Abstract
Targeting and down-regulation of ErbB2, a member of EGF receptor family, is regarded as one of the key aspect for cancer treatment because it is often overexpressed in breast and ovarian cancer cells. Although natural ligands for ErbB2 have not been found, unlike other ErbB receptors, EC-1, a 20-amino acid circular peptide, has been shown to bind to ErbB2 as an artificial ligand. Previously we showed EC-1 peptide did not induce the internalization of ErbB2 in SK-BR-3 cells. In this report, we designed divalent and multivalent forms of EC-1 peptide with the Fc portion of the human IgG and bionanocapsule modified with ZZ-tag on its surface to improve the interaction with ErbB2. These forms showed higher affinity to ErbB2 than that of EC-1 monomer. Furthermore, prominent endosomal accumulation of ErbB2 occurred in SK-BR-3 cells when stimulated with EC-Fc ligand multivalently displayed on the surface of the bionanocapsule, whereas SK-BR-3 cells as themselves displayed stringent mechanism against ErbB2 internalization without stimulation. The multivalent form of EC-1 peptide appeared to internalize ErbB2 more efficiently than divalent form did. This internalization was unaffected by the inhibition of clathrin association, but inhibited when the cholesterol was depleted which explained either caveolar or GPI-AP-early endocytic compartment (GEEC) pathway. Because of the lack of caveolin-1 expression, caveolar machinery may be lost in SK-BR-3 cell line. Therefore, it is suggested that the multivalent form of EC-1 induces the internalization of ErbB2 through the GEEC pathway.
Keywords: ErbB2, EC-1 peptide, internalization, multivalent display, bionanocapsule
Introduction
The epidermal growth factor receptor (EGFR) family consisting of ErbB1 (EGFR), ErbB2, ErbB3 and ErbB4 is known to play a crucial part in the development and progression of cancers. The signal for cell survival, differentiation and proliferation is achieved through both homo- and heterodimerization of the receptors [1, 2]. Anomalous increase in the expression of the ErbB2 is an identified factor for the development of several cancer types [1, 3] and high ErbB2 levels parallel the worse prognosis for breast cancer patients [4–6]. ErbB2 also associates with ErbB3 in the hetero dimerization and is considered the most active dimer that initiates cancers [7, 8]. ErbB2 is overexpressed in breast (30%) and ovarian (15–30%) cancer [9–11]. The myriad roles played by ErbB2 have made it one of the most preferred targets for the treatment of cancer. ErbB2 stringently internalizes efficiently recruiting back to the cell surface [12]. The mechanism of ErbB2 recycling and degradation is complex and appears to vary depending on the cell types. Various groups have shown that geldanamycin treatment efficiently internalizes the ErbB2 ligand [13–15]. Although the internalization of ErbB2 is enhanced when multivalently cross-linked with antibody such as anti-ErbB2 antibody sc-08 [13, 16–19], there is no known natural ligand that promotes the ErbB2 internalization till date. By phage display method, small peptides, which possess binding affinity to ErbB2, have been isolated [20–22]. EC-1 peptide, a cyclic 20-amino-acid-peptide, is also one of the artificial ErbB2 ligands isolated from random peptide library of phage display [23]. EC-1 peptide is considered to abolish the tyrosine phosphorylation of the intracellular domain of ErbB2 and to inhibit the proliferation of SK-BR-3 cells overexpressing ErbB2.
Previously, EC-1 peptide fused to GFP was shown to stimulate internalization of ErbB2 in SK-OV-3 cells whereas ErbB2 in SK-BR-3 cells retained on the surface [24]. Because anti-ErbB2 antibody sc-08 stimulated the internalization of ErbB2 in both cell lines, the mechanism of internalization stimulated by EC-1 peptide was considered quite different from that by anti-ErbB2 antibody sc-08. In this study, we designed multivalent form using EC-1 peptide fused to human IgG-Fc domain (EC-Fc) and bionanocapsule displaying ZZ-tag on the surface [25]. We then asked whether the multivalent forms stimulated the internalization of ErbB2 in SK-BR-3 cells.
