Summary
Gender bias and the role of sex hormones in autoimmune diseases are well established. In specific pathogen-free (SPF) non-obese diabetic (NOD) mice, females have 1.3–4.4 times higher incidence of Type 1 diabetes (T1D). Germ-free (GF) mice lost the gender bias (female/male ratio 1.1–1.2). Gut microbiota differed in males and females, a trend reversed by male castration confirming that androgens influence gut microbiota. Colonization of GF NOD mice with defined microbiota revealed that some, but not all, lineages overrepresented in male mice supported a gender bias in T1D. Although, protection of males did not correlate with blood androgen concentration, hormone-supported expansion of selected microbial lineages may work as a positive feedback mechanism contributing to the sexual dimorphism of autoimmune diseases. Gene expression analysis suggested pathways involved in protection of males from T1D by microbiota. Our results favor a two-signal model of gender bias, in which hormones and microbes together trigger protective pathways.
Introduction
Sexual dimorphism is a characteristic feature of many major autoimmune diseases(Fish, 2008; Whitacre, 2001). In most cases females have higher incidence and/or severity of the disease. Sex hormones play a pivotal role in the gender bias with a commonly accepted view that androgens are protective. Castration of males enhances disease progression in animal models of systemic lupus erythematosus (SLE)(Roubinian et al., 1978) and type 1 diabetes (T1D)(Makino et al., 1981), whereas castration of females has variable effects(Fitzpatrick et al., 1991; Makino et al., 1981). Supplementation of females with androgens leads to their protection from these diseases(Fox, 1992; Roubinian et al., 1978), and hormone therapy is used in human SLE patients(Kanda et al., 1997).
However, the hormonal influence on the sexual dimorphism of T1D in non-obese diabetic (NOD) mice appears to be sensitive to environmental influences: T1D incidence varies between institutions(Pozzilli et al., 1993) and even with time within the same institution (Table 1). Most importantly, germ-free (GF) NOD animals have much smaller differences in T1D incidence between genders: an independent rederivation of NOD/ShiLTJ mice into germ-free conditions resulted in remarkably similar incidence of T1D to that previously reported by our group (Table 1). Given the wide variation in T1D incidence and the female/male ratio of affected mice, these results lead to two conclusions: first, that the environmental settings and variations in commensal microbiota influence gender bias in NOD animals, and second, that the influence is likely affected by the composition of the microbiota. Thus, there likely exists an unknown interaction between known hormonal influences(Kovats and Carreras, 2008) and known microbial influences on T1D(Mathis and Benoist, 2012).
Table 1.
Gender bias of T1D in NOD colonies.
| Institution | Female (F) | Male (M) | Ratio F/M |
|---|---|---|---|
|
SPF Facilities
| |||
| U of Chicago | 60% | 25% | 2.4 |
| Yale 2008 (Wen et al., 2008) | 90% | 70% | 1.3 |
| Taconic 2009* | 80% | 50% | 1.6 |
| Harvard 2011 (Kriegel et al., 2011) | 50–60% | 10–15% | 4.4 |
| 2002** | 75% | 50% | 1.5 |
| 2003** | 85% | 60% | 1.4 |
| 2004** | 90% | 45% | 2.0 |
| 2005** | 80% | 40% | 2.0 |
| 2006** | 65% | 35% | 1.9 |
| 2007** | 100% | 65% | 1.5 |
| 2008** | 90% | 85% | 1.1 |
| 2009** | 90% | 65% | 1.4 |
|
| |||
|
Germ-Free Facilities
| |||
| Taconic 2008 (Wen et al., 2008) | 100% | 85% | 1.2 |
| U of Chicago 2012 | 95% | 84% | 1.1 |
Cumulative incidence of diabetes in NOD mice at 30 weeks of age.
Taconic Farms NOD incidence study. http://www.taconic.com/user-assets/Documents/Products%20and%20Services/Animal%20Models/NOD_Onset_2009.pdf
T1DR-T1D repository at The Jackson Laboratory. Data available at http://type1diabetes.jax.org/gqc_incidence_studies.html
Three models can explain these results. Linear model A suggests that hormones regulate the microbes (either through immune or metabolic mechanisms), and that microbes then activate the protective effector mechanisms. Linear model B suggests that microbes are regulators of hormonal metabolism, and that the hormones are the actual effectors. Finally, both microbiota and hormones could contribute in an additive fashion to the effector mechanisms (two-signal model C). To test these models, we used high throughput sequencing of male, female and castrated male microbiota, reconstitution of GF animals with candidate microbiota revealed by sequencing, and gene expression analysis of the pancreatic lymph nodes from GF and specific pathogen-free (SPF) males and females. Whereas the hormone-dependent differences in microbiota composition were readily detectable, not all microbes that expanded in males were capable of protecting them from T1D. Although colonized animals showed higher blood testosterone concentrations compared to GF animals, there was no strict correlation between the ability to induce testosterone and protection from T1D. Gene expression analysis and genetic data suggested that at least one protective mechanism was mediated by a pro-inflammatory cytokine, IFN-γ. Thus, our results favor a model, in which signals from both hormones and microbes are integrated for prevention of the disease development.
Results
Differences in microbial composition in males and females are driven by hormones
Hormonal regulation of the microbe-controlling mechanisms predicts that commensal composition should be different between the two genders. To test this, we compared the microbial communities in male and female mice before and after puberty. Mice were separated at 4 weeks and single-housed to prevent microbiota exchange. Bacterial DNA from the cecal contents of 4 week old (prepubescent) mice and 10–13 week old (post-pubescent) was subjected to high-throughput sequencing and subsequent annotation of the 16S rRNA encoding genes.
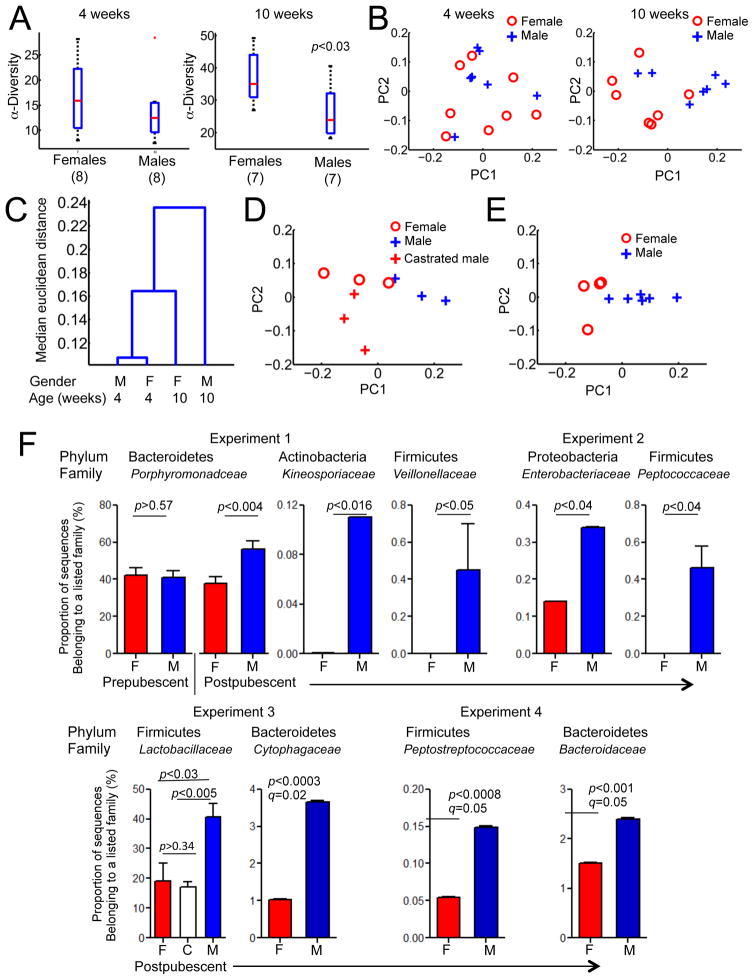
First, we asked whether the representation of different lineages of commensal microbes differed between these animals. The α-diversity (number of microbial species in a given sample) was not significantly different between the genders in 4 week-old mice. On the contrary, post-pubescent mice harbored gender-biased microbial communities (Figure 1A).
