Abstract
We report that several nanomaterials induced enhanced mineralization (increased numbers and larger areas of mineral nests) in MC3T3-E1 bone cells, with the highest response being induced by silver nanoparticles (AgNPs). We demonstrate that AgNPs altered microRNA expression resulting in specific gene expression associated with bone formation. We suggest that the identified essential transcriptional factors and bone morphogenetic proteins play an important role in activation of the process of mineralization in bone cells exposed to AgNPs.
Keywords: nanomaterials, bone cells, mineralization, miRNA regulation
Introduction
Bone is a living dynamic tissue and its constant rebuilding occurs through the combined action of osteoblast cells that generate bone and osteoclast cells that reabsorb it. Recently, a number of studies have focused on the growth of bone on nanostructural materials and the complex interactions between such materials and bone cells, both in vitro and in vivo[1, 2]. Because of their size and morphological properties, nanoscale materials can interact with cells and living tissues making them ideal vehicles for accelerated tissue regeneration and enhanced cellular proliferation [3, 4]. Moreover, these various shaped nanomaterials have been shown to specifically target and penetrate cells allowing delivery of genetic material [5]. In the present study, osteoblast cells (MC3T3-E1 cell line, derived from newborn mice calvaria) were incubated with various nanomaterials in order to study the effects of such structures on the biological activity of the cells, with a focus on mineralization rates. This type of cell was shown to be a model system for studying complex biological processes related to mineralization and bone formation of the osteoid matrix [6]. Additionally, they were found to readily interact with nanostructural materials and the observed effects could be translated to in vivo conditions with potentially positive benefits for the treatment of a number of bone related conditions, such as osteoporosis, or resorption of tissues. Here we report the in vitro effects of various types of nanomaterials (single walled carbon nanotubes – SWCNTs, hydroxyapatite nanoparticles – HAP, titanium dioxide nanoparticles – TiO2 and silver nanoparticles – AgNPs) on cell calcification and mineralization by MC3T3-E1 cells [7, 8], which display time-dependent and sequential expression of osteoblast characteristics similar to that found in in vivo bone tissue. The aforementioned nanomaterials were chosen due to their relevance to specific areas of nanoscience and their significant potential in bio-medical applications.
AgNPs with an average diameter of 23.0 ± 2.0 nm were used in this study [9]. AgNPs have been widely used as a biomaterial in a number of biomedical applications ranging from anti-microbial coatings to surface enhanced Raman Spectroscopy sensing [10, 11]. HAP is one of the most commonly used materials in bone graft because of its demonstrated ability to enhance mineral formation and promote bone calcification during in vitro and in vivo experiments [12, 13]. Although it has a high potential in bone regeneration, the HAP bio-reactivity when in a nanostructural form, is still not fully understood. TiO2 is one of the most corrosion – resistant materials that has been used intensively in orthopaedics, dentistry and a number of other applications [14]. Also it was previously shown that TiO2 nano-morphologically modified coatings can be used to reduce the adverse inflammatory effects of titanium implants and promote more advanced tissue healing following surgical procedures [14]. SWCNTs are superb one-dimensional nanostructures and their use in bio-medical applications range from highly accurate and sensitive biosensors [15], scaffolds for bone regeneration [16] and other bio-medical applications [17, 18]. Moreover, we have previously shown that carbon nanotubes can synergistically enhance the activity of various drugs [19]. In this work, the SWCNTs had a diameter ranging between 0.8 and 1.7 nm, average lengths of several microns and were prepared according to the methods presented in the Supporting Information section [20, 21].
In this work, we have demonstrated that these nanomaterials greatly enhance the level of the extracellular matrix formation and mineral deposition by MC3T3-E1 cells and the response was a function of the type of nanomaterials used. The most significant enhancement of cellular mineralization was observed when using AgNPs, followed by the HAP, TiO2 and SWCNTs. Interestingly, all the nanomaterials used in this study induced a higher mineralization in the MC3T3-E1 cells when compared to the control – untreated cells. Although the mechanism that governs the interactions between the nanomaterials and cells are not fully understood, it is thought that they may increase osteoblast cell cAMP production and intracellular calcium levels, affect DNA synthesis, alter collagen protein production and/or to be closely related to regulation of alkaline phosphatase (ALP) activity [22, 23].
Because nanomaterials have been observed to penetrate various sub-cellular compartments, including the nucleus, they may have the potential to alter normal biological processes. Given that the nanoparticles increased mineralization in these particular cells, they are expected to have affected the activity of ALP found on the bone cell membrane and which is considered an essential marker for bone cell proliferation and differentiation [7]. Alizarin red, a dye used to stain inorganic calcium deposition [24, 25], mineralized versus the unmineralized nodules [26], mineral deposition [27, 28], mineralized matrix [29] and mineralized bone nodule formation has been used extensively to study and quantify bone differentiation in model cell systems. In addition to alizarin red staining, electron diffraction spectroscopy (EDS), scanning electron microscopy (SEM), X-ray diffraction and Raman spectroscopy [30–33] have been used to examine and quantify mineralization of inorganic calcium formed by the bone cells. Nanostructural materials were shown to present negative cytotoxic effects due to the generation of reactive oxygen species, membrane disruption, disruption of protein or gene functions or DNA damaging [34]. Still, all the toxic effects induced by such nanoscale materials are mostly found to be dose dependent. As a result, a toxicological threshold has to be determined for each type of nanomaterials individually and has to take into consideration the shape, size, concentration, surface functionalization and the biological system to which they are exposed to. Most of the nanomedical procedures (ranging from cancer targeting and destruction to tissue engineering) involving the use of engineered nanomaterials, will have to be done at concentration values below the toxic threshold in order to limit any undesired toxic effects induced by the nanomaterials. The preliminary cytotoxicity studies (not presented here) performed for the nanoparticle concentrations used for the experiments presented in this manuscript, did not indicate the onset of any significant negative effects for the cells.