Materials and methods
Cell cultures
Human breast carcinoma derived cell lines, MCF-7, MDA-MB-453 and SK-BR-3, and human ovarian carcinoma cell line SK-OV-3 were from the ATCC (VA). SK-BR-3 and SK-OV-3 cells were grown at 37°C in RPMI-1640 medium supplemented with 10% FBS and MCF-7 in DMEM medium supplemented with 10% FBS under an atmosphere of 5% CO2 and MDA-MB-453 cells in Leibovitz’s L-15 medium supplemented with 10% FBS without conditioning CO2. All culture media were supplemented with 2 mM glutamine just before use.
Construction of expression plasmids
The expression plasmid for EC-Fc was constructed as follows. The DNA fragment coding human IgG-Fc domain was excised from the plasmid pBO593 [26] with restriction enzymes AgeIand NotIand then ligated at the 3′-end of a synthetic oligonucleotide coding for the EC-1 peptide. The resultant sequence coding EC-Fc was ligated to the 3′-end of the sequence coding the signal peptide derived from human RNase1 [27]. Finally, the sequence coding EC-Fc with a secretion signal peptide was cloned downstream of CMV promoter to construct the plasmid pBO854 with hygromycin resistant gene. Simultaneously, the expression vector for Fc domain without EC-1 moiety was constructed in the same procedure to construct the plasmid pBO853.
Preparation of EC-Fc protein
Chinese hamster ovary (CHO) cells were transfected with the expression plasmid pBO854 for EC-Fc protein. The transformed cells were cultured in the presence of 100 μg/ml Hygromycin B (Wako Pure-Chemicals, Osaka, Japan) for several weeks and cells stably expressing EC-Fc were pooled. Ten million of the transformed cells were seeded in 500 ml of CHO-S-SFMII (GIBCO, Carlsbad, CA, USA) containing 100 μg/ml hygromycin at 37°C for 5 days. At the end of culture, the medium was collected and centrifuged to remove precipitates. The supernatants were passed over a 1-ml column of Protein G agarose (Invitrogen, Carlsbad, CA, USA) equilibrated with PBS. The column was then washed with PBS and the bound protein was eluted with 0.1 M sodium phosphate buffer, pH 2.6. Fractions containing EC-Fc were readily neutralized with sodium phosphate buffer, pH 8.0 by adding 1/10 volume of each fraction. The buffer was then replaced with PBS using a PD-10 column (GE Healthcare, Piscataway, NJ). The preparation of Fc protein without EC-1 moiety was based on the same procedure.
Competition EIA to estimate Kd value
The value of dissociation constant (Kd) between the ligand and ErbB2 was estimated as previously described with slight modification [24, 28]. Briefly, each well of 96-well plate was coated with 200 ng of sErbB2 in 0.1 M sodium bicarbonate buffer, pH 8.6, overnight at 4°C followed by blocking with PBS containing 1% BSA. After the rinse with PBS containing 0.1% Tween 20 (PBST), EC-Fc and varying concentration of sErbB2, which were premixed 30 min before, was applied to the wells. After the incubation for 30 min at 25 °C, the wells were washed with PBST and 50 μl of protein A-HRP conjugate (Sigma-Aldrich) diluted to 1:10,000 was added and incubated for 1 hr. Wells were then washed eight times with PBST and 100 μl of OPD solution was added for the HRP reaction. After 15 min, the reaction was quenched by 50 μl of 0.5 M sulphuric acid and the absorbance at 492 nm of each well was measured by a microplate reader MTP-120 (Corona, Japan). Each experiment was performed in triplicate and the mean values and standard deviations were calculated.