Figure 1. Gender-related differences in commensal microbiota composition.
A. Alpha diversity of microbiota from 4 week-old and 10 week-old mice, grouped by gender, mean±SEM. Significance of changes in diversity between genders is reported using a non-parametric Mann-Whitney test.
B. Principal component analysis of microbiota diversity at the family level from 4 week-old (8 males, 8 females), and 10 week-old mice (7 males, 7 females).
C. Hierarchical clustering of 16S sequences of cecal samples from 4-week and 10-week old male and female mice were performed using the Euclidean distance metric and clustering by average linkage. The dendrogram shows the linkage points at increasing degree of dissimilarity.
D. Principal component analysis of microbiota diversity at the family level from 13 wk old littermate mice, 3 females, 3 males, and 3 castrated males.
E. Principal component analysis of microbiota diversity at the family level from 13 week old gnotobiotic littermate mice colonized with an SPF microbiota at birth.
F. Relative abundances of bacterial families between male and female mice. Mean and standard error is marked for each family, significance (p value) is calculated using one tailed t-test. q values obtained by Benjamini-Hochberg approach to adjust for false discovery rate were above 0.05 in most cases (not shown) except experiment 4. See also Figure S1.
Second, principal component analysis (PCA) was performed to determine whether differences in abundance of bacterial families in pre- and post-pubescent mice reflected gender-bias (Figure 1B). The first and second principal components, explaining ~75–80% of the total variance in both groups, only segregated the 10-week old mice by gender and not the 4-week-old mice. The comparison of post-pubescent males and females to prepubescent mice revealed that males deviated from the initially acquired microbiota with time, whereas the adult female microbiota stayed similar to the input microbiota of young mice (Figure 1C). We repeated the sequencing experiment with an independent group of animals and confirmed that postpubescent mice had more differences associated with gender compared to prepubescent mice (not shown).
To test whether removal of the androgen source by castration would drive male microbiota closer to female microbiota, we sequenced 16S rRNA genes from microbiota of male, female and castrated male littermates. Although the number of experimental animals was naturally limited by the size of the litter, the PCA analysis indicated that female and castrated male microbiota were closer to each other than to male microbiota (Figure 1D).
Finally, to eliminate possible adverse influences of the SPF environment on microbiota composition, we colonized germ-free mice with microbiota from an SPF female and compared male vs. female microbiota after puberty. PCA analysis again distinctly segregated male and female microbiota (Figure 1E). The first and second principal components explained ~90% of the total variance in these experiments.
Thus, it is fair to conclude that male microbiota composition deviates more from the input microbiota and that this deviation is indeed hormone-dependent.
Input microbiota defines the gender-specific changes in microbiota
An important question is whether male hormones support or inhibit specific microbial lineages in a reproducible manner, or whether the differences depend on the input microbiota (inherited from mother and coming from the environment). Thus, we compared the abundance of microbial families between adult males and females in four independent experiments. In the first experiment, an unbiased differential analysis of bacterial families showed an expansion of Porphyromonadaceae family (from the Bacteroidetes phylum) in males, which also corresponded to the largest coefficient in the first principal component of this experiment. Further unbiased screening of differences in abundance of bacterial families revealed Veillonellaceae and Kineosporiaceae to be also differentially abundant in males (Figure 1F). Interestingly, the second experiment where NOD mice from an independent set of parents were tested at 13 weeks of age revealed a very different overall commensal composition (Figure 1F and Figure S2) with Peptococcaceae and Enterobacteriaceae dominance in males. Furthermore, in the third experiment, males had more abundant Lactobacillaceae compared to females or castrated males (Figure 1F), which also corresponded to the largest coefficient in the first principal component of this experiment (Figure 1D). Finally, in the experiment with colonization of GF mice, unbiased differential analysis revealed additional new bacterial families differentially expressed between males and females, including Cytophagaceae Peptostreptococcaceae and Bacteroidaceae (Figure 1F).
Thus, the existence of a post-pubescent gender bias in microbial diversity and representation of individual families became evident. However, the lack of consistent microbial variability between all experiments suggests that this variability is likely dependent on the input microbiota. This finding offers an explanation for the variation in T1D gender bias between animal facilities(Pozzilli et al., 1993) (Table 1) and makes two important predictions: a. different microbes could be important for protection in males, and b. some of the changes may be driven by the same mechanisms but are functionally irrelevant for protection from diabetes. The only definitive way to test these predictions was to use a gnotobiotic approach.
Only selected microbes support gender bias in gnotobiotic NOD mice
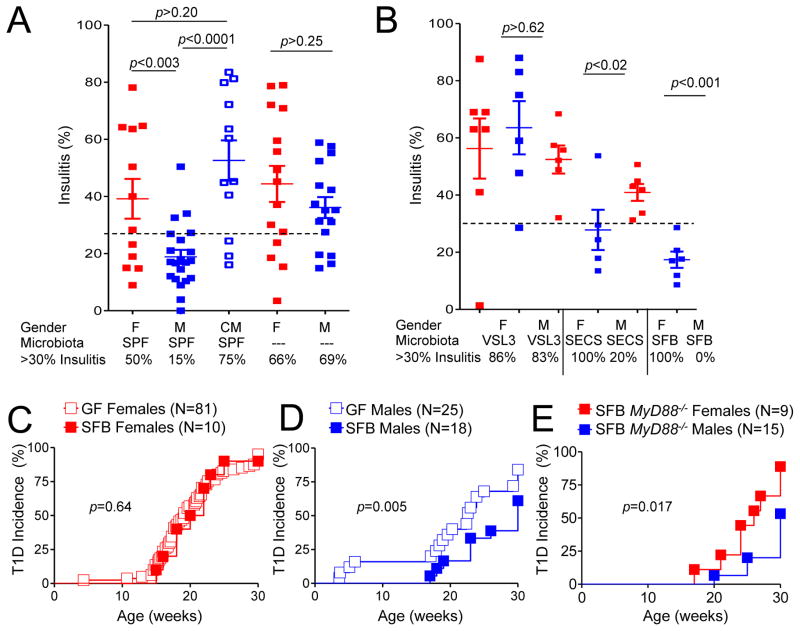
To address the requirement for specific microbiota for induction of gender bias, GF parents were inoculated with different microbes and bred within the isolators. Their progeny were either observed for T1D development or used at 12–13 weeks of age for analysis of the severity of mononuclear infiltration. The selection of microorganisms was based on the results of comparison of the male and female microbiota composition. The histopathology approach (Figure 2A,B) was adequate since it revealed a strong gender bias in specific-pathogen free (SPF) mice and a decrease in the gender bias in GF mice (Figure 2A,C) corresponding perfectly with the incidence of T1D in these groups. Representative images of islet infiltration are shown in Figure S2. First, we colonized GF mice with the probiotic mix VSL3(Calcinaro et al., 2005), which is enriched in Lactobacillaceae and Bifidobacteria, because Lactobacillaceae were enhanced in males in one of the sequencing experiments. However, VSL3 did not reduce the severity of histopathology in males or females (Figure 2B), if anything, it was enhanced by VSL3. Thus, VSL3 was not capable of inducing the gender bias compared to the undefined SPF community.
Figure 2. Influence of microbial lineages on the gender bias in T1D development in NOD mice.
A. Histopathology (% of islets with infiltrates beyond peri-insulitis) in 13 wk old mice from male and female SPF NOD mice, from males castrated at 4 wks of age and from GF male and female mice. Data are represented as mean insulitis score±SEM. 30% infiltration was chosen as an arbitrary threshold
B. Histopathology in 13 wk old gnotobiotic NOD mice reconstituted with indicated microbiota. Data are represented as mean insulitis score±SEM. Germ-free mice were reconstituted by natural acquisition of microbes from parents infected by gastric gavage during breeding.
C. Diabetes incidence in NOD GF female mice and in gnotobiotic NOD female mice monocolonized with SFB.
D. Diabetes incidence in NOD GF male mice and in gnotobiotic NOD male mice monocolonized with SFB. The differences between SFB-colonized and GF males were significant (p=0.005), as well as between SFB-colonized females and males (p=0.002).
