Abstract
FMS-like tyrosine kinase 3 (FLT3) normally functions in the survival/proliferation of hematopoietic stem/progenitor cells, but its constitutive activation by internal tandem duplication (ITD) mutations correlates with a poor prognosis in AML. The development of FLT3 tyrosine kinase inhibitors (TKI) is a promising strategy, but resistance that arises during the course of treatment caused by secondary mutations within the mutated gene itself poses a significant challenge. In an effort to predict FLT3 resistance mutations that might develop in patients, we used saturation mutagenesis of FLT3/ITD followed by selection of transfected cells in FLT3 TKI. We identified F621L, A627P, F691L and Y842C mutations in FLT3/ITD that confer varying levels of resistance to FLT3 TKI. Western blotting confirmed that some FLT3 TKI were ineffective at inhibiting FLT3 autophosphorylation and signaling through MAP kinase, STAT5 and AKT in some mutants. Balb/c mice transplanted with the FLT3/ITD Y842C mutation confirmed resistance to sorafenib in vivo but not to lestaurtinib. These results indicate a growing number of FLT3 mutations that are likely to be encountered in patients. Such knowledge, combined with known remaining sensitivity to other FLT3 TKI, will be important to establish as secondary drug treatments that can be substituted when these mutants are encountered.
Keywords: acute myeloid leukemia, mutant FLT3, drug resistance, tyrosine kinase inhibitors
INTRODUCTION
FMS-like tyrosine kinase 3 (FLT3) is a class III receptor tyrosine kinase that is expressed on early hematopoietic stem cells/progenitors and is vital for development of normal levels of mature myeloid and lymphoid cells.1, 2 FLT3 is normally activated by binding FLT3 ligand (FL), which prompts receptor dimerization and kinase activity with the subsequent activation of multiple downstream signaling pathways, including Ras/MAP kinase, AKT and STAT5.3–6 High coexpression of FLT3 with FL may lead to dysregulation of activity via autocrine or paracrine mechanisms that promote leukemogenesis, as has been observed in MLL-rearranged infant ALL.7–9 Mutation of FLT3 leads to constitutive kinase activation and transfection of these forms leads to transformation of cytokine-dependent cell lines to factor independence. FLT3 mutations represent one of the most common molecular perturbations in acute myeloid leukemia (AML), accounting for 30–35% of de novo cases.10–12 FLT3 mutations generally occur in the juxtamembrane (JM) domain or in the kinase domain (KD). The JM mutations factor in approximately 23% of newly diagnosed cases of AML and occur as in-frame internal tandem duplications (ITDs) of varying length, resulting in duplication of a sequence of typically 4–50 amino acids, often accompanied by a one or two amino-acid insert.10 The crystal structure of FLT3 shows that the JM domain functions as an autoinhibitory mechanism to regulate FLT3 kinase activity, and disruption by mutations destabilize its conformation.13 KD mutations constitute about 7–10% of AML cases and usually present as missense mutations of the activation loop, most commonly at D835.11, 12 Because of its proliferative stimulus and frequent mutation rate in AML, FLT3 has been deemed as a highly desirable target for modulation.
The impressive response of chronic myelogenous leukemia patients to BCR-ABL TKI generated enthusiasm for molecularly targeted therapies in other malignancies dependent on constitutively activated kinase signaling. However, the development of resistance to imatinib due to the acquisition of point mutations in BCR-ABL also foreshadows a similar outcome now being reported in AML patients expressing a FLT3/ITD mutation being treated with FLT3 TKI.14–16 Resistance mutations often decrease the affinity of a TKI for its target and necessitate the use of a structurally unrelated inhibitor if one is available. This expectation has led to investigations attempting to identify a spectrum of secondary mutations of FLT3/ITD in the laboratory, which confer resistance to FLT3 TKI prior to their emergence in the clinic. Several groups have employed various techniques to identify FLT3 resistance mutations.17–21 In contrast to the wide array of BCR-ABL resistance mutations, relatively few FLT3 resistance mutations have been identified, which may partially reflect the failure to achieve sufficient levels of inhibition of FLT3 signaling in many trials,22 In this study, we identified the F691L and Y842C mutations previously identified as well as two novel mutations, F621L and A627P, that cause resistance to select TKI. These results suggest that novel mutations arising in FLT3/ITD, perhaps by selection during the course of treatment with a TKI, may prove to be refractory to FLT3 mutant AML management using most TKIs and emphasize the need for development of FLT3 inhibitors that can overcome resistance due to mutations.
MATERIALS AND METHODS
Reagents and antibodies
Lestaurtinib, midostaurin, sunitinib, sorafenib and AC220 were purchased from LC Labs (Westchester, PA, USA). KW2449 was from Kyowa Hakko Kirin Co., Ltd. (Tokyo, Japan). AGS324 was provided by Aviv Gazit. Recombinant human interleukin-3 was purchased from Pepro Tech, Inc. (Rocky Hill, NJ, USA). FLT3 S-18 and STAT5 antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA, USA), 4G10 anti-phosphotyrosine antibody and recombinant protein A-agarose were from Upstate Biotechnology (Lake Placid, NY, USA) and CD135-phycqerythrin (PE)-conjugated and annexin V-PE antibodies were from BD Pharmingen (San Jose, CA, USA). PhosphoMAP kinase, phospho STAT5, phosphoAKT, MAP kinase and AKT antibodies were from Cell Signaling Technologies, Inc. (Beverly, MA, USA). Goat anti-mouse and goat anti-rabbit horseradish peroxidase antibodies and the enhanced chemiluminescence kit were from Amersham Biosciences (Arlington Heights, IL, USA).
DNA constructs and cells
BaF3 or TF-1 cells were cultured in RPMI medium supplemented with 1 ng/ml recombinant human interleukin-3 or 1 ng/ml of granulocyte-macrophage colony-stimulating factor, respectively. FLT3/ITD cells were established from a patient sample as previously described.23 FLT3 point mutations were generated by site-directed mutagenesis in the pBabe Neo vector containing FLT3/ITD complementary DNA using the QuickChange Site-Directed Mutagenesis (Stratagene, La Jolla, CA, USA) and used to transfect BaF3 cells using the Nucleofector II from Amaxa Biosystems (Walkersville, MD, USA). BaF3 cells were chosen for confirmation of resistance because we eventually wanted to transplant a resistant clone to syngeneic Balb/c mice and determine whether FLT3 TKI could overcome the mutation in vivo.
Selection for resistant clones
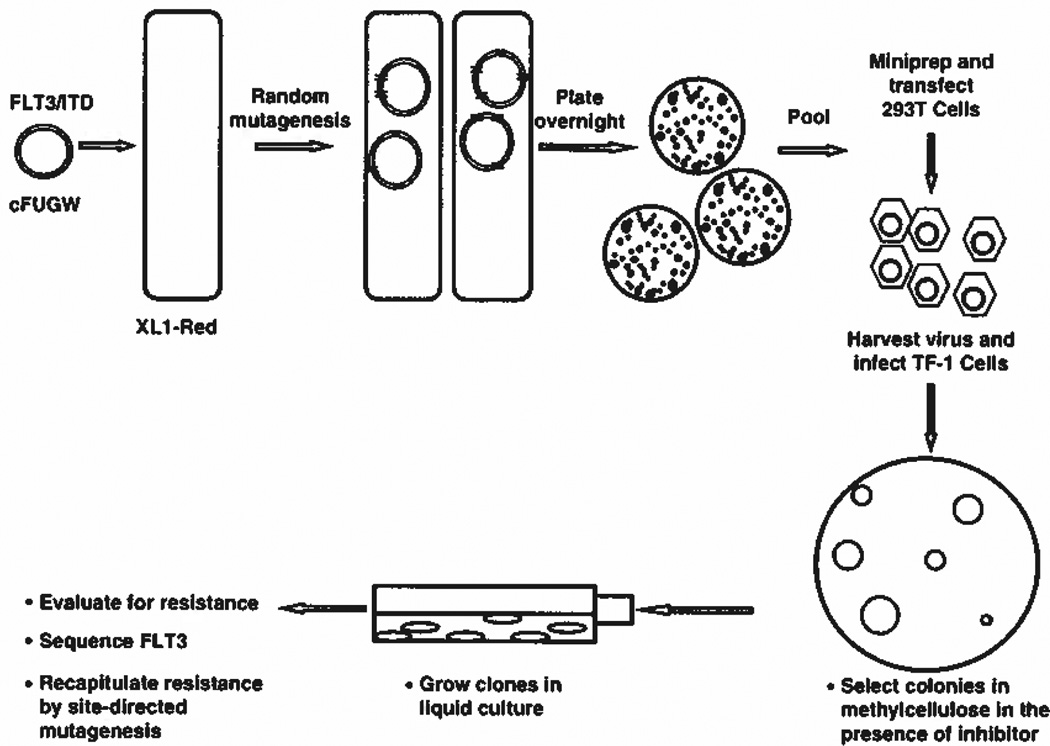
The cFUGW plasmid containing a FLT3/ITD complementary DNA was used to transform the XL1-Red mutator strain of E. coli as instructed by the manufacturer (Stratagene).24 Transformed cells were allowed to propagate overnight at 37 °C on agar plates containing ampicillin at which point plasmids were harvested and linearized by Not I digestion. The purified products were used for packaging in lentivirus in NIH293T cells. The viral supernatant was used to transduce granulocyte-macrophage colony-stimulating factor-dependent TF-1 cells. Cells were then plated in cytokine-deficient methylceliulose in the presence of a FLT3 TKI for 2 weeks at which point FLT3 in clones of interest was sequenced.