Preparation of multivalently displayed EC-1 peptide on bionanocapsule
To prepare the multivalent form of EC-1 peptide, EC-Fc proteins were displayed on the surface of bionanocapsule (BNC), which was composed of the recombinant surface antigen of hepatitis B virus [29]. To confer the high affinity for IgG Fc domain to BNC, the surface antigen was designed to fuse with the ZZ motif in the protein A derived from Staphylococcus aureus[25]. The resultant ZZ-tagged BNC (ZZ-BNC) was labelled with FITC by incubating with 1 mg/ml of FITC at 25°C for 15 min and the reaction mixture was further incubated at 4°C overnight in 0.1 M sodium bicarbonate buffer, pH 8.6. The molar ratio of FITC to ZZ-BNC was 110 to 1. After incubation, free FITC was removed by PD-10 column equilibrated with PBS. To prepare multivalently displayed form of EC-1 peptide, the molar ratio of ligand and BNC was optimized using Fc protein and human IgG (Sigma-Aldrich). To optimize the ratio FITC-labelled ZZ-BNC was mixed and incubated with Fc protein or human IgG at different ratio of 1:10, 1:20, 1:40, 1:60, 1:80 and 1:100 for 1 hr at 4°C. The precipitates formed were removed by centrifugation at 13,000 rpm for 5 min and the supernatant was transferred to a 96-black-well plate and fluorescence intensity at 530 nm of each well was measured by a microplate reader MTP-800 (Corona, Japan). Each experiment was performed in triplicate and the mean values and standard deviations were calculated. To characterize binding capacity of EC-Fc protein to ZZ-BNC, EC-Fc protein and ZZ-BNC were mixed in different ratios and incubated for 1 hr at 4°C. After centrifugation, the EC-Fc/BNC complex in the supernatant was immunoprecipitated with anti-HBsAg microbeads (Abott) by incubation overnight at 4°C. The beads were then washed three times in PBS followed by centrifugation for 30 sec at 4°C. Then the beads were suspended in Laemmli buffer containing β-mercaptoethanol, boiled for 5 min at 95°C, and subjected to SDS-PAGE and Western blot with anti-human IgG Fc antibody. As the quantitative control of Western blot, 125, 250 and 500 ng of EC-Fc was applied at the same time. Thus, the efficient ratio of EC-Fc and ZZ-BNC was determined and the multivalent form of EC-1 peptide was prepared as EC-Fc/BNC. BNC bound to Fc protein (Fc/BNC) was prepared simultaneously as a control.
Confocal microscopic observation of ErbB2 internalization
For confocal microscopic observation, cells were grown on 35 mm glass base dish (Iwaki Science Products, Japan). With the conditioned media prepared from CHO cells expressing EC-Fc or Fc proteins in HEPES buffered RPMI-1640 supplemented with 1% BSA, the target cells were incubated for 90 min at 37°C. The cells were fixed with 4% paraformaldehyde in PBS, permeabilized with 0.2% Triton X-100 and were blocked with 1% BSA or 10% FBS in PBS. Then the cells were washed with PBS and incubated with FITC-labelled anti-human IgG Fc antibody (Sigma-Aldrich) diluted 1:500 for 30 min at 37°C. Prior to the preparation of EC-Fc/BNC or Fc/BNC, BNC was labelled with RITC (Sigma-Aldrich). Cells were incubated with RITC-labelled EC-Fc/BNC or Fc/BNC for 1, 4 and 8 hrs. The cells were washed with PBS and were fixed with 4% paraformaldehyde in PBS and then permeabilized with 0.1% TritonX-100 for 5 min. The fixed and permeabilized cells were blocked with 1% BSA in PBS and incubated with anti-ErbB2 antibody sc-08 (Santa Cruz Biotechnology, CA) for 1 hr followed by Alexa 488–labelled anti-rabbit IgG (Molecular Probe Inc., OR) for 30 min. After washing the cells with PBS, the visualization were carried under a confocal microscope, LSM 510 Meta (Carl Zeiss, Germany) equipped with Argon laser and LSM software (Carl Zeiss, Germany). Argon laser with excitation laser line of 488 nm coupled with the band-pass filter of 505 nm was used for the fluorescence of FITC and Alexa 488. He–Ne with 543 nm laser line coupled with the band-pass filter of 560 nm for the fluorescence of RITC. For inhibition studies, Chlorpromzine (CPZ; Wako Pure-Chemicals, Japan) and Methyl β-cyclodextrin (mβCD) was procured from Sigma Aldrich. The percentage of colocalization was calculated from Manders’ overlapping coefficients (red overlapping green) using the JACoP plugin of ImageJ.