E. Diabetes incidence in MyD88-negative NOD gnotobiotic mice monocolonized with SFB.
F. N=the number of mice per group. p values for incidence were determined using Kaplan-Meier statistics, for histopathology by Student’s t-test. See also Figure S2.
In the second microbiota comparison experiment Enterobacteriacae family (phylum Proteobacteria) was found amplified in males (Figure 1F). Colonization of GF mice with a proteobacterium that was cultured from males (Figure S2B) and had 16S rRNA gene sequence ‘similar to E.Coli and Shigella’ (SECS), led to a clear decrease in histopathology in males (Figure 2B).
Finally, we tested whether segmented filamentous bacteria (SFB), known for their pro-inflammatory properties(Gaboriau-Routhiau et al., 2009; Ivanov et al., 2009) and reported to be associated with protection in SPF mice(Kriegel et al., 2011), would protect animals from T1D. The efficiency of colonization with SFB was comparable between the two genders (Figure S2), but there was no influence on T1D incidence in females (Figure 2C). The T1D incidence in males (Figure 2D) was significantly reduced compared to both GF males (p=0.005) and SFB-colonized females (p=0.002). Thus, under our experimental conditions, another single bacterium, SFB, was capable of supporting the gender bias.
Since both genders of MyD88-negative NOD mice are protected under normal conditions(Wen et al., 2008), it was likely that the protection offered by the gender bias mechanism is different from the protection offered by the microbiota in MyD88-negative mice. Thus, we colonized germ-free MyD88-deficient NOD mice with SFB (Figure 2E). Gender bias was preserved in MyD88-deficient mice colonized with SFB. The data argue that MyD88 is dispensable for the gender bias and that distinct bacteria trigger the two protective mechanisms.
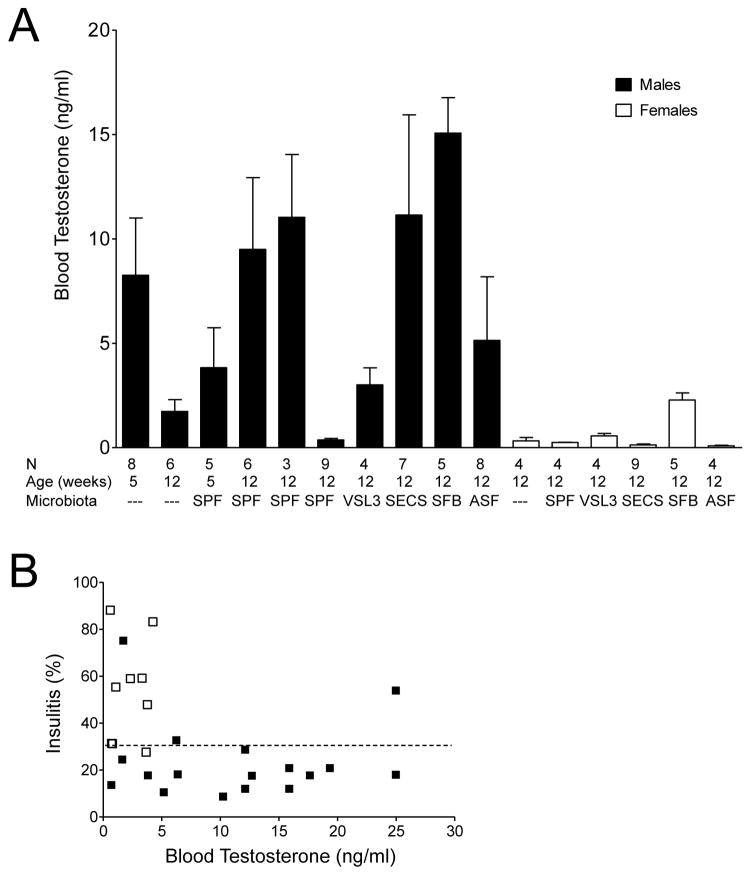
Microbiota elevates androgens to the threshold required for protection
Earlier studies have established that both testosterone(Makino et al., 1981) and microbes(Wen et al., 2008) were involved in the gender bias of T1D in NOD mice. Now that the gender-specific differences in microbiota were found to be androgen-dependent (Figure 1D), it was critical to determine whether the ability of microbiota to induce gender bias correlated with its ability to increase androgen concentration. Thus, we measured serum testosterone in SPF and GF mice, as well as in gnotobiotic mice. The elevated amounts of testosterone correlated with colonization with conventional microbiota and with SFB and SECS. At the same time colonization with VSL3 did not cause a rise in testosterone concentration (Figure 3A). Thus, it would seem that the linear model B supports the observed T1D progression. However, it was critical to determine whether a testosterone threshold necessary for protection could be reached and whether higher testosterone provided better protection. When the severity of islet infiltration was plotted against testosterone concentration in the same mice (Figure 3B), it became clear that after serum testosterone reached a certain concentration (less than 2 ng/ml), a further increase was not required for protection against T1D by microbes capable of providing a protective signal.
Figure 3. Microbiota and blood testosterone concentrations.
A. The blood testosterone concentrations of the 13 wk old GF, SPF and gnotobiotic males and females of indicated ages. Mean testosterone concentration±SEM. Blood samples were collected between 10 am and 12 pm. C- castrated and MC – mock-castrated males.
B. Islet histopathology in mice that demonstrated no gender bias (GF and VSL3 populated – open squares) and that demonstrated gender bias (SPF and SECS populated – black squares) plotted against blood testosterone concentrations. Data compiled from four experiments.
Gene expression analysis reveals possible signaling networks involved in the gender bias of T1D
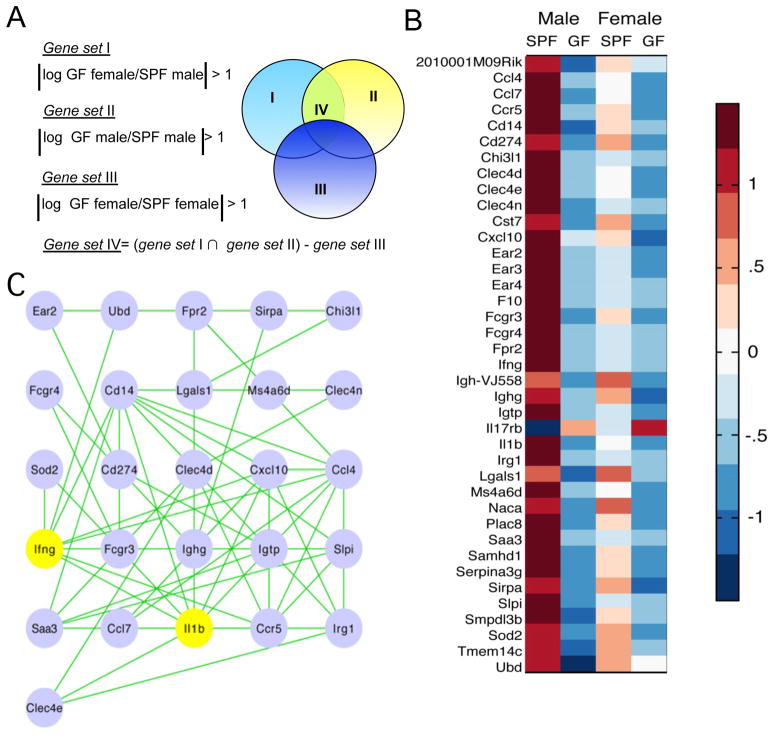
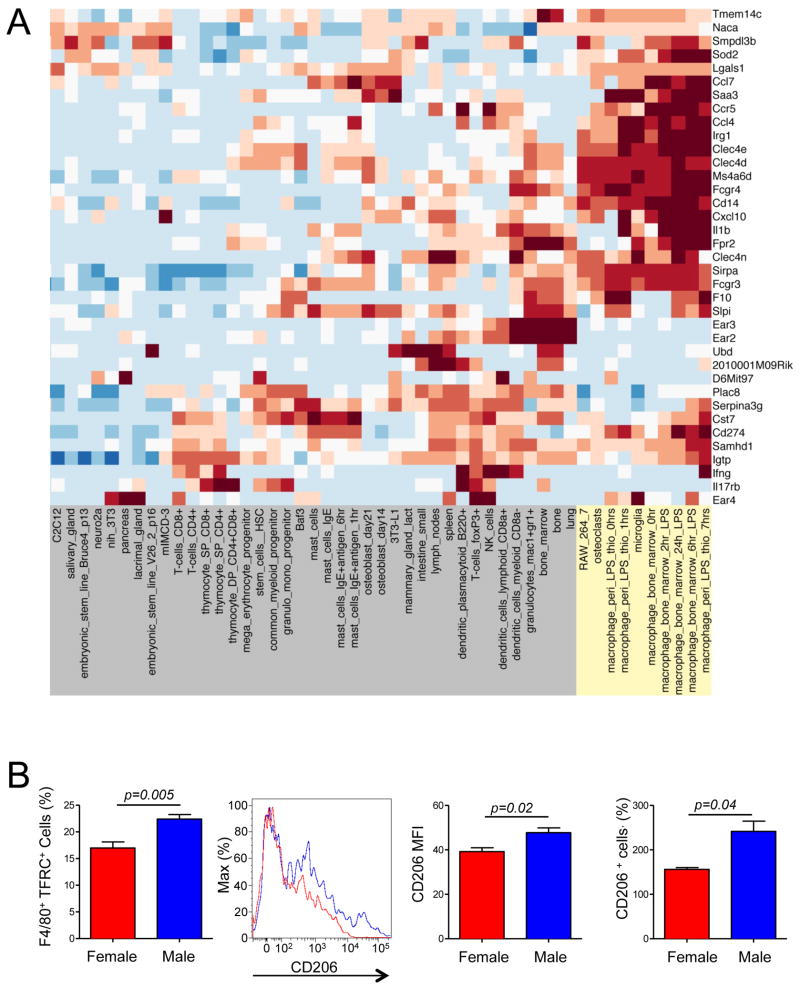