In addition to the observed phenotypic responses to the nanoparticles, we investigated the underlying molecular processes responsible for these observations. We chose to identify the genes associated with MC3T3 mineralization by evaluating microRNA (miRNA) regulation at various times subsequent to exposure of the cells to control media and control media supplemented with AgNPs. miRNAs comprise a family of 21–25 nucleotide non-coding small RNAs that regulate gene expression at the post-transcriptional level and participate in the regulation of almost every cellular process that has been investigated [35]. Currently, only a small number of miRNA target genes have been identified via direct experimentation. Instead, bioinformatic approaches, such as TargetScan, are used to determine the general principles governing miRNA target recognition and mechanism of action [36]. Recently, a study by Li et al.[37] detected miRNAs related to osteogenic differentiation induced by bone morphogenetic protein 2 (BMP2)-exposed pre-myogenic C2C12 cultured cells. They found that the miRNA predicted targets included the transcription factors Runx2 (Runt domain transcription factor), Msx2 (muscle segment 2 homeobox gene), Dlx3 (distal-less 3 homeobox gene), Smad1 (mothers against decapentaplegic transcription factor) and Smad5, as well as members of the Wnt/β-catenin pathway and BMPs and their receptors, and other signalling pathways that have been reported to promote osteogenesis. They concluded that BMP2 controls bone cell determination in this model in vitro cell system via a variety of different mechanisms. BMPs are multifunctional growth factors that play critical roles in embryonic development and cellular functions in postnatal and adult animals including acting as potent stimulators of bone formation [38].
Materials and methods
Nanoparticle synthesis and preparation
AgNPs (purity of 99.999 wt.%) with the average diameter of 23.0 ± 2.0 nm (Fig. S1) and the peak width at half-height of 7.1 ± 1.5 nm were prepared by borohydrate mediated reduction of silver nitrate and was based on the following protocol: in deionized water sodium borohydrate was introduced followed by sodium citrate followed by AgNO2 (drop wise) under slow stirring. Polyvinyl pyrrolidone was added to the solution and the mixture was stirred for 30 min. The resulting product was a golden yellowish in colour.
Single wall carbon nanotubes utilized in this work were synthesized on the bi-metallic catalyst system Fe-Co supported on MgO [20]. (Figs S2 and S3) HAP was purchased from Berkeley Advanced Biomaterials, Inc., (Berkeley, CA, USA) and they had a diameter of 20 ± 5 nm. The TiO2 nanoparticles were purchased from Nanostructures and Amorphous Materials, Inc. (Houston, TX, USA) and had a diameter of 20 ± 7 nm.
SEM images were obtained using a JEOL 7000F (Peabody, MA, USA) high-resolution scanning electron microscope coupled with an elemental analysis system EDAX and the Genesis Microanalysis software.
Cell culture
MC3T3 cells were purchased from ATCC, Inc. (Manassas, VA, USA) and maintained according to instructions provided. They were plated in 100 mm culture dishes at a density of 106/dish and supplemented by α– minimum essential medium with 10% foetal bovine serum (FBS) and 1% phosphatidylserine (PS) and incubated in 37 °C, 5% CO2 humidified incubator. Once at confluence, the cells were ethylenediaminetetraacetic acid trypsinized for further experiments. For experimental purposes, the cells were plated at a desired density in 24-well plates; 105/well and incubated for 24 hrs with 1 ml α-minimum essential medium with 10% FBS and 1% PS with or without nanoparticles (20 μg/ml). The nanoparticles of Ag, HAP, TiO2 and SWCNTs were all sonicated in the growth medium at a concentration of 20 μg/ ml and were introduced into the cell culture and incubated at 37°C in a 5% CO2 humidified incubator for 6 days until cells are confluent. The medium was changed every 48–72 hrs by aspirating half the volume and add 0.5 ml of fresh medium for each well. The cells were supplemented with the differentiation media as described in the following sections.
Characterization of the bone cells
Cells from the same passages were grown on 10 mm plastic cover slips at a density of (1 × 104 cells/dish) for the SEM images and supplemented with the growth medium as previously described. For the microscopic visualization, the cells were grown on 60 mm dishes under the same conditions and stained [tartrate-resistant acid phosphatase (TRACP) and ALP double-stain Kit] according to the manufacturer procedure.
Osteogenesis induction
The medium was aspirated and replaced by 1 ml of osteogenesis induction medium #1, containing approximately 99% cell culture medium, 0.02 mM/ml ascorbic acid 2 – phosphate solution and 1 mM/ml glycerol 2 – phosphate solution, this medium change corresponds to differentiation day 0 and was changed with 1 ml fresh osteogenesis induction medium #1 every 2–3 days.
On differentiation day 9, the medium was replaced by 1 ml fresh osteogenesis induction medium #2 by adding 5 nM/ml Melatonin solution to the osteogenesis induction medium #1, the medium was replaced by fresh osteogenesis induction medium #2 every 2–3 days.
Osteogenesis quantification assay
After 24 days the cells were fixed by 10% formaldehyde for 10 min. and washed 3 times for 5–10 min. each with 1× phosphate buffer saline and stained with an alizarin red stain solution by adding 400 μl to each well and incubated for 30 min. The stain was drained and the cells were washed three times with 1× phosphate-buffered saline (PBS) for 5–10 min. For the osteogenesis quantification 400 μl 10% acetic acid was added to each well and incubated for 30 min. with shaking to loosen the attached monolayer with the aid of a cell scraper; the cells and acetic acids were transferred to 1.5 ml microcentrifuge tubes and vortexed vigorously for 30 sec. The samples were heated to 85°C for 10 min. and transferred directly to ice for 5 min. for cooling. The samples were centrifuged at 20,000 ×g for 15 min. Four hundred microlitres of the supernatant was removed to 1.5 ml microcentrifuge tubes and 150 μl of ammonium hydroxide solution were added to each tube to neutralize the pH and insure it falls within the range of 4.1–4.5. Four hundred microlitres of the standard/sample were removed to the spectrophotometer cuvette and read at OD405 and the alizarin red stain concentration in each sample was plotted versus OD405. The alizarin red concentration in each sample was calculated according to the OD of the standard solution and the spectrophotometer was calibrated with a blank solution using 400 μl of 1× alizarin red S (ARS) dilution buffer.
