Summary
Radial glial progenitors (RGPs) are elongated epithelial cells which give rise to neurons, glia, and adult stem cells during brain development. RGP nuclei migrate basally during G1, apically using cytoplasmic dynein during G2, and undergo mitosis at the ventricular surface. By live imaging of in utero electroporated rat brain, we find that two distinct G2-specific mechanisms for dynein nuclear pore recruitment are essential for apical nuclear migration. The “RanBP2-BicD2” and “Nup133-CENP-F” pathways act sequentially, with Nup133 or CENP-F RNAi arresting nuclei close to the ventricular surface in a pre-mitotic state. Forced targeting of dynein to the nuclear envelope rescues nuclear migration and cell cycle progression, demonstrating that apical nuclear migration is not simply correlated with cell cycle progression from G2 to mitosis, but rather, is a required event. These results reveal that cell cycle control of apical nuclear migration occurs by motor recruitment, and identify a role for nucleus- and centrosome-associated forces in mitotic entry.
Radial glial progenitor (RGP) cells are precursors for the majority of neurons and glia in the vertebrate neocortex, as well as for adult stem cells (Götz and Huttner, 2005; Kriegstein and Alvarez-Buylla, 2009). RGPs are elongated epithelial cells which span the neural tube and developing cortex from the ventricular to the pial surface. They are highly proliferative (Noctor et al., 2001), but also serve as tracks for the migration of postmitotic neurons (Rakic, 1988). For these reasons, these cells play a uniquely important role in the development of the nervous system.
RGP cells also exhibit a distinctive and, until recently, largely mysterious form of cell-cycle dependent oscillatory nuclear movement known as interkinetic nuclear migration (INM) (Kosodo, 2012; Lee and Norden, 2012; Sauer, 1935; Spear and Erickson, 2012a; Taverna and Huttner, 2010). Mitotic divisions of RGP cells occur at the apical end of the cell, close to the ventricular surface of the developing neocortex (Figure 3A). The nuclei of RGP cells then ascend “basally” during G1, undergo S phase, and return apically to the ventricular surface during G2, where they again undergo mitosis. INM is a conserved form of behavior observed in multiple species and in the development of various tissues (Kishimoto et al., 2013), including mammalian and zebrafish neocortex and retina (Leung et al., 2011) and Drosophila imaginal disc (Meyer et al., 2011). The developmental purpose of this behavior is unknown, though it has been suggested that it contributes to cell fate regulation (Del Bene et al., 2008) or to maximize the packing density of proliferating cells (Kosodo, 2012).
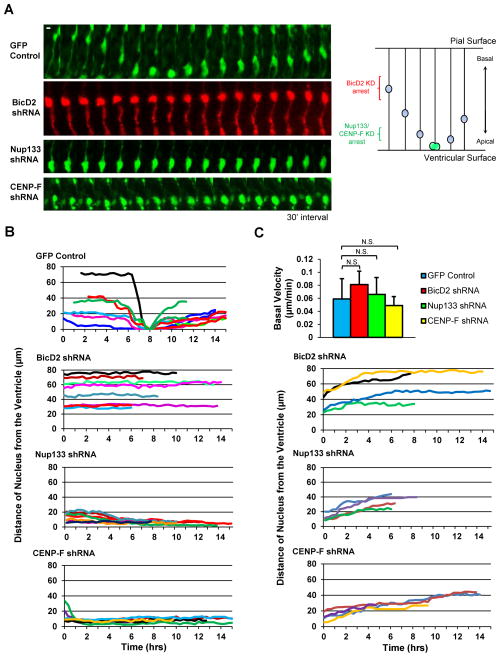
Figure 3. RNAi for Dynein NE Recruitment Factors Inhibits Apical nuclear migration.
A. E16 rat embryonic brains were subjected to in utero electroporation to express shRNAs corresponding to BicD2, Nup133, or CENP-F. Brain slices were placed into culture at E20 for live imaging over an 8–15 hr period. Control RGP cell is shown undergoing apical nuclear migration to the ventricular surface of the brain slice, followed by mitosis and basal migration. BicD2, Nup133, and CENP-F shRNAs each caused nuclear arrest. Model of INM depicted on right. Scale bar = 5 μm. B. Tracings of nuclei in Nup133, CENP-F, or BicD2 shRNA-expressing RGP cells show severe impairment of apical migration, with nuclei in BicD2 shRNA cells arresting further from the ventricular surface. C. No clear effect on basal nuclear migration was observed. Velocity is net distance/time. See also Figure S5 and Movie S1–S7.
The underlying mechanisms responsible for INM, its relationship to cell cycle progression, and the basis for spatial control of mitosis remained largely unaddressed until recently. We previously reported roles for microtubule motor proteins in INM (Tsai et al., 2005; 2010). By live imaging of the rat brain, we observed that centrosomes of RGP cells remain at the ventricular terminus throughout INM (Tsai et al., 2010). Microtubules were almost uniformly oriented with their minus ends directed toward the ventricular surface and their plus ends oriented basally. Consistent with this arrangement, we found that RNAi for the microtubule plus end-directed kinesin, KIF1A, specifically inhibited basal nuclear migration, whereas RNAi for cytoplasmic dynein and its regulator LIS1 specifically inhibited apical nuclear migration (Tsai et al., 2010). Another study found that inhibition of the dynein-cofactor dynactin interferes with apical, but stimulates basal nuclear migration in zebrafish retinal neuroepithelial cells (Del Bene et al., 2008). Roles for myosin II in INM in that system (Norden et al., 2009) and in basal nuclear migration in the embryonic mouse neocortex have also been reported (Schenk et al., 2009). No such role was detected in our own rat brain studies (Tsai et al., 2010), and the basis for the divergent results remains uncertain. A role for microtubules in the early stages of vertebrate brain development has also been supported by RNAi for diverse centrosomal and microtubule associated proteins (Ge et al., 2010; Kosodo et al., 2011; Yang et al., 2012).
Although centrosomes remain associated with nuclei during migration in a wide range of cell types, the centrosome-independent nuclear migration we have observed in rat brain RGP cells (Tsai et al., 2010) suggests that motors might act locally from the nuclear surface. Such a mechanism has been implicated in the transport of nuclei within mammalian myotubes (Cadot et al., 2012; Wilson and Holzbaur, 2012) and C. elegans hypodermal cells (Fridolfsson and Starr, 2010). In the latter case, cytoplasmic dynein is recruited to the nuclear envelope (NE) by a combination of nesprin and SUN proteins (the “LINC” complex), which together span the outer and inner NE (Fridolfsson et al., 2010; Starr and Fridolfsson, 2010). Members of these gene families have also been implicated in neuronal migration in the developing mouse brain using genetic and RNAi approaches (Zhang et al., 2009).
Additional mechanisms for dynein recruitment to the NE have been identified in G2 phase HeLa and U2OS cells. NE dynein has been reported to facilitate NE breakdown (NEB) (Beaudouin et al., 2002; Hebbar et al., 2008; Salina et al., 2002) and tether the nascent mitotic spindle to the NE for pole separation (Raaijmakers et al., 2012) and efficient chromosome capture (Bolhy et al., 2011; Jodoin et al., 2012; Splinter et al., 2010). Dynein is linked to the NE via interactions originating from two distinct nuclear pore components (Figure 1A–1B). The nucleoporin RanBP2 recruits BicD2, which, in turn, recruits both cytoplasmic dynein and its regulatory complex dynactin to the nuclear surface (Splinter et al., 2012; 2010). Nup133, another nucleoporin, independently recruits CENP-F (Bolhy et al., 2011). CENP-F, in turn, recruits NudE and NudEL, each of which bind directly to cytoplasmic dynein and its regulator LIS1 (Mckenney et al., 2010; Niethammer et al., 2000; Sasaki et al., 2000). This latter mechanism for dynein recruitment becomes active in late G2-prophase (Bolhy et al., 2011), though the extent of temporal overlap with the BicD2 pathway is uncertain.
