Abstract
Breast cancer (BC) is the most common malignancy of women in the developed world. To better understand its pathogenesis, knowledge of normal breast development is crucial, as BC is the result of disregulation of physiologic processes. The aim of this study was to investigate the impact of reproductive life stages on the transcriptional profile of the mammary gland in a primate model. Comparative transcriptomic analyses were carried out using breast tissues from 28 female cynomolgus macaques (Macaca fascicularis) at the following life stages: prepubertal (n = 5), adolescent (n = 4), adult luteal (n = 5), pregnant (n = 6), lactating (n = 3), and postmenopausal (n = 5). Mammary gland RNA was hybridized to Affymetrix GeneChip® Rhesus Macaque Genome Arrays. Differential gene expression was analyzed using ANOVA and cluster analysis. Hierarchical cluster analysis revealed distinct separation of life stage groups. More than 2,225 differentially expressed mRNAs were identified. Gene families or pathways that changed across life stages included those related to estrogen and androgen (ESR1, PGR, TFF1, GREB1, AR, 17HSDB2, 17HSDB7, STS, HSD11B1, AKR1C4), prolactin (PRLR, ELF5, STAT5, CSN1S1), insulin-like growth factor signaling (IGF1, IGFBP1, IGFBP5), extracellular matrix (POSTN, TGFB1, COL5A2, COL12A1, FOXC1, LAMC1, PDG-FRA, TGFB2), and differentiation (CD24, CD29, CD44, CD61, ALDH1, BRCA1, FOXA1, POSTN, DICER1, LIG4, KLF4, NOTCH2, RIF1, BMPR1A, TGFB2). Pregnancy and lactation displayed distinct patterns of gene expression. ESR1 and IGF1 were significantly higher in the adolescent compared to the adult animals, whereas differentiation pathways were overrepresented in adult animals and pregnancy-associated life stages. Few individual genes were distinctly different in postmenopausal animals. Our data demonstrate characteristic patterns of gene expression during breast development. Several of the pathways activated during pubertal development have been implicated in cancer development and metastasis, supporting the idea that other developmental markers may have application as biomarkers for BC.
Keywords: Breast development, Gene microarray, Breast cancer, Stem cell, Estrogen, Progesterone
Introduction
Major determinants of breast cancer (BC) risk include age, genetics, and reproductive history. Timing of reproductive milestones such as age at menarche and menopause, pregnancy, and lactation has an impact on breast development and thereby may alter the risk of developing BC later in life. Therefore, the study of breast development and differentiation across the lifespan may shed light on the origins of BC.
Mammary gland development may be separated into embryonic, adolescent, and adult phases, each of which is differentially regulated [1–4]. Embryonic branching of the breast is thought to occur without any hormonal requirement as it is not impaired in the absence of ER-alpha (ERα), ER-beta (ERβ), progesterone receptor (PGR), or the receptors for growth hormone (GH) or prolactin (PRLR). During adolescent branching, GH, insulin-like growth factor (IGF)-1, and ovarian estrogens as well as ERα are crucial; adult tertiary side-branching requires progesterone and its receptor [5].
All epithelial cells in the mammary gland are thought to originate from a common stem cell [6]. During differentiation from ERα negative stem cells to ERα positive progenitor cells and consecutively to basal- or luminal-restricted progenitors, these cells express different cell surface markers. Although unique stem cell markers have proven difficult to define, patterns of protein expression can distinguish mammary epithelial cells with stem-like or progenitor-like characteristics [7, 8].
As stem cells are undifferentiated and relatively resistant to apoptosis they may accrue DNA damage and mutations, making them susceptible to cancer initiation [9, 10]. The stem cell model of carcinogenesis proposes that BC originates in tissue stem or progenitor cells probably through deregulation of self-renewal pathways, resulting in BC cells with the defining stem cell properties of self-renewal and differentiation. Self-renewal drives tumorigenesis, whereas differentiation contributes to tumor phenotypic heterogeneity [11].
To investigate the impact of reproductive stages and their respective characteristic hormonal profiles on mammary gene expression in the human breast, one would have to take breast biopsies from healthy girls and women without any exogenous hormonal exposure such as hormonal contraception, fertility drugs, or postmenopausal hormone therapy. This is practically and ethically impossible. A unique model for studying these questions is the monkey model. Female cynomolgus macaques (Macaca fascicularis) have distinctive reproductive similarities to women, including a 28-day menstrual cycle, comparable ovarian hormone patterns, and natural ovarian senescence. Macaques have >95% overall genetic sequence identity to humans, including key genes involved in BC susceptibility [12]. Human and macaque mammary glands are similar in terms of microanatomy and development, sex steroid receptor expression, responses to exogenous hormones, and the development of a heterogeneous spectrum of hyperplastic and neoplastic lesions with aging [13–15].
The aim of this study was to identify the impact of reproductive stages and their respective characteristic hormonal profiles on normal breast development by comparing gene expression patterns of the mammary gland from young animals (prepubertal and adolescent), adults (premenopausal in luteal cycle phase and postmenopausal), and animals in pregnancy-associated life stages (pregnancy and lactation) in cynomolgus monkeys. By gaining a better understanding of breast development, we also aimed to identify new biomarkers of BC that might proceed to new approaches in targeted therapies.
Methods
Animal characteristics and life stage selection
Healthy control female cynomolgus macaques (Macaca fascicularis) were imported from the Institut Pertanian Bogor (Bogor, Indonesia). Diets fed were nutritionally complete for non-human primates, free of soy isoflavones, and providing 42% of calories from fat in order to model the North American diet. Animals were housed in social groups of at least 4 animals. All procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of the Wake Forest University School of Medicine, which is accredited by the Association for the Advancement and Accreditation of Laboratory Animal Care. The animals (total n = 28) were selected from four life stage groups: prepubertal (n = 5), adolescent (n = 4), adult luteal (n = 5), pregnant (n = 6), lactating (n = 3), and postmenopausal (n = 5). Ages were determined by dental eruption or known birth dates. Serum estradiol and progesterone levels were determined in the Yerkes Assay Services laboratory, Yerkes National Primate Research Center. Prepubertal and adolescent animals were of similar estimated age; the prepubertal stage was defined retrospectively, as referring to a sample taken more than 6 months prior to menarche. Adolescent animals were defined by menarche having occurred recently (within the preceding 6 months). Monkeys at adult luteal stage were in the luteal phase of the menstrual cycle, which was defined by endometrial and ovarian histology at necropsy. Pregnant monkeys were in the third trimester of gestation. Post-menopausal monkeys had undergone oophorectomy, which was performed at least 2.5 years prior to necropsy. Stages of breast development were confirmed by histopathology, using characteristics we have described previously [14, 16].
Tissue collection
Mammary gland, including associated fat and connective tissues, was collected either by breast biopsy or at necropsy. All prepubertal and adolescent samples and one sample from a lactating animal were collected by biopsy. Necropsy samples were only taken from animals from which they could be collected quickly (within 5 min of preplanned euthanasia). For biopsies, animals were anesthetized with ketamine (10–15 mg/kg) and buprenorphine (0.01 mg/kg), a 1.5-cm incision was made in the upper outer breast quadrant, and a small (0.4 gram) sample of mammary gland was removed. Biopsies were performed by an experienced veterinary surgeon. The incision was sutured, and the animals were monitored and given analgesia during recovery following IACUC-approved clinical procedures. Half of the biopsy sample was frozen; the other half was fixed at 4°C in 4% paraformaldehyde for 24 h, transferred to 70% ethanol, and then processed for histology using standard procedures. For necropsy samples, euthanasia was performed as part of unrelated IACUC-approved experimental protocols. Mammary tissues, ovaries, and uteri were removed, and fixed and frozen samples were collected as above. Each breast sample used for gene microarray analysis was evaluated by histology and mammary whole mount analysis to assure the presence of epithelial tissue with typical morphology for each developmental stage [17].
Gene microarray assays
Total RNA was extracted from frozen mammary samples using Tri Reagent (Molecular Research Center, Cincinnati, OH), purified using an RNeasy Mini kit (Qiagen, Valencia, CA, USA), and quantified using a Nanodrop ND-100 UV–VIS spectrophotometer (NanoDrop, Wilmington, DE). RNA intactness and quality were confirmed using an Agilent 2100 Bioanalyzer (Wilmington, DE). Only samples with an RNA integrity number (RIN) greater than 8.0 were used for hybridization. One microgram of total RNA from each sample was labeled using the GeneChip One-Cycle Target Labeling kit (Affymetrix, Inc., Santa Clara, CA) following the manufacturer’s protocol. RNA was then fragmented and hybridized to GeneChip Rhesus Macaque Genome Arrays (Affymetrix, Inc., Santa Clara, CA) for 17 h, prior to washing and scanning. Data were extracted from scanned images using GeneChip Operating Software (Affymetrix, Inc., Santa Clara, CA). RNA quality control and microarray assays were performed at Cogenics®, a Division of Clinical Data (Morrisville, NC; http://www.cogenics.com). One mammary gland sample from the prepubertal life stage group did not yield sufficient RNA for hybridization and was excluded, bringing the total number of arrays to 27 and the subset number of prepubertal animals to four.
Quantitative RT-PCR
RNA was reverse-transcribed using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Transcript levels for targets related to estrogen, androgen, and prolactin metabolism were measured using quantitative real-time polymerase chain reaction (qRT-PCR). Cynomolgus macaque, rhesus macaque, or human ABI Taqman primer probe assays were used as targets, and samples were normalized to endogenous GAPDH and ACTB using cynomolgus macaque-specific primer probes. Stock macaque mammary tissues and macaque BC tumors were run in triplicate on each plate as external calibrators. Reactions were performed with ABI PRISM® 7500 Fast Sequence Detection System (Applied Biosystems), and relative expression was determined using the ΔΔCt method calculated by ABI Relative Quantification 7500 Software v2.0.1.
Statistics
Microarray data analyses were performed using the GeneSifter software program (VizX Labs LLC, Seattle, WA, USA; http://www.genesifter.net). Intensity data were converted to a log2 scale, screened for heterogeneity among samples, and evaluated using supervised analysis of variance (ANOVA) and pairwise comparisons between treatments. Principal components analysis (PCA), pattern navigation, and cluster analysis were performed on data subsets filtered by expression threshold. All P values were corrected for multiple comparisons using the Benjamini and Hochberg method (Padj) [18], which derives a false discovery rate estimate from the raw P values [19]. Differences in gene numbers altered by each treatment were compared using a Fisher’s exact test.