Cells viability analysis and trypan blue assay
The cytotoxic effects of the nanomaterials were determined by trypan blue exclusion, a well known standard method to detect cell viability. The cells were cultured for 24 hrs with different nanomaterials at the concentrations of 20 μg/ml in the appropriate growth medium in a 48-well plate in a desired density. Then, the cells were dissociated with trypsin and transferred to 1.5 Eppendorf tubes and centrifuged. Finally, 25 μl of 1× trypan blue dye was added to each sample and incubated for less than 5 min. The viable cell number was counted using a haemacytometer, and the viability values were compared to the negative control. Figure S4 shows the proliferation rates of the cells unexposed to any nanomaterials.
Alizarin red staining
Cells were washed once with distilled water and then fixed with phosphate-buffered formalin for 20 min. The fixed cells were also washed once with distilled water and subsequently stained with 1% ARS solution for 5 min., the remaining dye was washed out twice with distilled water and the cells were washed once more and then dehydrated with ethanol.
ALP double staining
After washing with PBS, the cells were incubated with AgNPs as described above for three different time-points (6, 15, 24 days) and were fixed with the fixative solution [citrate buffer (pH 5.4) containing 60% acetone and 10% methanol] for 10 min. at room temperature. After fixation, the cells were washed thoroughly with PBS and incubated with the substrate solution for acid phosphatase for 30 min. at 37°C, for double staining of the ALP; the cells were incubated with the substrate solution for the ALP after washing and incubated for another 30 min. For the nuclear staining the cells were stained with methyl green and incubated for 5 min. at room temperature and washed three times with sterilized distilled water. The cells were visualized under the microscope.
Alkaline phosphatase activity assay
The bone cells were incubated with AgNPs as described above. Whole-cell extracts were collected at 0 day from the untreated cells and after 6 days of differentiation for the AgNPs treated cells, and incubated for 60 min. with 50 mM Tris-HCl (pH 8.8), 10 mM MgCl2 and 20 mM of p-nitrophenylphosphate, at 37°C. Following incubation the absorbance at 405 nm was measured spectrophotometrically to quantify the amount of p- nitrophenylphosphate produced. Total cell protein was assayed according to the method described by the commercial kit supplier (ALPase assay; Takara Bio, Inc., Shiga, Japan).
miRNA isolation
The cells were rinsed twice with cold 1× PBS and lysed in 600 μl RNA lysis/binding buffer (Ambion, Inc., Austin, TX, USA). miRNAs were isolated using mirVana™ miRNA isolation kit (Ambion, Inc.) that specifically capture small RNAs with lengths of less than 200 nucleotides. The RNAs were initially eluted in 100 μl nuclease-free water (Ambion, Inc.) and then dried using a Savant DNA 110 SpeedVac vacuum concentrator (Thermo Scientific, Pittsburgh, PA, USA) to increase RNA concentrations. RNA concentrations were determined using a NanoDrop 1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
PCR Array analysis of miRNA expression
Two hundred nanograms of enriched small RNAs were converted into cDNA using RT2 miRNA First Strand Kit (SABiosciences Corporation, Frederick, MD, USA). The cDNAs were mixed with 2× RT2 SYBR Green PCR Master Mix (SABiosciences Corporation) and dispersed into 384-well Mouse Genome miRNA PCR Array (MAM-3100E, SABiosciences Corporation) with 10 μl/well reaction mix. The PCR array contained a panel of primer sets for 376 mouse miRNAs, four small RNAs as the internal controls and four quality controls. The real-time qRT-PCR was performed on a 7900 real-time PCR system (Applied Biosystems, Inc., Foster, CA, USA) with the following cycling parameters: 95°C for 10 min., then 40 cycles of 95°C for 15 sec., 60°C for 30 sec. and 72°C for 30 sec. SYBR Green fluorescence was recorded from every well during the annealing step of each cycle. The threshold cycle (Ct) value of each sample was calculated with software SDS 2.3 (Applied Biosystems, Inc.). To calculate Cts, we set the threshold line as 0.15 and kept it the same across all of the analyses. The baseline was automatically defined by the software.
Normalization and statistical analysis
Normalization and statistical analysis of miRNA expression were conducted using SABiosciences Corporation’s Online PCR Array Data Analysis Web Portal. The ΔΔCt method was utilized to calculate the fold change (FC). Four genes, snoRNA251, snoRNA202, snoRNA142 and U6 in the PCR arrays, were used as endogenous controls to normalize each sample. To determine the effects of nanoparticle treatment, the formula: FC = 2^ [–(mean of ΔCt values of nanoparticle-treated samples at a specified time-point – mean of ΔCt values of control samples at the same time-point)] was used for up-regulated genes, while FC =−2^ (mean of ΔCt values of nanoparticle-treated samples at a specified time-point – mean of ΔCt values of control samples at the same time-point) was used for the down-regulation genes, and P-values were calculated using t-tests between nanoparticle-treated and control samples at each time-point to determine whether there was a significant difference for miRNA expression. To determine the miRNA response in untreated MC3T3 cell, the formula: FC = 2^ [−(mean of ΔCt values of samples after initiation of the MC3T3 cell culture – mean of ΔCt values of control samples at day 0)] was used for up-regulated genes, while FC =−2^ (mean of ΔCt values of samples after initiation of MC3T3 cell culture – mean of ΔCt values of control samples at day 0) was used for the down-regulations, and P-values were calculated using t-tests between day 0 samples and samples at each of the subsequent time-points to determine whether there is a significant difference for miRNA expression.