Growth inhibition and apoptosis
Cells were seeded in the presence or absence of inhibitor for 48 h with or without 1 ng/ml of interleukin-3. Cell proliferation/viability was measured as described using the MTT assay according to the manufacturer's instructions (Roche Applied Science, Indianapolis, IN, USA) or induction of apoptosis with annexin V/7-AAD (BD Biosciences, San Diego, CA, USA).25
Immunoprecipitation and western blotting
FLT3 was analyzed by performing immunoprecipitation, 5DS-PAGE and western blotting as previously described.26 Proteins were transferred to a polyvinyl diflouride membrane (Millipore, Bedford, MA, USA) and probed for FLT3 using S-18 and 4G10 phosphotyrosine antibodies. FLT3 was then detected on a Li-Cor imager using Odyssey software (both from Li-Cor Biosciences, Lincoln, NE, USA). Other proteins were detected in whole cell lysates using the indicated antibodies and a horseradish peroxidase-conjugated goat anti-rabbit secondary antibody followed by enhanced chemiluminescence.
Engraftment in Balb/c mice
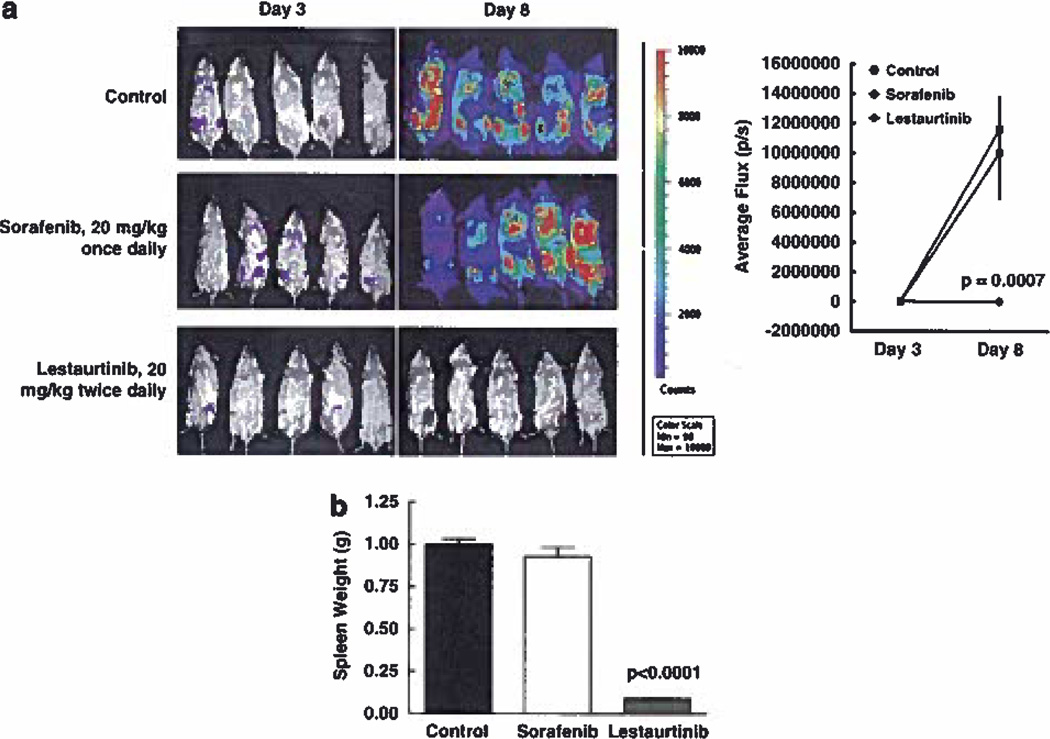
BaF3 FLT3/ITD Y842C cells were transfected with the L3GFP plasmid (a gift from Dr Linzhao Cheng of Johns Hopkins University) containing genes for luciferase and green fluorescent protein (GFP). For engraftment, 2 × 105 cells were injected by tail vein into syngeneic female Balb/c mice (Jackson Laboratories, Bar Harbor, ME, USA) (day 0). Starting 3 days later, mice were treated with 20mg/kg of lestaurtinib twice daily by subcutaneous injection, with 20 mg/kg sorafenib once daily or with a vehicle control. Mice were imaged by peritoneal injection of luciferin and visualized on an IVIS Spectrum imager (Caliper LifeSciences, Hopkinton, MA, USA) using Living Image software for analysis on days 3 and 8. All animal procedures were conducted in accordance with the policy of the Johns Hopkins University School of Medicine Animal Care and Use Committee.
Structural modeling of FLT3 TKI binding to resistance mutations Models of the FLT3 kinase complexed with lestaurtinib or midostaurin were constructed by superimposing the crystal structure of Lck kinase with staurosporine bound (Protein Data Bank (QJP)27 and superimposed on FLT3 (PDB code 1RJB).13 The resulting staurosporine placement relative to FLT3 serves as a suitable substitute for lestaurtinib and midostaurin binding owing to their related pharmacophore and the high homology of Lck with FLT3. Further confirmation was also provided by comparing lapatinib binding to EGFR (PDB code 1XKK).28 Likewise, FLT3 binding of sorafenib was modeled by superposing the FLT3 kinase structure on the raf kinase with sorafenib bound (PDB code 3GCS)29 and superimposed on the FLT3 crystal structure.13, 29 Images were generated and analyzed using PyMol.
RESULTS
Identification of resistance mutations
The scheme used to identify FLT3 resistance mutations is provided in Figure 1, and is based on a screen to identify resistance mutations in BCR-ABL.24 TF-1 cell colonies that grew in the presence of FLT3 TKI were picked for analysis. TF-1 cells do not normally express FLT3, so FLT3 was amplified by primers that covered the complementary DNA sequence and confirmed by sequencing. Cells were selected in methylceliulose at the minimum FLT3 TKI concentrations determined to inhibit growth of the parental FLT3/ITD cell colonies. These concentrations corresponded to 20 nm lestaurtinib, 20 nm midostaurin, 50 nm KW2449 and 100 nm AGS324 following a 2-week treatment. Despite efforts to reduce the frequency of clones generating resistance to FLT3 TKI caused by mechanisms unrelated to FLT3 mutations, only 9 out of 1500 clones analyzed harbored unique mutations of FLT3. Of the nine mutations, four were confirmed to confer resistance when site-directed mutagenesis was used to duplicate the isolated mutation in the FLT3/ITD backbone followed by transfection into BaF3 cells. The confirmed FLT3 TKI resistance mutations isolated were: F621L, A627P, F691L and Y842C. Approximately 600 colonies that were selected in the presence of lestaurtinib were sequenced and found not to contain resistance mutations. Of ~400 colonies that were selected in midostaurin, the A627P mutation was detected once as was the F691L mutation. Of 300 colonies sequenced from KW2449 treatment, the F621L mutation was identified once and the Y842C mutation 12 times. Of nearly 200 colonies grown in AGS324, the F621L mutation was selected once and the Y842C mutation was seen 23 times.
Figure 1.
