Abstract
HOX genes encode transcription factors that play a key role in morphogenesis and cell differentiation during embryogenesis of animals. Human neuroblastoma cells are known to be chemically induced to differentiate into neuronal or Schwannian cells. In the present study, we investigated the roles of HOX genes in differentiation of GOTO neuroblastoma cells into Schwannian cells.When GOTO cells were grown in the presence of 5-bromo-2′-deoxyuridine (BrdU), they increased the expressions of two HOX genes (HOXC6 and HOXC11) and marker genes for Schwannian cells (S100β and myelin basic protein). Forced expression of HOXC11 alone or both HOXC6 isoform 1 and HOXC11 induced the expression of S100β in GOTO cells. In transient transfection experiments, the overexpression of HOXC6 and HOXC11 transactivated the S100β promoter-reporter construct. Taken together, our results suggest that HOXC6 and HOXC11 are associated with differentiation of GOTO cells into Schwannian cells through the transcriptional activation of S100β gene.
Keywords: homeobox gene, HOXC6 - HOXC11, neuroblastoma, Schwannian cell
Introduction
Neuroblastoma is one of the most common paediatric solid tumours, which is derived from the sympathetic nervous system and often arises in the adrenal gland [1]. It is characterized by a variety of clinical malignancy, ranging from tumours which undergo spontaneous regression to those which metastasize to distant organs [2]. It is also known that neuroblastoma cells bi-directionally differentiate into neuronal or Schwannian cells in the presence of particular chemicals. Human neuroblastoma cell line GOTO, which was used in the present study, is induced to differentiate into Schwannian cells in the presence of 5-bromo- 2′-deoxyuridine (BrdU) and into neuronal cells in the presence of dibutyryl cyclic AMP (dbcAMP) [3–6]. GOTO cells treated with BrdU changed their morphology, from small and spindle-shape to larger, flattened and polygonal shape, and increased the expression of S100β protein, which is one of the markers for glial and Schwannian cells in nervous system. It has been suggested that the effect of BrdU on the GOTO cells is due to its incorporation into cellular DNA sequences during early S phase of the cell cycle, thus affecting specific target (switching loci) genes involved in cell differentiation [5, 6]. On the other hand, GOTO cells treated with dbcAMP extend neurite-like processes and increase neurofilament [3, 4].
HOX genes are one of the members of homeobox genes, and encode transcription factors. The genes contain a common sequence element of 180 bp, the homeobox, which codes for a highly conserved 60- amino-acid homeodomain [7]. The homeodomain is responsible for recognition and binding of sequencespecific DNA motifs, and cis-regulates the transcription of target genes [7]. Human genome contains 39 HOX genes present in four gene clusters named HOXA, HOXB, HOXC and HOXD, found on the separate chromosomes, 7, 17, 12 and 2, respectively [8]. Individual HOX genes on each four gene cluster are classified into 13 paralogous groups on the basis of their DNA homology and their positions on their respective chromosomes. HOX genes function as the conductors of morphogenesis and cell differentiation during embryogenesis of animals [9, 10]. During embryonic morphogenesis, HOX gene expressions are regulated in a precise spatiotemporal manner: the 3′-end genes of the clusters are expressed earlier and in more anterior domain than the genes localized in more 5′ positions. Mutational analysis in the mouse indicated that Hox genes play a crucial role in differentiation of nervous system [11, 12]. Further, there is evidence indicating that Hox genes control the renewal or commitment of stem cells or progenitor cells in hematopoiesis [13]. Recently, it has been found that HOX genes are also expressed in some normal adult organs in characteristic patterns [14, 15]. These suggest that HOX genes work as regulators of differentiation or maturation in organs and tissues of not only embryo but also postnatal body.
In this study, we attempted to determine whether HOX genes play a role in the differentiation of GOTO cells into Schwannian cells.