Prediction of target genes
The predicted target genes of differentially expressed miRNAs were obtained from TargetScan database (http://www.targetscan.org/). TargetScan predicts biological targets of miRNAs by searching for the presence of conserved 8 mer and 7 mer sites that match the seed region of each miRNA [39, 40]. TargetScan is a widely used database for miRNA target gene prediction and was demonstrated that its prediction matches best with the experimental data from proteomics [41].
Results
Figure 1 shows the experimental design used in this study. The cells were incubated with either control medium or with medium plus nanoparticles for 24 hrs. After removal of the nanoparticles, the cultures were further incubated with fresh medium. At confluence (day 6), all of the cultures were supplemented with Osteo I medium for 6 days and then supplemented with Osteo II medium and further cultured to day 24. In the cultures treated with AgNPs, in addition to mineralization, miRNAs expression was also analysed at 6, 15 and 24 days after the treatment with these nanoparticles.
Fig 1.
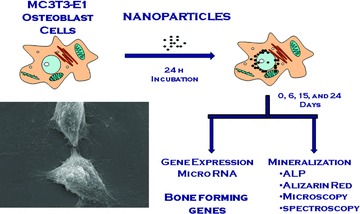
Diagram of the experimental design: MC3T3-E1 cells were incubated with nanomaterials for 24 hrs, after which the nanomaterials were removed. This point was considered as day 0. The cells were further incubated with fresh medium and Osteo I and/or Osteo II media for up to 24 days and the effects of the nanoparticles on miRNA expression and mineralization were assessed on days 6, 15 and 24.
During the 24 hr incubation process, the nanoparticles appear to have penetrated cells suggesting their possible location in the cytoplasm and nucleus. Figure 2 shows the optical images of the MC3T3-E1 cells before and after incubation with the four types of nanomaterials used in this study. It is clearly seen that most of the nanoparticles coalesce around the nucleus while some appeared to penetrate and accumulate inside the nucleus. As a result, they are expected to strongly interact with the various sub-cellular environments and possibly produce significant genetic and functional modifications.
Fig 2.
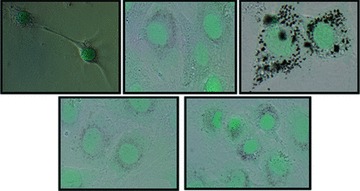
Representative photomicrograph showing the characteristic features of the bone cells stained with methyl green (A) control-unexposed to nanomaterials; (B) incubated with AgNPs; (C) SWCNTs; (D) HAP and (E) TiO2.
Effect of nanomaterials on cell mineralization in vitro
The formation of mineralized bone nodules is considered a marker for the final stages of osteoblast differentiation and can be analysed and quantified by staining fixed cell populations with ARS. To demonstrate the value of alizarin red staining in the observation of mineralized nodule formation, we have imaged all the conditions up to 24 days of culture. Elution and analysis of the ARS levels bound to the mineralized nodules (Fig. 3A–E) confirmed an enhanced matrix formation in the osteoblast cell cultures that were exposed to the four types of nanomaterials. The results showed that there was negligible staining for the control cells grown in Osteo I and II media (unexposed to any nanoparticles) with only few areas of weak alizarin red staining (Fig. 3A). The effect of enhanced mineralization varied with the type of nanomaterials, with the AgNPs having the most significant effect (Fig. 3F). Figure 3G and H shows the SEM and elemental analysis (EDS) of the mineralization nests, indicating the presence of Ca, P, O typical to mineralized structures.
Fig 3.
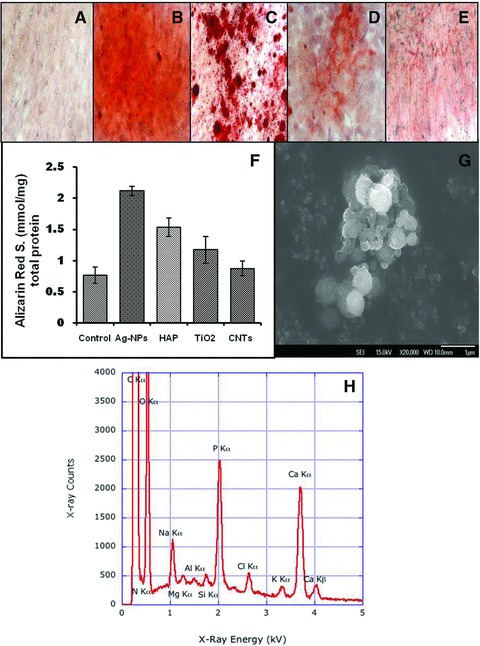
The mineralized nodule formation of osteoblasts in the presence of nanomaterials stained by alizarin red stain. All samples were incubated with Osteo I and II differentiation media (cf. ‘Materials and methods’): (A) cells without nanomaterials; (B) cells with AgNPs; (C) cells with HAP, (D) cells with TiO2; (E) cells with SWCNTs. (F) Effect of nanomaterials type on the concentration of ARS stain when osteoblastic bone cells were incubated in the presence of (20 μg/ml) of AgNPs, HAP, TiO2 and SWCNTs compared with the control samples. (G) SEM image of a mineralization nests and (H) elemental analysis of the mineral nests by EDS indicating the specific elements that are present in the mineral tissue. The experiments were assessed on day 24. Alizarin red concentrations were determined by comparing the samples OD405 with a standard sample of 2 mM of ARS diluted with 1× ARS dilution buffer and expressed as the concentration of the eluted ARS normalized by the standard protein.
Because the AgNPs induced the highest level of mineralization (increase in both area and intensity of mineralized nodules) when introduced into cell cultures, they were further used to investigate the kinetics of mineral formation at several time periods (6, 15 and 24 days). Figure 4A and B shows a significant increase in the level of alizarin red stain for the cells exposed to AgNPs when compared to the control and the trend was found to increase as the time progressed from 6 to 15 and 24 days of incubation. Figure 4B shows the actual stained Petri dishes clearly indicating that the cells exposed to AgNPs induced higher mineralization (darker red colour) throughout the 24 days as compared to the controls. Given its significant effect on mineralization, as compared to the other conditions, only the AgNPs were used in further studies to understand the role of the nanomaterials on ALP activity and gene expression.