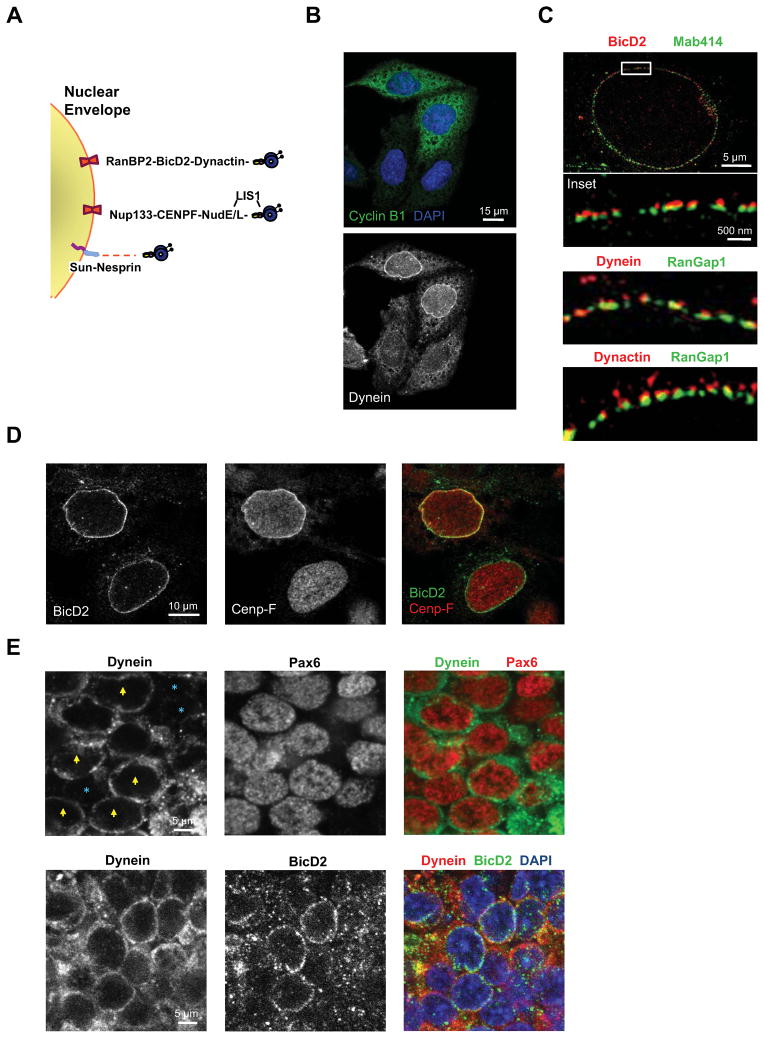
Figure 1. Mechanisms for cytoplasmic dynein recruitment to the nuclear envelope.
A. Diagram representing G2-specific NE dynein recruitment mechanisms via nucleoporins Nup133 and RanBP2. Dynein is also shown linked to the NE by SUN-nesprin complexes, a mechanism not known to be cell cycle regulated. B. Triple staining with anti-dynein, anti-Cyclin B1, and DAPI (DNA) showing NE dynein localization specifically in cyclin B1 expressing HeLa cells. C. 3D-Structured Illumination Microscopy (3D-SIM) revealing the association of BicD2, dynein, and dynactin with HeLa cell nuclear pores, marked using Mab414 and anti-RanGap1. D. HeLa cells were double-labeled with anti-BicD2 and anti-CENP-F antibodies to test for temporal overlap between the two cell cycle-dependent NE dynein recruitment mechanisms. All cells exhibiting CENP-F-positive NEs were also positive for BicD2, but only a fraction of BicD2-positive cells showed NE CENP-F staining. E. NE labeling in the ventricular zone (VZ) in E19 rat brain sections. Top row, NE dynein staining was seen in a subset of RGP cells (yellow arrowhead), but absent in others (blue asterisks). RGP cells were identified by Pax6 immunostaining. Bottom, dynein and BicD2 colocalize at the NE, with many BicD2-positive cells also positive for dynein. Confocal microscopy was used throughout unless otherwise stated. See also Figure S1, S2, and S4A.
We reasoned that, because apical INM is dynein-dependent and occurs during G2, related mechanisms might play a role in INM in the developing brain. The mer mouse, which has a missense mutation in Nup133, in fact, shows defects in early embryonic brain development, though the underlying mechanism was not explored (Lupu et al., 2008). We report here that interference with each of the G2-specific dynein recruitment pathways specifically inhibits apical, but not basal nuclear migration, providing evidence that these pathways are important in brain development and supporting a role for NE dynein in INM. Inhibition of the BicD2 and Nup133 pathways arrests nuclei early vs. late in apical migration, respectively, and in each case, in a premitotic state. Forced recruitment of dynein to the NE rescues apical migration and provides experimental evidence for spatial control of mitotic entry. These results provide insight into the role of microtubule motor proteins in INM, and the first clues into the mechanism for its cell cycle control.
Results
Relative roles of NE dynein recruitment factors in non-neural cells in vitro
As a basis for interpreting the relative roles of the three NE dynein recruitment mechanisms (Figure 1A) in vivo, we examined their temporal interrelationship further. The RanBP2-BicD2 and Nup133-CENP-F pathways (hereafter termed the “BicD2” and “Nup133” pathways, referring to the most upstream component targeted in this study) have each been implicated in G2-mediated force generation between the centrosome-centered microtubule array and the NE (Beaudouin et al., 2002; Bolhy et al., 2011; Salina et al., 2002; Splinter et al., 2010). The participation of Nup133 and RanBP2 in the two pathways suggested that dynein might directly localize to nuclear pores, rather than to the entire NE surface. Using 3D-structured illumination microscopy (3D-SIM), we found that dynein, dynactin, and BicD2 appeared as discrete puncta along the NE (Figure 1C). The puncta were significantly associated with the nuclear pores in G2 HeLa cells (Figure 1C and S1), supporting specific nuclear pore associations underlying dynein linkage to the NE.
We observed that CENP-F localization to the NE was restricted to prophase cells, identified by phosphohistone H3 (PH3) staining and DNA condensation, whereas BicD2 was observed at the NE in nearly all cyclin B1+ cells (not shown). This suggests that the BicD2 pathway becomes active prior to the Nup133 pathway. As a test of this possibility, we double-labeled HeLa cells with anti-CENP-F and anti-BicD2 antibodies (Figure 1D). We found only 21.4% ±5.7 of BicD2-positive nuclei to react with anti-CENP-F (n=107), whereas 100% of CENP-F-positive nuclei reacted with anti-BicD2 (n=64). These data support early G2 activation of the BicD2 pathway, and an additive role for the two pathways late in G2-prophase.
We also evaluated the distribution and function of nesprin-SUN complexes in nonneuronal cells. As judged by immunostaining, Nesprin-1 and -2 were present at the HeLa cell NE throughout the cell cycle (Figure S2A). Expression of a dominant negative KASH domain construct, RFP-KASH (Luxton et al., 2010), which interferes broadly with nesprin-SUN interactions, displaced Nesprin-1 and -2 from the NE, but had no detectable effect on dynein localization (Figure S2B–S2C). These results support a primary role for the Nup133 and BicD2 mechanisms in dynein NE recruitment in HeLa cells.
NE dynein recruitment in rat brain development
We also tested whether dynein could be detected at the NE in RGP cells in situ by immunostaining. The high levels of soluble and vesicular dynein in cytoplasm have historically made its localization difficult even in flat, non-neuronal cells. Using minimal fixation, however, we detected clear dynein staining at the nuclear rim in some, but not all, RPG cells in the ventricular zone of the E20 rat neocortex (Figure 1E and S4A). We also observed double-labeling of nuclei with anti-dynein and anti-BicD2 antibodies, supporting a potential role for the nonneuronal dynein recruitment mechanisms in RPG cells.
To test this possibility more directly, we expressed shRNAs for the NE dynein recruitment genes using in utero electroporation in E16 rat brain, each of which were found to reduce target expression levels in Rat2 cells (Figure S3). In E20 control rat brain, we observed extensive distribution of neurons throughout the outer regions of the developing neocortex, the intermediate zone (IZ) and cortical plate (CP; Figure 2A–2B). In contrast, cells expressing shRNAs for BicD2, Nup133, and CENP-F exhibited severe impairment in neuronal distribution, evidenced by an almost complete absence of cells in the IZ and CP regions. This effect was similar to that previously observed for LIS1 and dynein RNAi (Figure 2A) (Shu et al., 2004; Tsai et al., 2005), consistent with a role for the recruitment genes in dynein regulation. Triple mutant shRNAs for BicD2 and Nup133 had no effect on neuronal distribution (Figure S4B). RNAi targeting of BicD1, a BicD2 paralogue that participates in targeting of dynein to the Golgi apparatus, but has no role at the NE (Splinter et al., 2010), produced no effect on neuronal distribution (Figure 2A).