Quantitative RT-PCR data were analyzed using JMP (version 8.0.2, SAS Institute, Cary, NC). All relative expression data were evaluated for normality and homogeneity of variances among groups. Data were log-transformed and evaluated using ANOVA or the Kruskal–Wallis non-parametric test, then retransformed to original scale, and reported as fold change in prepubertal (control) group with 95% confidence interval. Multiple comparisons were made using Student’s T-test for each pair, with two-tailed significance level of 0.05.
Results
Animal characteristics and tissue histology
Characteristics of animals in the study are presented in Table 1. Breast tissue of prepubertal animals consisted primarily of immature ducts and terminal end buds. Adult-type lobular units were present in all other life stages; pregnant animals were distinguished by marked lobular proliferation, and lactating animals by pronounced secretory activity. Representative histologic images of the breast tissue at each life stage are shown in Fig. 1, including immunohistochemistry for the proliferation marker Ki67.
Table 1.
Characteristics of animals in the study
| Group | N | Age (year) | Body weight (kg) | Estradiol (pg/ml) | Progesterone (ng/ml) |
|---|---|---|---|---|---|
| Prepubertal | 4 | 2.8a | 1.9 ± 0.1 | 12.3 ± 8.5 | 1.1 ± 0.7 |
| Adolescent | 4 | 2.8b | 2.2 ± 0.3 | 4.6 ± 2.4 | 0.9 ± 0.7 |
| Adult (luteal phase) | 5 | 13.2 ± 5.8 | 3.5 ± 0.5 | 23.2 ± 15.6 | 2.2 ± 0.7 |
| Pregnant | 6 | 10.4 ± 2.8 | 3.3 ± 0.7 | 373.1 ± 151.3* | 5.2 ± 0.6* |
| Lactating | 3 | 8.1 ± 1.8 | 3.8 ± 2.0 | 8.6 ± 0.6 | 1.5 ± 1.0 |
| Postmenopausal | 5 | 18.4 ± 3.7 | 3.5 ± 0.9 | 3.1 ± 0.0 | 0.7 ± 0.7 |
Data are means ± standard deviation. Ages are estimated based on dentition
Different from all other groups at P < 0.05 (Tukey’s HSD test)
Sample collected greater than 6 months before menarche
Sample collected within 6 months after menarche
Fig. 1.
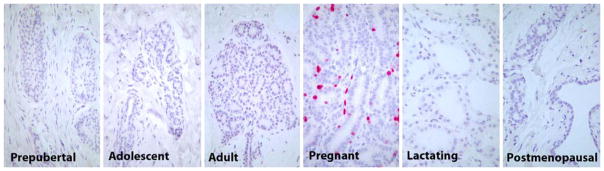
Photomicrographs of breast histology for each life stage studied. Objective magnification was the same (×40) for all images. Immunohistochemical stain for the proliferation marker Ki67; hematoxylin counterstain
Global gene expression profiles
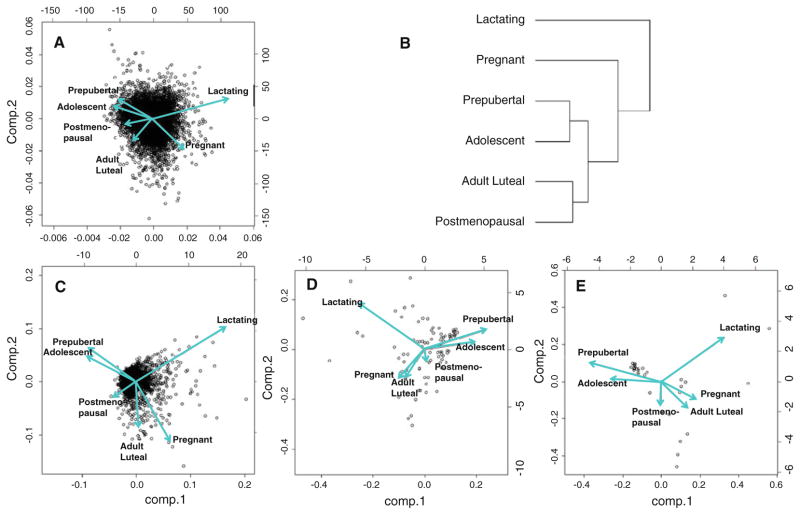
Principal components analysis and hierarchical clustering were initially used to evaluate mammary gland gene expression patterns across life stages (Fig. 2). Overall, 26,481 probe sets were detected at a quality >2 and threshold fold change (FC) >1.5. Among these probes, PCA showed divergent vectors for juvenile animals (prepubertal and adolescent), pregnancy-associated life stages (pregnant and lactating), and adult animals (adult luteal and postmenopausal). Corresponding clustering dendrograms showed clear separation of all groups, with closer associations between the following groups: prepubertal and adolescent; pregnant and lactating; and adult luteal and postmenopausal.
Fig. 2.
Principal components analysis (PCA) of gene expression profiles in the breast across life stages and corresponding hierarchical clustering dendrogram (b). Vector diagrams correspond to the following gene filters: all genes detected (quality > 2, n = 26,484) (a); genes significantly altered (ANOVA Padj < 0.05, quality > 2) with FC >1.2 (n = 1,964) (c), 2.0 (n = 89) (d), and 3.0 (n = 31) (e) in all groups combined versus prepubertal
Overall, 1,964 significantly differentially expressed probe sets were identified (ANOVA Padj < 0.05) for which FC > 1.2. The overall pattern of gene expression by PCA was similar regardless of the threshold for FC > 1.2, 2.0 and 3.0 at Padj < 0.05 for young animals. However, among the adult groups at higher FC, pregnant animals diverged from lactating animals and were more closely associated with adult luteal animals.
Most genes contributing to the difference between juvenile and adult animals were expressed in common by adult luteal, pregnant, and postmenopausal groups. However, most genes expressed by lactating animals were not shared by other groups. Between the two juvenile groups, no genes differed at a threshold of FC > 10, and there were only 137 differentially expressed genes at a threshold >1.8 (Padj < 0.05). Adult groups showed similar overall directional patterns in gene expression changes, but each differed in the number of transcripts altered from prepubertal subjects and the magnitude of gene expression changes. For example, adult luteal and postmenopausal profiles resulted in 60 (37 up-regulated and 23 down-regulated) and 55 (36 up-regulated and 19 down-regulated) altered transcripts relative to the prepubertal profile with FC > 10, respectively. Even more differences in the number of altered transcripts were apparent when comparing pregnancy-associated life stages versus prepubertal profiles with FC > 10 revealing 136 differences in pregnant animals (59 up-regulated and 77 down-regulated) and 437 differences in lactating animals (109 up-regulated and 328 down-regulated). Adult differences from the prepubertal group are shown graphically in Fig. 3.
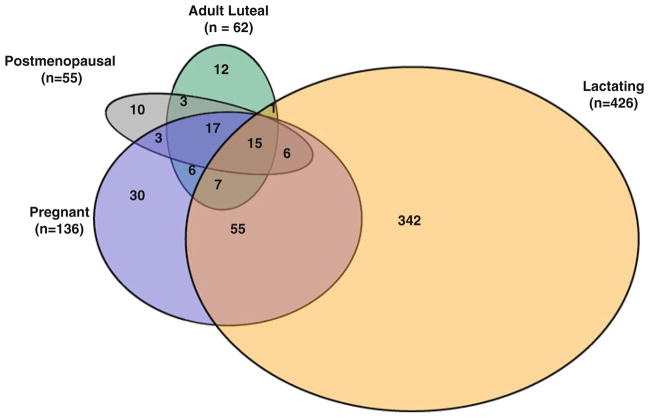
Fig. 3.
Venn diagram presenting overlapping genes within life stage groups being significantly different from prepubertal animals at threshold fold change >10 and Padj < 0.05. There are no genes in the adolescent group falling into this category
Ten genes were regulated in common in all adult life stage groups and differed from prepubertal animals at threshold >10 and Padj < 0.05 (Table 2). Among these differentially expressed genes, those associated with estrogen-regulated processes, proliferation, adhesion, and survival (TFF1, GREB1, NEK10, LRRN3, PPM1K, PGR, IGFBP1) decreased with advancing age, reaching their lowest levels during adult luteal phase (IGFBP1), pregnancy (TFF1, NEK10, PPM1K, LRRN3), lactation (PGR), and postmenopause (GREB1), respectively. Pairwise comparisons of gene expression between all groups revealed significant differences between prepubertal animals and all adult groups except postmenopausal. Among the four adult premenopausal life stage groups, significant differences were found for mammary LRRN3, GREB1, PPM1K, NEK10, and PGR gene expression (Padj ≤ 0.05). LRRN3 expression was significantly lower in pregnant than in postmenopausal breast tissue. PGR expression was significantly lower in lactating than in pregnant and adult luteal breast tissue. GREB1 expression was lower in lactating animals than in pregnant animals. Conversely, PPM1K and NEK10 gene expression were significantly lower in pregnant than in lactating breast tissue.
Table 2.
Gene expression microarray data
| Prepubertal (a) | Adolescent (b) | Adult luteal (c) | Pregnant (d) | Lactating (e) | Postmenopausal (f) | Pairwise comparisons Padj < 0.05 | |
|---|---|---|---|---|---|---|---|
| CSN1S1 | 1.00 | 2.40 | 35.54 | 43.76 | 101.24 | 22.66 | a vs. c-f b vs. c-e e vs. c, d |
| ELF5 | 1.00 | 1.22 | 10.59 | 9.16 | 12.87 | 2.35 | a, b, f vs. c-e |
| GREB1 | 1.00 | −1.42 | −13.58 | −12.76 | −27.89 | −29.17 | a vs. c-f b vs. c-e d vs. e |
| IGFBP1 | 1.00 | 1.34 | −89.23 | −86.31 | −83.32 | −19.67 | a vs. c-f b vs. c-e |
| IGHG1 | 1.00 | 1.11 | 42.67 | 36.76 | 15.45 | 34.62 | a vs. c-f b vs. c-e c vs. e |
| LRRN3 | 1.00 | −1.57 | −18.72 | −23.83 | −16.31 | −16.24 | a vs. c-f b vs. c-e d vs. f |
| NEK10 | 1.00 | −1.43 | −14.73 | −30.99 | −14.18 | −10.09 | a vs. c-f b vs. c-e d vs. e |
| PGR | 1.00 | −1.02 | −8.95 | −23.77 | −112.26 | −15.81 | a vs. c-f b vs. c-e e vs. c, d, f |
| PPM1K | 1.00 | −1.74 | −12.21 | −12.53 | −7.08 | −12.44 | a vs. c-f b vs. c-e d vs. e |
| TFF1 | 1.00 | 1.16 | −84.59 | −168.88 | −97.09 | −57.84 | a vs. c-f b vs. c-e |
Fold change in relative expression for adult life stage groups significantly different from prepubertal animals at threshold >10 and Padj < 0.05. Prepubertal and adolescent groups do not differ from each other
In contrast, the expression of three genes associated with glandular differentiation (CSN1S1, IGHG1, and ELF5) was significantly increased with advancing age when compared with prepubertal life stage, reaching their maximum during the luteal phase of the menstrual cycle (IGHG1) or lactation (CSN1S1, ELF5), respectively (Padj ≤ 0.05). Pairwise comparisons of differentiation gene expression revealed a significant difference between prepubertal and adult pre-menopausal or pregnancy-associated life stages (Padj ≤ 0.05), but again, not for postmenopausal subjects. The differentiation markers CSN1S1 and ELF5 peaked during lactation, differing from all other groups including pregnant mammary gland (Padj ≤ 0.05). Postmenopausal mammary ELF5 expression was significantly decreased when compared with the adult luteal and pregnancy-associated life stages (Padj ≤ 0.05).