Fig 4.
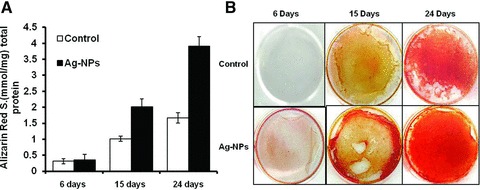
Effect of AgNPs on the concentration of alizarin red stain as a function of time, 105 cells were plated per 35 mm well with and without AgNPs (20 μg/ml) and incubated for 6, 15 and 24 days (A). The results were derived from three experiments, with six cultures for each variable in each experiment. Bars represent the concentration of the eluted ARS stain which is normalized with the standard dye. (B) Actual stained Petri dishes with alizarin red for control and the cell cultured with AgNPs for 6, 15 and 24 days.
Induction of alkaline phosphatase activity by AgNPs
ALP is an enzyme commonly used as a marker for the calcification level during the bone maturation process [42]. The effect of the AgNPs on ALP activity and level was measured at various points in time. The level of ALP was found to increase significantly when the cells were exposed to AgNPs, as compared to the control samples, confirming the increased level of mineralization presented in Figures 3 and 4. Figure 5 shows the increased level of ALP activity for the cells exposed to AgNPs for 24 hrs and further incubated in fresh medium for 6 days.
Fig 5.
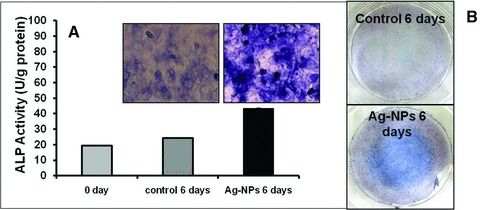
Effect of AgNPs on the ALP activity of MC3T3-E1 cells. (A) Microscopic images showing the enhanced level of ALP after 6 days after AgNP exposure. The cells were stained by ALP double staining and the level of the ALP enzyme was evaluated. (B) Actual images of the ALP stained Petri dishes for the control and the cells exposed to AgNPs and cultured for 6 days.
Discussions
We suggest that certain marker morphogenetic proteins and/or important transcriptional factors could be involved in the effects observed in cells exposed to AgNPs. In order to identify the target genes involved in activation of mineralization in response to AgNPs, miRNA microarray analysis was performed. This assay was recently used for identification of gene targets that are positive regulators of bone formation [34]. In our set of experiments, miRNA assessment allowed us to determine structural and regulatory genes required for bone formation that were significantly affected in control cultures of MC3T3-E1 cells, as well as cultures that were exposed to AgNPs. We chose to detect miRNA expression profiles on the same days that the phenotypic response to AgNPs was determined (Fig. 4). Tables 1 and 2 show selected up-regulated miRNAs and predicted gene targets, associated with particular miRNAs, in control cells and cells exposed to AgNPs at the three time-points. Although several BMPs were affected in the control cells due to the presence of osteogenesis differentiation media, there was a delay of several days in miRNA regulation of expression for a number of BMPs compared with the AgNPs exposed cultures (Tables 1 and 2). Thus, no BMP gene targets were detected on day 6 of culture in the controls, while a large number of these multifunctional bone forming growth factors were observed on day 6 in the AgNPs exposed cultures. Additionally, the exposure of cells to AgNPs not only resulted in the miRNA regulation of similar BMPs that were found in the control cultures, but also included several gene-target BMPs not found in the controls; for example BMP3, BMP6, BMP7 and BMP8B. BMP2 is a critical protein required to induce osteoblast differentiation [35] and the AgNPs were found to induce expression of miRNAs associated with the gene that is responsible for the production of this protein on day 6 of the experiment. We suggest that significant changes in miRNAs that are related to the regulation of BMPs in cells exposed to AgNPs play an important role in the process of mineralization in these cells. The data presented in Tables 1 and 2 support our hypothesis. We found that miRNA regulation of essential transcriptional factors involved in osteoblast formation exhibited a similar trend that was documented for BMPs. The up-regulation of several miRNAs related to transcription factors was delayed in control (unexposed) cells compared to AgNPs-exposed cells (Tables 1 and 2). Regulation of miRNAs that can affect Smad5 transcriptional factor appeared on day 6 in AgNPs-exposed cells but not in control cells. The number of transcriptional factors associated with bone formation such as Runx2, Dlx3, Msx2 were affected by correspondent miRNAs only in cells exposed to AgNPs. Runx2 is a common target of transforming growth factor β1 and BMP2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12 [43]. Lee and his colleagues also showed that Runx2 was a major target of BMP2 in pluripotent mesenchymal cells and that osteoblast specific gene expression was also dependent on the transcription factor Smad5, an upstream regulator of Runx2 [37, 44]. Our data (Tables 1 and 2) unequivocally indicated that AgNPs are not only responsible for the miRNA regulation of expression of Smad1/5 but also of Runx2. Both Runx2 and Smad1/5 are transcription factors essential for osteogenesis and activate bone-specific genes in a synergistic way. The difference in the regulation of BMPs and their transcription factors is most likely responsible for the differential Alizarin stain intensity observed in Figures 3 and 4.
Table 1.