Scheme used to identify FLT3/ITD resistance mutations. XL1-Red cells were transformed with the cFUGW plasmid carrying a human FLT3/ITD and grown overnight in agarose. The harvested DNA was used to transduce hematopoietic cells, which were then selected for resistance based on their ability to grow colonies in methylcellulose in the presence of a FLT3 TKI for 2 weeks. The resulting colonies were expanded in liquid culture growth medium, analyzed and sequenced for FLT3 mutations.
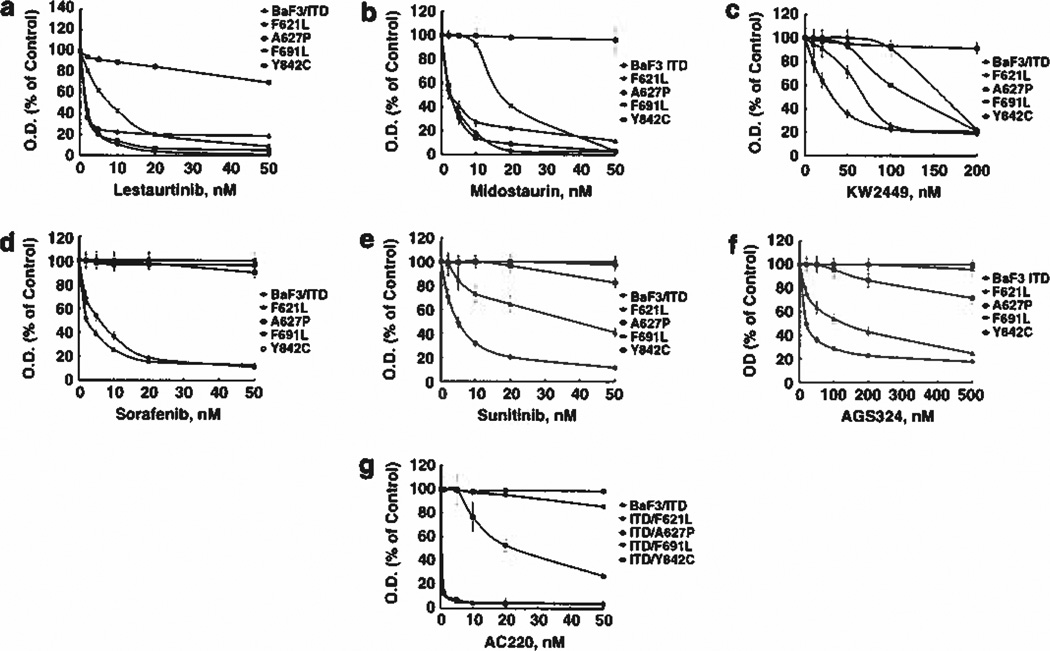
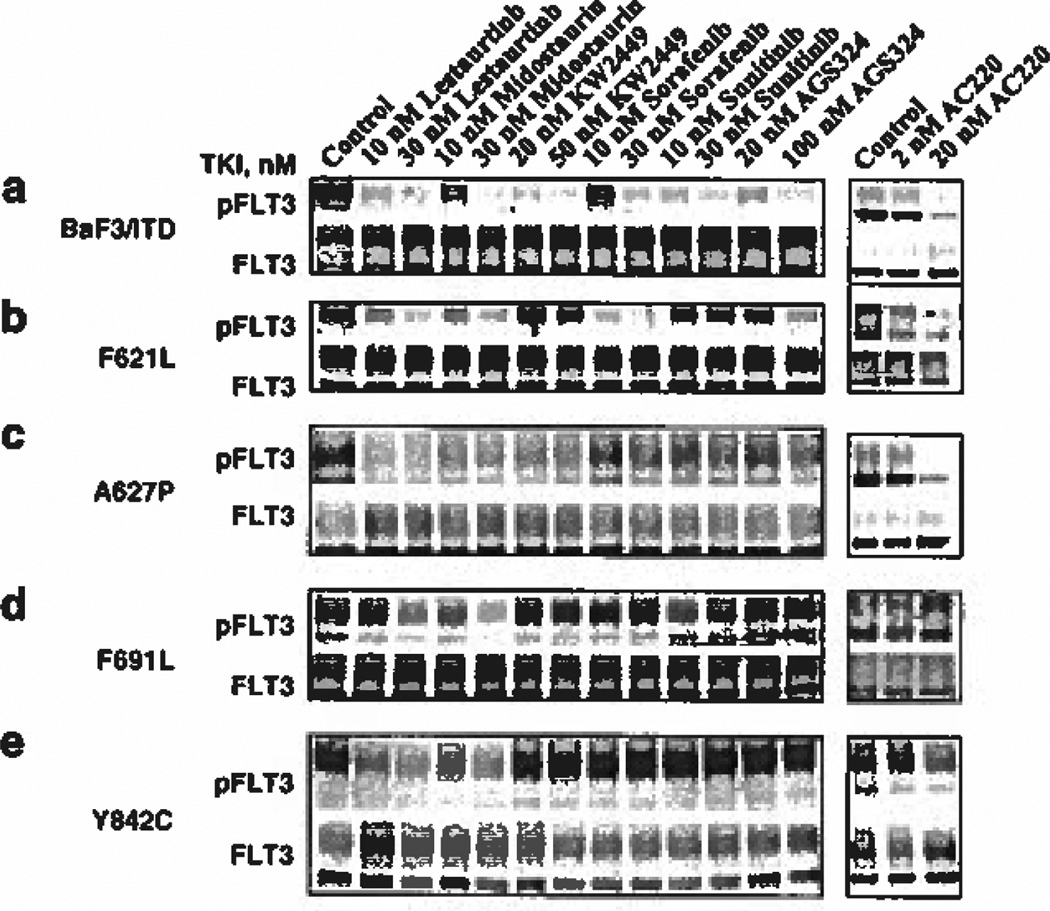
Each of the isolated mutations was then tested in BaF3 cells for the ability to engender resistance against six different FLT3 TKI as measured by MTT (growth/survival) assay. Lestaurtinib and midostaurin have undergone phase III clinical testing for treatment of FLT3 mutant AML. Lestaurtinib inhibited growth of BaF3 FLT3/ITD cells in the MTT assay with an IC50 of 2 nm (Figure 2a). The F621L and Y842C mutants displayed no resistance to this inhibitor, but the F691L mutant exhibited a fourfold increase in the IC50 (8 nm) to lestaurtinib. The A627P mutant resulted in such a large shift in the dose-response curve that the IC50 for lestaurtinib was not reached over the concentration range tested. Next, we examined the effect of lestaurtinib on FLT3 activation in each mutant directly by western blotting. Lestaurtinib demonstrated inhibitory activity on FLT3 autophosphorylation in all mutants, though some residual phosphorylation could be observed at 10 nm in the F621L and Y842C mutants (Figures 3a-e). The F691L mutant displayed resistance to lestaurtinib at 10 nm but was partially inhibited at 30 nm (Figure 3d). Thus, the western blotting results examining FLT3 inhibition by lestaurtinib for each mutant correlated well with the MTT data except for the A627P mutant, which showed sensitivity to inhibition of FLT3 phosphorylation by lestaurtinib but resistance to growth inhibition. Recently, it was shown that a mutation within BCR-ABL that inhibited its phosphorylation also led to inhibition of STAT5 phosphorylation but no effect on MAP kinase, AKT or other pathways resulted.30 This suggests a possible inherent property of the FLT3/ITD A627P conformation to act as a scaffold that permits substrate binding and activation that is compatible with reduced FLT3 kinase phosphorylation. Treatment of each resistant mutant with midostaurin revealed a similar pattern of resistance seen with lestaurtinib, with the F621L and Y842C mutants being as sensitive as the control BaF3/ITD cells (Figure 2b). Like the results obtained with lestaurtinib, the F691L mutant displayed partial resistance to midostaurin causing a fourfold increase in the IC50 (18 nm) for growth inhibition compared with BaF3/ITD cells (ICS0 of 4 nm). Like lestaurtinib, midostaurin was also ineffective at preventing proliferation in the A627P mutant. Midostaurin also exerted FLT3 inhibitory activity of each mutant, which mirrored the activity of lestaurtinib, with only the F691L mutant exhibiting partial resistance (Figures 3a-e). Thus, the MTT data are in agreement with the western blotting results, which show inhibition of FLT3 autophosphorylation by midostaurin associated with inhibition of proliferation of each mutant with the exception of the A627P mutant.
Figure 2.
