced degradation of p27Kip1, the proteasome inhibitor MG132 was added to the cells 2 hrs before harvest. p27Kip1 was then detected by Western blot analysis. It was found MG132 could not restore p27Kip1 protein expression in SGC7901/RPS13 cells (Fig. 7C), which suggested RPS13 did not promote p27Kip1 degradation. As revealed by real-time PCR, the mRNA level of p27Kip1 was decreased in SGC7901/RPS13 cells and increased in SGC7901/siRPS13 cells comparing to their corresponding control cells (Fig. 7D). Taken together, it was demonstrated that RPS13 could suppress p27Kip1 mRNA expression and thus down-regulate p27Kip1 in gastric cancer cells.
Discussion
RPs as basal organelle proteins in living cells play important roles in regulating protein biosynthesis to maintain cell survival. Besides accumulating evidence on the role of components of the translation apparatus, there was also evidence suggesting that a number of RPs have an extra-ribosomal capacity, for example regulation of cell proliferation, apoptosis and transformation, independent of their conventional role in translation [16, 17]. These findings were explained by a ‘one gene-dual function’ phenomenon, where a gene product can be multifunctional [18]. A previous study reported that the mitochondrial RP L41 (MRPL41) plays an important role in p53-dependent apoptosis and arrests the cell cycle by increasing the p21 (WAF1/CIP1) and p27Kip1 levels under the growth inhibitory conditions [19, 20]. In addition, it has been discovered that human RPL13a induces apoptosis, presumably by arresting cell growth in the G2/M phase of the cell cycle [21]. Furthermore, RPS29 was found to induce apoptosis in H520 cells by marginally increasing the release of apoptosis inducing factor and reducing the NF-κB dependent transcriptional activity [22]. Similar results have been found in RPL15 and RPS3 [23].
The present study demonstrated an extra-ribosomal function of RPS13 in gastric cancer. The expression and biological significance of RPS13 in human gastric cancer was firstly investigated.
It was found that RPS13 was expressed at a much higher level in gastric cancer tissues and cell lines than that in normal gastric mucosa and epithelial cell line. Other groups have reported over-expression of RPS13 in colon cancer and small cell lung cancer cells [1, 2]. They found that the corresponding mRNA of RPS13 was around 30-fold more abundant in the colon cancer cells than in normal colon [1] and about 50-fold more abundant in lung metastasis than in normal lung [2], which suggested RPS13 might be involved in the regulation of cancer metastasis. So we further analysed the relationship between RPS13 protein expression and clinicopathological characteristics of the gastric cancer specimens. The results revealed that RPS13 was up-regulated in gastric cancer but it did not correlate with patient's gender, age, tumour differentiation grade, TNM (Tumour, Lymph Node, Metastasis) stage and lymph node metastasis. We speculated that RPS13 might be involved in the development of gastric cancer and play important roles in tumorigenesis. To prove our hypothesis, we examined the effects of RPS13 on the growth and tumorigenesis of gastric cancer cells. It was revealed that RPS13 could promote the growth and tumorigenesis of gastric cancer cells. Previously, it has been reported that a close correlation was found between the expression level of RPS13 and the growth rate of two rat colon-carcinoma cell lines [1]. These data supported an important role of RPS13 in promoting cancer cell proliferation and tumorigenicity. It also suggested that RPS13 may serve as a useful biological marker of gastric cancer onset and can be used to evaluate the growth rate of tumour cells.
It has been reported that RPS13 took part in modulating subunit interactions in multiple steps of the translation cycle [24] and regulating the eIF4G-dependent translation through direct interactions with PDCD4 in senescent fibroblasts [25]. However, investigations on RPS13-deficient mutant revealed that RPS13 was not essential for cell growth [24]. Our previous work also indicated that overexpression of RPS13 did not alter the protein synthesis rates of gastric cancer cells [12]. Similarly, it was found that inducing exogenous RPS3a expression alone in NIH 3T3 cells did not increase protein synthetic activity [26]. It is accepted that, independent, non-coordinate changes in expression of an individual RP gene, or of a subset of RP genes, can occur under various cellular conditions, and have no direct association or correlation with protein synthetic activities per se [27, 28]. Therefore, we inferred that the stimulating effects of RPS13 on the growth and tumorigenesis of gastric cancer cells should be its extra-ribosomal function and have little relationship with protein synthetic activities.