Biotinylation assay for the internalization of ErbB2
Biotinylation was performed as described previously [24]. Briefly, 5×105 cells were plated and cultured in a 60-mm tissue culture dish with complete medium. At the 90% confluency, the cells were washed twice in Hanks’ balanced salt solution (HBSS) for 10 min at 4°C. Sulfo-NHS-SS-Biotin (Pierce Chemical) dissolved in HBSS at a concentration of 0.5 mg/ml was added to the cells at 4°C with mild shaking for 20 min. This reaction was repeated twice. The cells were washed with HEPES buffered RPMI supplemented with 1% BSA and 2 mM glutamine (RPMI-BSA) for 10 min at 4°C. Cells were incubated with either 30 μg/ml EC-Fc protein, or anti-ErbB2 monoclonal antibody sc-08 diluted to 1:100 in RPMI-BSA for 1 hr at 37°C. For the multivalent display studies the SK-BR-3 cells were incubated with 20, 100 and 200 nM of EC-Fc and 1, 5 and 10 nM of EC-Fc/BNC, where EC-Fc:BNC = 20:1. The cells incubated with anti-ErbB2 antibody sc-08 were washed and incubated for 1 hr at 37°C with HRP conjugated anti-mouse IgG (Cell Signaling Technology, MA) diluted to 1:400 with RPMI-BSA. The incubation was stopped by transferring the dishes back on ice, and the cells rinsed twice with HBSS. The biotin on the cell surface was cleaved under reducing condition of 20 mM DTT in 50 mM Tris-HCl, pH 8.7 containing 100 mM NaCl and 2.5 mM CaCl2 for 20 min at 4°C, which was repeated twice. The cells were then washed three times with HBSS, and lysed in the lysis buffer consisting of 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 10 mM NaF, 1 mM vanadate, and protease inhibitor cocktail (Sigma-Aldrich) by the incubation for 20 min at 4°C. The lysates were collected and sonicated twice, and cell extracts were clarified by centrifugation for 5 min at 4°C. Protein concentrations in the extracts were determined by BCA assay (Pierce Chemical). Twenty microlitres of avidin agarose (Sigma-Aldrich) was added to the extracts and incubated overnight at 4°C. After centrifugations for 30 sec at 4°C, the agarose was washed three times in the lysis buffer, suspended in Laemmli buffer supplemented with β-mercaptoethanol, heated 5 min at 95°C, and subjected to SDS-PAGE followed by Western blotting. Transferrin receptor internalization was taken as a control for the immunoprecipitation experiments because transferrin is constitutively replenished from the surface of the cell to the endosome and from recycling endosome to the cell surface.
Analysis of cellular uptake of EC-Fc/BNC
The receptor-mediated cellular uptake of EC-Fc/BNC was evaluated. The cells in 60-mm dish were washed twice with ice-cold PBS and incubated with 2 nM (10 μg/ml) of EC-Fc/BNC for 5 hrs at 4°C or 37°C. After incubation, the cells were washed twice with ice-cold PBS to remove unbound BNC and then collected by the treatment with 0.025% trypsin. After centrifugation at 6,000 rpm for 1 min the supernatant was discarded and the cell pellet was washed twice with ice-cold PBS. The cells were then lysed in the lysis buffer, further incubated for 20 min at 4°C and sonicated twice. The cell extracts were clarified by centrifugation for 5 min at 4°C. Twenty microlitres of anti-HBsAg microbeads suspension was added to the extracts and incubated overnight at 4°C. After centrifugations for 30 sec at 4°C, the beads were washed three times in PBS, suspended in Laemmli buffer supplemented with β-mercaptoethanol, heated for 5 min at 95°C, and subjected to SDS-PAGE followed by Western blotting.
Flow cytometric analysis of ErbB2 internalization
For flow cytometric analysis, the cells seeded in a 100-mm dish were washed with PBS and were collected with enzyme free cell dissociation buffer (GIBCO) after incubating at 37°C for 5 min. The dissociation reaction was stopped by adding serum to the sample. Five hundred thousand cells were suspended in 0.1% BSA, containing 0.1% sodium azide in PBS (PBSB). After suspended in PBSB supplemented with EC-Fc, EC-Fc/BNC, Fc and Fc/BNC the cells were subsequently incubated on ice for 30 min. For internalization experiment, the cells were fixed and then permeabilized with 0.2% Triton X-100 in PBS. Then washed with PBSB, the cells were incubated with anti-human Fc antibody labelled with FITC at a dilution of 1:500 in PBSB followed by incubation on ice for 30 min. The cells were then processed for flow cytometric analysis by a FACS Calibur (Becton, Dickinson and Company).