Growth of BaF3 FLT3/ITD resistance mutants in the presence of FLT3 tyrosine kinase inhibitors. Resistance was confirmed by performing site-directed mutagenesis of FLT3/ITD and nucleoporation in BaF3 cells. The resulting clones were grown in the absence of interleukin-3 and analyzed by treatment in increasing concentrations of FLT3 TKI for 2 days, after which growth inhibition was assessed using the MTT assay. Means are representative of at least three independent experiments. Cells expressing FLT3 resistance mutations were treated with (a) Lestaurtinib, (b) Midostaurin, (c) KW2449, (d) Sorafenib, (e) Sunitinib, (f) AGS324 or (g) AC220.
Figure 3.
FLT3 tyrosine kinase inhibitors fail to inhibit FLT3 phosphorylation in resistant mutants. BaF3 FLT3/ITD resistance mutants were treated with the indicated concentrations of FLT3 TKI for 1 h. Lysates (500 µg) were immunoprecipitated with FLT3 antibody (S-18) and probed for activation using 4G10 anti-phosphotyrosine antibody. The membrane was then stripped and reprobed for FLT3 expression. (a) BaF3 FLT3/ITD cells, (b) FLT3/ITD F621L cells, (c) FLT3/ITD A627P cells, (d) FLT3/ITD F691L cells and (e) FLT3/ITD Y842C cells.
KW2449 recently underwent testing in phase II clinical trials targeting mutant FLT3. MTT results show growth inhibition of BaF3/ITD cells with an IC50 of 25 nm, but each of the mutants demonstrated degrees of resistance to this inhibitor (Figure 2c). KW2449 IC50 values shifted by 2-fold for the F621L mutant (50 nm), by > 8-fold for the A627P mutant (> 200 nm), by 7-fold for the F691L mutant (170 nm) and by 4-fold for the Y842C mutant (100 nm). Western blotting shows that KW2449 is ineffective at inhibiting FLT3 autophosphorylation of the F621L, F691L and Y842C resistant mutants at concentrations that inhibit FLT3/ITD activation in the BaF3 cells but it was able to inhibit FLT3 phosphorylation in the A627P mutant (Figures 3a-e).
Sorafenib and sunitinib are TKI, both approved for use in renal cell carcinoma, that have activity against FLT3/ITD mutants and are in clinical trials for FLT3/ITD AML. Sorafenib inhibited proliferation of BaF3 FLT3/ITD-expressing cells with an IC50 of 8 nm, and was actually more potent at growth inhibition of the F621L cells (IC50 of 2 nm) (Figure 2d). The IC50 for growth inhibition by sorafenib was not reached at the concentrations tested for the F691L, A627P and Y842C mutants. In agreement with the MTT results, western blotting confirmed that FLT3 autophosphorylation was inhibited by sorafenib in the BaF3/ITD and F621L mutant cells but was resistant to inhibition in each of the other mutant cell lines (Figures 3a-e).
Sunitinib inhibited growth of BaF3/ITD cells with an IC50 of 5 nm, but partial or complete resistance was observed for each of the mutant cell lines. The IC50 shifted to 30 nm for the F621L mutant and to > 50 nm for each of the A627P, F691L and Y842C mutants. Western blotting confirmed that sunitinib inhibited FLT3/ITD autophosphorylation in the BaF3/ITD cells, but had little to no effect on FLT3 activation in any of the resistant mutants (Figures 3a-e).
AGS324 inhibited proliferation of BaF3/ITD cells in the MTT assay with an IC50 of 20 nm but the IC50 shifted to 100 nm in the F621L mutant and > 500 nm in the A627P, F691L and Y842C mutants. These results are in agreement with western blotting results, which show that AGS324 partially inhibited FLT3/ITD autophosphorylation in BaF3/ITD cells at 20 nm and completely at 200 nm (Figures 3a-e). The F621L mutant was not affected at 20 nm but at 200 nm was mostly inhibited. No FLT3 inhibitory activity by AGS324 was observed at either concentration tested in the A627P, F691L or Y842E mutants.
While this paper was under review, a study was published that reported several FLT3 mutations causing resistance to AC220 in vitro and in patients.14 AC220 is one of the newest and most selective FLT3 TKI to achieve clinical trial status for AML. In light of the importance of AC220, we tested it against the mutations identified in our screen. Figure 2g shows that AC220 inhibited growth of the F621L mutant as potently as it did against BaF3 FLT3/ITD cells (IC50 < 1 nm). However, A627P and F691L mutant cells were completely unresponsive to treatment with AC220. The Y842C mutation caused the IC50 to shift to ~20 nm. Likewise, the response of each mutant cell line to growth inhibition by AC220 was paralleled by the responsiveness of each mutation to inhibition of FLT3 autophosphorylation and downstream signaling pathways (Figures 3 and 4).
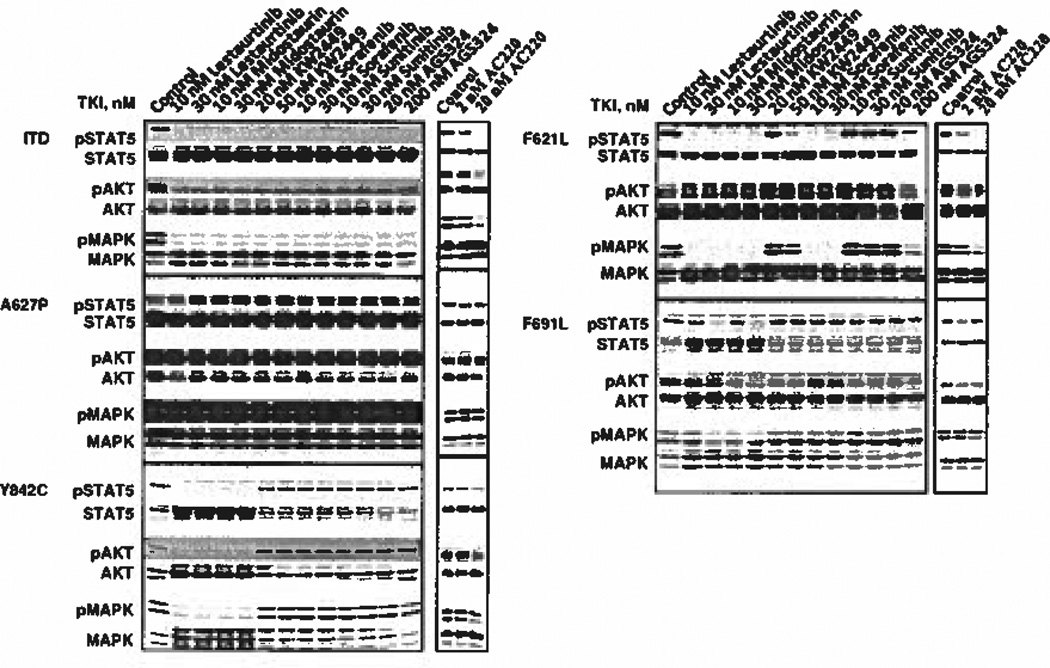
Figure 4.
FLT3 signaling remains intact in some resistant mutants when treated with a FLT3 tyrosine kinase inhibitor. BaF3 FLT3/ITD cells expressing the unmutated ITD, the F621L/ITD, the A627P/ITD, the F691L/ITD or the Y842C/ITD mutation were treated with the indicated concentrations of FLT3 TKI for 1 h. Cells were lysed in NP-40 buffer for 30 mins on ice, Lysates were separated on 8% SDS-PAGE gels, transferred to polyvinyl diflouride membranes using antibodies for pSTAT5, pAKT, pMAPK and their total protein counterparts and analyzed by enhanced chemiluminescence.