To address how RPS13 promoted gastric cancer cell growth, the effects of RPS13 on cell cycle progression were evaluated. It was found that the percentage of cells at S phase was increased in the presence of RPS13 overexpression whereas the percentage of cells at G1 phase was increased while RPS13 was down-regulated in gastric cancer cells. Subsequent analysis confirmed that RPS13 could accelerate G1 to S phase transition of gastric cancer cells. The progression from G1 to S phase is tightly regulated by cyclin D, cyclin E, CDK2, CDK4, p16INK4A and p27Kip1[29]. Among these regulators, only p27Kip1 was down-regulated by RPS13 in gastric cancer cells. p27Kip1 is highly expressed in G0 and early G1 phase and then decreases in late G1, at the transition into S phase. The decrease of p27Kip1 protein level in late G1 phase is caused by pro-teosome-mediated degradation [15]. It was demonstrated in the present study that RPS13 exerted no effects on p27Kip1 degradation, which was demonstrated by the failure of proteosome inhibitor to restore p27Kip1 expression in gastric cancer cells over-expressing RPS13. In addition, there was no interaction between RPS13 and p27Kip1 proteins (data not shown). Further studies revealed that RPS13 suppressed p27Kip1 mRNA expression in gastric cancer cells. However, how RPS13 inhibits p27Kip1 mRNA expression level and whether there are other mechanisms responsible for the stimulative effects of RPS13 on the growth of gastric cancer cells still need further investigation.
It is widely accepted that p27Kip1 functions as a tumour suppressor [15]. Clinical investigations revealed that p27Kip1 expression was lost in most cases of gastric carcinomas compared with normal gastric tissues and that loss of p27Kip1 was associated with poor prognosis [30, 31]. Moreover, it has been reported that expression of p27Kip1 in gastric cancers exhibited no significant correlation with tumour stage or lymph node status [32]. The present indicated no correlation of RPS13 with gastric cancer TNM stage and lymph node metastasis. These data suggested that suppression of p27Kip1 and up-regulation of RPS13 are early events in the developing process of gastric cancer. In support of this speculation, it was recently reported that p27Kip1 expression gradually dropped off when gastric mucosa lesions developed from atrophic gastritis to intestinal metaplasia, and to dysplasia [33]. Furthermore, the cyclooxygenase-2 inhibitor celecoxib caused growth inhibition of human gastric carcinoma cells as well as significantly increased expression of p27Kip1[34]. Considering there is currently no predictive marker for gastroen-terologists to pick out the patients with gastric precancerous lesions (including atrophic gastritis, intestinal metaplasia and dysplasia) who would eventually develop gastric cancer, it is meaningful to further examine the expression of RPS13 in gastric precancerous lesions and to assess the predictive value of RPS13 and p27Kip1 for gastric cancer.
In conclusion, the present study revealed overexpression of RPS13 in gastric cancer tissues. It was demonstrated that RPS13 could promote the growth and G1/S transition of gastric cancer cells through inhibiting p27Kip1 mRNA expression. Therefore, RPS13 is likely to play an important role in tumorigenesis of human gastric cancer and it could be a potential biomarker and therapeutic target for gastric cancer.
Acknowledgments
This work was supported by National Natural Sciences Foundation of China, No. 30670971.