Western blotting and image analysis
Proteins resolved on acrylamide gel were transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad, VA). The membrane was blocked with 10% skim milk in 10 mM Tris-HCl, pH 7.4, 150 mM NaCl containing 0.1% Tween-20 (TBST). The blots were then probed with a biotinylated polyclonal anti-human IgG antibody diluted to 1:1,000 (Dako, Denmark) or anti–preS-1 monoclonal antibody (Beacle, Japan) conjugated to HRP diluted to 1:15,000 in blocking buffer, followed by a HRP-Avidin (Zymed, CA) in TBST containing 10% skim milk. The HRP signal was developed with Western Lighting™ and Western Lighting Plus Chemiluminescence Reagent (Perkin Elmer, MA) and the intensity of the bands were visualized by LAS image 4000 (Fujifilm, Japan). Quantitative assessments of the relative intensity of the blots were performed with NIH ImageJ.
Results
Preparation of EC-Fc protein
EC-Fc fusion protein was designed as EC-1 peptide fused to the amino terminus of IgG Fc domain (Fig. 1A). EC-Fc protein secreted from recombinant CHO cells were immunoprecipitated with protein A-agarose. The bound proteins were analysed by Western blotting with anti-human IgG to detect the Fc domain. As shown in Figure 1B, EC-Fc was observed as a monomer at approximately 30 kDa under reducing condition, whereas dimer at approximately 60 kDa was observed together with the tetramer, hexamer and higher order of oligomeric forms under non-reducing condition in either Coomassie Brilliant Blue (CBB) staining or Western blotting. The Fc domain without EC moiety prepared as a control was also observed as monomer at approximately 30 kDa under reducing condition, whereas only dimer formed as a result of the S-S-bond in the hinge region was observed at approximately 60 kDa under non-reducing condition. No further oligomeric forms were observed for Fc. These results suggested that the hinge region in the Fc domain should be effective to prepare dimeric form of Fc and EC-Fc proteins through disulfide bonds. The oligomeric forms observed as tetramer, hexamer and higher order can be because of the intermolecular disulfide bonds between cysteine residues in each EC-1 peptide. It is worthwhile noticing that no trimers were observed probably because extra-disulfide linkages were formed only between the EC-1 moieties of each EC-Fc proteins. The reduced protein in the CBB staining and Western blotting indicated an increment in the theoretical size of the artificial ligands, Fc and EC-Fc, which could be attributed to the N-linked glycosylation of the Fc portion of the human IgG. In this context, the oligomers should have two EC-1 moieties as correctly cyclized peptide through disulfide bonds in a molecule (Fig. 1C). The majority of EC-Fc was considered divalent form of EC-1 peptide even if the oligomeric forms such as hexamer and higher oligomer were formed because high oligomerization should precipitate EC-Fc and prevent it from working as ligand of ErbB2. For the susceptibility to the reducing conditions, EC-Fc was treated with DTT at various concentrations ranging from 0.5 to 5 mM at 4°C and 25°C (Fig. 1D). As the result, no order of disulfide bonds appeared to be present in the sensitivity to the reduction reagent in oligomeric forms because the reduction did not stop at a dimer even in low concentration of DTT. The specific binding affinity of EC-Fc to sErbB2 was evaluated by competition EIA (Fig. 1E). Scatchard plot revealed a high affinity binding site with a Kd of 26 nM, whereas a low affinity site was estimated to be 360 μM.
Fig 1.