Materials and methods
Cells and cell culture
Human neuroblastoma cell line GOTO was obtained from Health Science Research Resources Bank (Sennan, Japan). This cell line is derived from neuroblastoma arising from the adrenal gland [16]. The cells were grown on tissue culture dishes in a 1: 1 (v/v) mixture of Dulbecco's modified Eagle's minimum essential medium and Ham's F12 medium (DME/F12) supplemented with 10% of foetal bovine serum (FBS). The cells were cultured in a CO2 incubator (37°C, 5% CO2 and 95% air). After 24 hrs subculture, cells were maintained in DME/F12 containing 10% FBS with or without 5 μg/ml of 5-bromo-2′-deoxyuridine (BrdU, Sigma Chemical, St. Louis, MO, USA) or 1 mM dibutyryl cyclic AMP (dbcAMP, Sigma Chemical) for 1–9 days with a change of medium every 24 hrs.
Construction of HOX gene expression plasmid vectors and transfection
The HOXC11 genomic DNA, including the full coding region and intron, was obtained by the PCR amplification using the primers (Forward primer: 5′-ACGATGTTTAACTCGGTCAAC- 3′; reverse primer: 5′-TTACAGCAGAGGATTTCCCGA- 3′). The PCR product was TA-cloned into pCR2.1 plasmid vector (Invitrogen, Burlington, Canada) by using TOPO-TA cloning kit (Invitrogen) (pCR2.1-HOXC11). The pCR2.1-HOXC11 was digested with HindIII and XhoI, and subcloned into the multi-cloning site of pCDNA3.1Hyg(-) (Invitrogen) which had been digested with HindIII and XhoI (pCDNA3.1Hyg(-)-HOXC11). The expression plasmid vectors of HOXC6 isoform 1 and isoform 2 were constructed according to the following procedures. Total RNA was extracted from GOTO cells treated with BrdU. Three micrograms of total RNA sample was subjected to cDNA synthesis for 2 hrs at 37°C in 50 μl of reaction mixture containing 4 U/μl of Molony murine leukemia virus reverse transcriptase, 10 mM dithiothreitol, 0.5 mM MgCl2, 0.5 μM dNTP, and 2 μM random primer. PCR amplification of cDNA for full length of HOXC6 isoform 1 or isoform 2 was performed in 50 μl of reaction mixture containing 1 μl of cDNA sample, 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 0.125U/μl of LA-Taq polymerase (Takara, Ohtsu, Japan), and the primer set for isoform 1 or isoform 2 (HOXC6 isoform 1 forward primer: 5_- AGGTAAAGGCAAAGGGATGAAT-3_; reverse primer: 5_- CCTGGTCACTCTTTCTGCTTCTTA-3_ and HOXC6 isoform 2 forward primer: 5_- GCCGTATGACTATGGATCTAATTC- 3_; reverse primer: 5_- CCTGGTCACTCTTTCTGCTTCT- 3_). The PCR products were TA-cloned into pCR2.1 plasmid vector in the same manner as HOXC11 (pCR2.1-HOXC6 isoform 1 or isoform 2). Next, cDNA fragments of HOXC6 isoform 1 or isoform 2 with HindIII-recognition sequence at 5_-terminus and XbaI- recognition sequence at 3_-terminus were amplified from pCR2.1- HOXC6 isoform 1 or isoform 2 by using the primer pair (isoform 1 forward primer: 5_-CGCAAGCTTAAAGGGATGAATTCCTACTTC- 3_; reverse primer: 5_-CGCTCTAGACTCTTTCTGCTTCTCCTCTTC- 3_ or isoform 2 forward primer: 5_-CGCAAGCTTGACATGCTCTCAAACTGCAGACA- 3_; reverse primer: 5_-CGCTCTAGAACTCTTTCTGCTTCTCCTCTTC- 3_). The cDNA fragment was digested with both HindIII and XbaI restriction enzymes, and subcloned into the HindIII-XbaI site of pCDNA3.1Neo-myc- His(A) (pCDNA3.1Neo-myc-His(A)-HOXC6 isoform 1 or pCDNA3.1Neo-myc-His(A)-HOXC6 isoform 2). The sequences of pCDNA3.1Hyg(-)-HOXC11, pCDNA3.1Neomyc- His(A)-HOXC6 isoform 1 and pCDNA3.1Neo-myc- His(A)-HOXC6 isoform 2 were confirmed by automatic DNA sequencing using ABI PRISM 377 sequencer (Applied BioSystems, Foster City, CA, USA).