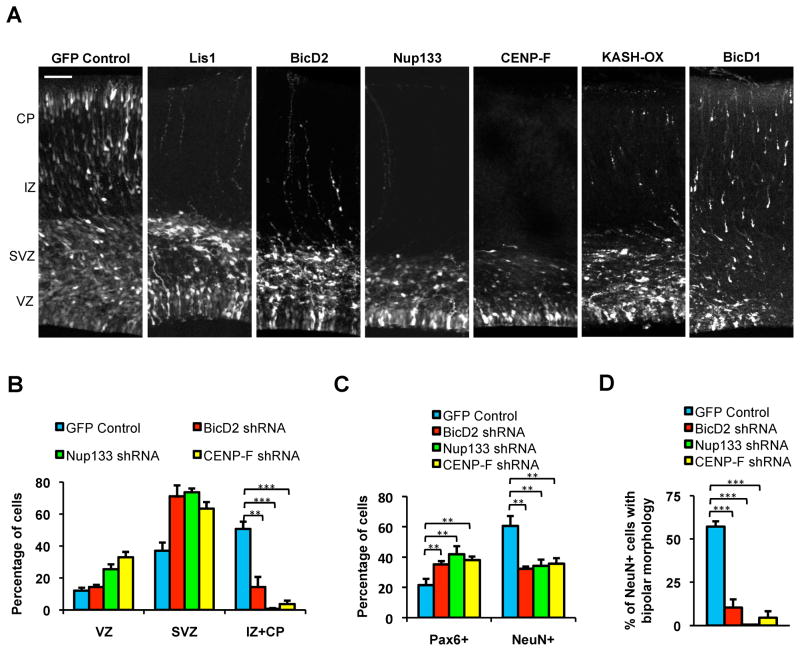
Figure 2. Inhibition of dynein NE recruitment mechanisms affects overall neuronal migration in embryonic rat brain.
A. E16 rat embryonic brains were subjected to in utero electroporation with the pRNAT vector expressing shRNAs corresponding to the genes noted along with a fluorescent reporter, or with an RFP-KASH construct (n=3 brains per condition). Brain tissue was fixed and sectioned at E20. Expression of BicD2, Nup133, or CENP-F shRNAs resulted in a marked reduction in distribution of electroporated cells throughout the intermediate zone (IZ) and cortical plate (CP), comparable to the effects of LIS1 shRNA. KASH expression resulted in an intermediate cell redistribution phenotype. No clear effect was observed for BicD1. Scale bar = 50 μm. B. Quantification of transfected cells within the VZ, subventricular zone (SVZ), and IZ+CP show an increase in Nup133, CENP-F, and BicD2 shRNA-expressing cells in the SVZ, and a decrease in the IZ+CP. C. Quantification of Pax6+ (RGP) and NeuN+ (neuronal) cells in brain electroporated with BicD2, Nup133, or CENP-F shRNA shows an increase in RPG cells and a decrease in neurons. D. Quantification of NeuN+ neurons that exhibited a migratory, bipolar morphology supports a loss in migrating neurons. *P<0.05; **P<0.01; ***P<0.001; N.S., Not Significant; Error bars = S.D. (throughout legends). See also Figure S3, S4B, and S5A.
BicD2, Nup133, and CENP-F RNAi each caused a reduction in the number of postmitotic neurons as judged using the neuronal marker, NeuN (Figure 2C). Of the NeuN-positive cells, most were located in the SVZ and had a multipolar morphology, though a few bipolar cells could also be detected in the lower IZ in the case of BicD2 and CENP-F RNAi (Figure 2D). Together, these results suggested that passage through the stages of neurogenesis and migration, including the multipolar-to-bipolar transition, was inhibited. RNAi for BicD2, Nup133, and CENP-F each caused an increase in the percentage of RGP cells as determined by Pax6, a marker specific for RGP cells (Figure 2C). These observations suggested a failure in the initial stages of neurogenesis.
To test the role of nesprin-SUN LINC complexes, we used in utero electroporation in E16 rat brain to express the dominant negative RFP-KASH construct. By E20, the nesprin fragment could be detected at the nuclear surface in many transfected cells (Figure S5A). The fraction of neuronal precursor cells in the outer regions of the developing neocortex was decreased compared to the control (Figure 2A), consistent with a role for nesprins in rodent brain development (Zhang et al., 2009).
Effects on INM in RGP Cells
Together, these results suggested contributions for both the BicD2 and Nup-133 NE dynein recruitment pathways during neurogenesis and migration. To test the role of dynein recruitment factors specifically during INM, we imaged RGP cells in live brain slices four days after in utero electroporation. Nuclei in control cells exhibited clear migration in the apical and basal directions, with mitosis occurring at the ventricular surface (Figure 3A–3B and Movie S1). In contrast, BicD2 RNAi caused specific inhibition of apical, but not basal nuclear migration with nuclei arrested at least 30 μm from the ventricular surface throughout the recording period (Figure 3C, Movie S2, and Movie S5). None of the nuclei in cells subjected to BicD2 RNAi reached the ventricular surface.
Nup133 and CENP-F RNAi each also severely inhibited apical INM (Figure 3A–3B and Movie S3–S4). However, nuclei in both cases arrested much closer to the ventricular surface than observed following BicD2 RNAi. Nuclei in either Nup133 or CENP-F shRNA- expressing cells accumulated within 10 μm of the ventricle, with none successfully reaching it (Fig. 3B). Apical movement could still be detected in some nuclei prior to complete arrest. Basal migration occurred at normal rates in another subset of these cells, arguing against a role for the Nup133 pathway in this phase of INM (Figure 3C and Movie S6–S7).
The live behavior was supported by analysis of fixed brain tissue as a function of time following in utero electroporation with shRNAs. Progressive changes in the overall distribution of RGP cell somata could be observed between 2 and 4 days following in utero electroporation with BicD2, Nup133, and CENP-F shRNAs (not shown). For BicD2 RNAi, the distribution of nuclei shifted to increasing distances from the ventricle, with enrichment of cell bodies at >30 μm from the ventricular surface by four days (Figure 4A–4B). In striking contrast, Nup133 shRNA-expressing cell bodies gradually accumulated toward the ventricular surface, 70% within 10 μm, and many as close as 2–5 μm (Figure 4B–4C). The majority of cells, even those closely approaching the ventricle, still showed a detectable “endfoot” between the soma and ventricular surface, which often appeared swollen by the encroaching nucleus. CENP-F RNAi caused similar effects, though the concentration of nuclei near the ventricular surface was not as extreme (Figure 4).
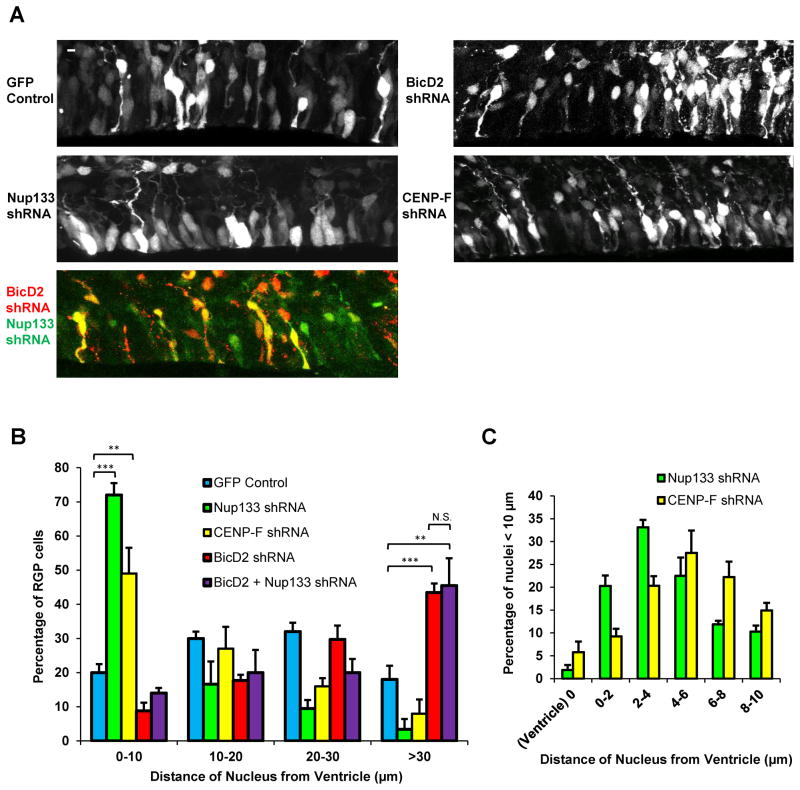
Figure 4. RNAi-induced arrest of nuclear migration early vs. late during apical INM.