A secondary goal of this study was to identify signaling pathways that are significantly altered across life stages (threshold > 1.5; ANOVA Padj < 0.00001). Overall, 4,962 differently regulated genes covering 10 KEGG pathways and 23 relevant gene ontology cohorts were identified. We focused on pathways related to androgen and estrogen metabolism, mammary stem cells, and profiles relevant for BC development.
Sex steroid receptors and steroid metabolism
Gene expression differences by array for the major receptors of the estrogen, progesterone, prolactin, and androgen (ESR1, PGRB, PRLR, AR), and key enzymes in steroid metabolism are shown in Table 3. ESR1 and PGRB decreased in the transition from prepuberty to adolescence, and AR gene expression was maximal during adolescence after which it significantly decreased (Padj ≤ 0.05). ESR1 expression had its minimum during postmenopause, whereas AR and most strikingly PGRB expression were lowest during lactation. Lower gene expression for ESR1, PGRB, and AR during lactation was significant relative to other life stages (Padj ≤ 0.05). PGRB was less than <1/100th of prepubertal levels during lactation. Expression of PRLR significantly increased with maturity in comparison with the prepubertal life stage, reaching its maximum during pregnancy where it was significantly higher than in lactating and postmenopausal breast (Padj ≤ 0.05).
Table 3.
Gene expression array data for sex steroid receptors and metabolism
| Prepubertal (a) | Adolescent (b) | Adult luteal (c) | Pregnant (d) | Lactating (e) | Postmenopausal (f) | Pairwise comparisons Padj < 0.05 | |
|---|---|---|---|---|---|---|---|
| ESR1 | 1.00 | −1.34 | −5.01 | −4.79 | −3.36 | −5.10 | a vs. b-f b vs. c-f c vs. e |
| PRLR | 1.00 | 1.26 | 3.20 | 4.51 | 1.88 | 1.61 | a vs. b-f b vs. d c vs. e d vs. e, f |
| PGRB | 1.00 | −1.02 | −8.95 | −23.77 | −112.26 | −15.81 | a, b vs. c-f e vs. c, d, f |
| AR | 1.00 | 1.33 | −2.10 | −2.72 | −7.54 | −1.18 | a vs. b-f b vs. c-e c vs. e d vs. e, f e vs. f |
| MKI67 | 1.00 | 1.06 | 1.08 | 1.03 | 1.18 | −1.17 | a vs. b-f c vs. d, e d vs. e, f e vs. f |
| STS | 1.00 | −1.17 | −1.23 | −1.31 | −1.43 | 1.32 | a vs. b-f b, c, f vs. d, e |
| HSD11B1 | 1.00 | 1.53 | −1.21 | −2.04 | −1.35 | −1.05 | a vs. b-f d vs. b, c, f |
| HSD17B7 | 1.00 | −1.12 | −1.21 | −1.84 | −1.53 | −1.10 | a vs. b-f b, f vs. d, e c vs. d |
| HSD17B2 | 1.00 | 1.03 | 4.66 | 4.92 | 3.11 | 3.31 | a vs. b-f b vs. c-e |
| AKR1C4 | 1.00 | −1.56 | −1.34 | −2.93 | −3.14 | −1.52 | a vs. b-f b vs. d c, f vs. d, e |
Up- and down-regulated overlapping genes within life stage groups significantly different from the prepubertal group at P <0.05. Pairwise comparisons at Padj < 0.05 were made, and the relevant results are presented in the text
Five genes involved in C21, C19, and C18 steroid formation were differently expressed relative to prepubertal gene profiles (AKR1C4, STS, HSD11B1, HSD17B2, and HSD17B7; Table 3). AKR1C4 and STS decreased during the pubertal transition and were lower in all adult groups. Expression of three genes (STS, HSD11B1, and HSD17B7) coding for enzymes associated with the formation of more potent steroid hormones was significantly decreased with maturity when compared with prepubertal animals, reaching their minimum during pregnancy (HSD11B1, HSD17B7) and lactation (STS), respectively (Padj ≤ 0.05). However, for these genes, postmenopausal mammary gene expression significantly increased to levels comparable to adolescent and adult luteal life stages for STS, HSD11B1, and HSD17B7 (Padj ≤ 0.05). Expression of two genes (HSD17B2 and AKR1C4) coding for enzymes associated with steroid hormone inactivation to less-potent metabolites was either significantly increased (HSD17B2) or decreased (AKR1C4) with advancing age when compared with prepubertal life stage, respectively (Padj ≤ 0.05; Table 3).
Markers of stem cells and differentiation
Life stage effects on genes related to mammary stem cells and differentiation were evaluated using gene ontology analysis (stem cell division n = 9; stem cell differentiation n = 33). Genes differentially regulated at FC > 1.2 versus prepubertal (Padj = 0.00001) were initially used for this analysis (n = 4,955). Herein, 8 genes were found to be statistically significant (P = 0.001) for stem cell regulation (DICER1, IGF1, LIG4, KLF4, NOTCH2, RIF1, BMPR1A, and TGFB2). In addition, we analyzed our data set for gene expression patterns of stem cell markers presented by LaMarca and Rosen (ELF5, CD29, CD24, CD61, CD44, ALDH1, BRCA1, GATA3, FOXA1, STAT5, and POSTN) [6], and we also included the proliferation marker MKI67 in this category (Table 4).
Table 4.
Mammary progenitor cell markers
| Prepubertal (a) | Adolescent (b) | Adult luteal (c) | Pregnant (d) | Lactating (e) | Postmenopausal (f) | Pairwise comparisons Padj < 0.05 | |
|---|---|---|---|---|---|---|---|
| ELF5 | 1.00 | 1.22 | 10.59 | 9.16 | 12.87 | 2.35 | a, b, f vs. c-e |
| CD24 | 1.00 | 1.09 | 1.66 | 1.21 | 3.97 | 1.59 | a vs. b-f e vs. b-d, f |
| STAT5 | 1.00 | 1.17 | 2.81 | 5.96 | 1.68 | 1.14 | a vs. b-f b vs. d c, f vs. d, e d vs. e |
| ALDH1 | 1.00 | 1.10 | 1.38 | −1.36 | 1.47 | 1.19 | a vs. b-f b vs. d d vs. e, f |
| DICER | 1.00 | −1.23 | −1.25 | −2.25 | −4.02 | −1.20 | a vs. b-f b, c vs. d, e d vs. e, f e vs. f |
| RIF1 | 1.00 | −1.37 | −1.29 | −2.44 | −4.49 | −1.44 | a vs. b-f b, c vs. d, e e vs. f |
| BMPR1A | 1.00 | −1.12 | −1.14 | −1.56 | −1.56 | −1.16 | a vs. b-f b vs. d c, f vs. d, e |
| TGFB2 | 1.00 | −1.40 | −1.17 | −3.99 | −6.23 | −1.22 | a vs. b-f b, c, f vs. d, e |
| CD44 | 1.00 | −1.09 | −1.30 | −2.99 | −5.09 | −1.15 | a vs. b-f b vs. d, e c vs. e |
| CD29 | 1.00 | 1.00 | −1.24 | −1.85 | −4.86 | −3.86 | a vs. b-f b, c, f vs. d, e |
| FOXA1 | 1.00 | −1.04 | −1.76 | −2.28 | −2.42 | −1.44 | a vs. b-f b, c, f vs. d, e |
| KLF4 | 1.00 | 1.05 | 1.61 | −2.76 | −4.19 | 1.51 | a vs. b-f b vs. c-e c, f vs. d, e |
| NOTCH2 | 1.00 | 1.17 | 1.19 | −1.58 | −2.37 | 1.11 | a vs. b-f b, c vs. d, e d vs. e, f e vs. f |
| LIG4 | 1.00 | −1.12 | 1.37 | −2.03 | −4.09 | 1.12 | a vs. b-f b vs. c-e c vs. d, e d vs. e, f e vs. f |
| BRCA1 | 1.00 | 1.11 | 1.06 | −1.18 | −1.68 | −1.22 | a vs. b-f e vs. b-d, f |
| POSTN | 1.00 | 1.16 | −10.06 | −28.27 | −92.92 | −9.17 | a vs. b-f b vs. d-f c, d vs. e e vs. f |
| IGF1 | 1.00 | 1.28 | −2.46 | −2.22 | −8.22 | −2.28 | a vs. b-f b vs. d-f e vs. c, d e vs. f |
| CD61 | 1.00 | −1.07 | 1.20 | −1.20 | 1.27 | −1.20 | a vs. b-f e vs. b-d, f |
Up- and down-regulated overlapping genes within life stage groups being significantly different from prepubertal animals at P < 0.05. Pairwise comparisons at Padj < 0.05 were made, and the relevant results are presented in the text
A distinct pattern of decreased gene expression for putative stem cell markers and regulators of differentiation was seen during pregnancy or lactation when compared to juvenile and adult animals; diminished expression was seen for DICER1, RIF1, BMPR1A, TGFB2, LIG4, KLF4, NOTCH2, CD44, FOXA1, and CD29 (Padj ≤ 0.05). In contrast, mammary gene expression of the differentiation and stem cell markers ELF5, CD24, STAT5, and ALDH1 increased with advancing age peaking during pregnancy or lactation (Padj ≤ 0.05). Other potential stem cell markers presented a heterogeneous gene expression profile across life stages (KLF4, POSTN, NOTCH2, IGF1, BRCA1, LIG4, and CD61) with an increase during puberty followed by a decrease during pregnancy-associated life stages (KLF4, POSTN, NOTCH2, IGF1, BRCA1, and LIG4) and postmenopause (POSTN, IGF1, and BRCA1; Padj ≤ 0.05). MKI67 gene expression remained relatively stable across life stages.