These expression plasmid vectors and the control vectors (pCDNA3.1Hyg(-) and pCDNA3.1Neo-myc-His(A)) were transfected into GOTO cells by lipofection method using Lipofectamine and PLUS reagent (Invitrogen).
RNA extraction and cDNA preparation
Total cellular RNA was extracted from cultures of GOTO cells with Trizol reagent (Invitrogen) according to the manufacturer's instruction. For exclusion of contaminated genomic DNA, 50 μg of total RNA was treated with DNase [14]. For preparation of cDNA for RT-duplex PCR analysis, 3 μg of DNase-treated total RNA was subjected to cDNA synthesis for 2 hrs at 37°C in 50 μl of reaction mixture containing 4 U/μl Moloney murine leukemia virus reverse transcriptase (Invitrogen), 7.5 mM dithiothreitol, 0.5 mM MgCl2, 0.5 μM dNTP, 2 μM random primer. For real-time PCR, 1 μg of DNase-treated total RNA was subjected to cDNA synthesis in 100 μl of reaction mixture containing Taq Man RT buffer (Applied Biosystems), 5.5 mM MgCl2, 500 μM dNTP, 2.5 μM random hexamers, 0.4 U/μl RNase inhibitor and 1.25 U/μl MultiScribe reverse transcriptase. The reverse transcription reaction was performed sequentially for 10 min at 25°C, for 30 min at 48°C, and for 5 min at 95°C.
Real-time reverse-transcription PCR (RT-PCR) analysis for HOX genes
Real-time RT-PCR assays were carried out by using ABI PRISM 7900HT (Applied Biosystems) with SYBR-green fluorescence under the same condition as described in our previous report [14]. The primer sets for amplification of 39 HOX genes and β-actin gene were listed in our previous report [14].
Semi-quantitative RT-duplex PCR analysis for neuron- or Schwann cell-related genes
PCR amplification of cDNA was performed in 20 μl of reaction mixture containing 2 μl of cDNA sample, 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 2.5 U/ml of Taq polymerase (Wako, Tokyo, Japan) and two kinds of primer sets (10 nM of neuron- or Schwann cell-related gene, for example, S100β gene and 2 nM of β-actin gene as an internal control) by using Gene Amp PCR System 2400-R (Perkin–Elmer, Norwalk, CT, USA). Co-amplification of the specific gene and β-actin gene was achieved in a single reaction mixture. Primer pairs used for the RTduplex PCR were as follows: S100β, forward primer (5_- GCCACGAGTTCTTTGAACATGA-3_) and reverse primer (5_-TGATGCAGGCCGTTAAAACA-3_); myelin basic protein (MBP), forward primer (5_-GGACTGTCCCTGAGCAGATT- 3_) and reverse primer (5′-AATCCCTTGTGAGCCGATTT- 3_); HOXC6 isoform 1, forward primer (5_- AAAGGGATGAATTCCTACTTC-3_) and reverse primer (5_- CTCTTTCTGCTTCTCCTCTTC-3_); HOXC6 isoform 1 and 2, forward primer (5_-GACATGCTCTCAAACTGCAGACA- 3) and reverse primer (5_-CTCTTTCTGCTTCTCCTCTTC- 3_); HOXC11, forward primer (5_-ATCCCAGCTCGTCCGGTTCAGCCCA- 3_) and reverse primer (5_-GTCCGTCAGGTTCAGCATC- 3_); β-actin, forward primer (5_-TCTACAATGAGCTGCGTGTGGCTCC-3_ or 5_-ACCTCATGAAGATCCTCACCGAGCG- 3_) and reverse primer (5(-AGGAAGGAAGGCTGGAAGAGTGCCT-3_ or 5_- AGGAAGGAAGGCTGGAAGAGTGCCT-3_).