To test for general effects of dynein NE recruitment genes on nuclear position, shRNAs for BicD2, Nup133, and CENP-F were expressed in E16 rat embryonic brains, which were then fixed and imaged at E20 (n=3 brains per condition). A. Distribution of RGP nuclei. Control nuclei were distributed at a range of distances from the ventricular surface. Nuclei of BicD2 RNAi expressing cells accumulated relatively far from the ventricular surface. In contrast, nuclei of Nup133 and CENP-F RNAi expressing cells accumulated close to this site. Double knockdown of BicD2 and Nup133 resulted in a nuclear distribution similar to that for BicD2 RNAi alone. Scale bar = 5 μm for all panels. B. Quantification of nuclear distances. RGP cells were identified morphologically and by Pax6 staining, and distance was measured from the ventricular surface to the closest (bottom) edge of the nucleus. Nup133 and CENP-F RNAi caused a marked accumulation of nuclei close to the ventricular surface, which peaked at ~5 μm (panel C).
Together the live and fixed tissue imaging results are consistent with the sequential activation of the two mechanisms in nonneuronal cells (Figure 1). In this view, BicD2 RNAi causes nuclear arrest in RGP cells early during apical migration, whereas Nup133 or CENP-F RNAi results in late nuclear arrest. In support of this possibility, knockdown of Nup133 along with BicD2 did not produce a severer phenotype than that for BicD2 knockdown alone (Figure 4A–B).
Finally, expression of the RFP-KASH dominant negative had no detectable effect on either basal or apical nuclear migration (Figure S5B). Nonetheless, this construct severely impeded subsequent glial-guided neuronal migration (Figure S5C). This result is consistent with a previous report of defective neuronal migration (Zhang et al., 2009), but argues that nesprins are not involved in INM in RGP cells. Our data, therefore, identify two G2-specific mechanisms for NE dynein recruitment in these cells, and a later role for the SUN-Nesprin LINC complex in migrating neurons.
Near-apically arrested RGP cells are in a premitotic state
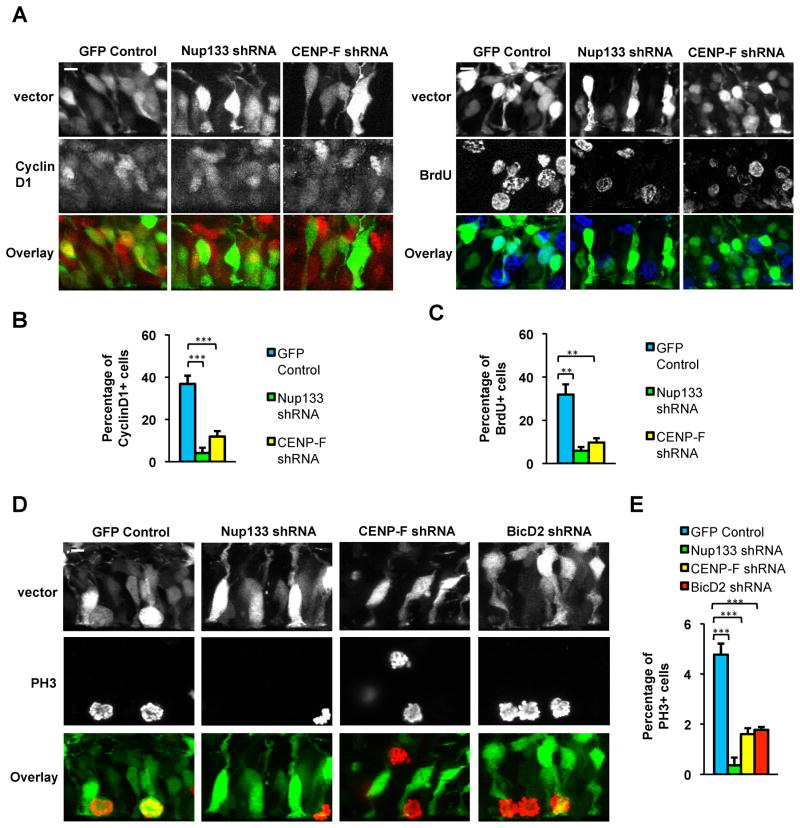
In cells subjected to Nup133 and CENP-F RNAi, the accumulation of nuclei near, but not at the ventricular surface suggested an arrest late in the cell cycle. Anti-cyclin D1 immunostaining and BrDU pulse labeling revealed a pronounced decrease in the fraction of Nup133 or CENP-F shRNA expressing cells in G1 or S phase, respectively, consistent with a late G2 arrest (Figure 5A–5C). The vast majority of Nup133 and CENP-F knockdown cells, however, were almost completely negative for PH3, an early mitotic indicator (Figure 5D–5E). Also, there was no evidence of ectopic mitotic events in cells subjected to RNAi for Nup133 or CENP-F. Rather, the few cells that entered mitosis did so at the ventricular surface. A similar decrease in the number of PH3+ cells was observed with BicD2 RNAi (Figure 5D–5E). These results strongly suggest that RGP nuclei must reach the ventricular surface to enter mitosis.
Figure 5. Nuclei in apically arrested RGP cells fail to enter mitosis.
E16 rat embryonic brains were electroporated with vectors expressing shRNAs for Nup133, CENP-F, or BicD2. Sections were stained at E20 with cell cycle markers and scored for the percent of positive RGP nuclei (n=3 brains per condition). A–C. Nup133 and CENP-F RNAi dramatically reduced the percentage of nuclei positive for the G1 marker Cyclin D1 or for the S-phase marker BrdU (following 15 min pulse labeling). D–E. Substantial decreases in the fraction of anti-phosphohistone H3 (PH3) positive nuclei were observed under all RNAi conditions, with an almost completely loss resulting from Nup133 RNAi. Scale bars = 5 μm for all panels.
Centrosome behavior
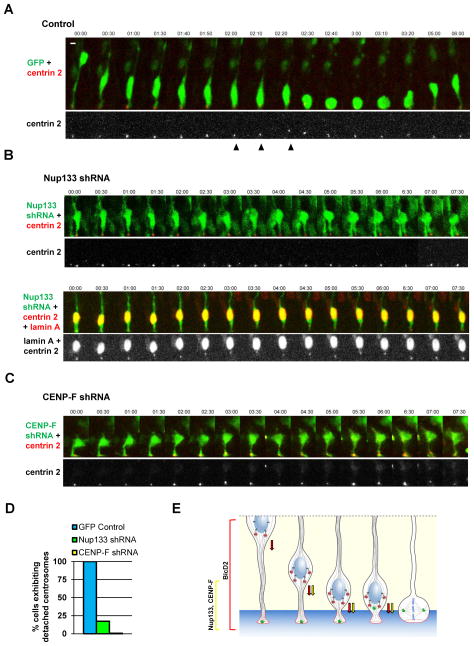
The nature of a potential trigger for mitotic entry in RGP cells once apical migration is complete remains uncertain. A promising candidate is the centrosome, which remains at the ventricular surface throughout the cell cycle, but is ultimately required in spindle assembly and can promote timely mitotic entry (Hachet et al., 2007; Portier et al., 2007). A recent study in chick neural tube and embryonic mouse neocortex reported that centrosomes can depart from the ventricular surface and travel basally to meet the apically migrating nucleus (Spear and Erickson, 2012b). NEB occurred soon after, and the soma then migrated to the ventricular surface. We observed striking, but a more limited range of centrosome motility in embryonic rat brain (Figure 6A and S7A). This behavior occurred in all RGP cells examined (n=9) and involved a brief, short-range departure of the centrosome from the ventricular terminus of the RGP cell (5 μm avg; 8 μm maximum) to meet the apically migrating nucleus. Intriguingly, the distance traveled is similar to that between Nup133 or CENP-F RNAi-arrested nuclei and the ventricular terminus of the RGP cell. The centrosome “hopping” behavior was completely inhibited in all CENP-F depleted cells examined (n=5), and in 83% of Nup133 depleted cells (n=18; Figure 6B–6D). In the few cases where centrosome departure occurred, multiple futile attempts to reach the nucleus could be seen (Figure 6C and S7B), each followed by return of the centrosome towards the ventricular surface.