FLT3 signaling in resistant mutants remains intact in the presence of FLT3 TKI
In light of the evidence that many AML patients expressing mutant FLT3 acquire TKI resistance that cannot be accounted for by mutations within FLT3 itself, we examined FLT3 signaling pathways to determine whether resistance to FLT3 inhibition correlated with retention of FLT3 signaling pathways in the mutants identified in the present study. Figure 4a shows that in BaF3/ITD cells, each FLT3 TKI that was tested inhibited FLT3/ITD autophosphorylation and this resulted in termination of signaling through STAT5, AKT and MAP kinase pathways. Against the F621L mutant (Figure 4b), all three signaling pathways were completely inhibited by lestaurtinib, midostaurin and sorafenib, whereas KW2449 and AGS324 could only effect partial inhibition of downstream signaling events even at the higher concentrations. Sunitinib exhibited almost no inhibitory activity against any of the signaling pathways in this FLT3 mutant. The A627P mutant displayed strong activation of MAP kinase and AKT but a rather weak phosphorylated STAT5 signal (Figure 4c). Despite inhibition of FLT3/ITD autophosphorylation caused by treatment with several of the TKI, all pathways remained activated in the presence of each FLT3 TKI. This supports the idea that other mechanisms related to mutation at 627 contribute to resistance, similar to published results in which MAP kinase remained fully activated despite inhibition of FLT3/ITD A627E autophosphorylation by midostaurin.31 The FLT3/ITD F691L mutant displayed a low level of resistance to both lestaurtinib and midostaurin as was evident in the ability of these two FLT3 TKI to inhibit downstream signaling pathways only at the higher 30 nm concentrations but not at 10 nm (Figure 4d). Otherwise, STAT5, AKT and MAP kinase phosphorylation was largely unaffected when this mutant was treated even with high concentrations of KW2449, sorafenib, sunitinib or AGS324. Downstream signaling of the FLT3/ITD Y842C mutant showed a high level of resistance to treatment with KW2449, sorafenib, sunitinib and AGS324, but remained sensitive to lestaurtinib and midostaurin (Figure 4e). Figure 4e shows that the ability of this mutation to signal through STAT5, AKT and MAP kinase in the presence of TKI mirrored the results seen when FLT3 activation was probed. Lestaurtinib and midostaurin blocked all three signaling pathways in this mutant, which was consistent with the effects seen on FLT3 autophosphorylation.
FLT3/ITD Y842C causes resistance in vivo
The FLT3/ITD Y842C mutation produced high-level resistance to KW2449, sorafenib, sunitinib and AGS324 in vitro but remained sensitive to lestaurtinib and midostaurin. We evaluated this mutation further in vivo by transplanting luciferase-positive cells in syngeneic Balb/c mice and treating with a FLT3 TKI that was ineffective in vitro (sorafenib) and one that was effective (lestaurtinib). Bioluminescence emanating from transplanted luc + cells upon intraperitoneal injection of luciferin was used to quantify the level of FLT3/ITD engraftment in mice. Figure 5 shows that injection of BaF3 FLT3/ITD Y842C luc + cells in mice led to engraftment in the bone marrow that was visible by bioluminescence by day 3 (Figure 5a). Drug or lestaurtinib vehicle treatment of each group began at day 3 and continued until day 8. Bioluminescence increased dramatically in the control group from a mean flux of 22 800 photons/s counts on day 3 to 11 670 000 by day 8. As expected, based on the in vitro data, twice daily administration with lestaurtinib led to a significant reduction in the progression of the FLT3/ITD Y842C cells (P = 0.0007). In contrast, engraftment continued unabated in mice treated with sorafenib to levels similar to the control group (day 8 mean level of 10 000 000 photons/s). Figure 5b shows that lestaurtinib also significantly reduced mean spleen weights compared with control and sorafenib-treated mice (0.09g vs 1.02g and vs 0.93 g, P < 0.0001 vs both).
Figure 5.
FLT3/ITD Y842C confers resistance in vivo, (a) BaF3 FLT3/ITD Y842C luc + cells were transplanted via tail vein in syngeneic Balb/c mice on day 0. Bioluminescence was monitored by peritoneal injection of luciferin on day 3 to monitor engraftment. Treatment with sorafenib (20 mg/kg, once daily), lestaurtinib (20 mg/kg, twice daily) or lestaurtinib vehicle control began on day 3 and continued until day 8 to assess FLT3 TKI activity, (b) The treatments described in (a) were continued until day 15 when mice in the control group became sick at which point spleen weights were measured in all groups. Values expressed are the means ± s.e.m.
DISCUSSION
A number of FLT3 TKI have progressed to clinical trials for FLT3 AML32 Management of AML, however, is complicated by the fact that it is a multi-mutational disease with typically 8–10 somatic mutations per AML patient. This results in FLT3 inhibition alone often being insufficient to induce cell death of most FLT3-mutated AML blasts.33, 34 In contrast, early stage chronic myeloid leukemia is driven primarily by expression of the oncogenic BCR-ABL fusion protein, and targeting BCR-ABL kinase activity has been remarkably successful as a strategy to improve survival in chronic myelogenous leukemia patients.35–38 However, selection for point mutations in BCR-ABL conferring resistance via reduced binding is a mechanism that reduces efficacy, leading to relapse.39, 40
While nearly 100 mutations have been clinically described in chronic myelogenous leukemia patients that are at least partially resistant to imatinib,41 FLT3 TKI monotherapy has not selected for nearly as many mutations. There are a number of reasons for this with the major ones likely being: (1) a lack of success of first-generation FLT3 TKI in achieving thorough inhibition of FLT3 kinase activity. As more recent FLT3 TKI are more successful in vivo, the frequency of selecting for resistance mutations is increasing;14–16 (2) in FLT3/ITD AML, it appears there are 8–10 other mutations that contribute to leukemogenesis and so resistance is easily selected for via pathways that have nothing to do with resistance to inhibiting FLT3. Nevertheless, resistance mutations are likely to have an increasingly important role as FLT3 TKI that achieve targeted levels of inhibition are developed.
Identification of FLT3 resistance mutations is still at an early stage. One of the first in vitro screens identified resistance to midostaurin at four residues within the ATP-binding cleft of FLT3/ITD17 These included substitutions at A627, N676, F691 and G697 that conferred varying levels of resistance. A later study also identified mutations at N676 or F691 as mediators of resistance to midostaurin or sorafenib, respectively, and selection in SU5614 led to mutations mapping exclusively to TKD2.18 A recent saturation mutagenesis screen found mutations at D835, Y842 and F691 that caused resistance to AC220, and the authors also detected D835 and F691 in FLT3 mutant AML patients treated with AC220, in effect further validating the in vitro screening protocols.14
One of the most frequent FLT3/ITD resistance mutations discovered to date is the selection within the ITD allele of point mutations in the second half of the KD (TKD2). Activating point mutations within the KD of FLT3 occur in the absence of an ITD mutation at a rate of 7–10% in AML patients, and some of these are insensitive to some FLT3 TKI.42 Thus, the acquisition of such a mutation in a FLT3/ITD allele confers TKI resistance. The most common activating point mutations occur on the activation loop and they respond with differential sensitivity to various FLT3 TKI, though they all respond to both lestaurtinib and midostaurin. Emerging data are now implicating the acquisition of point mutations in TKD2 within a FLT3/ITD allele as a major source of resistance to sorafenib or AC220.14–16
Different FLT3 TKI appear to affect both the frequency of selection for resistance mutations and the residues affected. Compared with other FLT3 TKI, lestaurtinib and midostaurin are effective against more known FLT3 mutations, especially those occurring on the activation loop; in fact, no resistance mutations occurring in the second half of the KD have been reported in patients as being resistant to these two agents. In contrast, many of the mutations located on or near the activation loop show widely varying sensitivity to different FLT3 TKI. In this regard, lestaurtinib and midostaurin bear similarity to nilotinib and dasatanib, while the remaining FLT3 TKI more closely resemble imatinib. For example, treatment with imatinib generates numerous mutations throughout BCR-ABL that confer imatinib resistance, but only a handful of resistance mutations emerge while patients are on nilotinib or dasatanib, and these tend to be dominated by the T315I substitution.41 This has caused some to postulate that lestaurtinib and midostaurin may act as class I kinase inhibitors, which target the activated FLT3 conformation, while other FLT3 TKI act as class II inhibitors that bind to inactive FLT3, in analogy to structural studies of BCR-ABL in complex with imatinib43–45 With both FLT3/ITD and BCR-ABL, far more point mutations have been identified that are capable of conferring resistance to class II TKI compared with class I inhibitors. Thus, one might propose that FLT3/ITD AML patients who develop a FLT3 KD mutation that causes resistance to a class II inhibitor might still respond to a class I inhibitor. In fact, the A848P mutation within the FLT3/ITD allele that was selected for in one AML patient while being treated with sunitinib and sorafenib was shown to still be responsive to midostaurin in vitro.46 Another characteristic of FLT3 TKI that might be important in limiting the number of resistance mutations that emerge during treatment relates to the degree of selectivity of the inhibitor. Broad-spectrum inhibitors such as lestaurtinib would be expected to produce more toxkity at high concentrations in AML patients, but such inhibitors might reduce the number of resistance mutations that emerge during treatment compared with a more selective inhibitor.