To address the question of which signaling pathways may be affected by both gender and microbiota, we compared gene expression patterns in the pancreatic lymph nodes (PLN), critical for T1D development (Gagnerault et al., 2002; Hoglund et al., 1999; Turley et al., 2005). Only one group out of four (two genders in SPF and GF conditions, respectively) was protected from T1D – the SPF males. Thus, we performed a series of set operations to identify genes that were specifically regulated by microbes in males (Figure 4A). First, genes with similar expression between SPF males and GF females were removed, followed by removal of similarities between SPF and GF males, and, finally, genes that are different between SPF and GF females were removed from the list. The resulting expression pattern of about 40 genes (Figure 4B) was organized into a network of signaling pathways using the STRING database(Szklarczyk et al., 2010). The signatures of the two specific pathways became apparent: interferon-γ (IFN-γ) and interleukin-1β (IL-1β) (Figure 4C). Since PLNs are complex organs, we asked what cell type was critical for the gene expression pattern that we have detected. For that, we generated a gene abundance profile comparing the expression of 40 genes with the gene expression patterns from a panel of 96 cell types from the BioGPS murine RNA Gene Expression Atlas (http://biogps.gnf.org). Hierarchical clustering associated this expression pattern with the monocyte-macrophage cell lineage (Figure 5A and Figure S3). Additionally, an up-regulation of Chitinase-3-like protease 1 gene (Chi3l-1) suggested a more abundant presence of the tolerogenic ‘alternatively activated’ or M2 macrophages(Gordon and Taylor, 2005; Loke et al., 2002; Raes et al., 2002) in SPF males. The representation of macrophages with M2 surface markers was compared between SPF males and females by staining of PLN cells liberated by collagenase. More M2 type macrophages [as determined by the numbers of cells positive for transferrin receptor (Becker et al., 2012; Johnston et al., 2012) and CD206 (Gordon and Martinez, 2010; Stein et al., 1992) and by intensity of CD206 expression] were found in the male mice (Figure 5B).
Figure 4. Analysis of the changes in gene expression driven by microbes and gender.
A. SPF and GF NOD males and females were used as donors of the PLN. Of the four groups only one (SPF males) is protected from T1D. The logic of arrival at the gene set IV, specific to this group, is shown.
B. Heat-map of expression of the genes from set IV. The intensity of the color corresponds to the strength of expression relative to the mean expression across all conditions.
C. Gene set IV organized in a network using the STRING database. Genes encoding IFN-γ and IL-1β are highlighted in yellow.
Figure 5.
A. Many genes belonging to set IV (Figure 4 A) have a signature characteristic of macrophages (highlighted in yellow) as determined by comparison to BioGPS murine RNA Gene Expression Atlas data. The intensity of the color corresponds to the strength of expression relative to the mean expression across all cell types. The complete expression map can be found in Figure S3.
B. Enhanced presence of alternatively activated macrophages in age-matched adult male SFP mice compared to SPF female mice. CD11b+F4/80+ macrophages were stained with antibodies to transferrin receptor (TFRC, left panel) or to CD206. An example of CD206 staining is shown along with quantitation of both CD206+ cell number and the mean fluorescence intensity (MFI). 3 to 5 mice per group were used in these experiments. Data representative of 2–3 independent experiments. Data are represented as mean cell number or MFI±SEM.
Available gene-targeting data support the involvement of the discovered network in male protection from T1D
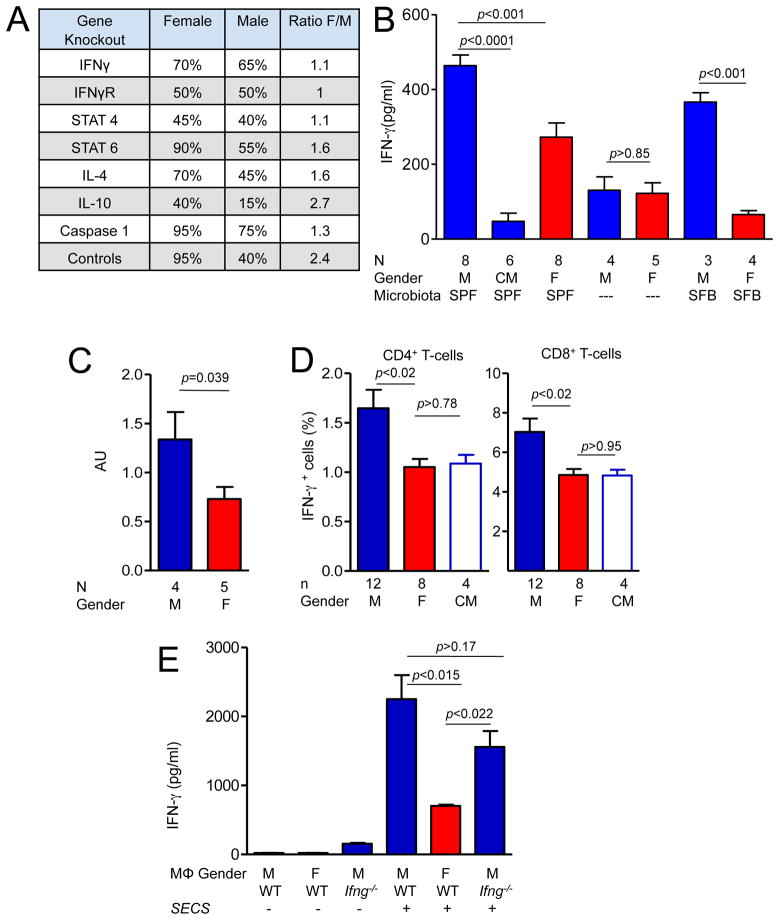
The best support for importance of the gene expression data comes from genetic ablation experiments. Publicly available reports (compilation of the data based on the Type 1 Diabetes Resource, T1DR, at the Jackson Laboratory) on male and female incidence in the relevant gene-targeted mice showed that the loss of IFN-γ signaling removes the gender bias from T1D (Figure 6A). NOD mice with targeted deletions of genes encoding IFN-γ, IFN-γ-receptor-1, and of the downstream signaling molecule STAT4, all had similar incidence of T1D in females and males at 30 weeks of age. During the same time period in the same facility, NOD mice negative for IL12, IL10 and IL4 continued to exhibit gender-biased autoimmunity (Figure 6A). Genetic deletion of Casp1, which is necessary for the cleavage of inactive pro-IL-1β (and of pro-IL-18), has been reported to lead to higher incidence in males compared to males derived from the same back-crossing experiment (Schott et al., 2004). The female incidence was not affected by the lack of Caspase-1. These results strongly support our findings of potential involvement of IFN-γ and IL-1β in the gender-biased protection from T1D.