Figure 6. Pre-mitotic centrosome dynamics.
E16 rat embryonic brains were co-transfected by in utero electroporation with cDNAs encoding shRNAs along with DsRed-centrin2 to image centrosomes. Brain slices were placed in culture at E20 for live imaging of RGP cells. Time-lapse images were generated at time intervals indicated at top in min. A. Centrosomes expressing control shRNA vector were retained at the ventricular surface throughout INM until 2:00–2:20 min, at which time the centrosome (arrowhead) departs to meet the soma. Following contact, the centrosome and soma migrate together to the ventricular surface (2:40) and the cell continues through INM (3:20). B. Time lapse recordings of Nup133 shRNA co-electroporated with centrin2 (top) and with centrin2 plus lamin A (bottom). In 83% of Nup133 depleted cells, the centrosome remained at the ventricular surface throughout the recording. In 17% of cases (bottom), centrosomes jumped towards the nucleus as in control cells, but then returned alone to the ventricular surface. The nuclei in both Nup133 examples showed only limited mobility throughout the recording. The lower example shows that the NE remained intact. C. CENP-F shRNA resulted in similar nuclear and centrosome arrest to that seen in the majority of Nup133 RNAi cases. D. Quantification of centrosome behavior shows departure from ventricular surface in all control cells, but in few to no Nup133 and CENP-F shRNA-expressing cells. E. Model for apical INM. Dynein is shown sequentially recruited to RGP cell nuclear pores via the BicD2 and Nup133 pathways. Arrows represent BicD2- (red) and Nup133- (yellow) mediated dynein pulling forces exerted on the NE. The centrosome (green) remains at the ventricular surface of the RGP cell throughout INM, but then moves towards the nucleus just prior to mitosis. Scale bars = 5 μm for all panels. See also Figure S7A–S7B.
Rescue of Apical INM and Cell Cycle Progression Defects by BicD2 Expression
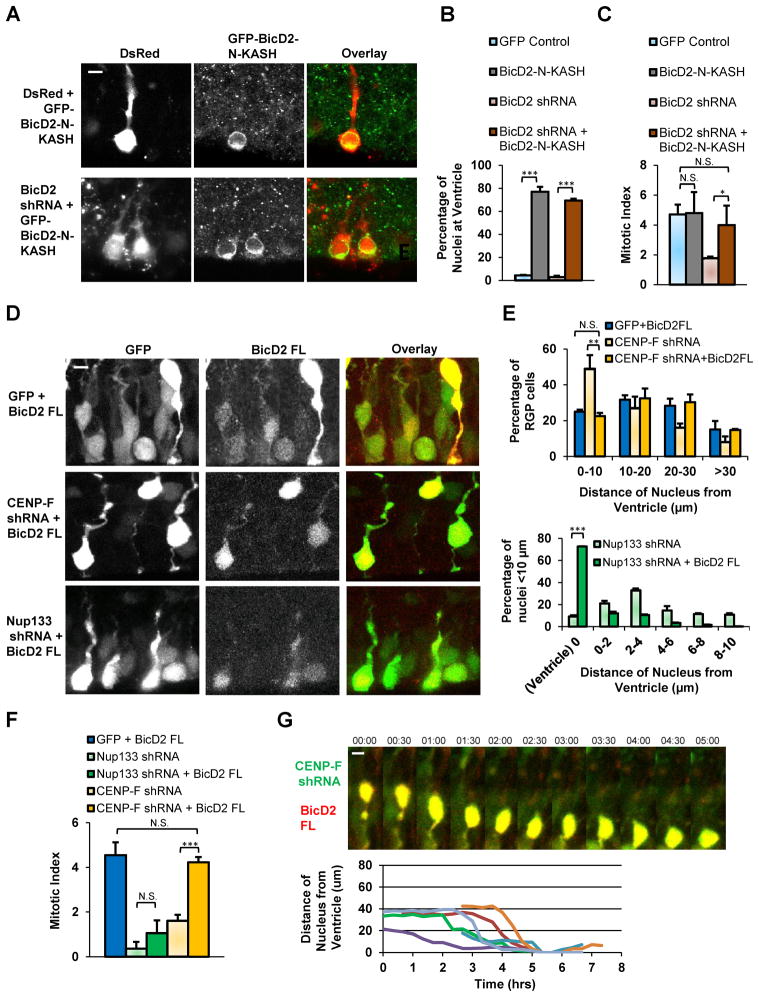
The effects of Nup133, CENP-F, and BicD2 inhibition on apical INM and mitotic entry suggest an important role in these processes for dynein forces generated at the NE. To test if dynein targeting to this site is sufficient to drive apical INM, we used a previously characterized fusion construct consisting of the dynein-interacting N-terminal portion of BicD2 fused to the nesprin-3 KASH domain (Splinter et al., 2010). This construct lacks the C-terminal BicD2 site responsible for G2-specific NE targeting, and uses the KASH domain in its place for constitutive, cell cycle independent NE dynein targeting. Our own current data reveal KASH domain targeting to the RPG cell NE without effect on INM (Figure S5). BicD2-N-KASH expression in RPG cells alone or in combination with BicD2 shRNA caused a dramatic accumulation of most transfected somata at the ventricular surface (Figure 7A–7B and S7C). Mitotic cells, as determined by PH3 staining, were still restricted to the ventricular surface, but so were a large number of nonmitotic nuclei. Overall mitotic index in the transfected cells was near that in controls. These results together suggest that cell cycle progression persists in the BicD2-N-KASH-expressing cells, but that excess recruitment of NE dynein may overpower the kinesin-generated forces involved in basal INM during G1.
Figure 7. Rescue of apically arrested nuclei by targeting of NE dynein.
E16 rat embryonic brains were transfected with GFP-BicD2-N-KASH as a general NE dynein targeting approach. Alternatively, full length BicD2 was expressed to enhance dynein targeting to apically migrating RGP nuclei. Brain slices were fixed and sectioned at E20 (n = 3 brains per condition). A. Nuclei of cells expressing GFP-BicD2-N-KASH alone or in combination with BicD2 shRNA showed marked accumulation at the ventricular surface. Note that GFP-BicD2-N-KASH decorates the NE. B. Quantification of transfected RGP nuclei located at the ventricular surface. C. Sections were also stained for PH3 and percentage of PH3+ cells is compared with control and BicD2 RNAi conditions (from Figure 5E). D. RGP cells expressing full length BicD2 alone or in combination with CENP-F or Nup133 shRNA. E. Quantification of nuclear distances from the ventricular surface to the bottom of the nucleus. Top, nuclear distances of cells expressing either full length BicD2, CENP-F shRNA (from Figure 4B), both. Bottom, distances of nuclear located ≤ 10 μm from ventricular surface in cells expressing Nup133 shRNA alone (from Figure 4C) or in combination with full length BicD2. F. Quantification of PH3+ cells among RGP cells transfected with full length BicD2 or, in combination with, Nup133 or CENP-F shRNA. Percentages of PH3+ cells are compared to Nup133 and CENP-F RNAi conditions (from Figure 5E). G. Live recording and tracings of RGP cells co-expressing full length BicD2 and CENP-F shRNA undergoing apical nuclear migration completely to the ventricular surface. Scale bar = 5 μm. See also Figure S4C, S6, and S7C–S7D.