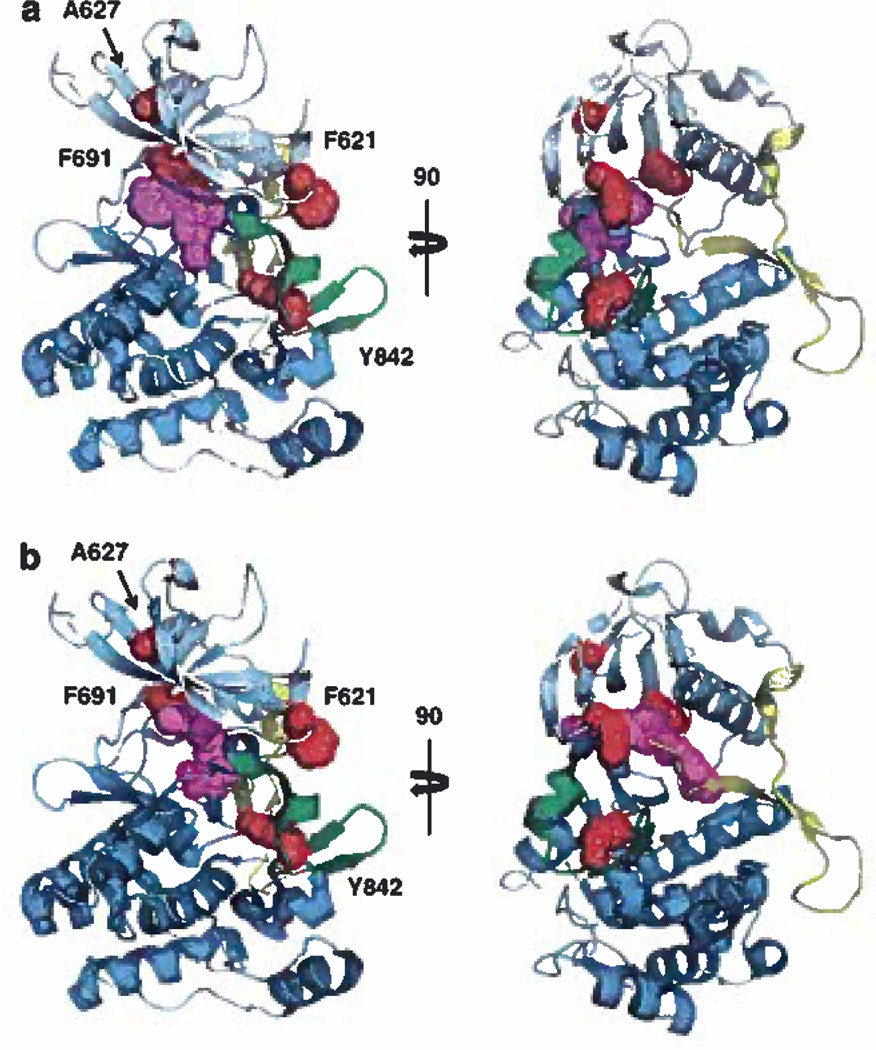
Modeling of FLT3 with inhibitors bound shows that F691 is the only residue in this study likely to make direct contact with FLT3 TKI (Figure 6). This position is analogous to the ‘gatekeeper’ T315 residue in BCR-ABL, which when mutated to isoleucine, confers pan-resistance to BCR-ABL TKI, including imatinib, nilotinib and dasatinib, by inhibiting drug binding. Our results suggest that mutation to leucine (F691L) probably would not result in steric hindrance because of the smaller size of the substituted side chain. However, mutation to leucine may result in weaker interactions with inhibitors, thus decreasing their affinity for FLT3.
Figure 6.
Structural modeling of FLT3 TKI with resistant mutants. (a) Lestaurtinib/midostaurin (magenta) binding to FLT3 was based on staurosporine bound to Lck kinase and superimposed on the FLT3 crystal structure. FLT3 TKI resistance mutations (red) were identified in the ATP-binding deft (F621L, A627P or F691L) or the activation loop (Y842C). The N-terminus is shown in pale cyan, the C-terminus in cyan, the juxtamembrane in yellow (partially removed for greater clarity) and the activation loop in green, (b) Sorafenib (magenta) binding to FLT3 was based on its atomic coordinates in complex with p38 Map Kinase. Images were generated using PyMol and are shown as orthogonal views and views rotated 90°.
Although F621 and A627 appear unlikely to make direct contact with FLT3 TKI in these models, both of these residues lie within the nucleotide-binding loop and their mutation may affect transitions from active to inactive forms. In fact, mutation to proline at position A627, located at the roof of the ATP-binding cleft, is predicted to restrict a backbone phi conformation in the β1-β2 region that is normally allowed by alanine. The F621L mutation displayed the lowest level of TKI resistance, which may not be surprising considering that F621 is further outside of the cleft and that leucine is less bulky than phenylalanine. Figure 6 shows that neither staurosporine nor sorafenib is likely to contact F621, which is consistent with the biological data showing inhibition of kinase activity in this mutation.
Y842 is located on the activation loop of the carboxy terminal kinase lobe, so assessing the effects of a residue so far removed from the ATP-binding site is complicated by the lack of crystal structures of different activated FLT3 conformations. However, the activation loops in related kinases adopt a wide range of conformations. In the autoinhibited state, the unphosphorylated activation loop is folded between the amino- (N) and carboxy- (C) terminal kinase lobes and prevents ATP binding by contacting the JM-binding motif that is embedded between the N and C lobes. In activated kinases, the tyrosine-phosphorylated activation loop swings outward, thus permitting access of ATP and substrate to the active site. The Y842C mutation likely alters the ensemble of activation loop conformations such that ATP can still enter the nucleotide-binding site but sorafenib is less able to align itself in its preferred orientation. Structural modeling shows that sorafenib does in fact extend further into the ATP cleft than midostaurin and occupies an adjacent site within FLT3. Sorafenib also clashes with the JM domain in a way that midostaurin does not.
The FLT3/ITD Y842C mutant might pose significant challenges to treatment with some TKI because it was highly resistant to both sunitinib and sorafenib. It is important to note that five AML patients have been identified with activating mutations of FLT3 Y842C or N841.47, 48 This repeated occurrence strongly suggests that these mutations are likely to be more frequently encountered as a mechanism of resistance to select FLT3 TKI monotherapy (such as sorafenib and sunitinib). The FLT3/ITD A627P mutation was atypical in that it imparted resistance to growth inhibition to all inhibitors despite the finding that FLT3 autophosphorylation was inhibited by several TKI, similar to a report that an AML patient who was on midostaurin progressed due to outgrowth of a leukemic clone bearing a FLT3/ITD A627E mutation.31 In that report, FLT3 autophosphorylation was inhibited by midostaurin but MAP kinase phosphorylation remained intact. When we combined lestaurtinib with the MEK inhibitor, U0126, a modest suppression of the FLT3/ITD A627E mutant cells resulted, which indicates that the AKT and/or STAT5 pathways activated by FLT3/ITD are still providing some survival/proliferation advantages. These findings point to additional mechanisms of resistance, both related and unrelated to FLT3 mutations, as has been reported.20, 49–51 The relative rate of occurrence of different mechanisms of resistance in patients treated on FLT3 TKI is not known, but mutations are being detected at a greater rate as successful inhibition of FLT3 kinase activity is achieved. Until recently, only three resistance mutations had been reported, while AML patients were being treated with a TKI and only the FLT3/ITD N676K mutation actually imparted resistance to inhibition of FLT3 phosphorylation by midostaurin.52 While this manuscript was in preparation, additional resistance mutations were identified clinically in FLT3 mutant AML patients treated with the FLT3 TKI sorafenib or AC220. The recently detected mutations include substitutions at D835 and one of the mutations identified in our screen, F691L, all of which occurred as secondary mutations within the FLT3/ITD allele. The increasing number of AML patients who are found to express FLT3 resistance mutations following TKI treatment provides compelling evidence that mutations such as FLT3/ITD F621L, A627P, F691L and Y842C found in the present screen and others may lead to significant therapeutic challenges. As the list continues to grow, a knowledge of which FLT3 TKI are still active against these mutations will be required to better manage the emergence of resistance cases. The results presented here indicate that some FLT3 TKI will be less effective against several FLT3 resistance mutants and that even lestaurtinib and midostaurin, which target the greatest range of mutations, will exhibit reduced potency against some clones. Thus, there is a need to profile FLT3/ITD mutations for their ability to confer resistance to FLT3 TKI and a need to expand the repertoire of inhibitors available for clinical use against emerging TKI-resistant clones.