The luminal progenitor cell population is characterized by the cell surface marker CD29lo CD24+ CD61+ and the differentiated luminal cell population by CD29lo CD24+ CD61−. Pairwise comparisons revealed a significant increase in CD24 gene expression in lactating mammary gland in comparison with other life stages, whereas CD29 gene expression was significantly lower in pregnant and lactating mammary gland compared to young animals (Padj ≤ 0.05). CD61 gene expression was significantly higher in adult luteal and lactating and lower in pregnant and postmenopausal mammary gland in comparison with prepubertal subjects (Padj ≤ 0.05). Expression of four genes associated with cell differentiation, repair mechanisms, and apoptosis (DICER1, RIF1, BMPR1A, and TGFB2) was significantly decreased with advancing age when compared with prepubertal animals (Padj ≤ 0.05).
Expression of six genes associated with cell differentiation was significantly increased during adult life stages when compared with juvenile animals (LIG4, KLF4, NOTCH2, ELF5, STAT5, ALDH1; Padj ≤ 0.05). Within adult premenopausal life stages, no significant differences in expression of DICER1, IGF1, LIG4, KLF4, NOTCH2, RIF1, BMPR1A, TGFB2, STAT5, or ALDH1 were observed. Surprisingly, postmenopausal mammary gene expression did not differ significantly from the adolescent expression pattern, with the exception of IGF1 gene expression that was significantly higher during adolescence than in any other life stage examined (Padj ≤ 0.05). Within pregnancy-associated life stages, DICER1, IGF1, LIG4, NOTCH2, STAT5, CD29, BRCA1, and POSTN gene expression was significantly lower in breast tissue of lactating animals relative to pregnant animals (Padj ≤ 0.05). The opposite was true for CD24 and CD61 (Padj ≤ 0.05). Genes DICER1, IGF1, LIG4, NOTCH2, KLF4, RIF1, BMPR1A, TGFB2, CD29, CD44, FOXA1, BRCA1, and POSTN exhibited lower expression in lactating compared to postmenopausal breast tissue (Padj ≤ 0.05). The opposite was true for ELF5, CD24, STAT5, ALDH1, and CD61 gene expression (Padj ≤ 0.05).
Extracellular matrix signaling
Life stage effects on genes related to extracellular matrix signaling were evaluated using gene ontology analysis (extracellular matrix organization n = 179). Genes differentially regulated at FC > 1.2 versus prepubertal (Padj = 0.00001) were initially used for this analysis (n = 42). We selected eight significantly differently regulated genes (Table 5).
Table 5.
Extracellular matrix signaling
| Prepubertal (a) | Adolescent (b) | Adult luteal (c) | Pregnant (d) | Lactating (e) | Postmenopausal (f) | Pairwise comparisons Padj < 0.05 | |
|---|---|---|---|---|---|---|---|
| POSTN | 1.00 | 1.16 | −10.06 | −28.27 | −92.92 | −9.17 | a vs. b-f b vs. d-f c, d vs. e e vs. f |
| TGFB1 | 1.00 | 1.06 | −2.46 | −4.90 | −14.89 | −1.30 | a vs. b-e b vs. d, e e vs. c, d, f |
| COL5A2 | 1.00 | 1.15 | −3.78 | −4.27 | −20.15 | −2.68 | a vs. b-f b vs. d, e e vs. c, d, f |
| COL12A1 | 1.00 | −1.05 | −2.49 | −5.53 | −10.87 | −1.77 | a vs. b-f b vs. d, e e vs. c, d, f |
| FOXC1 | 1.00 | 2.07 | 9.15 | 4.49 | −1.74 | 2.89 | a vs. b-f b, c vs. d, e e vs. d, f |
| LAMC1 | 1.00 | −1.08 | −1.61 | −3.77 | −8.55 | −1.26 | a vs. b-f d vs. b, c e vs. b-d |
| PDGFRA | 1.00 | −1.12 | −1.76 | −2.79 | −2.40 | −1.49 | a vs. b-f b vs. d, e c vs. d |
| TGFB2 | 1.00 | −1.49 | −1.36 | −3.73 | −6.23 | −1.76 | a vs. b-f b, c, f vs. d, e |
Up- and down-regulated overlapping genes within life stage groups being significantly different from prepubertal animals at P < 0.05. Pairwise Comparisons at Padj < 0.05 were made, and the relevant results are presented in the text
Overall, gene expression (POSTN, TGFB1, COL5A2, COL12A1, LAMC1, PDGFRA, TGFB2) was decreased during all life stages in comparison with prepubertal animals reaching a nadir during lactation (TGFB1, PDGFRA, LAMC1, COL12A1, COL5A2; Padj ≤ 0.05). POSTN in particular was expressed most abundantly in juveniles, with a tenfold lower expression in cycling or postmenopausal adults and a 30- to 90-fold lower expression in pregnant and lactating animals, respectively. However, there was a significant initial increase in TGFB1, POSTN, and COL5A2 gene expression during adolescence (Padj ≤ 0.05). In contrast, FOXC1 was significantly up-regulated in adulthood in comparison with prepuberty (Padj ≤ 0.05), with lower expression in lactation.
Breast cancer–associated genes
Next, we selected genes that have been associated with BC development, described in the 70-gene Amsterdam signature that forms the basis for the MammaPrint® assay [20]. Out of 50 genes represented in the MammaPrint® assay, 21 probes were significantly differently regulated at Padj ≤ 0.05 in our data set, representing six genes (Table 6).
Table 6.
Genes related to BC
| Prepubertal (a) | Adolescent (b) | Adult luteal (c) | Pregnant (d) | Lactating (e) | Postmenopausal (f) | Pairwise comparisons Padj < 0.05 | |
|---|---|---|---|---|---|---|---|
| SERF1A | 1.00 | −1.55 | −1.90 | −4.46 | −6.18 | −1.46 | a vs. b-f b, c vs. d, e e vs. f |
| GTM3 | 1.00 | −1.20 | −1.76 | −1.62 | −3.07 | −1.42 | a vs. b-f e vs. b-d, f |
| PECI | 1.00 | −1.32 | −1.64 | −2.33 | −1.80 | −1.17 | a vs. b-f b vs. d, e e vs. f |
| IGFB5 | 1.00 | 1.47 | −1.15 | −2.72 | −8.67 | −1.69 | a vs. b-f b vs. c-e c vs. d, e d vs. e e vs. f |
| TGFB3 | 1.00 | 1.27 | −1.20 | −1.86 | −5.58 | −1.34 | a vs. b-f e vs. b-d, f |
| CENPA | 1.00 | 1.45 | 2.08 | 1.63 | −2.16 | −1.45 | a vs. b-f c, d vs. e, f e vs. f |
Up- and down-regulated genes shared by adult life stage groups that were significantly different from prepubertal group at Padj < 0.01. Pairwise comparisons at Padj < 0.05 were made, and the relevant results are presented in the text
In general, mammary expression of SERF1A, GTM3, and PECI genes decreased significantly with advancing age in comparison with prepubertal life stage, reaching their minimum during pregnancy (PECI) and lactation (SERF1A, GTM3), respectively (Padj ≤ 0.05).
However, gene expression of IGFBP5 and TGFB3 revealed a significant increase during adolescence, followed by a similar significant decrease in advanced life stages, also reaching a minimum during lactation (Padj ≤ 0.05). In contrast, CENPA gene expression was significantly increased in adolescent, adult luteal, and pregnant mammary glands when compared with prepubertal subjects and was only significantly decreased during lactation and postmenopause (Padj ≤ 0.05). Lactating mammary gland presented a distinct gene expression pattern since pairwise analysis revealed a significant difference between lactating and young and adult life stages for SERF1A, IGFBP5, GTM3, and TGFB3, respectively (Padj ≤ 0.05). There was a significant difference between lactating and pregnant mammary gene expression for GSTM3, IGFBP5, TGFB3, and CENPA (Padj ≤ 0.05).
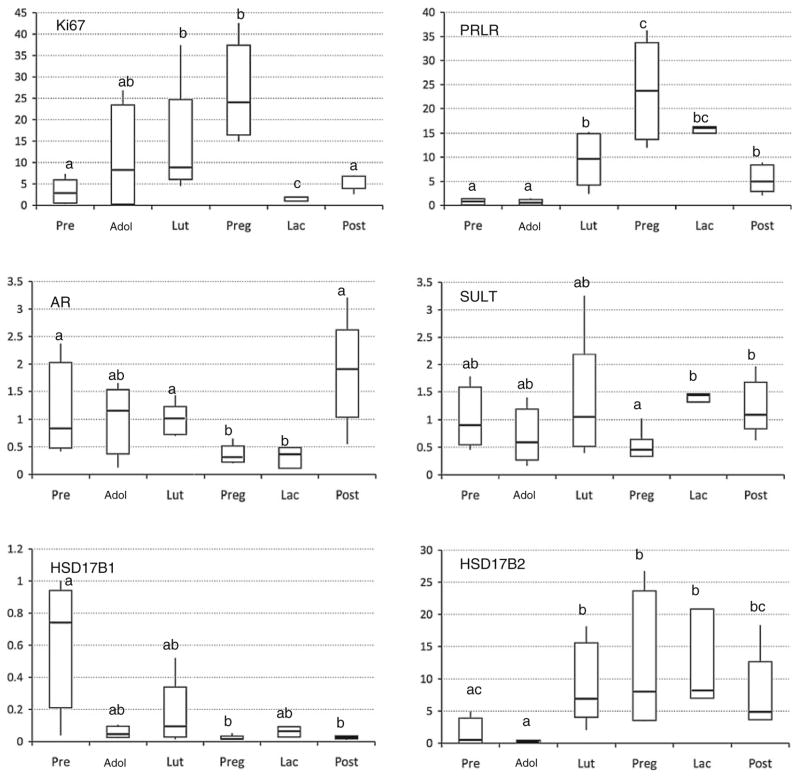
Quantitative RT-PCR
Quantitative RT-PCR was also used to assess expression levels of 14 key genes associated with estrogen, androgen, and prolactin signaling and metabolism, which had shown differential expression by microarray. Six of these showed significant fold change in expression in adulthood compared to prepuberty (Fig. 4). The genes were MKI67, PRLR, AR, SULT, HSD17B1, and HSD17B2. We found that expression profile of these genes among different life stages was similar to that shown with microarray. Consistent with our microarray results, MKI67 and PRLR were up-regulated during adulthood and pregnancy. Expression of AR was down-regulated in pregnancy and lactation. Expression of SULT mRNA, encoding for the enzyme that sulfates and thereby deactivates estrone, was diminished during pregnancy, relative to other groups. Expression of HSD17B1, which encodes a major estrogen biosynthetic enzyme converting estrone to estradiol, was highest in prepubertal animals. Conversely, mRNA for the enzyme catalyzing the reverse reaction (HSD17B2) was more abundant in adult life stages.