We also expressed RNAi-insensitive BicD2, which rescued the effects of BicD2 RNAi (Figure S4C). We found, in addition, partial or extensive rescue of Nup133 or CENP-F RNAi. In the former case, BicD2 expression rescued apical nuclear migration but not mitosis, as evidenced by a dramatic accumulation of PH3-negative nuclei at the ventricular surface (Figure 7D–7F and S7D). This observation could be explained by a general mitotic defect in Nup133-depleted cells, which we confirmed using Rat2 fibroblasts (Figure S6). In marked contrast, BicD2 expression in CENP-F depleted RPG cells, rescued apical nuclear migration (Figure 7G) and mitotic index, with RGP cells distributed normally throughout the ventricular zone (Figure 7D–7F and S7D). These results reveal that INM can be completely rescued by forced recruitment of dynein to the NE.
Discussion
We previously found that INM functions by long-range kinesin- and dynein-mediated nuclear transport along an array of uniformly directed microtubules (Tsai et al., 2010). The lack of direct centrosome involvement in transporting nuclei suggested that the motor proteins act from the nucleus, a relatively unusual mechanism in vertebrates. We now provide evidence that dynein, in particular, is present at the nuclear surface, and that RNAi for genes involved in dynein recruitment to the NE inhibit INM. Because these mechanisms are G2-specific (Bolhy et al., 2011; Splinter et al., 2010), our results provide important insight into another long-standing question, the mechanisms responsible for INM cell cycle control.
Role of Dynein NE Recruitment Factors
Dynein is observed at the NE in nonneuronal cells specifically during G2 (Beaudouin et al., 2002; Salina et al., 2002). We obtained the first microscopic evidence that dynein, dynactin, and BicD2 associate with nuclear pores, as is predicted from the published role for Nup133 and BicD2 in dynein recruitment. These results strongly support the specificity of dynein localization to the NE, and help identify the pores as a form of dynein cargo. Colocalization experiments further indicated sequential activation of the two mechanisms, with BicD2 activated before Nup133/CENP-F during G2 in nonneuronal cells. Although we observed nesprins at the NE throughout the HeLa cell cycle, there was no effect of a nesprin KASH dominant negative domain (Luxton et al., 2010) on dynein recruitment. This was despite clear decoration of the NE by the fragment itself. Together, these data indicate little, if any, role for nesprins in dynein recruitment to the HeLa NE.
A role for nuclear pore-mediated dynein and its G2-recruitment pathways in INM was supported by colocalization of dynein with BicD2 at the NE in RGP cells. The high levels of soluble and vesicular dynein in cytoplasm have made its localization problematic even in flat, nonneuronal cells, and analysis in brain tissue is further compounded by the lack of useful G2 markers. Dynein colocalization with BicD2 at the RGP NE provides the first in situ evidence for G2-specific dynein localization to this site.
Functional analysis in brain
Functional data in support of a role for NE dynein in INM was obtained using RNAi for Nup133, CENP-F, and BicD2. shRNA expression in each case produced strong inhibition of INM in RGP cells in embryonic rat brain as judged both by live imaging and analysis of fixed tissue. The effects of RNAi were clearly specific for apical vs. basal migration, further supporting a role for dynein in nuclear migration toward the ventricular surface of the brain.
These data strongly confirm a role for dynein in apical INM. Furthermore, they support a role for a specific dynein subfraction that associated with the NE vs. other forms of cellular cargo. Because Nup133 depletion might affect aspects of nuclear function, we performed RNAi for CENP-F, which produced very similar defects in INM. This protein has a well-established role in kinetochore function, acting in recruitment of dynein and some of its cofactors (Vergnolle and Taylor, 2007). Nonetheless, we observed no evidence for accumulation of CENP-F knockdown cells in mitosis. This result suggests, therefore, that inhibition of apical INM is sufficiently severe to predominate in the CENP-F RNAi phenotype.
We note that, in addition to the effects of BicD2, Nup133, and CENP-F RNAi on INM, neurons also accumulated in the subventricular zone of the developing rat brain. These observations suggest that the dynein NE recruitment genes participate not only in INM, but in subsequent aspects of neuronal migration as well. Whether these effects also reflect roles in NE dynein recruitment remains to be determined.
In contrast to these results, the KASH dominant negative fragment caused no detectable effect on INM as monitored live, consistent with the lack of cell cycle-dependent NE dynein recruitment we find in nonneuronal cells. However, KASH expression blocked neuronal migration in the IZ of the embryonic rat brain. These results are consistent with neuronal defects observed in nesprin and SUN knockout mice (Zhang et al., 2009), but identify a specific switch in dynein NE recruitment mechanism during neuronal differentiation.
The sequential effects of BicD2 vs. CENP-F/Nup133 RNAi provide further evidence for an unexpected level of complexity in the control of INM. Consistent with sequential activation of the two dynein NE recruitment pathways in nonneuronal cells, we observed BicD2 RNAi to arrest nuclei relatively far from the ventricular surface. This result implies that BicD2 recruits sufficient levels of NE-associated dynein to complete only the initial part of the apical migration process. The Nup133 recruitment pathway, in contrast, seems to fine-tune apical INM, and act at a critical late stage in cell cycle and developmental progression. We note that this analysis could not be readily conducted in the mer mutant mouse due to early lethality, but suggest that NE dynein recruitment could explain the reported reduction in neurogenesis in the neural tube (Lupu et al., 2008).
Our results provide evidence that G2-dependent dynein NE recruitment may be generally important in vertebrate cell behavior. We note that in nonneuronal cells, the role of NE dynein is nonessential (Beaudouin et al., 2002; Salina et al., 2002). In contrast, in RGP cells, the BicD2 and Nup133 mechanisms each appear to be essential both for apical migration, and for cell cycle progression. Conceivably, it is for developmental situations such as this that these mechanisms have been conserved during evolution.
Centrosome behavior and its role in mitotic entry
The purpose and mechanism by which nuclei of the RGP cells undergo mitosis only at the ventricular surface are unknown. In earlier work, we found that LIS1 and dynein RNAi arrest nuclei at a range of positions relative to the ventricular surface, (Tsai et al., 2005; 2010), but unlike nonneuronal cells, the arrested RGP nuclei were nonmitotic. Our current results add strong additional support for positional control of mitotic entry in these cells. Disruption of the Nup133 pathway, especially by CENP-F RNAi, is particularly intriguing in this regard as it results in arrest of nuclei within a few μm of the ventricular surface with little or no evidence of progression into mitosis. These results suggest that a mitotic entry signal must be restricted to the extreme apical terminus of the cell.
Although centrosomes of RGP cells remain at the apical terminus throughout INM, recent analysis of chicken neural tube has revealed instances of centrosome departure from the ventricular surface just prior to mitosis (Spear and Erickson, 2012b). Contact with the nucleus appeared to initiate NEB, suggesting that the trigger for mitotic entry might be associated with the centrosome. We observed dissociation of the centrosome from the apical terminus of the RGP cell when nuclei reached a distance of 3–8 um from this site, a distance much smaller than those reported in chick neural tube. Centrosomes then moved rapidly from the ventricular terminus of the RGP cells to meet the nucleus. After contact, the soma progressed to the ventricular surface. Intriguingly, Nup133 or CENP-F RNAi greatly decreased the frequency of centrosome “hopping” events, and, when they did occur, the centrosome returned alone to the ventricular surface.
Our results together reveal centrosome departure from the ventricular surface to be a standard aspect of late INM behavior. We argue, furthermore, that centrosome hopping must depend, at least in part, on NE-associated dynein forces as indicated by the inhibitory effects of Nup133 and CENP-F RNAi on centrosome behavior (Figure 6E). The additional force from Nup133-mediated dynein recruitment may be needed to pull the apically bound centrosome towards the nucleus. How the centrosome-nucleus complex completes migration to the ventricular surface after contact is uncertain, but could involve restoring force generated from the apical cell cortex.
Targeting dynein to the NE is sufficient for nuclear migration to the ventricular surface
It is uncertain why two successive mechanisms should be required for dynein recruitment to the NE. At the least, it suggests that forces generated by either one alone are insufficient to complete apical migration. To test this possibility and to gain further insight into the role of nuclear and centrosome transport in mitotic entry and progression, we attempted to target excess dynein to the NE. We used the BicD2 N-terminal domain to target dynein to the NE via the nesprin-3 KASH domain. Expression of this construct had a potent effect on RGP cells, dramatically increasing the number of non-mitotic nuclei at the ventricular surface. These results suggest that increased NE dynein can overcome even the opposing kinesin-3 generated forces (Tsai et al., 2010) responsible for G1 basal nuclear migration.