Luciferase reporter assay
The luciferase reporter plasmid containing the S100β promoter upstream of the firefly luciferase gene was constructed by inserting DNA fragments into the HindIII and XhoI sites of pGL3-basic (Promega, Madison, WI, USA). S100β promoter was PCR-amplified from genomic DNA of human umbilical vein endothelial cells with the primers as follows: four kinds of forward primers, S100β (-1002) (5_-TGGGAAACCCTTTCACGGCCACTT-3_); S100β (-773) (5_-CTTCCTCAGGCTGGCTGGAGGC-3_); S100bβ (-589) (5(-CACCCGTTTTAGTCCCTGCAGACA-3_); S100β (-388) (5′-TCAGCCTGTACTTGGAAGCTGCTTG-3′) and a reverse primer (5_-CTTCCTTGTCTCACCCCTTCCTGG- 3_). The lucifearse reporter constructs containing four kinds of fragments from the S100β promoter region were named pGL3-basic-S100β(-1002/+69), pGL3-basic-S100β (-773/+69), pGL3-basic-S100β(-589/+69), and pGL3- basic-S100β(-388/+69), respectively. GOTO cells were seeded onto 12-well plates (1 × 104/well). After 48 hrsincubation, medium was replaced with serum-free DME/F12, and the GOTO cells were 3 hrs-transiently transfected with the firefly luciferase reporter plasmids (1 μg/well), Renilla luciferase expression plasmid vector (pRLCMV, Promega) (3 ng/well) as a control vector to normalize transfection efficiencies, and HOX gene expression plasmid vectors (0.5 μg/well). After the transfection, the cells were washed with DME/F12 medium and incubated for an additional 48 hrs. Luciferase assay was then performed by using a dual luciferase reporter assay system (Promega) according to the manufacturer's instructions. Luciferase activity was measured with Berthold Mini Limat LB950L (Berthold, Tokyo, Japan).
Results
Induction of differentiation of GOTO cells into Schwannian cell-like cells
First, we confirmed that GOTO cells differentiated into Schwannian cell-like cells by the treatment with 5-bromo-2′-deoxyuridine (BrdU). GOTO cells were cultured in the presence of 5 μg/ml of BrdU for 1–9 days, and their expression levels of S100β and myelin basic protein (MBP) as markers of Schwannian cells were detected by RT-duplex PCR. As shown in Figure 1, when GOTO cells were treated with BrdU, S100β and MBP were detected after 3 day- and 5 day-incubation, respectively, and their expression levels were maintained till day 9. When GOTO cells were incubated in the presence of 1 mM dibutyryl cyclic AMP (dbcAMP) which was known to induce differention into neuronal cells, the expression level of neither S100β nor MBP was affected.
1.
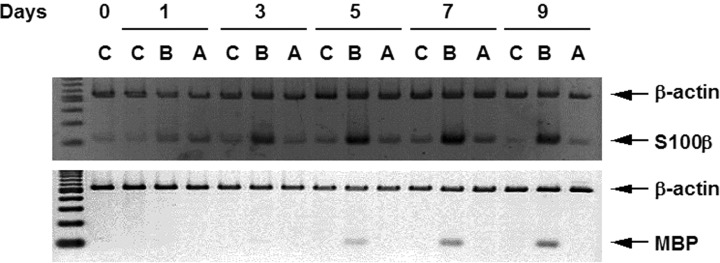
Expressions of S100β and myelin basic protein genes in GOTO cells grown in the presence of BrdU. Total RNA was extracted from the cells cultured in the presence of 5 μg/ml of BrdU (B), 1 mM dbcAMP (A) or no chemicals (C) for the indicated period (days). The total RNA was reverse-transcribed, and a duplex PCR with primers for S100β or MBP and for β-actin was performed. PCR products were electrophoresed and stained with ethidium bromide.
Changes in HOX gene expressions of GOTO cells by the treatment with BrdU
Next, we examined chronological changes of the expression of 39 HOX genes in GOTO cells treated with BrdU. Non-treated GOTO cells showed the expression of 8 HOX genes, HOXC4, C6, C9, C10, D8, D9 and D13, of 39 genes (data not shown). The BrdU-treatment altered the expression of two genes, HOXC6 and C11. The treatment showed no change in the expression of the other 37 HOX genes. The expression of HOXC6 in GOTO cells treated with BrdU was increased up to twofold on day 3 compared to non-treated ones, and then gradually decreased (Fig. 2). The expression of HOXC11 rapidly increased (twofold on day 1 and fourfold on day 3 compared to non-treated cells) and remained high till day 9 when the cells were treated with BrdU (Fig. 2).