Fig. 4.
Gene expression measurements taken by qRT-PCR. Box plots of the 6 out of 13 genes examined with significant overall ANOVA at P < 0.05. Significant pairwise differences are indicated by differing symbols. P values for pairwise differences are <0.01 (MKI67, PRLR, AR, and 17BHSD2), <0.03 (17BHSD1), or <0.05 (SULT)
Discussion
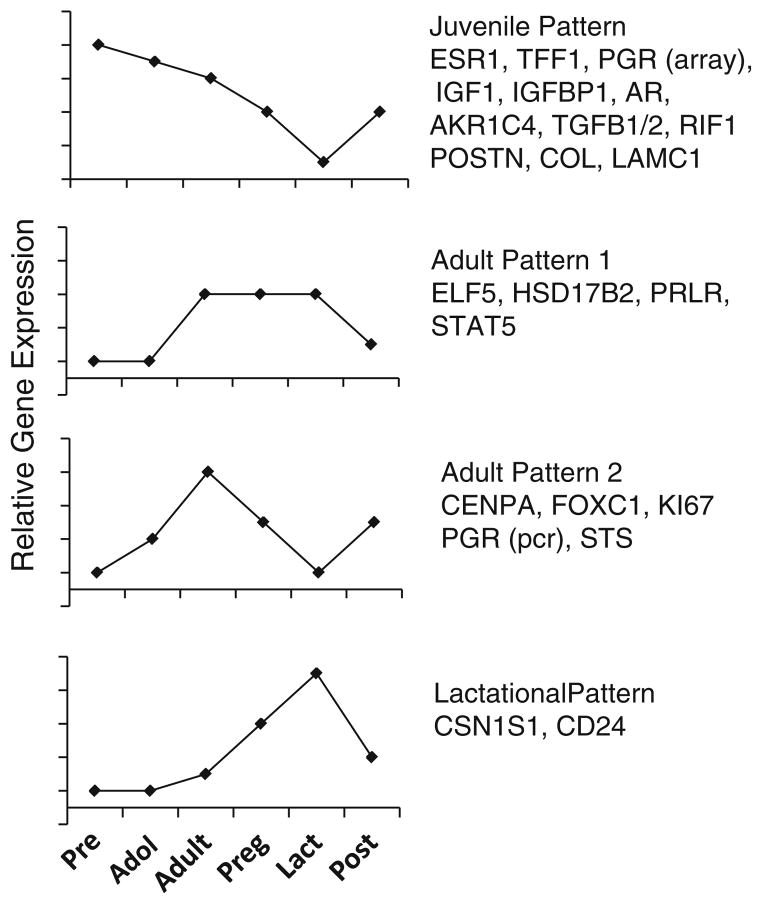
Our data demonstrate distinct life stage-specific patterns of gene expression during breast development. Simplified examples of expression patterns are shown in Fig. 5. With respect to mammary development during puberty, our findings support previous observations that GH operating via IGF-1, ovarian estrogens, and the respective receptors are crucial for normal breast development [2].
Fig. 5.
Schematic illustration of predominant patterns of gene expression from the microarray data. Y axis is not to scale
Estrogen-regulated genes such as ESR1 and IGF1 were found to be up-regulated during adolescence. The increase in IGF1 was not accompanied by an increase in IGFBP1 that would hamper its proliferative activity by protein binding. Other estrogen-regulated genes such as GREB1, TFF1, and PGR(B) were elevated during pubertal stages and decreased with advancing age. GREB1 is an ERα target gene and ERα coactivator that regulates estrogen-induced proliferation in BC cells. It is overexpressed in ERα-positive BC by 3.5-fold compared to ERα-negative BC [21]. TFFs are associated with mucin-secreting epithelial cells and contribute to mucosal defense and healing. TFF1 is expressed in approximately 50% of human breast tumors and enhances anchorage-independent growth, increased cell migration, and invasion [22]. As an estrogen-induced protein, it indicates likely responsiveness to endocrine treatment [23]. Progesterone via its receptor PGR is essential for mammary lobuloalveolar development. In virgin mammary gland, the A subtype of PGR dominates [24]. The PGR B signaling pathway shares regulatory cross-talk with other pathways such as RANKL [25]. The ratio of PGR isoforms may have a regulatory effect during development [26, 27]. PRLR expression has implications beyond prolactin per se; placental lactogen and primate growth hormones bind the PRLR [28, 29], which results in the activation of various signaling pathways including Jak2/Stat5 [30, 31], Shc/Grb2/Ras/Raf/Mek/MapK [32–34], and PKB/PI3 K [35–37].
Pregnancy-associated life stages markedly differed from other life stages. Prolactin is a major driver of development during pregnancy both directly and through stimulation of ovarian progesterone production. These hormones induce rapid and global proliferation of epithelial cells within the ductal epithelium and developing alveoli. During the second half of pregnancy, the cells of the alveoli differentiate and polarize to form the secretory alveolar epithelium, capable of milk production and secretion during lactation. Withdrawal of ovarian and placental progesterone brings about the onset of secretory activation [38]. Accordingly, PRLR gene expression reached its maximum during pregnancy and remained high during lactation in our study. Similarly, gene expression of CSN1S1, one of the predominant phosphoproteins in milk, tremendously increased during lactation. However, production of caseins is also stimulated by cortisol [37]. Enzyme HSD11B1 catalyzes the reduction of cortisone to active cortisol, and not surprisingly, we found that HSD11B1 gene expression paralleled the increased production of milk proteins.
During postmenopause, expression of various genes came back to adult luteal level (STS, HSD11B1, HSD17B7, HSD17B2, TFF1, GREB1, CSN1S1, IGFBP1, NEK10, IGHG1, LRRN3, PPM1K, PGR, ESR1, PRLR, AR). However, since the animals in this study were imported from abroad where they had lived in a breeding colony, it is likely that they had been pregnant before. Therefore, the gene “thumb print” of past pregnancy may be still present in our adult luteal subgroup.
AR gene expression decreased across life stage. AR rapidly modulates the expression of genes involved in proliferation and differentiation. Only recently, short-term androgen treatment has been shown to diminish the estrogen-induced proliferative response of the breast in rhesus monkeys [39, 40]. Furthermore, there is emerging evidence that the androgen-signaling pathway plays a critical role in breast carcinogenesis, independent of ER. In humans, although hyperandrogenemia is thought to be a risk factor for BC, expression of AR in BC is associated with lower tumor burden and favorable differentiation [41].
Breast tissue and mammary cancer cells possess the enzymatic systems necessary for local biosynthesis of steroid hormones, including enzymes involved in both the activation (STS, HSD11B1, HSD17B7) and inactivation (HSD17B2, AKR1C4) of hormones. The increase in ovarian serum estrogens during puberty and even more during pregnancy was accompanied by a decrease in mammary STS and HSD17B7 and an increase in HSD17B2 favouring the formation of estrone, estrone sulfate, and estriol. This control mechanism may protect the mammary gland from excessive estrogenic exposure. Indeed, HSD17B2 has been shown to be the predominant HSD17B subtype in normal breast tissue, whereas in cancerous tissue, the ratio of HSD17B1 to HSD17B2 changes, leading to a higher amount of locally formed potent estradiol [42]. Similarly, STS activity is significantly higher in cancerous in comparison with normal breast tissue [43].
There is evidence for a hierarchical model in which all types of epithelial cells in the mammary gland originate from a common multipotent stem cell [6]. In this model, ERα-negative stem cells (CD24+/CD29hi) undergo asymmetric division to give rise to undifferentiated, ERα-positive progenitor cells. These multipotent progenitor cells may also differentiate into basal-restricted or luminal-restricted progenitors and alveolar-restricted lineages (CD61+). During pregnancy, prolactin-mediated GATA3 may contribute to alveolar cell development, whereas ELF5, also a target of prolactin signaling, establishes the secretory alveolar lineage [44].
Across life stages, we found a significant increase in CD24 and ALDH1 gene expression, which was highest during lactation; this seems to indicate an amplification of uncommitted progenitor cells during breast development but may also indicate a lack of specificity for these putative stem cell markers in the context of the mature breast. Further differentiation to CD61-expressing luminal progenitors mainly occurred during adult luteal and lactating life stages. We also found an increase in ELF5 gene expression across life stages, reaching a maximum during adult luteal and lactating mammary gland, respectively, thus paralleling CD61 and PRLR gene expression. As mentioned above, GATA3 regulates mammary cell fate at multiple time points throughout mammary gland development including embryonic stage. Thus, it is not surprising that we did not find a significant impact of the life stages examined when using prepubertal animals as a reference of comparison (data not shown).
The association of ALDH1 and BRCA1, which has well-established roles in DNA repair and chromosome stability, has been investigated by Wicha et al. [45]. They suggest that BRCA1 plays a role in the differentiation of ALDH1-positive/ER-negative stem/progenitor cells into ERα-positive luminal epithelial cells. A loss of BRCA1 function may therefore cause a block in epithelial cell differentiation and the expansion of undifferentiated, ER-negative stem cells. BRCA1 expression was highest early in life and reduced in pregnancy, lactation, and postmenopausal phases, with a nadir during lactation.
There is a variety of stem cell markers generally associated with cell differentiation, repair mechanism, and apoptosis, which displayed different gene expression patterns across life stages. The majority of stem cell markers analyzed revealed a decrease in gene expression immediately with the transition into puberty (DICER1, RIF1, BMPR1A, TGFB2, LIG4, KLF4, NOTCH2, FOXA1), while others decreased after puberty (IGF1, KLF4, POSTN, NOTCH2, LIG4). Some of them have been investigated in BC. For example, DICER1 is thought to be involved in tumorigenesis. Its down-regulation may be related to the metastatic spread of tumors [46]. The IGF pathway has also been linked to mammary carcinogenesis in animal models. Higher IGF1 circulating levels significantly increased cancer risk, including premenopausal BC [47]. Other genes that were differentially expressed in our animals have uncertain relevance. For example, KLF4 may act as a transcriptional activator or repressor depending on the promoter context and/or cooperation with other transcription factors. There is no consensus on whether KLF4 functions as a tumor suppressor or an oncogene in BC [48–50]. Similarly, the role of STAT5A/B in BC is complex. In early stages of BC, STAT5A/B may promote malignant transformation and enhance growth of the tumor. This is in contrast to established BC, where STAT5A/B may mediate the critical cues for maintaining differentiation [51]. TGFB2 as part of the TGF-beta pathway is generally antiproliferative in epithelial cells but can also play either a tumor-suppressing or a tumor-promoting role in human breast carcinogenesis, depending on age and pathologic features of prognostic significance [52]. LIG4 encoding the protein DNA ligase IV that is involved in double-strand break repair has been shown either to have no significant relationship to BC risk [53, 54] or to be associated with a decrease in BC risk [55].