Figure 6.
A. Manipulation of sexual dimorphism of T1D in NOD mice by gene knock-outs (Chatenoud et al., 2001; Serreze et al., 2001; Serreze et al., 2000) as reported by T1DR or by an original paper for Casp1−/− mice(Schott et al., 2004).
B. IFN-γ expression in the PLNs of males and females in GF, SPF and SFB-monocolonized NOD mice. Mean cytokine concentration±SEM. Tissues were collected from 13 week-old mice, homogenized and used for IFN-γ-specific ELISA. c - SPF males castrated at 4 weeks of age.
C. IFN-γ-specific RNA expression in 12–13 week old SPF males and females was compared by RT- quantitative PCR. AU-arbitrary units. Mean±SEM.
D. The percentage of IFN-γ-positive cells among CD4+ and CD8+ T cells from the PLNs of male, female and castrated male (c) NOD mice. Mean±SEM.
E. Peritoneal macrophages from female, male and IFN-γ-negative male mice were pretreated or not with heat-inactivated SECS bacteria overnight before addition of G9C8 T cells and their cognate peptide. IFN-γ was measured by ELISA in the supernatants after overnight culture. Data from a representative experiment of three independent experiments. Mean±SEM.
N= mice per group. p values were determined by Student’s t-test. See also Figure S4.
IFN–γ production in regional lymph nodes is enhanced in NOD males
To further study the importance of IFN–γ in mediating the gender bias in NOD mice, we tested its expression in the pancreatic lymph nodes by measuring the amounts of protein and IFN-γ-encoding mRNA (Figure 6B, C). Higher concentrations of IFN-γ were found in SPF males compared to SPF females, and its amounts in GF mouse PLNs were much lower (Figure 6B). The same was true for the SFB-monocolonized mice (Figure 6B). The amounts of IFN-γ in the male PLNs measured at 12 weeks of age were severely reduced after castration at 4 weeks of age (Figure 6B). IFN-γ-specific RNA expression measured by quantitative PCR was significantly higher in PLN of the postpubescent males (Figure 6C).
We then asked whether there were more IFN-γ-producing cells among CD4+ and CD8+ T cells in the PLNs of male mice. Lymph node cells were activated with phorbol myristate acetate (PMA) and ionomycin in vitro and stained for surface markers and intracellular IFN-γ. Male PLNs (as well as mesenteric nodes, but not spleens, Figure S4) contained significantly more IFN-γ-producing Tcells than PLNs of female and castrated male mice (Figure 6D). Enhanced production of IFN-γ by T cells in males could result from direct signaling by hormones and/or microbes, or reflect the influence of these signals on antigen-presenting cells. Since most of the microbiota-dependent gene expression differences between males and females pointed at macrophage-monocyte as a critical cell lineage (Figure 5A), we asked whether macrophages from males and females were equally capable of eliciting an IFN-γ response from T cells. Peritoneal macrophages from male, female and Ifng−/− male mice were either pretreated with heat-inactivated SECS bacteria or not, washed and insulin-specific CD8+ T cells from G9C8 T cell receptor transgenic mice(Varanasi et al., 2012) were added along with a cognate peptide. Only macrophages exposed to bacteria elicited IFN-γ from the T cells, and male macrophages were clearly more efficient in doing so than female macrophages (Figure 6E).
In sum, our results show that despite expansion of multiple microbial lineages in male NOD mice, only some lineages can provide necessary signals to host cells that also receive a second signal from androgens. Together these signals control the gender bias in T1D development in NOD mice.
Discussion
The importance of the intestinal commensal microbiota for development of T1D has been clearly demonstrated in NOD mice lacking the MyD88 signaling adaptor(Wen et al., 2008). Importantly, GF NOD mice used in these experiments lose the commonly observed gender bias (enhanced T1D development in females). Thus, it became clear that hormones and microbiota interact to modify the course of the disease progression (Figure S5). We sought to test the three models explaining the mechanisms behind the gender bias. Sequencing of 16S rRNA genes from male and female mice revealed that the microbiota can be gender-biased and that the adult female microbiota is more similar to the microbiota of prepubescent mice of both genders than the male microbiota. Thus, puberty affects the male microbiota composition, which becomes less diverse than the female microbiota. Importantly, comparison of male, female and castrated male microbiota demonstrated that sex hormones rather than X-chromosome-associated factors were important for the change in microbiota composition.
To test whether microbial deviations seen in male mice were meaningful (capable of protecting colonized mice), we used a gnotobiotic approach. If the first model was correct, one would expect microbes enhanced in male NOD mice to be protective in both male and female mice. However, mice mono-colonized with SECS proteobacteria demonstrated a protective effect in males only. Similarly, mono-colonization with SFB led to gender-biased protection measured by T1D incidence and islet pathology. These observations did not fit well with the model in which hormones regulated microbes and microbes were the effectors challenging autoimmunity. If that hypothesis were true, we would expect the protective effect to become unbiased in terms of gender.
Interestingly, VSL3 has been previously reported to be protective in female NOD mice upon continuous application via oral gavage(Calcinaro et al., 2005). It is possible that VSL3 works in concert with the complex SPF microbiota to elicit protection, but such a protection could be mediated by a non-gender-biased mechanism. SFB have been previously associated with protection in female SPF NOD mice(Kriegel et al., 2011). The disparity of the results could be due to an interaction of SFB with other members of the microbial community (absent in our facility) to provide non-gender-biased protection, or by variation in SFB strains themselves.
These experiments also demonstrated that not all androgen-driven changes in microbiota are meaningful in terms of supporting the gender bias in T1D, and provide an additional argument in favor of the gnotobiotic experimental approach. Finally, they also made it clear that many microbial lineages are capable of supporting the gender bias. In fact, our colony of NOD mice was SFB-free but exhibited the gender bias.
Although SPF and some gnotobiotic mice had higher blood testosterone concentrations compared to females and GF mice, it was likely that a certain threshold of testosterone concentration should be achieved before it is able to aid bacteria in eliciting protection from T1D. A general argument in favor of the threshold requirement is that testosterone amounts in males are highly variable during the day (Bartke et al., 1973). Moreover, GF male rats have slightly higher testosterone amounts compared to SPF male rats(Snyder and Wostmann, 1985), and blood testosterone concentrations in GF males entering puberty at 5 weeks of age were higher compared to SPF males, suggesting that microbiota can also negatively regulate testosterone. In addition to these arguments, we have previously reported(Wen et al., 2008) that gnotobiotic male NOD mice carrying the Altered Schaedler’s Flora (ASF)(Dewhirst et al., 1999) microbial consortium did not show a reduction in T1D incidence (78% at 30 weeks, compared to 84% in germ-free males) and the incidence curves were not significantly different (p=0.48), very similar to a recently published experiment (Markle et al., 2013), in which ASF does not significantly protect gnotobiotic NOD males from T1D. However, ASF was perfectly capable of increasing blood testosterone concentration in associated male mice to an average of 5 ng/ml. Thus, a consortium with a poor ability to induce the gender bias supports enhancement of testosterone.
Plotting islet pathology vs. testosterone concentration suggested that when the threshold is achieved, it is the nature of the microbial stimulus that matters, once again supporting the two-signal hypothesis and suggesting that androgen enhancement by microbiota is not enough to explain how the gender bias works.
The ability of microbes to regulate hormones and of hormones to change microbial diversity cannot be simply discarded and must be taken into account. In a modified gender bias model hormone enhancement and maintenance of a gender-biased microbiota provide a regulatory feedback loop needed to maintain the gender bias of T1D protection. It is also likely that in complex microbial communities the functions of hormone enhancement and provision of regulatory signals to the immune system could be divided between different members of the community.
What are the protective mechanisms that are induced by hormones and microbes? Gene-expression analysis has revealed possible signaling mechanisms that are involved in the process. Two points need to be made clear: a. these mechanisms are likely just the tip of the iceberg of gender bias, and b. not all connections shown by this experiment are necessarily meaningful. Moreover, the differences detected by microarray analysis may reflect both microbiota-induced changes and changes induced by a different course of T1D development.