2.
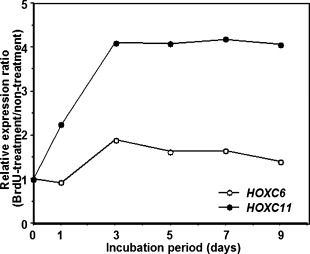
Chronological changes in expressions of HOXC6 and HOXC11 genes in GOTO cells grown in the presence of BrdU. GOTO cells were grown in the same condition as Fig. 1. Total RNA treated with DNase was subjected to quantitative real-time RT-PCR. The expression levels of HOX genes were determined as the ratio of the HOX gene to the internal reference gene (β-actin). Data are represented as the relative ratio of the HOX gene expression level of GOTO cells treated with BrdU to that of non-treated GOTO cells.
Identification of isoform of HOXC6 of which expression level was elevated by BrdU
It is known that there are two kinds of isoforms in HOXC6 transcripts [17]. To determine which isoform was increased in GOTO cells treated with BrdU, we designed two kinds of PCR primer pairs: One was able to amplify only isoform 1 and the other was able to do both isoform 1 and isoform 2. By using the primer pairs and RNA extracted from GOTO cells treated with BrdU, RT-PCR analysis was performed. As shown in Figure 3, both isoforms were originally expressed in GOTO cells, and isoform 1 was increased by the BrdU-treatment. However, no significant increase of isoform 2 was found because the increase of isoform 1 plus 2 was not higher than that of isoform 1 alone.
3.
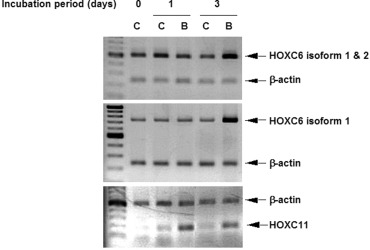
Expressions of HOXC6 isoform 1 and 2 in GOTO cells grown in the presence of BrdU. Total RNA was extracted from the cells grown in the presence of BrdU (B) or in the absence of BrdU (C) for the indicated periods. The expressions of HOXC6 isoform 1, isoform 2 and HOXC11 were analyzed by RT-duplex PCR.
Changes in expressions of differentiation-related genes by the transduction of HOXC6 and/or HOXC11 gene
To clarify whether HOX genes of which expression was increased by BrdU were associated with differentiation of GOTO cells into Schwannian cell-like cells, we analyzed the expressions of the differentiation- related genes such as S100β and MBP in GOTO cells transduced with HOXC6 and/or HOXC11 genes. The GOTO cells transiently transfected with both HOXC6 isoform 1 and HOXC11 expression vector showed high expression of S100β gene (Fig. 4). Forced expression of only HOXC11 gene also induced the expression of S100β gene although its level was relatively low compared to that by the transfection with both HOXC6 isoform 1 and HOXC11 gene (Fig. 4). The transduction of HOXC6 and/or HOXC11 gene did not alter the expression of MBP genes (Fig. 4).
4.
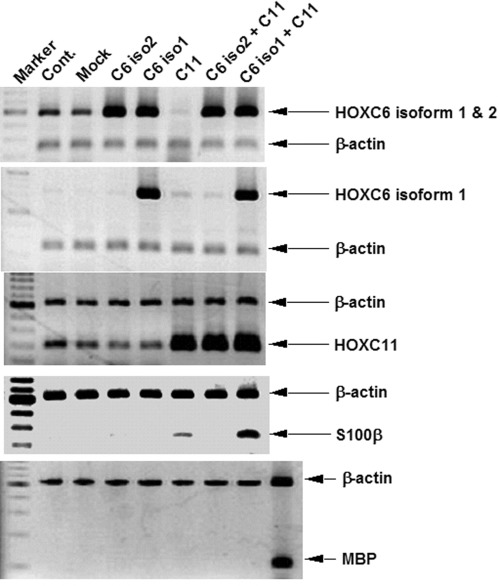
Expressions of S100β and MBP in GOTO cells transfected with HOXC6 and/or HOXC11 genes. Total RNA was extracted from the cells 2 days after the transfection of HOXC6- and/or HOXC11-encoding plasmid vectors. The expressions of genes present here were analyzed by RT-duplex PCR. The sample shown in the right lane at the bottom panel was derived from RNA of GOTO cells treated with BrdU for 9 days.