ACKNOWLEDGEMENTS
We are grateful to Dr Linzhao Cheng (Johns Hopkins University) for providing us with the L3GFP vector used to visualize FLT3/ITD engraftment and to members of the lab for numerous thoughtful discussions. This work was supported by grants from the NCI (CA90770 and CA90668), Leukemia and Lymphoma Society and Giant Food Pediatric Cancer Research Fund. DS is also supported by the Kyle Haydock Professorship.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
ABW designed experiments, performed research, analyzed data and wrote the manuscript; LL and BN performed research; ML analyzed data; PB and DL analyzed data and wrote the manuscript; DS designed experiments, supervised the project, analyzed data and wrote the manuscript.
REFERENCES
- 1.Small D, Levenstein M, Kim E, Carrow C, Amin S, Rockwell P, et al. STK-1, the human homolog of Flk-2/Flt-3, is selectively expressed in CD34 + human bone marrow cells and is involved in the proliferation of early progenitor stem cells. Proc Natl Acad Sci USA. 1994;91:459–463. doi: 10.1073/pnas.91.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosnet O, Schiff C, Pebusque MJ, Marchetto S, Tonnelle C, Toiron Y, et al. Human FLT3/FLK2 gene: cDNA cloning and expression in hematopoietic cells. Blood. 1993;82:1110–1119. [PubMed] [Google Scholar]
- 3.Lavagna-Sevenier C, Marchetto S, Birnbaum D, Rosnet O. FLT3 signaling in hematopoietic cells involves CBL, SHC and an unknown P115 as prominent tyrosine-phosphorybted substrates. Leukemia. 1998;12:301–310. doi: 10.1038/sj.leu.2400921. [DOI] [PubMed] [Google Scholar]
- 4.Rosnet O, Buhring HJ, deLapeyriere O, Beslu N, Lavagna C, Marchetto S, et al. Expression and signal transduction of the FLT3 tyrosine kinase receptor. Acta Haematol. 1996;95:218–223. doi: 10.1159/000203881. [DOI] [PubMed] [Google Scholar]
- 5.Lyman SD, James L, Vanden Bos T, de Vries P, Brasel K, Gliniak B, et al. Molecular cloning of a ligand for the flt3/flk-2 tyrosine kinase receptor: a proliferative factor for primitive hematopoietic cells. Cell. 1993;75:1157–1167. doi: 10.1016/0092-8674(93)90325-k. [DOI] [PubMed] [Google Scholar]
- 6.Zhang S, Broxmeyer HE. Flt3 ligand induces tyrosine phosphorylation of gab1 and gab2 and their association with shp-2, grb2, and PI3 kinase. Biochem Biophys Res Commun. 2000;277:195–199. doi: 10.1006/bbrc.2000.3662. [DOI] [PubMed] [Google Scholar]
- 7.Brown P, Levis M, Shurtleff S, Campana D, Downing J, Small D. FLT3 inhibition selectively kills childhood acute lymphoblastic leukemia cells with high levels of FLT3 expression. Blood. 2005;105:812–820. doi: 10.1182/blood-2004-06-2498. [DOI] [PubMed] [Google Scholar]
- 8.Armstrong SA, Staunton JE, Silverman LB, Pietere R, den Boer ML, Minden MD, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 9.Zheng R, Levis M, Piloto O, Brown P, Baldwin BR, Gorin NC, et al. FLT3 ligand causes autocrine signaling in acute myeloid leukemia cells. Blood. 2004;103:267–274. doi: 10.1182/blood-2003-06-1969. [DOI] [PubMed] [Google Scholar]
- 10.Nakao M, Yokota S, Iwai T, Kaneko H, Horlike S, Kashima K, et al. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia. 1996;10:1911–1918. [PubMed] [Google Scholar]
- 11.Abu-Duhier FM, Goodeve AC, Wilson GA, Care RS, Peake IR, Reilly JT. Identification of novel FLT-3 Asp835 mutations in adult acute myeloid leukaemia. Br J Haematol. 2001;113:983–988. doi: 10.1046/j.1365-2141.2001.02850.x. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto Y, Kiyoi H, Nakano Y, Suzuki R, Kodera Y, Miyawaki S, et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood. 2001;97:2434–2439. doi: 10.1182/blood.v97.8.2434. [DOI] [PubMed] [Google Scholar]
- 13.Griffith J, Black J, Faerman C, Swenson L, Wynn M, Lu F, et al. The structural basis for autoinhibition of FLT3 by the juxtamembrane domain. Mol Cell. 2004;13:169–178. doi: 10.1016/s1097-2765(03)00505-7. [DOI] [PubMed] [Google Scholar]
- 14.Smith CC, Wang Q, Chin CS, Salerno S, Damon LE, Levis MJ, et al. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature. 2012;485:260–263. doi: 10.1038/nature11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alvarado Y, Kantarjian H, Ravandi F, Luthra R, Borthakur G, Manero GG, et al. FLT3 inhibitor treatment in FLT3-mutated AML is associated with development of secondary FLT3-TKD mutations. Blood. 2011;118:1493. [Google Scholar]
- 16.Zhang W, Konopleva M, Jacamo RO, Borthakur G, Chen W, Cortes JE, et al. Acquired point mutations of TKD are responsible for sorafenib resistance in FLT3-ITD mutant AML. Blood. 2011:118. [Google Scholar]
- 17.Cools J, Mentens N, Furet P, Fabbro D, Clark JJ, Griffin JD, et al. Prediction of resistance to small molecule FLT3 inhibitors: implications for moleculariy targeted therapy of acute leukemia. Cancer Res. 2004;64:6385–6389. doi: 10.1158/0008-5472.CAN-04-2148. [DOI] [PubMed] [Google Scholar]
- 18.von Bubnoff N, Engh RA, Aberg E, Sanger J, Peschel C, Duyster J. FMS-like tyrosine kinase 3-internal tandem duplication tyrosine kinase inhibitors display a nonoverlapping profile of resistance mutations in vitro. Cancer Res. 2009;69:3032–3041. doi: 10.1158/0008-5472.CAN-08-2923. [DOI] [PubMed] [Google Scholar]
- 19.Bagrintseva K, Schwab R, Kohl TM, Schnittger S, Eichenlaub S, Ellwart JW, et al. Mutations in the tyrosine kinase domain of FLT3 define a new molecular mechanism of acquired drug resistance to PTK inhibitors in FLT3-ITD transformed hematopoietic cells. Blood. 2004;103:2266–2275. doi: 10.1182/blood-2003-05-1653. [DOI] [PubMed] [Google Scholar]
- 20.Zhou J, Bi C, Janakakumara JV, Liu SC, Chng WJ, Tay KG, et al. Enhanced activation of STAT pathways and overexpression of survivin confer resistance to FLT3 inhibitors and could be therapeutic targets in AML. Blood. 2009;113:4052–4062. doi: 10.1182/blood-2008-05-156422. [DOI] [PubMed] [Google Scholar]
- 21.Stolzel F, Steudel C, Oelschlagel U, Mohr B, Koch S, Ehninger G, et al. Mechanisms of resistance against PKC412 in resistant FLT3-ITD positive human acute myeloid leukemia cells. Ann Hematol. 89:653–662. doi: 10.1007/s00277-009-0889-1. [DOI] [PubMed] [Google Scholar]
- 22.Prescott H, Kantarjian H, Cortes J, Ravandi F. Emerging FMS-like tyrosine kinase 3 inhibitors for the treatment of acute myelogenous leukemia. Expert Opin Emerg Drugs. 2011;16:407–423. doi: 10.1517/14728214.2011.568938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tse KF, Allebach J, Levis M, Smith BD, Bohmer FD, Small D. Inhibition of the transforming activity of FLT3 internal tandem duplication mutants from AML patients by a tyrosine kinase inhibitor. Leukemia. 2002;16:2027–2036. doi: 10.1038/sj.leu.2402674. [DOI] [PubMed] [Google Scholar]
- 24.Azam M, Latek RR, Daley GQ. Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell. 2003;112:831–843. doi: 10.1016/s0092-8674(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 25.Levis M, Allebach J, Tse KF, Zheng R, Baldwin BR, Smith BD, et al. A FLT3-targeted tyrosine kinase inhibitor is cytotoxic to leukemia cells in vitro and in vivo. Blood. 2002;99:3885–3891. doi: 10.1182/blood.v99.11.3885. [DOI] [PubMed] [Google Scholar]