Regulatory genes (transcriptional factors) and selected structural genes regulated by overexpression of miRNAs in control cells grown in the presence of osteogenesis I and II growth media
| Day of exposure | miRNA related to putative structural genes | Putative structural genes (gene targets of miRNAs) | miRNA related to putative transcription factors | Putative transcription factors (gene targets of miRNAs) |
|---|---|---|---|---|
| 6 | None | None | None | None |
| 15 | mir-874 | BMP1 | mir-133; mir-142–5p | Dlx3 |
| mir-142–5p; | BMP2 | mir-300; let-7d | Smad1 | |
| mir-17 | BMPR1B | mir-17 | Smad5 | |
| mir-101a | BMPR2 | |||
| mir-17 | CRIM1 | |||
| mir-17 | ||||
| 24 | mir-345–5p | BMP2 | mir-497 | Smad5 |
| mir-497 | BMPR1A | |||
| mir-361 | BMPR2 | |||
| mir-497 | BMP8A |
BMP1, BMP2, BMP8A: bone morphogenetic protein 1, 2, 8A, respectively; BMPR1A, BMPR1B: bone morphogenetic protein receptor, type 1A, type 1B, respectively; BMPR2: bone morphogenetic protein receptor, type 2; CRIM1: cysteine-rich transmembrane BMP regulator 1; Dlx3: Distal-less 3 homeobox gene; Smad1 and Smad5: mothers against decapentaplegic transcription factor.
Table 2.
Regulatory genes (transcriptional factors) and selected structural genes regulated by overexpression of miRNAs in cells exposed to osteogenesis I and II growth media in presence of AgNPs
| Day of exposure | miRNA related to putative structural genes | Putative structural genes (gene targets of miRNAs) | miRNA related to putative transcription factors | Putative transcription factors (gene targets of miRNAs) |
|---|---|---|---|---|
| 6 | mir-374 mir-374 mir-29b mir-721 mir-721 mir-295; mir-374 | BMP2 BMP3 BMPR1A BMPR1B BMPR2 CRIM1 | mir-130a; mir-130b | Smad5 |
| 15 | mir-124 mir-325 mir-16 mir-331–3p; mir-763 mir-503; mir-29b; mir-29c; mir-16; mir-130a; mir-130b; mir-721; mir-381 mir-466d-3p; mir-101b; mir-124 mir-291a-5p; mir-742; mir-153; mir-106b; mir-130a; mir-130b; mir-721; mir-93; mir-25; mir-92b; mir-19a; mir-361; mir-351; mir-381 mir-466d-5p; mir-760; mir-19a mir-335–5p; mir-23b; mir-16; mir-106b; mir-93; mir-18a; mir-295; mir-302b | BMP6 BMP7 BMP8A BMP8B BMPR1A BMPR1B BMPR2 BMPER CRIM1 | mir-325; mir-30b, mir-217; mir-466d-3p; mir-23b mir-381 mir-19a; mir-29c mir-182; mir-291–5p; mir-30b mir-130a; mir-130b; mir-721; mir-224; mir-19a; mir-106b; mir-93; mir-16; mir-291a-5p; mir-23b | Runx2 Msx2 Dlx3 Smad1 Smad5 |
| 24 | mir-141 | BMP1 | mir-384-p mir-384p mir-486 | Runx2 Dlx3 Smad5 |
Runx2: Runt domain transcription factor; Msx2: muscle segment 2 homeobox gene. See Table 1 for description of the other genes and transcription factors.
The miRNA studies clearly show that the AgNPs have significant effects on bone-specific genes or transcriptional factors regulation through miRNAs inducing an enhanced formation of bone morphogenic proteins with positive effects on the overall minerals deposited by the cells. Although in this work we mostly focused on the analysis of regulation of the genes and the transcriptional factors affecting bone formation, the AgNPs were also found to affect a number of genes that are related to stress, cellular assembly and organization, cellular function and maintenance, cell to cell signalling and interaction, protein synthesis and degradation, and carbohydrate metabolism (data not shown here). The mechanisms by which the nanoparticles are inducing these effects are still under investigation using a combination of genomic, proteomic and bioinformatics approaches. Further investigation of the ‘cross-talk’ between transcriptional factors and BMPs in bone cells is important in order to understand the processes by which nanoparticles, especially AgNPs, enhance bone formation. To our knowledge, these are the first data indicating that a large number of engineered nanomaterials play a significant role in bone mineralization at the genomic level and suggesting that other nanoparticles not evaluated in this study may also alter mineralization in these cells in a similar manner. Although the use of nanomaterials for bone generation has been previously reported, the major goal for these studies was mostly to create nontoxic 3D nano-based scaffolds for cellular proliferation and tissue growth. In this work we show that the uptake of the nanoparticles by bone cells was found to have a positive effect on the overall mineralization level of these cells. As a result, these results can be the foundation of a bone regeneration technological platform, in which antibody targeted nanoparticles can be delivered in vivo to the osteoblast cells only and increase their corresponding level of mineral formation. Such approaches could find excellent applications in addressing medical conditions such as osteoporosis, bone death or resorption without the need for surgical interventions.
Conclusions
A thorough evaluation of the effects due to various nanomaterials on cell calcification and bone matrix formation in MC3T3-E1 bone cells is very important, given the newly found nanoparticles–cells interactions and the nanoparticles-induced enhanced extracellular bone matrix formation. In this study, it was found that several nanomaterials induced calcification of MC3T3-E1 cells with the highest response being induced by the AgNPs. The enhanced magnitude of the mineralization and ALP expression in the cells incubated with AgNPs was significantly greater than the levels of the enzyme analysed in the cells treated with the other nanomaterials studied (SWCNTs, TiO2 and HAP). Alizarin red staining is still the standard method to visualize mineralization in osteogenic cell cultures. In the present study the number of the nodules formed in the culture of bone cells treated with AgNPs was higher than those induced by the other nanomaterials and untreated control cells. In addition to these phenotypic responses, we found that the exposure of cells to AgNPs affected genes responsible for osteogenic differentiation. The miRNA analysis indicated that miRNA regulation and expression of essential transcriptional factors and BMPs played an important role in activating the process of mineralization in bone cells exposed to AgNPs. To our knowledge, this is the first report that describes the regulation of a specific developmental activity by a nanomaterial resulting in a beneficial outcome.
Acknowledgments
Financial support from the Arkansas Science and Technology Authority (ASTA) grant #08-CAT-03 and the U.S. Army Telemedicine and Advanced Research Center is highly appreciated.