HOXC6 and HOXC11 activate transcription from the S100β promoter
We generated four kinds of luciferase reporter constructs to assess whether S100β promoter region could mediate transcriptional activation by HOXC6 and/or HOXC11. The reporter constructs contained fragments ranging from −1002 to +69, from −773 to +69, from −589 to +69 or from −388 to +69 of the S100β genomic sequence. GOTO cells were transiently co-transfected with the reporter plasmid and HOX gene expression plasmid vector (HOXC6 and/or HOXC11). As shown in Figure 5, HOXC6 isoform 1 or HOXC11 increased the basal reporter activity in every reporter whereas HOXC6 isoform 2 did not. Co-transfection of HOXC6 (isoform 1 or 2) and HOXC11 led to the maximum activation of the three kinds of reporters which contained a fragment of one of the promoter regions (−773/+69, −589/+69 or −388/+69). In case of using the reporter containing a fragment of the promoter region −1002/+69, the activation level by co-transfection of HOXC6 isoform 2 and HOXC11 was lower than that by co-transfection of HOXC6 isoform 1 and HOXC11.
5.
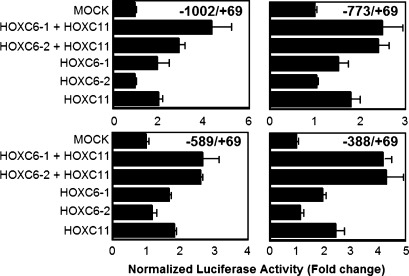
Activation of S100β promoter by HOXC6 and HOXC11. GOTO cells were co-transfected with the S100β promoter- reporter constructs (pGL3-basic-S100β(−1002/+69), pGL3-basic-S100β(−773/+69), pGL3-basic-S100β(−589/+69), or pGL3-basic-S100β(−388/+69)), the control plasmid for transfection efficiency (pRL-CMV) and the effector plasmid (pcDNA3.1Hyg(−)-HOXC11, pCDNA3.1Neo-myc-His(A)-HOXC6 isoform 1 or pCDNA3.1Neo-myc-His(A)-HOXC6 isoform 2). Twenty-four hours after the transfection, luciferase activity was measured.We assigned a value 1 to the activity of cells transfected with mock plasmid vectors (relative luciferase activity). Columns and bars represent the mean value and SD from three independent experiments.
Discussion
It is known that human neuroblastoma GOTO cells differentiate into Schwannian cells by the treatment with BrdU [5, 6].To confirm this phenomenon, we first examined the alteration of the expressions of marker genes of Schwannian cells such as S100β and myelin basic protein by the treatment with BrdU. It was confirmed that the BrdU-treatment induced the expressions of S100β and MBP in GOTO cells. By using this in vitro differentiation model, we attempted to understand the roles of HOX genes in differentiation of GOTO cells into Schwannian cells.
During differentiation into Schwannian cells by BrdU, the expression of HOXC6 was transiently increased to the peak on day 3, and the expression of HOXC11 was increased up to fourfold against the non-treated cells on day 3 and remained till day 9. To test whether the increased expressions of these HOX genes induce differentiation into Schwannian cells, we transiently transfected GOTO cells with the HOX gene expression vectors. As a result, the co-transfection of HOXC6 isoform 1 and HOXC11 elevated the expression level of endogenous S100β gene to a highest level whereas any transfection of HOXC6 and/or HOXC11 did not induce the expression of MBP. Thus, the increased expression of HOXC6 isoform 1 and HOXC11 is likely to result in induction of the expression of S100β at least during the differentiation into Schwannian cells.We could think of some possibilities why the transfection of HOXC6 and HOXC11 did not induce the expression of MBP. One possibility is that these HOX gene products were not involved in the transcription of MBP gene. Another possibility is the difference in timing of assay. Although the expression of MBP was first detected on day 5 after the BrdU-treatment, the MBP expression was analyzed 2 days after the transfection of HOX gene encoding plasmid vectors. Therefore, it might have taken longer time for MBP gene to be induced by the transfection of HOXC6 and HOXC11 encoding vectors.