- 26.Tse KF, Mukherjee G, Small D. Constitutive activation of FLT3 stimulates multiple intracellular signal transducers and results in transformation. Leukemia. 2000;14:1766–1776. doi: 10.1038/sj.leu.2401905. [DOI] [PubMed] [Google Scholar]
- 27.Zhu X, Kim JL, Newcomb JR, Rose PE, Stover DR, Toledo LM, et al. Structural analysis of the lymphocyte-specific kinase Lck in complex with non-selective and Src family selective kinase inhibitors. Structure. 1999;7:651–661. doi: 10.1016/s0969-2126(99)80086-0. [DOI] [PubMed] [Google Scholar]
- 28.Wood ER, Truesdale AT, McDonald OB, Yuan D, Hassell D, Dickerson SH, et al. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004;64:6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- 29.Simard JR, Grutter C, Pawar V, Aust B, Wolf A, Rabiller M, et al. High-throughput screening to identify inhibitors which stabilize inactive kinase conformations in p38alpha. J Am Chem Soc. 2009;131:18478–18488. doi: 10.1021/ja907795q. [DOI] [PubMed] [Google Scholar]
- 30.Grebien F, Hantschel O, Wojcik J, Kaupe I, Kovacic B, Wyrzucki AM, et al. Targeting the SH2-kinase interface in Bcr-Abl inhibits leukemogenesis. Cell. 2011;147:306–319. doi: 10.1016/j.cell.2011.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breitenbuecher F, Markova B, Kasper S, Carius B, Stauder T, Bohmer FD, et al. A novel molecular mechanism of primary resistance to FLT3-kinase inhibitors in AML. Blood. 2009;113:4063–4073. doi: 10.1182/blood-2007-11-126664. [DOI] [PubMed] [Google Scholar]
- 32.el-Shami K, Stone RM, Smith BD. FLT3 inhibitors in acute myeloid leukemia. Expert Rev Hemotol. 2008;1:153–160. doi: 10.1586/17474086.1.2.153. [DOI] [PubMed] [Google Scholar]
- 33.Knapper S, Mills KI, Gilkes AF, Austin SJ, Walsh V, Burnett AK. The effects of lestaurtinib (CEP701) and PKC412 on primary AML blasts: the induction of cytotoxicity varies with dependence on FLT3 signaling in both FLT3-mutated and wild-type cases. Blood. 2006;108:3494–3503. doi: 10.1182/blood-2006-04-015487. [DOI] [PubMed] [Google Scholar]
- 34.Siendones E, Barbarroja N, Torres LA, Buendia P, Velasco F, Dorado G, et al. Inhibition of Flt3-activating mutations does not prevent constitutive activation of ERK/Akt/STAT pathways in some AML cells: a possible cause for the limited effectiveness of monotherapy with small-molecule inhibitors. Hematol Oncol. 2007;25:30–37. doi: 10.1002/hon.805. [DOI] [PubMed] [Google Scholar]
- 35.Kurzrock R, Talpaz M. The molecular pathology of chronic myelogenous leukaemia. Br J Haematol. 1991;79(Suppl 1):34–37. doi: 10.1111/j.1365-2141.1991.tb08116.x. [DOI] [PubMed] [Google Scholar]
- 36.Druker BJ. Current treatment approaches for chronic myelogenous leukemia. Cancer J. 2001;7(Suppl 1):S14–S18. [PubMed] [Google Scholar]
- 37.Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 38.O’Brien SG, Guilhot F, Larson RA, Grathman I, Baccarani M, Cervantes F, et al. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2003;348:994–1004. doi: 10.1056/NEJMoa022457. [DOI] [PubMed] [Google Scholar]
- 39.Apperley JF. Part I. mechanisms of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol. 2007;8:1018–1029. doi: 10.1016/S1470-2045(07)70342-X. [DOI] [PubMed] [Google Scholar]
- 40.O’Hare T, Deininger MW, Eide CA, Clackson T, Druker BJ. Targeting the BCR-ABL signaling pathway in therapy-resistant Philadelphia chromosome-positive leukemia. Clin Cancer Res. 2011;17:212–221. doi: 10.1158/1078-0432.CCR-09-3314. [DOI] [PubMed] [Google Scholar]
- 41.Soverini S, Hochhaus A, Nicolini FE, Gruber F, Lange T, Saglio g, et al. BCR-ABL kinase domain mutation analysis in chronic myeloid leukemia patients treated with tyrosine kinase inhibitors: recommendations from an expert panel on behalf of European LeukemiaNet. Blood. 2011;118:1208–1215. doi: 10.1182/blood-2010-12-326405. [DOI] [PubMed] [Google Scholar]
- 42.Clark JJ, Cools J, Curley DP, Yu JC, Lokker NA, Giese NA, et al. Variable sensitivity of FLT3 activation loop mutations to the small molecule tyrosine kinase inhibitor MLNS18. Blood. 2004;104:2867–2872. doi: 10.1182/blood-2003-12-4446. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y, Gray NS. Rational design of inhibitors that bind to inactive kinase conformations. Nat Chem Biol. 2006;2:358–364. doi: 10.1038/nchembio799. [DOI] [PubMed] [Google Scholar]
- 44.Cowan-Jacob SW, Fendrich G, Floersheimer A, Furet P, Liebetanz J, Rummel G, et al. Structural biology contributions to the discovery of drugs to treat chronic myelogenous leukaemia. Acta Crystallogr D Biol Crystaltogr. 2007;63:80–93. doi: 10.1107/S0907444906047287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weisberg E, Roesel J, Furet P, Bold G, Imbach P, Florsheimer J, et al. Antileukemic effects of novel first- and second-generation FLT3 inhibitors: structure-affinity comparison. Genes Cancer. 2010;1:1021–1032. doi: 10.1177/1947601910396505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Bubnoff N, Rummelt C, Menzel H, Sigl M, Peschel C, Duyster J. Identification of a secondary FLT3/A848P mutation in a patient with FLT3-ITD-positive blast phase CMML and response to sunitinib and sorafenib. Leukemia. 2010;24:1523–1525. doi: 10.1038/leu.2010.122. [DOI] [PubMed] [Google Scholar]
- 47.Kindler T, Breitenbuecher F, Kasper S, Estey E, Giles F, Feldman E, et al. Identification of a novel activating mutation (Y842C) within the activation loop of FLT3 in patients with acute myeloid leukemia (AML) Blood. 2005;105:335–340. doi: 10.1182/blood-2004-02-0660. [DOI] [PubMed] [Google Scholar]
- 48.Jiang J, Paez JG, Lee JC, Bo R, Stone RM, DeAngelo DJ, et al. Identifying and characterizing a novel activating mutation of the FLT3 tyrosine kinase in AML. Blood. 2004;104:1855–1858. doi: 10.1182/blood-2004-02-0712. [DOI] [PubMed] [Google Scholar]
- 49.Mony U, Jawad M, Seedhouse C, Russell N, Pallis M. Resistance to FLT3 inhibition in an in vitro model of primary AML cells with a stem cell phenotype in a defined microenvironment. Leukemia. 2008;22:1395–1401. doi: 10.1038/leu.2008.125. [DOI] [PubMed] [Google Scholar]
- 50.Sato T, Yang X, Knapper S, White P, Smith BD, Galkin S, et al. FLT3 ligand impedes the efficacy of FLT3 inhibitors in vitro and in vivo. Blood. 2011;117:3286–3293. doi: 10.1182/blood-2010-01-266742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshimoto G, Miyamoto T, Jabbarzadeh-Tabrizi S, Iino T, Rocnik JL, Kikushige Y, et al. FLT3-ITD up-regulates MCL-1 to promote survival of stem cells in acute myeloid leukemia via FLT3-ITD specific STAT5 activation. Blood. 2009;114:5034–5043. doi: 10.1182/blood-2008-12-196055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heidel F, Solem FK, Breitenbuecher F, Lipka DB, Kasper S, Thiede MH, et al. Clinical resistance to the kinase inhibitor PKC412 in acute myeloid leukemia by mutation of Asn-676 in the FLT3 tyrosine kinase domain. Blood. 2006;107:293–300. doi: 10.1182/blood-2005-06-2469. [DOI] [PubMed] [Google Scholar]