Conflict of interest
The authors confirm that there are no conflicts of interest.
Supporting Information
TEM analysis of the AgNPs used for this study.
Fig. S2 TEM image of SWCNTs utilized in this research.
Fig. S3 Raman spectrum of SWCNTs collected with the 633 nm laser excitation of the full frequency domains showing all the corresponding vibrational modes. The inset represents a magnified area of the RBM region.
Fig. S4 Overall proliferation rates of pre-osteoblasts in culture. Cell proliferation was assessed by direct counting the cells at various time points and showed that the growth rates reached a plateau after 12 days. The cells were plated in 35 mm dishes with an initial density of 200,000 cells and the medium was regularly changed every 2-3 days.
References
- 1.Usui Y, Aoki K, Narita N, et al. Carbon nanotubes with high bone-tissue compatibility and bone-formation acceleration effects. Small. 2008;4:240–6. doi: 10.1002/smll.200700670. [DOI] [PubMed] [Google Scholar]
- 2.Oh S, Daraio C, Chen LH, et al. Significantly accelerated osteoblast cell growth on aligned TiO2 nanotubes. J Biomed Mater Res A. 2005;78:97–103. doi: 10.1002/jbm.a.30722. [DOI] [PubMed] [Google Scholar]
- 3.Wen HC, Lin YN, Jian SR, et al. Observation of growth of human fibroblasts on Silver. J Phys: Conf Ser. 2007;61:445–9. [Google Scholar]
- 4.Zhang L, Webster TJ. Nanotechnology and nanomaterials: promises for improved tissue regeneration. Nanotoday. 2009;4:66–80. [Google Scholar]
- 5.Dobson J. Gene therapy progress and prospects: magnetic nanoparticle-based gene delivery. Gene Ther. 2006;13:283–7. doi: 10.1038/sj.gt.3302720. [DOI] [PubMed] [Google Scholar]
- 6.Zhang JC, Li XX, Xu SJ, et al. Effects of rare earth ions on proliferation, differentiation and function expression of cultured osteoblasts in vitro. Prog Nat Sci. 2004;14:404–9. [Google Scholar]
- 7.Dean DD, Schwartz Z, Bonewald L, et al. Matrix vesicles produced by osteoblast-like cells in culture become significantly enriched in proteoglycan-degrading metalloproteinases after addition of beta-glycerophosphate and ascorbic acid. Calcif Tissue Int. 1994;54:399–08. doi: 10.1007/BF00305527. [DOI] [PubMed] [Google Scholar]
- 8.Matsumoto T, Igarashi C, Takeuchi Y, et al. Stimulation by 1,25-dihydroxyvitamin D3 of in vitro mineralization induced by osteoblast-like MC3T3-E1 cells. Bone. 1991;12:27–32. doi: 10.1016/8756-3282(91)90051-j. [DOI] [PubMed] [Google Scholar]
- 9.Mahmood M, Casciano DA, Mocan T, et al. Cytotoxicity and biological effects of functional nanomaterials delivered to various cell lines. J Appl Toxicol. 2010;30:74–83. doi: 10.1002/jat.1475. [DOI] [PubMed] [Google Scholar]
- 10.Wen HC, Lin YN, Jian SR, et al. Observation of growth of human fibroblasts on silver nanoparticles. J Phys. 2007;61:445–9. [Google Scholar]
- 11.Doering WE, Nie S. Single-molecule and single-nanoparticle SERS: examining the roles of surface active sites and chemical enhancement. J Phys Chem B. 2002;106:311–7. [Google Scholar]
- 12.Botelho CM, Brooks RA, Best SM, et al. Human osteoblast response to silicon-substituted hydroxyapatite. J Biomed Mater Res A. 2006;79:723–30. doi: 10.1002/jbm.a.30806. [DOI] [PubMed] [Google Scholar]
- 13.Chang YL, Stanford CM, Keller JC. Calcium and phosphate supplementation promotes bone cell mineralization: implications for hydroxyapatite (HA)-enhanced bone formation. J Biomed Mater Res. 2000;52:270–8. doi: 10.1002/1097-4636(200011)52:2<270::aid-jbm5>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 14.Liao H, Wurtz T, Li J. Influence of titanium ion on mineral formation and properties of osteoid nodules in rat calvaria cultures. J Biomed Mater Res. 1999;47:220–7. doi: 10.1002/(sici)1097-4636(199911)47:2<220::aid-jbm12>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 15.Chen RJ, Bangsaruntip S, Drouvalakis KA, et al. Noncovalent functionalization of carbon nanotubes for highly specific electronic biosensors. PNAS. 2003;100:4984–9. doi: 10.1073/pnas.0837064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi X, Sitharaman B, Pham QP, et al. In vitro cytotoxicity of single-walled carbon nanotube/biodegradable polymer nanocomposites. J Biomed Mater Res. 2007;86A:813–23. doi: 10.1002/jbm.a.31671. [DOI] [PubMed] [Google Scholar]
- 17.Zhang D, Yi C, Zhang J, et al. The effects of carbon nanotubes on the proliferation and differentiation of primary osteoblasts. Nanotechnology. 2007;18:475102. doi: 10.1007/978-1-60761-579-8_5. [DOI] [PubMed] [Google Scholar]
- 18.Magrez A, Kasas S, V Salicio, et al. Cellular toxicity of carbon-based nanomaterials. Nano Lett. 2006;6:1121–5. doi: 10.1021/nl060162e. [DOI] [PubMed] [Google Scholar]
- 19.Mahmood M, Karmakar A, Fejleh A, et al. Synergistic enhancement of cancer therapy using a combination of carbon nanotubes and anti-tumor drug. Nanomedicine. 2009;4:883–93. doi: 10.2217/nnm.09.76. [DOI] [PubMed] [Google Scholar]
- 20.Dervishi E, Li Z, Watanabe F, et al. Thermally controlled synthesis of single-wall carbon nanotubes with selective diameters. J Mater Chem. 2009;19:3004–12. [Google Scholar]
- 21.Dervishi E, Li Z, Xu Y, et al. The influence of Fe–Co/MgO catalyst composition on the growth properties of carbon nanotubes. Part Sci and Tech. 2009;27:222–37. [Google Scholar]