References
- 1.Denis MG, Chadeneau C, Lecabellec MT, et al. Over-expression of the S13 riboso-mal protein in actively growing cells. Int J Cancer. 1993;55:275–80. doi: 10.1002/ijc.2910550218. [DOI] [PubMed] [Google Scholar]
- 2.Zhang L, Cilley RE, Chinoy MR. Suppression subtractive hybridization to identify gene expressions in variant and classic small cell lung cancer cell lines. J Surg Res. 2000;93:108–19. doi: 10.1006/jsre.2000.5957. [DOI] [PubMed] [Google Scholar]
- 3.Kondoh N, Wakatsuki T, Ryo A, et al. Identification and characterization of genes associated with human hepatocellular car-cinogenesis. Cancer Res. 1999;59:4990–6. [PubMed] [Google Scholar]
- 4.Ganger DR, Hamilton PD, Klos DJ, et al. Differential expression of metallopanstim-ulin/S27 ribosomal protein in hepatic regeneration and neoplasia. Cancer Detect Prev. 2001;25:231–6. [PubMed] [Google Scholar]
- 5.Vaarala MH, Porvari KS, Kyllönen AP, et al. Several genes encoding ribosomal proteins are over-expressed in prostate-cancer cell lines: confirmation of L7a and L37 over-expression in prostate-cancer tissue samples. Int J Cancer. 1998;78:27–32. doi: 10.1002/(sici)1097-0215(19980925)78:1<27::aid-ijc6>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 6.Ohkia A, Hu Y, Wang M, et al. Evidence for prostate cancer-associated diagnostic marker-1: immunohistochemistry and in situ hybridization studies. Clin Cancer Res. 2004;10:2452–8. doi: 10.1158/1078-0432.ccr-03-0170. [DOI] [PubMed] [Google Scholar]
- 7.Bee A, Ke Y, Forootan S, et al. Ribosomal protein l19 is a prognostic marker for human prostate cancer. Clin Cancer Res. 2006;12:2061–5. doi: 10.1158/1078-0432.CCR-05-2445. [DOI] [PubMed] [Google Scholar]
- 8.Lü B, Xu J, Zhu Y, et al. Systemic analysis of the differential gene expression profile in a colonic adenoma-normal SSH library. Clin Chim Acta. 2007;378:42–7. doi: 10.1016/j.cca.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 9.Pogue-Geile K, Geiser JR, Shu M, et al. Ribosomal protein genes are overexpressed in colorectal cancer: isolation of a cDNA clone encoding the human S3 ribosomal protein. Mol Cell Biol. 1991;11:3842–9. doi: 10.1128/mcb.11.8.3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Lan M, Shi YQ, et al. Differential display of vincristine-resistance-related genes in gastric cancer SGC7901 cell. World J Gastroenterol. 2002;8:54–9. doi: 10.3748/wjg.v8.i1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Y, You H, Liu F, et al. Differentially expressed gene profiles between mul-tidrug resistant gastric adenocarcinoma cells and their parental cells. Cancer Lett. 2002;185:211–8. doi: 10.1016/s0304-3835(02)00264-1. [DOI] [PubMed] [Google Scholar]
- 12.Shi Y, Zhai H, Wang X, et al. Ribosomal proteins S13 and L23 promote multidrug resistance in gastric cancer cells by suppressing drug-induced apoptosis. Exp Cell Res. 2004;296:337–46. doi: 10.1016/j.yexcr.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 13.Guo XY, Shi YQ, Zhai HH, et al. Prokaryotic expression and purification of RPS13 and preparation of polyclonal antibody against RPS13. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2007;23:363–6. (in Chinese with English Abstract.) [PubMed] [Google Scholar]
- 14.Liang J, Pan Y, Zhang D, et al. Cellular prion protein promotes proliferation and G1/S transition of human gastric cancer cells SGC7901 and AGS. FASEB J. 2007;21:2247–56. doi: 10.1096/fj.06-7799com. [DOI] [PubMed] [Google Scholar]
- 15.Vervoorts J, Lüscher B. Post-translational regulation of the tumor suppressor p27(KIP1) Cell Mol Life Sci. 2008;65:3255–64. doi: 10.1007/s00018-008-8296-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wool IG. Extraribosomal functions of ribo-somal proteins. Trends Biochem Sci. 1996;21:164–5. [PubMed] [Google Scholar]
- 17.Lai MD, Xu J. Ribosomal proteins and col-orectal cancer. Curr Genomics. 2007;8:43–9. doi: 10.2174/138920207780076938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naora H. Involvement of ribosomal proteins in regulating cell growth and apoptosis: translational modulation or recruitment for extraribosomal activity? Immunol Cell Biol. 1999;77:197–205. doi: 10.1046/j.1440-1711.1999.00816.x. [DOI] [PubMed] [Google Scholar]