For other so-called BC stem cell markers, only sparse data are available so far. RIF1 contributes to ATM-mediated protection against DNA damage. There is some evidence that chromosome rearrangements such as RIF1 play an important role in BC development [56]. There are some links between BC and the transcriptional factor FOXA1, [57] NEK10, [58], and NOTCH2 [59].
The IGHG1 protein has been shown to be down-regulated in BC, [60] whereas ELF5 was expressed at higher levels in BC cells than normal epithelial cells [61]. GATA3 has been shown to drive invasive BC cells to undergo reversal of epithelial–mesenchymal transition, leading to the suppression of cancer metastasis [62]. To date, there are no data available on BC and BMPR1A, or LRRN3.
Extracellular matrix (ECM) molecules may provide important clues regarding the differentiation and biologic behavior of normal and neoplastic breast tissue. The high expression of POSTN seen in the adolescent breast in this study is interesting in light of the observation that POSTN overexpression in BC is associated with the development of bony metastases [63]. It may that the abnormal re-expression of this developmentally expressed protein contributes to the bone tropism of metastatic BC. Variations in other ECM molecules are less profound and may be explained at least in part by the relative proportions of stroma to glandular tissue in the tissues examined.
Diagnostic molecular tools have the potential to help clinicians decide how to treat a growing population of patients with early-stage BC. The MammaPrint® assay, developed in 2001 at the Netherlands Cancer Institute in Amsterdam, provides an expression profile of breast tumors for BC prognosis and prediction. The signature includes 70 relevant genes covering the hallmarks of cancer: cell cycle, metastasis, angiogenesis, and invasion [20]. In our data set, we found six genes covered by the MammaPrint® to be significantly regulated across life stages. GSTM3, PECI, TGFB3, IGFBP5, and SERF1A gene expression was reduced with advancing age, whereas CENPA gene expression increased until pregnancy followed by a significant decrease during lactation and postmenopause. Glutathione S-transferases such as GSTM3 metabolize a broad range of xenobiotics and carcinogens [64]. PECI is an auxiliary enzyme that catalyzes an isomerisation step required for the beta-oxidation of unsaturated fatty acids [65]. Expression of SERF1A, IGFB5, and the centromeric protein CENPA [66] is down-regulated in tumor tissue [20]. The function of SERF1A has not been elucidated yet.
This is the first study to investigate gene expression profiles in normal breast tissue across life stages. Our data demonstrate distinct patterns of gene expression during breast development and may shed light on the profound effect of reproductive life history on BC risk.
There has not been performed a comparable study in humans so far. In the mouse model, comparative gene expression analysis has focused on pregnancy, lactation, and involution after weaning [67–69]. Broader assessments across the lifespan have not been reported. In the murine gland, PCA across pregnancy, lactation, and involution revealed three distinct gene expression profiles with the majority of genes being associated with the “proliferation” profile during late pregnancy, stable expression during lactation, and diminished expression during involution [68]. Gene ontology analysis showed that biological processes such as ion transport, calcium-mediated signaling, transferase activity, and cell proliferation were mostly represented by the dominant “proliferation” PCA [68]. When focusing on single gene groups, adipocyte specific genes (fat specific protein 27, resistin, adiponectin, CAP, perilipin, hormone stimulated lipase, mitochondrial, dicarboxylic amino acid transporter) were shown to decline throughout pregnancy and early lactation, while milk protein genes (e.g., various caseins, mucin 1, a-lactalbumin, butyrophilin, xanthine oxireductase, parathyroid hormone related protein) increased over the time period. These findings are in agreement with our data. Genes associated with the regulation of glucose entry and utilization for lactose synthesis were mostly up-regulated at parturition (e.g., glucose transporter 1, citrate synthase, citrate transporter, ATP citrate lyase). Similarly, many genes that determine the nature of the fatty acids in the triglyceride were up-regulated at secretory activation [67, 70]. In their model of secretory activation, Rudolph et al. suggested a positive regulatory role for prolactin and the PRLR signaling pathway via Akt1 (fatty acid synthesis, lactose synthesis) and STAT5 (protein synthesis), while progesterone was thought to act as a negative regulator via direct prolactin inhibition and indirectly by IGFBP5, TGFB2, and Wnt5b signaling [68]. Similarly, our data showed a constantly high expression of PRLR, STAT5, and ELF5 during pregnancy and lactation, while the expression of the negative regulator TGFB2 and IGFBP5 was lowest during lactation.
The study described here has its limitations. Since this is an exploratory study design, many questions regarding single gene function cannot be answered. Furthermore, as mentioned above, since adult animals in this study were obtained as adults from a breeding colony, they are likely multiparous. The tissues examined were composed of a variety of cell types, and the proportions of tissues varied across the lifespan. Thus, some effects, such as the relatively high expression of collagens in the juvenile stages, likely represent the preponderance of fibroblasts in the tissue sampled.
Most interestingly, a variety of genes including BC stem cell genes currently discussed as potential biomarkers are expressed already early in life. Thus, one might speculate that, firstly, some developmental markers presented here may also serve as biomarkers in BC; secondly, most genes expressed later in life when BC risk is higher have already been expressed during pubertal breast development suggesting a “thumb print” of life events in breast tissue, which, thirdly, leads to the hypothesis that exposure to agents early in life (hormones, environmental and dietary factors, etc.) may alter the gene expression profile of the breast, thereby altering BC risk later in life.
Conclusion
Our data demonstrate distinct patterns of gene expression during breast development. Several of the pathways activated during pubertal development have been implicated in cancer development and metastasis, suggesting that developmental signals may have application as biomarkers for BC in later life. These data also illuminate imprinting effects in early life, as many genes associated with early breast differentiation persisted into later life. Possibly, therapeutic interventions early in life may alter BC risk later in life.
Acknowledgments
The authors are grateful for the technical support of Ms. Hermina Borgerink, Ms. Jean Gardin, Ms. Lisa O’Donnell, and Mr. Joseph Finley and also for the work of students Ms. Sara Dillon, Ms. Amelia Hubbard, and Mr. Russell O’Donnell. This study has been supported by the German Research Foundation Grants STU 469/2-1 and STU 469/3-1 (to PS), a German Society of Obstetrics and Gynecology Grant (to PS), and National Institutes of Health Grants R01 AT00639-06 (to JMC), RO3 AG18170 (to TCR), R01 R01AG017864 (to JKW), and P40 RR 021380 (to JDW).
Abbreviations
- AKR1C4
Aldo-keto reductase family 1, member C4
- AKT1
v-akt Murine thymoma viral oncogene homolog 1
- ALDH1
Aldehyde dehydrogenase 1 family, member A1
- AR
Androgen receptor
- BC
Breast cancer
- BMPR1A
Bone morphogenetic protein receptor, type 1A
- BRCA1
Breast cancer 1
- CAP
Cyclase-associated protein
- CD24, CD29, CD44, CD61
Cluster designation antigens 24, 29, 44, and 61
- CENPA
Centromeric protein A
- COL12A1
Collagen, type XII, alpha-1
- COL5A2
Collagen, type V, alpha-2
- CSN1S1
Casein alpha S1
- DICER1
Dicer 1, ribonuclease type III
- ELF5
E74-like factor 5
- ERα
Estrogen receptor alpha (protein)
- ERβ
Estrogen receptor beta (protein)
- ESR1
Estrogen receptor alpha (gene)
- ESR2
Estrogen receptor beta (gene)
- FOXA1
Forkhead box A1
- FOXC1
Forkhead box C1
- GATA3
Glutamyl-tRNA amidotransferase subunit A binding protein 3
- GH
Growth hormone
- GREB1
Gene regulated by estrogen in breast cancer 1
- GTM3
Glutathione s-transferase mu 3
- HSD11B1
11-beta-hydroxysteroid dehydrogenase type 1
- HSD17B2
17-beta-hydroxysteroid dehydrogenase type 2
- HSD17B7
17-beta-hydroxysteroid dehydrogenase type 7
- IGF1
Insulin-like growth factor 1
- IGFBP1
Insulin-like growth factor–binding protein 1
- IGFBP5
Insulin-like growth factor–binding protein 5
- IGHG1
IgG heavy-chain locus
- Jak2
Janus kinase 2
- KLF4
Kruppel-like factor 4
- LAMC1
Laminin, gamma-1
- LIG4
DNA ligase IV
- LRRN3
Leucine-rich repeat protein, neuronal, 3
- MKI67
Proliferation-related antigen Ki67
- NEK10
Never in mitosis gene a–related kinase 10
- NOTCH2
Notch gene homolog 2
- PCA
Principal components analysis
- PDGFRA
Platelet-derived growth factor receptor, alpha
- PECI
Peroxisomal D3, D2-enoyl-CoA isomerase
- PGR
Progesterone receptor
- PGRB
Progesterone receptor B
- POSTN
Periostin
- PPM1K
Protein phosphatase, PP2C domain-containing, 1K
- PRLR
Prolactin receptor
- RANKL
Receptor activator of NF-kappa-B ligand
- RIF1
RAP1 interacting factor homolog
- SERF1A
Small EDRK-rich factor 1A
- STAT5
Signal transducer and activator of transcription 5
- STS
Steroid sulfatase
- SULT
Sulfotransferase
- TFF1
Trefoil factor 1
- TGFB1
Transforming growth factor, beta-1
- TGFB2
Transforming growth factor, beta-2
- TGFB3
Transforming growth factor, beta-3
- WNT5B
Wingless-related MMTV integration site 5B
Contributor Information
Petra Stute, Email: petra.stute@insel.ch, Department of Gynecologic Endocrinology and Reproductive Medicine, University Women’s Hospital, Effingerstrasse 102, 3010 Berne, Switzerland.
Sonja Sielker, Arrows Biomedical Deutschland GmbH, Munster, Germany.
Charles E. Wood, Department of Pathology/Section on Comparative Medicine, Wake Forest University School of Medicine, Winston-Salem, NC 27157-1040, USA
Thomas C. Register, Department of Pathology/Section on Comparative Medicine, Wake Forest University School of Medicine, Winston-Salem, NC 27157-1040, USA
Cynthia J. Lees, Department of Pathology/Section on Comparative Medicine, Wake Forest University School of Medicine, Winston-Salem, NC 27157-1040, USA
Fitriya N. Dewi, Department of Pathology/Section on Comparative Medicine, Wake Forest University School of Medicine, Winston-Salem, NC 27157-1040, USA
J. Koudy Williams, Department of Pathology/Section on Comparative Medicine, Wake Forest University School of Medicine, Winston-Salem, NC 27157-1040, USA.