Genetic experiments usually provide the most definitive results. In the case where the microbiota is involved as an epigenetic factor influencing disease development, carefully controlled experiments are needed. Since the role of IFN-γ in the gender bias has been supported by simultaneous observation (at the same time and in the same facility) of the three genetically modified mouse strains relevant to IFN-γ signaling with mice carrying targeted mutations that did not support the gender bias, it is very likely that IFN-γ is central to at least one of the protective mechanisms.
Furthermore, lymph nodes of male NOD mice produced more IFN-γ compared to female lymph nodes and also contained more IFN-γ producing T cells How could excessive IFN-γ be involved in protection from T1D? One possibility is that this cytokine negatively regulates T helper cells required to activate B cells, which are clearly needed for the development of T1D (Serreze et al., 1998; Silveira et al., 2002). The other option is that IFN-γ works as a self-limiting factor inducing cell death in activated T cells(Liu and Janeway, 1990), including in T1D (Qin et al., 2004; Sobel et al., 2002).
It also appears that macrophages are already ‘imprinted’ by androgens in vivo and with the help of microbial signals stimulate T cells to produce more IFN-γ. The nature of imprinted cellular signals is not yet known and warrants further investigation, however, our findings support the two-signal model of gender bias.
Several other genes showed multiple connections in the gene interaction network However, the targeted deletion of Cd14(Kloting et al., 2004) and of tlr4(Dong et al., 2012) have preserved the gender bias, indicating that although CD14 was strongly expressed in SPF males this enhancement was not meaningful. The other cytokine revealed by gene expression analysis was IL-1β, also a proinflammatory cytokine. The role of IL-1β is still unclear, because IL-1-receptor requires MyD88 for signaling, whereas gender bias seems to be MyD88-independent It is, however, possible that several MyD88-dependent and -independent mechanisms controlled by hormones exist to support the gender bias of autoimmunity. The involvement of Caspase-1 in gender bias(Schott et al., 2004) also points at the inflammasome involvement.
It remains to be determined whether the mechanisms that are induced in a gender-biased manner can be used for treatment of non-gender-biased diseases. This is especially important because T1D in humans is not gender-biased, although the disease itself is likely a constellation of diseases that needs to be stratified to reveal the bias. Finally, it is essential to determine if gender bias in other autoimmune disorders is dependent on microbiota.
Experimental procedures
Mice
NOD/ShiLtJ (The Jackson Laboratory, Bar Harbor, ME) mice were kept under SPF and GF conditions at the University of Chicago Animal Resource Center. GF status was monitored by aerobic and anaerobic fecal cultures and PCR amplification of bacterial 16S rRNA genes from fecal DNA as previously described (Kane et al., 2011).
Gnotobiotic NOD mice were derived from GF mice by introduction of a specified bacterial community via gastric gavage to the parents in a separate isolator. Bacteria were transferred to the progeny naturally from the mother, and the efficiency of colonization of the progeny has been confirmed by PCR for 16S rRNA genes specific for the colonizing lineages. VSL3 mix containing Bifidobacterium breve, B. longum, B. infantis, Lactobacillus acidophilus, L. plantarum, L. casei L. bulgaricus and Streptococcus thermophilus was a generous gift from Dr. Claudio De Simone (VSL Pharmaceuticals, Inc, Gaithersburg, Maryland). ASF (Dewhirst et al., 1999) was obtained from Taconic Farms (Hudson, NY). SFB were kept frozen as cecal contents obtained from SFB monocolonized mice, defrosted and used to colonize GF mice. SECS bacterium was introduced to GF male and female mice at the time of weaning by gavage of 100μL of overnight cultures.
Surgery
Gonads were excised from 4 week old males anesthesized with ketamine and xylozine combination (100mg/kg and 5 mg/kg respectively) using the Change-A-Tip handheld cauterizer (Bovie Medical Corporation, Clearwater, FL). Mock-operated mice had an incision made followed by application of the wound clips.
Diabetes
Diabetes development was monitored by weekly testing of urine glucose with Diastix strips (Bayer, Elkhart, IN). At least 200 islets per group of animals were scored using pancreatic sections cut at 40μm intervals and graded as follows: 0, no visible infiltration; I, periinsulitis; II, insulitis (Wen et al., 2008).
Detection of IFN-γ
Pancreatic lymph nodes (2–10 mg) homogenized in cold PBS with protease inhibitors (Roche, Indianapolis, IN), spun for 5 min at 10,000 rpm at 4°C, diluted to 0.4mg of tissue/ml were assayed (50μl per sample) using an IFN-γ ELISA kit (BD Biosciences, San Diego, CA).
RNA Isolation and reverse transcription-PCR (RT-PCR)
One microgram of total RNA recovered from pancreatic lymph nodes of 10 week old mice by homogenizing samples in Trizol (Invitrogen, Carlsbad, CA) was reverse transcribed using SuperScript III First-Strand kit (Invitrogen) and Oligo-dT primers. SYBR Green (Bio-Rad, Hercules, CA) real-time PCR was performed in 20μL reactions using 2μL of cDNA. The following primers were used: GAPDH sense 5′ – AACGACCCCTTCATTGAC-3′, GAPDH antisense 5′-TCCACGACAT ACTCAGCAC-3′, IFN-γ sense 5′-AACGCTACACACTGCATCT-3′, IFN-γ antisense 5′-GAGCTCATTGAATGCTTGG-3′. AB StepOnePlus system (Applied Biosystems, Foster City CA) and StepOne software were used. For IFN-γ intracellular staining, lymph node and spleen cells were activated in vitro with PMA and Ionomycin for 4 hours, permeabilized and stained as described (Kriegel et al., 2011).
Flow Cytometry analysis
Lymph nodes and spleens from 10–13 week old mice were manually disrupted and either used as suspensions or incubated with Collagenase (0.2mg/mL) (Type II, Invitrogen) and DNAse (0.15mg/mL) (Roche) for 30′ at 37°C and passed through a nylon mesh to release macrophages. Cells were stained with directly conjugated antibodies to TFRC-APC (Transferrin receptor), F4/80-PerCP, and CD11b-PE or CD11b-PECy7 in combination with anti-CD206-PE, CD4-PeCy7 (all from eBioscience, San Diego, CA) and CD8-Pacific Blue (Biolegend, San Diego, CA). Anti-mouse IFN-γ XMG1.5-APC antibodies were from BD Biosciences. Cells were analyzed using a FACS Canto or LSR-II flowcytometers (BD Biosciences), and the data were analyzed with FlowJo software (V. 9.6.1, Tree Star, Ashland, OR). For macrophage analysis all gates were established such that 1–2% of cells stained with isotype controls were positive, dead cells were excluded by calcein blue staining (eBioscience).
T cell activation in vitro
Peritoneal macrophages were isolated four days after i.p. administration of 1.5 mL of thioglycolate (Difco Laboratories, Detroit MI). Macrophages were plated in Click’s medium at a density of 5×104 cells per well of a 96 well flat bottom plate, allowed to settle and attach and washed with PBS to remove non-adherent cells. Macrophages were stimulated for 18 hours with heat killed SECS at a ratio of approximately 1:25. Wells were washed with Click’s medium prior to addition of 7.5×104 purified G9C8 T cells and 3μg/ml of cognate peptide InsB15–23.
Testosterone measurements
Serum testosterone concentrations were determined using a rat/mouse testosterone ELISA kit (IBL America, Minneapolis, MN).
Statistical Analysis
Statistical analysis of histology scoring, serum testosterone concentration, and IFN-γ concentration was performed using Prism 5 (GraphPad). Results are expressed as means ± SEM. The statistical difference between two groups was determined by Student’s t test. For multiple groups the statistical difference was determined using the one-way analysis of variance (ANOVA). T1D incidence data was analyzed by Kaplan-Meier using Prism 5 (GraphPad). A p-value < 0.05 was considered statistically significant.