- 22.Montjovent MO, Burri N, Silke M, et al. Fetal bone cells for tissue engineering. Bone. 2004;35:13323–31. doi: 10.1016/j.bone.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 23.Kawazoe Y, Shiba T, Nakamura R, et al. Induction of calcification in MC3T3-E1 cells by inorganic polyphosphate. J Dent Res. 2004;83:613–8. doi: 10.1177/154405910408300806. [DOI] [PubMed] [Google Scholar]
- 24.Bellows CG, Aubin JE, Heersche JN, et al. Mineralized bone nodules formed in vitro from enzymatically released rat calvaria cell populations. Calcif Tissue Int. 1986;38:143–154. doi: 10.1007/BF02556874. [DOI] [PubMed] [Google Scholar]
- 25.Gerstenfeld LC, Chipman SD, Glowacki J, et al. Expression of differentiated function by mineralizing cultures of chicken osteoblasts. Dev Biol. 1987;122:49–60. doi: 10.1016/0012-1606(87)90331-9. [DOI] [PubMed] [Google Scholar]
- 26.Bellows CG, Aubin JE, Heersche JN. Physiological concentrations of glucocorticoids stimulate formation of bone nodules from isolated rat calvaria cells in vitro. Endocrinology. 1987;121:1985–92. doi: 10.1210/endo-121-6-1985. [DOI] [PubMed] [Google Scholar]
- 27.Marsh ME, Munne AM, Vogel JJ, et al. Mineralization of bone-like extracellular matrix in the absence of functional osteoblasts. J Bone Miner Res. 1995;10:1635–43. doi: 10.1002/jbmr.5650101105. [DOI] [PubMed] [Google Scholar]
- 28.Owen TA, Aronow M, Shalhoub V, et al. Progressive development of the rat osteoblast phenotype in vitro: reciprocal relationships in expression of genes associated with osteoblast proliferation and differentiation during formation of the bone extracellular matrix. J Cell Physiol. 1990;143:420–30. doi: 10.1002/jcp.1041430304. [DOI] [PubMed] [Google Scholar]
- 29.Chen D, Chen H, Feng JQ, et al. Osteoblastic cell lines derived from a transgenic mouse containing the osteocalcin promoter driving SV40 T-antigen. Mol Cell Diff. 1995;3:193–212. [Google Scholar]
- 30.Gregory CA, Grady GW, Peister A, et al. An Alizarin red-based assay of mineralization by adherent cells in culture: comparison with cetylpyridinium chloride extraction. Anal Biochem. 2004;329:77–84. doi: 10.1016/j.ab.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 31.Breen EC, Ignotz RA, McCabe L, et al. TGF beta alters growth and differentiation related gene expression in proliferating osteoblasts in vitro, preventing development of the mature bone phenotype. J Cell Physiol. 1994;160:323–35. doi: 10.1002/jcp.1041600214. [DOI] [PubMed] [Google Scholar]
- 32.Katayama UK, Miyamoto T, Koshihara T. Interleukin-4 enhances in vitro mineralization in human osteoblast-like cells. Biochem Biophys Res Commun. 1992;189:1521–6. doi: 10.1016/0006-291x(92)90248-j. [DOI] [PubMed] [Google Scholar]
- 33.Rey C, Kim HM, Gerstenfeld L, et al. Structural and chemical characteristics and maturation of the calcium phosphate crystals formed during the calcification of the organic matrix synthesized by chicken osteoblasts in cell culture. J Bone Miner Res. 1995;10:1577–88. doi: 10.1002/jbmr.5650101020. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Ali SF, Dervishi E, et al. Cytotoxicity effects of graphene and single-wall carbon nanotubes in neural phaeochromocytoma-derived PC12 cells. ACS Nano. 2010;4:3181–6. doi: 10.1021/nn1007176. [DOI] [PubMed] [Google Scholar]
- 35.Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol. 2009;10:141–8. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 36.Chen T. The role of microRNA in chemical carcinogenesis. J Env Sci Health C. 2010;28:1–36. doi: 10.1080/10590501.2010.481477. [DOI] [PubMed] [Google Scholar]
- 37.Li Z, Hassan MQ, Volinia S, et al. A microRna signature for a BMP-induced osteoblast lineage commitment program. Proc Natl Acd Sci USA. 2008;105:13906–11. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004;22:233–41. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- 39.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 40.Grimson A, Farh KK, Johnston WK, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baek D, Villén J, Shin C, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beck GR, Sullivan EC, Moran E, et al. Relationship between alkaline phosphatase levels, osteopontin expression and mineralization in differentiating MC3T3–E1 osteoblasts. J Cell Biochem. 1998;68:269–80. doi: 10.1002/(sici)1097-4644(19980201)68:2<269::aid-jcb13>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 44.Lee KS, Kim HJ, Li QL, et al. Runx2 is a common target of transforming growth factor beta 1 and bone morphogenetic protein 2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol Cell Biol. 2000;20:8783–92. doi: 10.1128/mcb.20.23.8783-8792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TEM analysis of the AgNPs used for this study.
Fig. S2 TEM image of SWCNTs utilized in this research.
Fig. S3 Raman spectrum of SWCNTs collected with the 633 nm laser excitation of the full frequency domains showing all the corresponding vibrational modes. The inset represents a magnified area of the RBM region.
Fig. S4 Overall proliferation rates of pre-osteoblasts in culture. Cell proliferation was assessed by direct counting the cells at various time points and showed that the growth rates reached a plateau after 12 days. The cells were plated in 35 mm dishes with an initial density of 200,000 cells and the medium was regularly changed every 2-3 days.