Janice D. Wagner, Department of Pathology/Section on Comparative Medicine, Wake Forest University School of Medicine, Winston-Salem, NC 27157-1040, USA
Ulrich Stefenelli, Services-In-Statistics, Würzburg, Germany.
J. Mark Cline, Department of Pathology/Section on Comparative Medicine, Wake Forest University School of Medicine, Winston-Salem, NC 27157-1040, USA.
References
- 1.Monaghan P, Perusinghe NP, Cowen P, Gusterson BA. Peripubertal human breast development. Anat Rec. 1990;226:501–508. doi: 10.1002/ar.1092260412. [DOI] [PubMed] [Google Scholar]
- 2.Russo J, Hu YF, Yang X, Russo IH. Developmental, cellular, and molecular basis of human breast cancer. J Natl Cancer Inst Monogr. 2000;27:17–37. doi: 10.1093/oxfordjournals.jncimonographs.a024241. [DOI] [PubMed] [Google Scholar]
- 3.Hovey RC, Trott JF. Morphogenesis of mammary gland development. Adv Exp Med Biol. 2004;554:219–228. doi: 10.1007/978-1-4757-4242-8_19. [DOI] [PubMed] [Google Scholar]
- 4.Hens JR, Wysolmerski JJ. Key stages of mammary gland development: molecular mechanisms involved in the formation of the embryonic mammary gland. Breast Cancer Res. 2005;7:220–224. doi: 10.1186/bcr1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sternlicht MD, Kouros-Mehr H, Lu P, Werb Z. Hormonal and local control of mammary branching morphogenesis. Differentiation. 2006;74:365–381. doi: 10.1111/j.1432-0436.2006.00105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LaMarca HL, Rosen JM. Minireview: hormones and mammary cell fate—what will I become when I grow up? Endocrinology. 2008;149:4317–4321. doi: 10.1210/en.2008-0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Booth BW, Boulanger CA, Smith GH. Alveolar progenitor cells develop in mouse mammary glands independent of pregnancy and lactation. J Cell Physiol. 2007;212:729–736. doi: 10.1002/jcp.21071. [DOI] [PubMed] [Google Scholar]
- 8.Wagner KU, Boulanger CA, Henry MD, Sgagias M, Hennighausen L, Smith GH. An adjunct mammary epithelial cell population in parous females: its role in functional adaptation and tissue renewal. Development. 2002;129:1377–1386. doi: 10.1242/dev.129.6.1377. [DOI] [PubMed] [Google Scholar]
- 9.Hilakivi-Clarke L. Nutritional modulation of terminal end buds: its relevance to breast cancer prevention. Curr Cancer Drug Targets. 2007;7:465–474. doi: 10.2174/156800907781386641. [DOI] [PubMed] [Google Scholar]
- 10.Molyneux G, Regan J, Smalley MJ. Mammary stem cells and breast cancer. Cell Mol Life Sci. 2007;64:3248–3260. doi: 10.1007/s00018-007-7391-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lacroix M. Significance, detection and markers of disseminated breast cancer cells. Endocr Relat Cancer. 2006;13:1033–1067. doi: 10.1677/ERC-06-0001. [DOI] [PubMed] [Google Scholar]
- 12.Magness CL, Fellin PC, Thomas MJ, Korth MJ, Agy MB, Proll SC, Fitzgibbon M, Scherer CA, Miner DG, Katze MG, Iadonato SP. Analysis of the Macaca mulatta transcriptome and the sequence divergence between Macaca and human. Genome Biol. 2005;6:R60. doi: 10.1186/gb-2005-6-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cline JM, Soderqvist G, von Schoultz B, Skoog L. Regional distribution of proliferating cells and hormone receptors in the mammary gland of surgically postmenopausal macaques. Gynecol Obstet Invest. 1997;44:41–46. doi: 10.1159/000291407. [DOI] [PubMed] [Google Scholar]
- 14.Cline JM, Wood CE. The mammary glands of macaques. Toxicol Pathol. 2008;36:130S–141S. doi: 10.1177/0192623308327411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stute P, Wood CE, Kaplan JR, Cline JM. Cyclic changes in the mammary gland of cynomolgus macaques. Fertil Steril. 2004;82:1160S–1170S. doi: 10.1016/j.fertnstert.2004.04.035. [DOI] [PubMed] [Google Scholar]
- 16.Wood CE, Hester JM, Cline JM. Mammary gland development in early pubertal female macaques. Toxicol Pathol. 2007;35(6):795–805. doi: 10.1080/01926230701584213. [DOI] [PubMed] [Google Scholar]
- 17.Cline JM. Assessing the mammary gland of nonhuman primates: effects of endogenous hormones and exogenous hormonal agents and growth factors. Birth Defects Res B Dev Reprod Toxicol. 2007;80:126–146. doi: 10.1002/bdrb.20112. [DOI] [PubMed] [Google Scholar]
- 18.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B M1. 1995;57:289–300. [Google Scholar]
- 19.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 20.van ‘t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 21.Ghosh G, Thompson DA, Weigel RJ. PDZK1 and GREB1 are estrogen-regulated genes expressed in hormone-responsive breast cancer. Cancer Res. 2000;60:6367–6375. [PubMed] [Google Scholar]
- 22.Amiry N, Kong X, Muniraj N, Kannan N, Grandison PM, Lin J, Yang Y, Vouyovitch CM, Borges S, Perry JK, Mertani HC, Zhu T, Liu D, Lobie PE. Trefoil factor-1 (TFF1) enhances oncogenicity of mammary carcinoma cells. Endocrinology. 2009;150(10):4473–4483. doi: 10.1210/en.2009-0066. [DOI] [PubMed] [Google Scholar]
- 23.Mathelin C, Tomasetto C, Rio MC. Trefoil factor 1 (pS2/TFF1), a peptide with numerous functions. Bull Cancer. 2005;92:773–781. [PubMed] [Google Scholar]
- 24.Conneely OM, Mulac-Jericevic B, Lydon JP. Progesterone-dependent regulation of female reproductive activity by two distinct progesterone receptor isoforms. Steroids. 2003;68:771–778. doi: 10.1016/s0039-128x(03)00126-0. [DOI] [PubMed] [Google Scholar]
- 25.Mulac-Jericevic B, Lydon JP, DeMayo FJ, Conneely OM. Defective mammary gland morphogenesis in mice lacking the progesterone receptor B isoform. PNAS. 2003;100:9744–9749. doi: 10.1073/pnas.1732707100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buck K, Vanek M, Groner B, Ball RK. Multiple forms of prolactin receptor messenger ribonucleic acid are specifically expressed and regulated in murine tissues and the mammary cell line HC11. Endocrinology. 1992;130:1108–1114. doi: 10.1210/endo.130.3.1537278. [DOI] [PubMed] [Google Scholar]
- 27.Clarke LA, Wathes DC, Jabbour HN. Expression and localization of prolactin receptor messenger ribonucleic acid in red deer ovary during the estrous cycle and pregnancy. Biol Reprod. 1997;57:865–872. doi: 10.1095/biolreprod57.4.865. [DOI] [PubMed] [Google Scholar]
- 28.Freeman ME, Kanyicska B, Lerant A, Nagy G. Prolactin: structure, function, and regulation of secretion. Physiol Rev. 2000;80:1523–1631. doi: 10.1152/physrev.2000.80.4.1523. [DOI] [PubMed] [Google Scholar]
- 29.Bole-Feysot C, Goffin V, Edery M, Binart N, Kelly PA. Prolactin (PRL) and its receptor: actions, signal transduction pathways and phenotypes observed in PRL receptor knockout mice. Endocr Rev. 1998;19:225–268. doi: 10.1210/edrv.19.3.0334. [DOI] [PubMed] [Google Scholar]
- 30.Gouilleux F, Wakao H, Mundt M, Groner B. Prolactin induces phosphorylation of Tyr694 of Stat5 (MGF), a prerequisite for DNA binding and induction of transcription. EMBO J. 1994;13:4361–4369. doi: 10.1002/j.1460-2075.1994.tb06756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X, Robinson GW, Gouilleux F, Groner B, Hennighausen L. Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. PNAS. 1995;92:8831–8835. doi: 10.1073/pnas.92.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erwin RA, Kirken RA, Malabarba MG, Farrar WL, Rui H. Prolactin activates Ras via signaling proteins SHC, growth factor receptor bound 2, and son of sevenless. Endocrinology. 1995;136:3512–3518. doi: 10.1210/endo.136.8.7628388. [DOI] [PubMed] [Google Scholar]
- 33.Das R, Vonderhaar BK. Activation of raf-1, MEK and MAP kinase in prolactin responsive mammary cells. Breast Cancer Res Treat. 1996;40:141–149. doi: 10.1007/BF01806209. [DOI] [PubMed] [Google Scholar]
- 34.Piccoletti R, Bendinelli P, Maroni P. Signal transduction pathway of prolactin in rat liver. Mol Cell Endocrinol. 1997;135:169–177. doi: 10.1016/s0303-7207(97)00204-9. [DOI] [PubMed] [Google Scholar]
- 35.Fresno Vara JA, Caceres MA, Silva A, Martin-Perez J. Src family kinases are required for prolactin induction of cell proliferation. Mol Biol Cell. 2001;12(7):2171–2183. doi: 10.1091/mbc.12.7.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tessier C, Prigent-Tessier A, Ferguson-Gottschall S, Gu Y, Gibori G. PRL antiapoptotic effect in the rat decidua involves the PI3 K/protein kinase B-mediated inhibition of caspase-3 activity. Endocrinology. 2001;142(9):4086–4094. doi: 10.1210/endo.142.9.8381. [DOI] [PubMed] [Google Scholar]
- 37.Oakes SR, Rogers RL, Naylor MJ, Ormandy CJ. Prolactin regulation of mammary gland development. J Mammary Gland Biol Neoplasia. 2008;13:13–28. doi: 10.1007/s10911-008-9069-5. [DOI] [PubMed] [Google Scholar]
- 38.Neville MC, McFadden TB, Forsyth I. Hormonal regulation of mammary differentiation and milk secretion. J Mammary Gland Biol Neoplasia. 2002;7:49–66. doi: 10.1023/a:1015770423167. [DOI] [PubMed] [Google Scholar]
- 39.Zhou J, Ng S, Adesanya-Famuiya O, Anderson K, Bondy CA. Testosterone inhibits estrogen-induced mammary epithelial proliferation and suppresses estrogen receptor expression. FASEB J. 2000;14:1725–1730. doi: 10.1096/fj.99-0863com. [DOI] [PubMed] [Google Scholar]