To assess whether HOXC6 and/or HOXC11 transcriptionally modulate S100β expression, co-transfection experiments were carried out in GOTO cells by using these HOX-encoding plasmid vectors and Luciferase reporter plasmid vectors. Luciferase reporter constructs include the promoter region of S100β (−1002 to +69) or its deleted region (−773 to +69, −589 to +69 or −388 to +69). In the experiments using each reporter, luciferase activity increased when HOXC6- and/or HOXC11-encoding vectors were co-transfected. Maximal luciferase activity was generated when both HOXC6 and HOXC11 were transduced. The promoter region within −1002 and −1 contained five putative HOX-binding motif (TAAT) and the most proximal site of promoter region (−388 to −1) also contained two HOX-binding sites [18]. As the co-transfection of HOXC6 (isoform 1 or 2) and HOXC11maximally increased luciferase activity of every reporter construct, two HOX-binding sites present in the most proximal region of promoter may be essential to activate the transcription of S100β by both HOXC6 and HOXC11. In the experiment using the reporter containing the promoter region (−1002 to −1), the co-transfection of HOXC6 isoform 1 and HOXC11 activated more efficiently the transcription of S100β than that of HOXC6 isoform 2 and HOXC11. This suggests that the promoter region within −1002 and −774, which includes one putative HOX-binding site, may be involved in the transactivation of S100β by HOXC6 isoform 1 but not that by isoform 2. The ability of HOXC6 isoform 1 co-operated with HOXC11 to increase the transcriptional activity of S100β appears to be tempered by the presence of the promoter region between −1002 and −774. Thus we estimate that HOXC6 and HOXC11 recognize the possible promoter region of S100β although we need further study to demonstrate that HOXC6 and HOXC11 directly bind to the putative DNA-binding sites and activate the transcription of S100β.
It was reported that HOXC6, but not HOXC11, was one of the HOX genes whose expressions were upregulated in human neuroblastoma cells differentiating into neuronal cells by retinoic acid or dbcAMP [19, 20].We also obtained the data that GOTO cells increased the expression of neuron-specific enolase and HOXC6 but not HOXC11 when they were treated with dbcAMP (unpublished data). Taken together, HOXC6 appears to be associated with differentiation into both Schwannian and neuronal cells whereas HOXC11 appears to be associated more specifically with differentiation into Schwannian cells.
Acknowledgments
The authors wish to thank Ms. Masako Yanome for her help in preparing the manuscript.
References
- 1.Reynolds CP, Seeger RC. Neuroblastoma. In: Haskell CM, editor. Cancer treatment. New York: WB Saunders; 2000. pp. 1214–36. [Google Scholar]
- 2.Brodeur GM, Nakagawara A. Molecular basis of clinical heterogeneity in neuroblastoma. Am J Pediatr Hematol Oncol. 1992;14:111–6. doi: 10.1097/00043426-199205000-00004. [DOI] [PubMed] [Google Scholar]
- 3.Murakami T, Ohmori H, Gotoh S, Tsuda T, Ohya R, Akiya S, Higashi K. Down modulation of N-myc, heatshock protein 70, and nucleolin during the differentiation of human neuroblastoma cells. J Biochem. 1991;110:146–50. doi: 10.1093/oxfordjournals.jbchem.a123533. [DOI] [PubMed] [Google Scholar]
- 4.Tsuneishi S, Sano K, Nakamura H. Serum depletion increases the neurofilament protein mRNA levels in a neuroblastoma cell line, GOTO. Brain Res Mol Brain Res. 1993;17:119–28. doi: 10.1016/0169-328x(93)90080-9. [DOI] [PubMed] [Google Scholar]