In contrast, overexpression of BicD2 alone had no apparent effect on INM in control cells. However, it partially or completely rescued the effects of BicD2, Nup133 or CENP-F RNAi. In the latter case, BicD2 restored apical migration as well as mitotic index, with all mitotic figures aligned at the ventricular surface. BicD2 expression drove nuclei to the ventricular surface in Nup133 shRNA-expressing cells, though mitotic index was still severely reduced. These results are consistent with a general mitotic defect (Figure S6) and suggest additional non-dynein roles for this nucleoporin, such as assembly of the Y-complex (Walther et al., 2003). Together the restorative effects of BicD2 expression argue that sufficient levels of dynein recruitment to the NE could sustain apical nuclear migration independent of the recruitment mechanism. Rescue of Nup133 or CENP-F RNAi by BicD2 also suggests that excess levels of the latter protein must target properly to the G2 NE.
Model for Graded Increase in Dynein Forces During Apical INM
Our data provides striking evidence for the differential roles of two NE dynein recruitment mechanisms in INM, and indicate that the individual mechanisms are each inadequate to complete this basic process. Nonetheless, BicD2 overexpression restored INM in shRNA-expressing RGP cells. This observation argues that BicD2-mediated dynein recruitment can, in principal, support INM. If so, then normal NE BicD2 expression or activity may be inadequate for INM. The need for a second NE dynein recruitment mechanism suggests a requirement for greater forces as the nucleus of the RGP cell approaches the ventricular surface of the brain (Figure 6E). This could be to overcome a higher density of nuclei in this region, or the resistance of the cell cortex as the nucleus attempts to enter the narrower endfoot region of the cell.
Conclusions
Our results provide the first evidence on the mechanism for cell cycle control of INM. Blocking apical migration of the RGP nucleus, even within 1–5 um of the ventricular surface, blocked mitotic entry, suggesting that a mitotic entry signal is tightly restricted to the ventricular terminus of the cell. Why a mechanism has evolved to ensure this behavior is unknown. We speculate that ectopic cell divisions must interfere in some substantial way with proper establishment of neurogenesis. Nucleokinesis at the ventricular surface provides access of the condensing chromosomes to the centrosomes, which assemble the mitotic spindle, and to apical cues associated with the cell cortex, which are likely important in controlling fate through spindle orientation. The specific consequences of ectopic mitosis remain an important question for further study. What can be concluded from the current study is that altered INM reduces RGP proliferation. The consequences on brain size seem evident, and may have important implications for understanding and controlling brain developmental disease.
Experimental Procedures
Full protocols continued under Extended Experimental Procedures.
Cell culture, transfection, and immunostaining
HeLa cells were cultured in DMEM supplemented with 10% FBS. Effectene reagent (Qiagen) was used for plasmid transfection and cells were analyzed after 2 days. For immunostaining, cells were washed in PBS, fixed in −20°C methanol, 10mM EGTA (10 min), permeabilized in PBS, 0.1% Triton X-100 (5 min) and stained in PBS, 0.05% Tween supplemented with donkey serum. For better visualization, cells stained for dynein, dynactin, BicD2 or CENP-F were incubated for 1 hour in Nocodazol (10μM) prior to fixation (Bolhy et al., 2011; Jodoin et al., 2012; Raaijmakers et al., 2012; Splinter et al., 2010). For 3D-SIM, cells were plated on high performance coverslips (0,170 ± 0.005 mm, ZEISS), stained as described previously and mounted in ProLong® Gold antifade reagent (Molecular Probes).
In utero electroporation and live imaging
Plasmids were transfected by intraventricular injection in embryonic rats in utero, followed by electroporation as described previously (Tsai et al., 2005). Animals were maintained according to protocols approved by the Institutional Animal Care and Use Committee at Columbia University. Coronal slices for live imaging were prepared 4 days after electroporation and imaged at intervals of 10 min for 8–15 hrs as described previously (Tsai et al., 2010). For BrDU labeling experiments, BrDU (Sigma-Aldritch) was injected at 50 mg/kg body weight intraperitoneally 20 min prior to embryo harvest.
Immunostaining of brain slices
Rat embryos were perfused transcardially with chilled saline and 4% paraformaldehyde (PFA) (EMS, wt/vol) and then incubated in 4% PFA overnight. Brain slices were sectioned coronally on a Vibratome (Leica microsystems). Brain slices were washed with PBS and stained in PBS, 0.3% triton-X100 supplemented with donkey serum. Primary antibodies were incubated overnight at 4°C and secondary antibodies were incubated for 2 hrs at room temperature. For BrDU immunostaining, brain slices were first incubated in 2N HCl for 25 min at 37°C and then washed in PBS prior to antibody incubation. For dynein and BicD2 immunostaining, rat brains were sectioned coronally on a Vibratome prior to fixation and incubated in Nocodazol (10μM) for 2 hrs for better visualization (Bolhy et al., 2011; Jodoin et al., 2012; Raaijmakers et al., 2012; Splinter et al., 2010). Brain slices were then fixed in methanol at −20°C for 20 min. Immunostaining proceeded as described above.
Supplementary Material
Highlights.
Nuclear Pore Recruitment of Dynein in RGPs Mediates G2 Apical Nuclear Migration.
Two-step Dynein Recruitment Required for Mitotic Entry and Neurogenesis.
RNAi for Late Recruitment Factors Interferes with Centrosome Motility.
Forced Dynein Recruitment Restores Nuclear Migration and Cell Cycle Progression.
Acknowledgments
We thank Drs. G. Gundersen, G. Luxton, H. Worman, C. Ostlund, and A. Laufer for advice and reagents; the Rockefeller Univ. BIRC for 3D-SIM imaging; and the Foundation des Treilles for providing a venue for stimulating scientific discussions. This project was supported by NIH HD40182 and GM102347 to RBV; ARC and ANR-12-BSV2-0008-01 to VD; NWO ALW-VIC1 to AA; and AHA postdoctoral fellowships to TN and ADB.