SFB quantitative PCR (qPCR) comparison
Snap-frozen cecal content was placed in lysis buffer and transferred into autoclaved cryotube containing 500 μL of 0.1-mm zirconium-silica beads (Biospec Products, Bartlesville, OK) and 20% SDS (210 μL), bead beaten for 2 min using a Mini-Beadbeater (Biospec, Bartlesville, OK), followed by phenol-chloroform extraction of DNA, as described (Turnbaugh et al., 2009). SYBR Green real-time PCR was performed using a dilution of sample that corresponded to 20ng of original cecal content per well using SFB-specific primers: SFB736 forward 5′ GACGCTGAGG CATGAG AGCAT-3′ and SFB884 reverse 5′-GACGGCACGGATTGTTATTCA-3′ (Barman et al., 2008). Cecum contents from GF mice served as negative controls.
Culturing and classification of SECS
Fecal samples from NOD male mice from our colony were homogenized using plastic pestles in 0.5% tergital 0.5%BSA PBS buffer followed by serial dilution and plating on LB and MacConkey agar, followed by incubation at 37 °C for 20 hours. A single colony from a MacConkey plate was subcultured and frozen stocks prepared in 10% Glycerol and stored at −80°C for long term storage. This clone was classified by colony PCR by amplification and sequencing of its 16S rDNA using universal bacterial primers 8F: 5′-AGAGTTTGATCCTGGCTCAG-3′, and 1392R: 5′-ACGGGCGGTGTGTAC-3′. The bacterium was classified using the Michigan State University Ribosomal Database Project classifier function (http://rdp.cme.msu.edu/)
Gene expression analysis
PLNs were isolated from 9–10 week old GF and SPF male and female NOD mice (3 mice per group). RNA was extracted using guanidinium-cesium chloride gradient ultracentrifugation (Chirgwin et al., 1979). RNA quality was assessed using agarose gel electrophoresis and the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto CA). Samples were analyzed using Illumina MouseRef 8 BeadChip (Illumina, San Diego, CA) array at the Functional Genomics Facility core at the University of Chicago.
Sets of genes with expression changes greater than 1 log-fold were identified with and without permutation testing. The combinatorial gene selection model identified a signature of 39 genes (gene set IV) regulated in a male-specific and germ-dependent manner, which were examined in the context of known and predicted gene-gene interactions using the STRING database (Szklarczyk et al., 2010) and identified a large connected sub-network with Il1b and Ifng serving as hubs based on number of interacting gene partners.
To determine cell-specific expression pattern by our gene signature, a gene abundance profile was generated using a panel of 96 different cell types and conditions available from the BioGPS murine RNA Gene Expression Atlas (http://biogps.gnf.org) (Lattin et al., 2008; Wu et al., 2009). Microarray data were deposited in GEO (Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/) database under accession number GSE49467.
Molecular Identification of Bacteria using 16S Sequencing
16S sequencing of samples from 4,10, and 13 week old mice
Cecal contents of male and female mice were collected using sterile instruments into cryovials and snap frozen in liquid nitrogen until processing. Samples were collected from three age groups of mice: prepubescent newly-weaned 4 week old mice, post-pubescent 10 week old mice and another group of 13 week old mice. DNA was extracted from cecal samples, and the V4 region of the 16S rRNA gene was amplified and sequenced on a 454 Genome Sequencer FLX Titanium platform (Roche Diagnostics and Beckman Coulter Genomics). Sequencing was performed at Argonne National Laboratory and at Research and Testing Laboratory (RTL), Lubbock, TX, as previously described (Dowd et al., 2008). On average 10,000 sequences per sample were acquired. Libraries were separated by exact matches to barcode tags and deposited to MG-RAST. The standard MG-RAST processing pipeline was used to (a) remove artificial replicate sequences produced by sequencing artifacts; (b) remove contaminant mouse sequences using DNA level matching to the mouse genome; (c) filter sequence reads based on length and on the number of ambiguous base calls with default settings. Sequences were deposited in Metagenomics Analysis Server (MG-RAST, http://metagenomics.anl.gov) database under the following accession numbers: mgp1501, mgp1470, mgp2995, mgp4244.
The alpha diversity was calculated using the antilog of the Shannon diversity index. The Shannon diversity index is an abundance-weighted average of the logarithm of the relative abundances of annotated species. The species-level annotations are from all the annotation source databases used by MG-RAST. Statistical analysis was carried out using the MATLAB (version 7.12.0.635) software package. The Mann–Whitney test was used to test if two alpha diversity distributions have equal medians, against the alternative that they do not have equal medians. Data is visualized by boxplots. In each box, the central red line is the median (50th percentile), the (upper and lower) edges of the box are the 75th and 25th percentiles, the error bars extend to the last datum within 1.5 of the interquartile range of the upper and lower quartiles, and outliers are plotted individually.
Family level PCA analysis and hierarchical clustering was performed in MATLAB. The difference between proportions of reads mapped to specific families in males and females was tested using a one-sided T-test. We controlled for multiple hypothesis testing using the q-value method by adjusting the p-value to reflect the false discovery rate (Benjamini Y. and Hochberg, 1995; Storey, 2002). Hierarchical clustering of 16S sequencing of cecal samples from 4-week and 10-week old male and female mice were performed using the Euclidean distance metric and clustering by average linkage. We standardized the data by calculating the median count of microbes at the family level for all samples in a group. The hierarchical clustering analysis allowed us to visualize similarity between pair-wise comparisons of the four groups. The results are displayed in a dendrogram, which shows the linkage points at increasing degree of dissimilarity.
Supplementary Material
Acknowledgments
We thank Dr. Claudio De Simone for his help with VSL3, Anuradha Nadimpalli and Sarah Owens for help with sequencing sample preparations, and Joseph Pickard for editing the manuscript. AC is supported by NIH grant AI082418, by JDRF grant 17-2011- 519. AC and DA are supported by NIH/NIDDK Digestive Disease Research Core Center grant DK42086. LY and MB were supported by Molecular and Cellular Biology training grant (T32 GM007183) and Clinical translational science award TL1 training grant (TL1TR000432), respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, Salzman N. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun. 2008;76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke A, Steele RE, Musto N, Caldwell BV. Fluctuations in plasma testosterone levels in adult male rats and mice. Endocrinology. 1973;92:1223–1228. doi: 10.1210/endo-92-4-1223. [DOI] [PubMed] [Google Scholar]
- Becker L, Liu NC, Averill MM, Yuan W, Pamir N, Peng Y, Irwin AD, Fu X, Bornfeldt KE, Heinecke JW. Unique proteomic signatures distinguish macrophages and dendritic cells. PLoS One. 2012;7:e33297. doi: 10.1371/journal.pone.0033297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. journal of the Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- Calcinaro F, Dionisi S, Marinaro M, Candeloro P, Bonato V, Marzotti S, Corneli RB, Ferretti E, Gulino A, Grasso F, et al. Oral probiotic administration induces interleukin-10 production and prevents spontaneous autoimmune diabetes in the non-obese diabetic mouse. Diabetologia. 2005;48:1565–1575. doi: 10.1007/s00125-005-1831-2. [DOI] [PubMed] [Google Scholar]
- Chatenoud L, Salomon B, Bluestone JA. Suppressor T cells--they’re back and critical for regulation of autoimmunity! Immunol Rev. 2001;182:149–163. doi: 10.1034/j.1600-065x.2001.1820112.x. [DOI] [PubMed] [Google Scholar]
- Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Dewhirst FE, Chien CC, Paster BJ, Ericson RL, Orcutt RP, Schauer DB, Fox JG. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl Environ Microbiol. 1999;65:3287–3292. doi: 10.1128/aem.65.8.3287-3292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Qi D, Yang L, Huang Y, Xiao X, Tai N, Wen L, Wong FS. TLR4 regulates cardiac lipid accumulation and diabetic heart disease in the nonobese diabetic mouse model of type 1 diabetes. American journal of physiology. Heart and circulatory physiology. 2012;303:H732–742. doi: 10.1152/ajpheart.00948.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd SE, Wolcott RD, Sun Y, McKeehan T, Smith E, Rhoads D. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP) PLoS One. 2008;3:e3326. doi: 10.1371/journal.pone.0003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish EN. The X-files in immunity: sex-based differences predispose immune responses. Nat Rev Immunol. 2008;8:737–744. doi: 10.1038/nri2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick F, Lepault F, Homo-Delarche F, Bach JF, Dardenne M. Influence of castration, alone or combined with thymectomy, on the development of diabetes in the nonobese diabetic mouse. Endocrinology. 1991;129:1382–1390. doi: 10.1210/endo-129-3-1382. [DOI] [PubMed] [Google Scholar]
- Fox HS. Androgen treatment prevents diabetes in nonobese diabetic mice. J Exp Med. 1992;175:1409–1412. doi: 10.1084/jem.175.5.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Gagnerault MC, Luan JJ, Lotton C, Lepault F. Pancreatic lymph nodes are required for priming of beta cell reactive T cells in NOD mice. J Exp Med. 2002;196:369–377. doi: 10.1084/jem.20011353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- Hoglund P, Mintern J, Waltzinger C, Heath W, Benoist C, Mathis D. Initiation of autoimmune diabetes by developmentally regulated presentation of islet cell antigens in the pancreatic lymph nodes. J Exp Med. 1999;189:331–339. doi: 10.1084/jem.189.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LK, Rims CR, Gill SE, McGuire JK, Manicone AM. Pulmonary macrophage subpopulations in the induction and resolution of acute lung injury. American journal of respiratory cell and molecular biology. 2012;47:417–426. doi: 10.1165/rcmb.2012-0090OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda N, Tsuchida T, Tamaki K. Testosterone suppresses anti-DNA antibody production in peripheral blood mononuclear cells from patients with systemic lupus erythematosus. Arthritis Rheum. 1997;40:1703–1711. doi: 10.1002/art.1780400921. [DOI] [PubMed] [Google Scholar]
- Kane M, Case LK, Kopaskie K, Kozlova A, MacDearmid C, Chervonsky AV, Golovkina TV. Successful transmission of a retrovirus depends on the commensal microbiota. Science. 2011;334:245–249. doi: 10.1126/science.1210718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloting N, Kloting I, Jack RS. CD14 triggers autoimmune Type 1 diabetes in the NOD mouse. Diabetologia. 2004;47:151–152. doi: 10.1007/s00125-003-1251-0. [DOI] [PubMed] [Google Scholar]
- Kovats S, Carreras E. Regulation of dendritic cell differentiation and function by estrogen receptor ligands. Cell Immunol. 2008;252:81–90. doi: 10.1016/j.cellimm.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci U S A. 2011;108:11548–11553. doi: 10.1073/pnas.1108924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattin JE, Schroder K, Su AI, Walker JR, Zhang J, Wiltshire T, Saijo K, Glass CK, Hume DA, Kellie S, Sweet MJ. Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome Res. 2008;4:5. doi: 10.1186/1745-7580-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Janeway CA., Jr Interferon gamma plays a critical role in induced cell death of effector T cell: a possible third mechanism of self-tolerance. J Exp Med. 1990;172:1735–1739. doi: 10.1084/jem.172.6.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loke P, Nair MG, Parkinson J, Guiliano D, Blaxter M, Allen JE. IL-4 dependent alternatively-activated macrophages have a distinctive in vivo gene expression phenotype. BMC Immunol. 2002;3:7. doi: 10.1186/1471-2172-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Kunimoto K, Muraoka Y, Katagiri K. Effect of castration on the appearance of diabetes in NOD mouse. Jikken Dobutsu. 1981;30:137–140. doi: 10.1538/expanim1978.30.2_137. [DOI] [PubMed] [Google Scholar]
- Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, von Bergen M, McCoy KD, Macpherson AJ, Danska JS. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- Mathis D, Benoist C. The influence of the microbiota on type-1 diabetes: on the threshold of a leap forward in our understanding. Immunol Rev. 2012;245:239–249. doi: 10.1111/j.1600-065X.2011.01084.x. [DOI] [PubMed] [Google Scholar]
- Pozzilli P, Signore A, Williams AJ, Beales PE. NOD mouse colonies around the world--recent facts and figures. Immunol Today. 1993;14:193–196. doi: 10.1016/0167-5699(93)90160-M. [DOI] [PubMed] [Google Scholar]
- Qin HY, Chaturvedi P, Singh B. In vivo apoptosis of diabetogenic T cells in NOD mice by IFN-gamma/TNF-alpha. International immunology. 2004;16:1723–1732. doi: 10.1093/intimm/dxh173. [DOI] [PubMed] [Google Scholar]
- Raes G, De Baetselier P, Noel W, Beschin A, Brombacher F, Hassanzadeh GhG. Differential expression of FIZZ1 and Ym1 in alternatively versus classically activated macrophages. J Leukoc Biol. 2002;71:597–602. [PubMed] [Google Scholar]
- Roubinian JR, Talal N, Greenspan JS, Goodman JR, Siiteri PK. Effect of castration and sex hormone treatment on survival, anti-nucleic acid antibodies, and glomerulonephritis in NZB/NZW F1 mice. J Exp Med. 1978;147:1568–1583. doi: 10.1084/jem.147.6.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott WH, Haskell BD, Tse HM, Milton MJ, Piganelli JD, Choisy-Rossi CM, Reifsnyder PC, Chervonsky AV, Leiter EH. Caspase-1 is not required for type 1 diabetes in the NOD mouse. Diabetes. 2004;53:99–104. doi: 10.2337/diabetes.53.1.99. [DOI] [PubMed] [Google Scholar]
- Serreze DV, Chapman HD, Post CM, Johnson EA, Suarez-Pinzon WL, Rabinovitch A. Th1 to Th2 cytokine shifts in nonobese diabetic mice: sometimes an outcome, rather than the cause, of diabetes resistance elicited by immunostimulation. J Immunol. 2001;166:1352–1359. doi: 10.4049/jimmunol.166.2.1352. [DOI] [PubMed] [Google Scholar]
- Serreze DV, Fleming SA, Chapman HD, Richard SD, Leiter EH, Tisch RM. B lymphocytes are critical antigen-presenting cells for the initiation of T cell-mediated autoimmune diabetes in nonobese diabetic mice. J Immunol. 1998;161:3912–3918. [PubMed] [Google Scholar]
- Serreze DV, Post CM, Chapman HD, Johnson EA, Lu B, Rothman PB. Interferon-gamma receptor signaling is dispensable in the development of autoimmune type 1 diabetes in NOD mice. Diabetes. 2000;49:2007–2011. doi: 10.2337/diabetes.49.12.2007. [DOI] [PubMed] [Google Scholar]
- Silveira PA, Johnson E, Chapman HD, Bui T, Tisch RM, Serreze DV. The preferential ability of B lymphocytes to act as diabetogenic APC in NOD mice depends on expression of self-antigen-specific immunoglobulin receptors. Eur J Immunol. 2002;32:3657–3666. doi: 10.1002/1521-4141(200212)32:12<3657::AID-IMMU3657>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Snyder DL, Wostmann BS. Comparison of blood chemistry and hormone levels from conventional and germfree rats. Prog Clin Biol Res. 1985;181:95–98. [PubMed] [Google Scholar]
- Sobel DO, Han J, Williams J, Yoon JW, Jun HS, Ahvazi B. Gamma interferon paradoxically inhibits the development of diabetes in the NOD mouse. Journal of autoimmunity. 2002;19:129–137. doi: 10.1006/jaut.2002.0604. [DOI] [PubMed] [Google Scholar]
- Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J Exp Med. 1992;176:287–292. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD. A direct approach to false discovery rates. Journal of the Royal Statistical Society. 2002;64:479–498. [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2010;39:D561–568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turley SJ, Lee JW, Dutton-Swain N, Mathis D, Benoist C. Endocrine self and gut non-self intersect in the pancreatic lymph nodes. Proc Natl Acad Sci U S A. 2005;102:17729–17733. doi: 10.1073/pnas.0509006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varanasi V, Avanesyan L, Schumann DM, Chervonsky AV. Cytotoxic mechanisms employed by mouse T cells to destroy pancreatic beta-cells. Diabetes. 2012;61:2862–2870. doi: 10.2337/db11-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitacre CC. Sex differences in autoimmune disease. Nat Immunol. 2001;2:777–780. doi: 10.1038/ni0901-777. [DOI] [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, 3rd, Su AI. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.