- 5.Sugimoto T, Kato T, Sawada T, Horii Y, Kemshead JT, Hino T, Morioka H, Hosoi H. Schwannian cell differentiation of human neuroblastoma cell lines in vitro induced by bromodeoxyuridine. Cancer Res. 1988;48:2531–7. [PubMed] [Google Scholar]
- 6.Tsunamoto K, Todo S, Imashuku S, Kato K. Induction of S 100 protein by 5-bromo-2'-deoxyuridine in human neuroblastoma cell lines. Cancer Res. 1988;48:170–4. [PubMed] [Google Scholar]
- 7.Levine M, Hoey T. Homeobox proteins as sequencespecific transcription factors. Cell. 1988;55:537–40. doi: 10.1016/0092-8674(88)90209-7. [DOI] [PubMed] [Google Scholar]
- 8.Ruddle FH, Bartels JL, Bentley KL, Kappen C, Murtha MT, Pendleton JW. Evolution of Hox genes. Annu Rev Genet. 1994;28:423–42. doi: 10.1146/annurev.ge.28.120194.002231. [DOI] [PubMed] [Google Scholar]
- 9.Gehring WJ, Hiromi Y. Homeotic genes and the homeobox. Annu Rev Genet. 1986;20:147–73. doi: 10.1146/annurev.ge.20.120186.001051. [DOI] [PubMed] [Google Scholar]
- 10.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 11.Lufkin T, Dierich A, LeMeur M, Mark M, Chambon P. Disruption of the Hox-1.6 homeobox gene results in defects in a region corresponding to its rostral domain of expression. Cell. 1991;66:1105–19. doi: 10.1016/0092-8674(91)90034-v. [DOI] [PubMed] [Google Scholar]
- 12.Chisaka O, Capecchi MR. Regionally restricted developmental defects resulting from targeted disruption of the mouse homeobox gene hox-1.5. Nature. 1991;350:473–9. doi: 10.1038/350473a0. [DOI] [PubMed] [Google Scholar]
- 13.Abramovich C, Humphries RK. Hox regulation of normal and leukemic hematopoietic stem cells. Curr Opin Hematol. 2005;12:210–6. doi: 10.1097/01.moh.0000160737.52349.aa. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi Y, Hamada J, Murakawa K, Takada M, Tada M, Nogami I, Hayashi N, Nakamori S, Monden M, Miyamoto M, Katoh H, Moriuchi T. Expression profiles of 39 HOX genes in normal human adult organs and anaplastic thyroid cancer cell lines by quantitative realtime RT-PCR system. Exp Cell Res. 2004;293:144–53. doi: 10.1016/j.yexcr.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 15.Yahagi N, Kosaki R, Ito T, Mitsuhashi T, Shimada H, Tomita M, Takahashi T, Kosaki K. Position-specific expression of Hox genes along the gastrointestinal tract. Congenit Anom. 2004;44:18–26. doi: 10.1111/j.1741-4520.2003.00004.x. [DOI] [PubMed] [Google Scholar]
- 16.Sekiguchi M, Oota T, Sakakibara K, Inui N, Fujii G. Establishment and characterization of a human neuroblastoma cell line in tissue culture. Jpn J Exp Med. 1979;49:67–83. [PubMed] [Google Scholar]
- 17.Jegalian BG, De Robertis EM. Homeotic transformations in the mouse induced by overexpression of a human Hox3.3 transgene. Cell. 1992;71:901–10. doi: 10.1016/0092-8674(92)90387-r. [DOI] [PubMed] [Google Scholar]
- 18.Gehring WJ, Qian YQ, Billeter M, Furukubo-Tokunaga K, Schier AF, Resendez-Perez D, Affolter M, Otting G, Wuthrich K. Homeodomain-DNA recognition. Cell. 1994;78:211–23. doi: 10.1016/0092-8674(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 19.Manohar CF, Furtado MR, Salwen HR, Cohn SL. Hox gene expression in differentiating human neuroblastoma cells. Biochem Mol Biol Int. 1993;30:733–41. [PubMed] [Google Scholar]
- 20.Manohar CF, Salwen HR, Furtado MR, Cohn SL. Upregulation of HOXC6, HOXD1, and HOXD8 homeobox gene expression in human neuroblastoma cells following chemical induction of differentiation. Tumour Biol. 1996;17:34–47. doi: 10.1159/000217965. [DOI] [PubMed] [Google Scholar]