Abbreviations
- RGP
Radial Glial Progenitor
- INM
Interkinetic Nuclear Migration
- NE
Nuclear Envelope
- NEB
Nuclear Envelope Breakdown
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beaudouin J, Gerlich D, Daigle N, Eils R, Ellenberg J. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell. 2002;108:83–96. doi: 10.1016/s0092-8674(01)00627-4. [DOI] [PubMed] [Google Scholar]
- Bolhy S, Bouhlel I, Dultz E, Nayak T, Zuccolo M, Gatti X, Vallee R, Ellenberg J, Doye V. A Nup133-dependent NPC-anchored network tethers centrosomes to the nuclear envelope in prophase. J Cell Biol. 2011;192:855–871. doi: 10.1083/jcb.201007118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadot B, Gache V, Vasyutina E, Falcone S, Birchmeier C, Gomes ER. Nuclear movement during myotube formation is microtubule and dynein dependent and is regulated by Cdc42, Par6 and Par3. EMBO Rep. 2012;13:741–749. doi: 10.1038/embor.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bene F, Wehman AM, Link BA, Baier H. Regulation of Neurogenesis by Interkinetic Nuclear Migration through an Apical-Basal Notch Gradient. Cell. 2008;134:1055–1065. doi: 10.1016/j.cell.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridolfsson HN, Starr DA. Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J Cell Biol. 2010;191:115–128. doi: 10.1083/jcb.201004118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridolfsson HN, Ly N, Meyerzon M, Starr DA. UNC-83 coordinates kinesin-1 and dynein activities at the nuclear envelope during nuclear migration. Dev Biol. 2010;338:237–250. doi: 10.1016/j.ydbio.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Frank CL, Calderon de Anda F, Tsai LH. Hook3 interacts with PCM1 to regulate pericentriolar material assembly and the timing of neurogenesis. Neuron. 2010;65:191–203. doi: 10.1016/j.neuron.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Hachet V, Canard C, Gönczy P. Centrosomes Promote Timely Mitotic Entry in C. elegans Embryos. Dev Cell. 2007;12:531–541. doi: 10.1016/j.devcel.2007.02.015. [DOI] [PubMed] [Google Scholar]
- Hebbar S, Mesngon MT, Guillotte AM, Desai B, Ayala R, Smith DS. Lis1 and Ndel1 influence the timing of nuclear envelope breakdown in neural stem cells. J Cell Biol. 2008;182:1063–1071. doi: 10.1083/jcb.200803071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jodoin JN, Shboul M, Sitaram P, Zein-Sabatto H, Reversade B, Lee E, Lee LA. Human Asunder promotes dynein recruitment and centrosomal tethering to the nucleus at mitotic entry. Mol Biol Cell. 2012;23:4713–4724. doi: 10.1091/mbc.E12-07-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto T, Fugo K, Kiyokawa T. Intracellular position of G2/M-phase nuclei in neoplastic and non-neoplastic pseudostratified glands suggests the occurrence of interkinetic nuclear migration. Med Mol Morphol. 2013 doi: 10.1007/s00795-013-0026-z. [DOI] [PubMed] [Google Scholar]
- Kosodo Y. Interkinetic nuclear migration: beyond a hallmark of neurogenesis. Cell Mol Life Sci. 2012;69:2727–2738. doi: 10.1007/s00018-012-0952-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosodo Y, Suetsugu T, Suda M, Mimori-Kiyosue Y, Toida K, Baba SA, Kimura A, Matsuzaki F. Regulation of interkinetic nuclear migration by cell cycle-coupled active and passive mechanisms in the developing brain. Embo J. 2011;30:1690–1704. doi: 10.1038/emboj.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A. The Glial Nature of Embryonic and Adult Neural Stem Cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HO, Norden C. Mechanisms controlling arrangements and movements of nuclei in pseudostratified epithelia. Trends Cell Biol. 2012;23:141–150. doi: 10.1016/j.tcb.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Leung L, Klopper AV, Grill SW, Harris WA, Norden C. Apical migration of nuclei during G2 is a prerequisite for all nuclear motion in zebrafish neuroepithelia. Development. 2011;138:5003–5013. doi: 10.1242/dev.071522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupu F, Alves A, Anderson K, Doye V, Lacy E. Nuclear Pore Composition Regulates Neural Stem/Progenitor Cell Differentiation in the Mouse Embryo. Dev Cell. 2008;14:831–842. doi: 10.1016/j.devcel.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GWG, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear Arrays of Nuclear Envelope Proteins Harness Retrograde Actin Flow for Nuclear Movement. Science. 2010;329:956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mckenney RJ, Vershinin M, Kunwar A, Vallee RB, Gross SP. LIS1 and NudE induce a persistent dynein force-producing state. Cell. 2010;141:304–314. doi: 10.1016/j.cell.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer EJ, Ikmi A, Gibson MC. Interkinetic Nuclear Migration Is a Broadly Conserved Feature of Cell Division in Pseudostratified Epithelia. Curr Biol. 2011;21:485–491. doi: 10.1016/j.cub.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Niethammer M, Smith DS, Ayala R, Peng J, Ko J, Lee MS, Morabito M, Tsai LH. NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron. 2000;28:697–711. doi: 10.1016/s0896-6273(00)00147-1. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- Norden C, Young S, Link BA, Harris WA. Actomyosin Is the Main Driver of Interkinetic Nuclear Migration in the Retina. Cell. 2009;138:1195–1208. doi: 10.1016/j.cell.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portier N, Audhya A, Maddox PS, Green RA, Dammermann A, Desai A, Oegema K. A Microtubule-Independent Role for Centrosomes and Aurora A in Nuclear Envelope Breakdown. Dev Cell. 2007;12:515–529. doi: 10.1016/j.devcel.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JA, van Heesbeen RGHP, Meaders JL, Geers EF, Fernandez-Garcia B, Tanenbaum ME, Medema REH. Nuclear envelope-associated dynein drives prophase centrosome separation and enables Eg5-independent bipolar spindle formation. Embo J. 2012;31:4179–4190. doi: 10.1038/emboj.2012.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- Salina D, Bodoor K, Eckley DM, Schroer TA, Rattner JB, Burke B. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 2002;108:97–107. doi: 10.1016/s0092-8674(01)00628-6. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Shionoya A, Ishida M, Gambello MJ, Yingling J, Wynshaw-Boris A, Hirotsune S. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 2000;28:681–696. doi: 10.1016/s0896-6273(00)00146-x. [DOI] [PubMed] [Google Scholar]
- Sauer F. MITOSIS IN THE NEURAL TUBE. J Comp Neurol. 1935:1–29. [Google Scholar]
- Schenk J, Wilsch-Bräuninger M, Calegari F, Huttner WB. Myosin II is required for interkinetic nuclear migration of neural progenitors. Proc Natl Acad Sci USA. 2009;106:16487–16492. doi: 10.1073/pnas.0908928106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu T, Ayala R, Nguyen MD, Xie Z, Gleeson JG, Tsai LH. Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron. 2004;44:263–277. doi: 10.1016/j.neuron.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Spear PC, Erickson CA. Interkinetic nuclear migration: A mysterious process in search of a function. Develop Growth Differ. 2012a;54:306–316. doi: 10.1111/j.1440-169X.2012.01342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear PC, Erickson CA. Apical movement during interkinetic nuclear migration is a two-step process. Dev Biol. 2012b;370:33–41. doi: 10.1016/j.ydbio.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter D, Razafsky DS, Schlager MA, Serra-Marques A, Grigoriev I, Demmers J, Keijzer N, Jiang K, Poser I, Hyman AA, et al. BICD2, dynactin, and LIS1 cooperate in regulating dynein recruitment to cellular structures. Mol Biol Cell. 2012;23:4226–4241. doi: 10.1091/mbc.E12-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter D, Tanenbaum ME, Lindqvist A, Jaarsma D, Flotho A, Yu KL, Grigoriev I, Engelsma D, Haasdijk ED, Keijzer N, et al. Bicaudal D2, Dynein, and Kinesin-1 Associate with Nuclear Pore Complexes and Regulate Centrosome and Nuclear Positioning during Mitotic Entry. PLoS Biol. 2010;8:e1000350. doi: 10.1371/journal.pbio.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Fridolfsson HN. Interactions Between Nuclei and the Cytoskeleton Are Mediated by SUN-KASH Nuclear-Envelope Bridges. Annu Rev Cell Dev Biol. 2010;26:421–444. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna E, Huttner WB. Neural Progenitor Nuclei IN Motion. Neuron. 2010;67:906–914. doi: 10.1016/j.neuron.2010.08.027. [DOI] [PubMed] [Google Scholar]
- Tsai JW, Chen Y, Kriegstein AR, Vallee R. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J Cell Biol. 2005;170:935–945. doi: 10.1083/jcb.200505166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JW, Lian WN, Kemal S, Kriegstein AR, Vallee RB. Kinesin 3 and cytoplasmic dynein mediate interkinetic nuclear migration in neural stem cells. Nat Neurosci. 2010;13:1463–1471. doi: 10.1038/nn.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnolle MAS, Taylor SS. Cenp-F Links Kinetochores to Ndel1/Nde1/Lis1/Dynein Microtubule Motor Complexes. Current Biology. 2007;17:1173–1179. doi: 10.1016/j.cub.2007.05.077. [DOI] [PubMed] [Google Scholar]
- Walther TC, Alves A, Pickersgill H, Loïodice I, Hetzer M, Galy V, Hülsmann BB, Köcher T, Wilm M, Allen T, et al. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 2003;113:195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- Wilson MH, Holzbaur ELF. Opposing microtubule motors drive robust nuclear dynamics in developing muscle cells. J Cell Sci. 2012;125:4158–4169. doi: 10.1242/jcs.108688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y-T, Wang C-L, Van Aelst L. DOCK7 interacts with TACC3 to regulate interkinetic nuclear migration and cortical neurogenesis. Nat Neurosci. 2012;15:1201–1210. doi: 10.1038/nn.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, Han M. SUN1/2 and Syne/Nesprin-1/2 Complexes Connect Centrosome to the Nucleus during Neurogenesis and Neuronal Migration in Mice. Neuron. 2009;64:173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.