- 40.Dimitrakakis C, Zhou J, Wang J, Belanger A, LaBrie F, Cheng C, Powell D, Bondy C. A physiologic role for testosterone in limiting estrogenic stimulation of the breast. Menopause. 2003;10:292–298. doi: 10.1097/01.GME.0000055522.67459.89. [DOI] [PubMed] [Google Scholar]
- 41.Park S, Koo J, Park HS, Kim JH, Choi SY, Lee JH, Park BW, Lee KS. Expression of androgen receptors in primary breast cancer. Ann Oncol. 2010;21:488–492. doi: 10.1093/annonc/mdp510. [DOI] [PubMed] [Google Scholar]
- 42.Vihko P, Herrala A, Härkönen P, Isomaa V, Kaija H, Kurkela R, Pulkka A. Control of cell proliferation by steroids: the role of 17HSDs. Mol Cell Endocrinol. 2006;248:141–148. doi: 10.1016/j.mce.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 43.Chetrite GS, Cortes-Prieto J, Philippe JC, Wright F, Pasqualini JR. Comparison of estrogen concentrations, estrone sulfatase and aromatase activities in normal, and in cancerous, human breast tissues. J Steroid Biochem Mol Biol. 2000;72:23–27. doi: 10.1016/s0960-0760(00)00040-6. [DOI] [PubMed] [Google Scholar]
- 44.Oakes SR, Naylor MJ, Asselin-Labat ML, Blazek KD, Gardiner-Garden M, Hilton HN, Kazlauskas M, Pritchard MA, Chodosh LA, Pfeffer PL, Lindeman GJ, Visvader JE, Ormandy CJ. The Ets transcription factor Elf5 specifies mammary alveolar cell fate. Genes Dev. 2008;22:581–586. doi: 10.1101/gad.1614608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu S, Ginestier C, Charafe-Jauffret E, Foco H, Kleer CG, Merajver SD, Dontu G, Wicha MS. BRCA1 regulates human mammary stem/progenitor cell fate. Proc Natl Acad Sci USA. 2008;105:1680–1685. doi: 10.1073/pnas.0711613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grelier G, Voirin N, Ay AS, Cox DG, Chabaud S, Treilleux I, Léon-Goddard S, Rimokh R, Mikaelian I, Venoux C, Puisieux A, Lasset C, Moyret-Lalle C. Prognostic value of Dicer expression in human breast cancers and association with the mesenchymal phenotype. Br J Cancer. 2009;101:673–683. doi: 10.1038/sj.bjc.6605193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen W, Wang S, Tian T, Bai J, Hu Z, Xu Y, Dong J, Chen F, Wang X, Shen H. Phenotypes and genotypes of insulin-like growth factor 1, IGF-binding protein-3 and cancer risk: evidence from 96 studies. Eur J Hum Genet. 2009;17:1668–1675. doi: 10.1038/ejhg.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Foster KW, Frost AR, McKie-Bell P, Lin CY, Engler JA, Grizzle WE, Ruppert JM. Increase of GKLF messenger RNA and protein expression during progression of breast cancer. Cancer Res. 2000;60:6488–6495. [PubMed] [Google Scholar]
- 49.Miller KA, Eklund EA, Peddinghaus ML, Cao Z, Fernandes N, Turk PW, Thimmapaya B, Weitzman SA. Kruppel-like factor 4 regulates laminin alpha 3A expression in mammary epithelial cells. Journal of Biological Chemistry. 2001;276:42863–42868. doi: 10.1074/jbc.M108130200. [DOI] [PubMed] [Google Scholar]
- 50.Pandya AY, Talley LI, Frost AR, Fitzgerald TJ, Trivedi V, Chakravarthy M, Chhieng DC, Grizzle WE, Engler JA, Krontiras H, Bland KI, LoBuglio AF, Lobo-Ruppert SM, Ruppert JM. Nuclear localization of KLF4 is associated with an aggressive phenotype in early-stage breast cancer. Clin Cancer Res. 2004;10:2709–2719. doi: 10.1158/1078-0432.ccr-03-0484. [DOI] [PubMed] [Google Scholar]
- 51.Tan SH, Nevalainen MT. Signal transducer and activator of transcription 5A/B in prostate and breast cancers. Endocr Relat Cancer. 2008;15:367–390. doi: 10.1677/ERC-08-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Figueroa JD, Flanders KC, Garcia-Closas M, Anderson WF, Yang XR, Matsuno RK, Duggan MA, Pfeiffer RM, Ooshima A, Cornelison R, Gierach GL, Brinton LA, Lissowska J, Peplonska B, Wakefield LM, Sherman ME. Expression of TGF-beta signaling factors in invasive breast cancers: relationships with age at diagnosis and tumor characteristics. Breast Cancer Res Treat. 2009;121:727–735. doi: 10.1007/s10549-009-0590-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.García-Closas M, Egan KM, Newcomb PA, Brinton LA, Titus-Ernstoff L, Chanock S, Welch R, Lissowska J, Peplonska B, Szeszenia-Dabrowska N, Zatonski W, Bardin-Mikolajczak A, Struewing JP. Polymorphisms in DNA double-strand break repair genes and risk of breast cancer: two population-based studies in USA and Poland, and meta-analyses. Hum Genet. 2006;119:376–788. doi: 10.1007/s00439-006-0135-z. [DOI] [PubMed] [Google Scholar]
- 54.Jakubowska A, Gronwald J, Menkiszak J, Górski B, Huzarski T, Byrski T, Tołoczko-Grabarek A, Gilbert M, Edler L, Zapatka M, Eils R, Lubiński J, Scott RJ, Hamann U. BRCA1-associated breast and ovarian cancer risks in Poland: no association with commonly studied polymorphisms. Breast Cancer Res Treat. 2010;119:201–211. doi: 10.1007/s10549-009-0390-5. [DOI] [PubMed] [Google Scholar]
- 55.Kuschel B, Auranen A, McBride S, Novik KL, Antoniou A, Lipscombe JM, Day NE, Easton DF, Ponder BA, Pharoah PD, Dunning A. Variants in DNA double-strand break repair genes and breast cancer susceptibility. Hum Mol Genet. 2002;11:1399–1407. doi: 10.1093/hmg/11.12.1399. [DOI] [PubMed] [Google Scholar]
- 56.Howarth KD, Blood KA, Ng BL, Beavis JC, Chua Y, Cooke SL, Raby S, Ichimura K, Collins VP, Carter NP, Edwards PA. Array painting reveals a high frequency of balanced translocations in breast cancer cell lines that break in cancer-relevant genes. Oncogene. 2007;27:3345–3359. doi: 10.1038/sj.onc.1210993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ademuyiwa FO, Thorat MA, Jain RK, Nakshatri H, Badve S. Expression of Forkhead-box protein A1, a marker of luminal A type breast cancer, parallels low Oncotype DX 21-gene recurrence scores. Mod Pathol. 2010;23:270–275. doi: 10.1038/modpathol.2009.172. [DOI] [PubMed] [Google Scholar]
- 58.Ahmed S, Thomas G, Ghoussaini M, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41:585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee SH, Jeong EG, Yoo NJ, Lee SH. Mutational analysis of NOTCH1, 2, 3 and 4 genes in common solid cancers and acute leukemias. APMIS. 2007;115:1357–1363. doi: 10.1111/j.1600-0463.2007.00751.x. [DOI] [PubMed] [Google Scholar]
- 60.Kabbage M, Chahed K, Hamrita B, Guillier CL, Trimeche M, Remadi S, Hoebeke J, Chouchane L. Protein alterations in infiltrating ductal carcinomas of the breast as detected by non-equilibrium pH gradient electrophoresis and mass spectrometry. J Biomed Biotechnol. 2008;2008:564127. doi: 10.1155/2008/564127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He J, Pan Y, Hu J, Albarracin C, Wu Y, Dai JL. Profile of Ets gene expression in human breast carcinoma. Cancer Biol Ther. 2007;6:76–82. doi: 10.4161/cbt.6.1.3551. [DOI] [PubMed] [Google Scholar]
- 62.Yan W, Cao QJ, Arenas RB, Bentley B, Shao R. GATA3 inhibits breast cancer metastasis through the reversal of epithelial-mesenchymal transition. J Biol Chem. 2010;285:14042–14051. doi: 10.1074/jbc.M110.105262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sasaki H, Yu CY, Dai M, Tam C, Loda M, Auclair D, Chen LB, Elias A. Elevated serum periostin levels in patients with bone metastases from breast but not lung cancer. Breast Cancer Res Treat. 2003;77(3):245–252. doi: 10.1023/a:1021899904332. [DOI] [PubMed] [Google Scholar]
- 64.Pearson WR, Vorachek WR, Xu SJ, Berger R, Hart I, Vannais D, Patterson D. Identification of class-mu glutathione transferase genes GSTM1-GSTM5 on human chromosome 1p13. Am J Hum Genet. 1993;53:220–233. [PMC free article] [PubMed] [Google Scholar]
- 65.Geisbrecht BV, Zhang D, Schulz H, Gould SJ. Characterization of PECI, a novel monofunctional delta-3, delta-2-enoyl-CoA isomerase of mammalian peroxisomes. J Biol Chem. 1999;274:21797–21803. doi: 10.1074/jbc.274.31.21797. [DOI] [PubMed] [Google Scholar]
- 66.Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 67.Rudolph MC, McManaman JL, Hunter L, Phang T, Neville MC. Functional development of the mammary gland: use of expression profiling and trajectory clustering to reveal changes in gene expression during pregnancy, lactation, and involution. J Mammary Gland Biol Neoplasia. 2003;8(3):287–307. doi: 10.1023/b:jomg.0000010030.73983.57. [DOI] [PubMed] [Google Scholar]
- 68.Lemay DG, Neville MC, Rudolph MC, Pollard KS, German JB. Gene regulatory networks in lactation: identification of global principles using bioinformatics. BMC Syst Biol. 2007;1:56. doi: 10.1186/1752-0509-1-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao H, Huang M, Chen Q, Wang Q, Pan Y. Comparative gene expression analysis in mouse models for identifying critical pathways in mammary gland development. Breast Cancer Res Treat. 2011 doi: 10.1007/s10549-011-1650-8. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 70.Anderson SM, Rudolph MC, McManaman JL, Neville MC. Key stages in mammary gland development. Secretory activation in the mammary gland: it’s not just about milk protein synthesis! Breast Cancer Res. 2007;9(1):204. doi: 10.1186/bcr1653. [DOI] [PMC free article] [PubMed] [Google Scholar]