- 19.Kim MJ, Yoo YA, Kim HJ, et al. Mitochondrial ribosomal protein L41 mediates serum starvation-induced cell-cycle arrest through an increase of p21(WAF1/CIP1) Biochem Biophys Res Commun. 2005;338:1179–84. doi: 10.1016/j.bbrc.2005.10.064. [DOI] [PubMed] [Google Scholar]
- 20.Yoo YA, Kim MJ, Park JK, et al. Mitochondrial ribosomal protein L41 suppresses cell growth in association with p53 and p27Kip1. Mol Cell Biol. 2005;25:6603–16. doi: 10.1128/MCB.25.15.6603-6616.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen FW, Davies JP, Ioannou YA. Differential gene expression in apoptosis: identification of ribosomal protein 23K, a cell proliferation inhibitor. Mol Genet Metab. 1998;64:271–82. doi: 10.1006/mgme.1998.2718. [DOI] [PubMed] [Google Scholar]
- 22.Khanna N, Sen S, Sharma H, et al. S29 ribosomal protein induces apoptosis in H520 cells and sensitizes them to chemotherapy. Biochem Biophys Res Commun. 2003;304:26–35. doi: 10.1016/s0006-291x(03)00532-1. [DOI] [PubMed] [Google Scholar]
- 23.Jang CY, Lee JY, Kim J. RpS3, a DNA repair endonuclease and ribosomal protein, is involved in apoptosis. FEBS Lett. 2004;560:81–5. doi: 10.1016/S0014-5793(04)00074-2. [DOI] [PubMed] [Google Scholar]
- 24.Cukras AR, Green R. Multiple effects of S13 in modulating the strength of inter-subunit interactions in the ribosome during translation. J Mol Biol. 2005;349:47–59. doi: 10.1016/j.jmb.2005.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang MJ, Ahn HS, Lee JY, et al. Up-regulation of PDCD4 in senescent human diploid fibroblasts. Biochem Biophys Res Commun. 2002;293:617–21. doi: 10.1016/S0006-291X(02)00264-4. [DOI] [PubMed] [Google Scholar]
- 26.Naora H, Takai I, Adachi M, Naora H. Altered cellular responses by varying expression of a ribosomal protein gene: sequential coordination of enhancement and suppression of ribosomal protein S3a gene expression induces apoptosis. J Cell Biol. 1998;141:741–53. doi: 10.1083/jcb.141.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrari S, Manfredini R, Tagliafico E, et al. Noncoordinated expression of S6, S11, and S14 ribosomal protein genes in leukemic blast cells. Cancer Res. 1990;50:5825–8. [PubMed] [Google Scholar]
- 28.Seshadri T, Uzman JA, Oshima J, et al. Identification of a transcript that is down-regulated in senescent human fibroblasts. Cloning, sequence analysis, and regulation of the human L7 ribosomal protein gene. J Biol Chem. 1993;268:18474–80. [PubMed] [Google Scholar]
- 29.Yu Y, Kovacevic Z, Richardson DR. Tuning cell cycle regulation with an iron key. Cell Cycle. 2007;6:1982–94. doi: 10.4161/cc.6.16.4603. [DOI] [PubMed] [Google Scholar]
- 30.Kim DH. Prognostic implications of cyclin B1, p34cdc2, p27(Kip1) and p53 expression in gastric cancer. Yonsei Med J. 2007;48:694–700. doi: 10.3349/ymj.2007.48.4.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gamboa-Dominguez A, Seidl S, Reyes-Gutierrez E, et al. Prognostic significance of p21WAF1/CIP1, p27Kip1, p53 and E-cadherin expression in gastric cancer. J Clin Pathol. 2007;60:756–61. doi: 10.1136/jcp.2006.038976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milne AN, Sitarz R, Carvalho R, et al. Molecular analysis of primary gastric cancer, corresponding xenografts, and 2 novel gastric carcinoma cell lines reveals novel alterations in gastric carcinogenesis. Hum Pathol. 2007;38:903–13. doi: 10.1016/j.humpath.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 33.Anagnostopoulos GK, Stefanou D, Arkoumani E, et al. Immunohistochemical expression of cell-cycle proteins in gastric precancerous lesions. J Gastroenterol Hepatol. 2008;23:626–31. doi: 10.1111/j.1440-1746.2007.05219.x. [DOI] [PubMed] [Google Scholar]
- 34.Liu H, Huang P, Xu X, et al. Anticancer effect of celecoxib via COX-2 dependent and independent mechanisms in human gastric cancers cells. Dig Dis Sci. 2009;54:1418–24. doi: 10.1007/s10620-008-0510-9. [DOI] [PubMed] [Google Scholar]