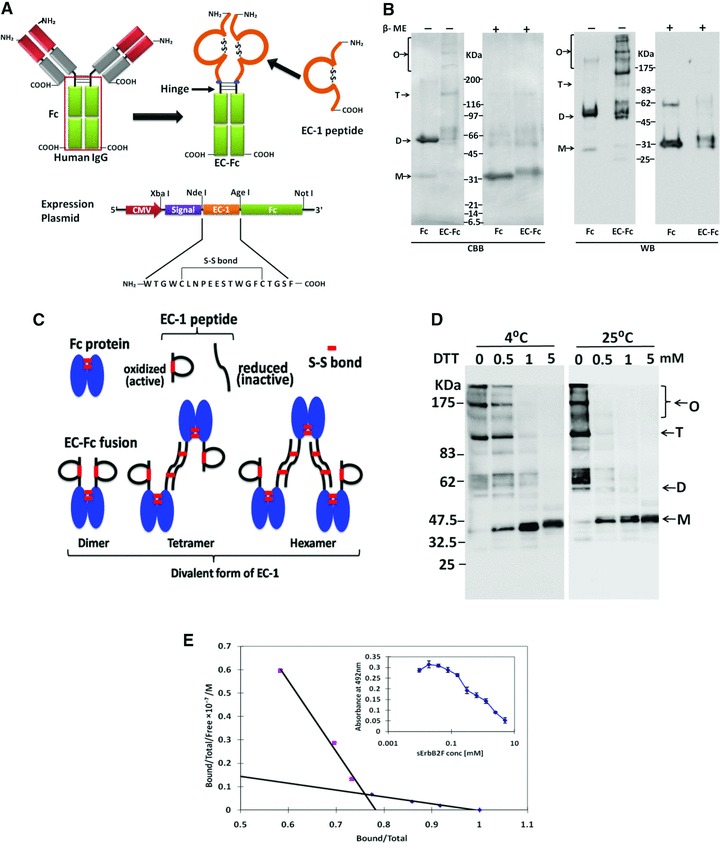
Construction and preparation of EC-Fc. (A) Schematic diagrams of EC-Fc. EC-1 peptide was fused to the human IgG Fc domain containing hinge region, so that the fusion protein EC-Fc should form dimer. The secretional expression of EC-Fc was designed with the signal peptide derived from human RNase I under the control of CMV promoter. The unique restriction enzyme sites Xba I, Nde I, Age I and Not I were used to construct the expression plasmid. (B) EC-Fc (theoretical MW = 30.0 kDa) or IgG Fc (theoretical MW = 27.6 kDa) was produced, purified with protein A-column and subjected to SDS-PAGE and Western blotting. Anti-human IgG Fc antibody recognized the purified EC-Fc without significant degradation. β-ME: betamercaptoethanol; CBB: Coomassie Brilliant Blue staining; WB: Western blotting. (C) Reduction of EC-Fc oligomeric form with DTT treatment. WB: Western blotting; M: monomer; D: dimer; T: tetramer; O: oligomer (B, C). Molecular weight markers range from 6.5 to 200 kDa (B, C). (D) Schematic representation of the probable dimer, tetramer and hexamer formation of EC-Fc and dimer formation of Fc through disulphide linkages. (E) Evaluation of the affinity of EC-Fc for ErbB2 by competitive EIA. Kd values between EC-Fc and sErbB2F were estimated from the Scatchard plots.
Internalization of ErbB2 was induced by EC-Fc in SK-BR-3 cells
Because the specific binding of EC-Fc to ErbB2 was confirmed, we then checked uptake of ErbB2 upon incubation with EC-Fc. For this assay, surface biotinylated cells were prepared and used. After incubation with EC-Fc, the biotin-labelled molecules retained on the cell surface were stripped off by the reducing reagent, whereas the molecules internalized were kept labelled with biotin so that they should be immunoprecipitated for the further analysis. The anti-ErbB2 antibody sc-08, which is known to induce internalization of ErbB2 in either SK-OV-3 cells or SK-BR-3 cells [13, 34], was used as a positive control. When evaluated on SK-BR-3 cells, EC-Fc was found to induce the internalization of ErbB2 (Fig. 2A). This internalization was estimated approximately 50% of that induced by anti-ErbB2 antibody sc-08. Internalized ErbB2 was observed when SK-OV-3 cells were treated with EC-Fc (Fig. 2B). In SK-OV-3 cells, the internalized ErbB2 induced by EC-Fc was almost equivalent with that induced by antibody sc-08. EC-Fc at 1 μM showed internalization of ErbB2 equivalent to that induced by anti-ErbB2 antibody sc-08 at 6.67 nM. This internalization was dependent on the doses of EC-Fc in a similar manner of monovalent EC-1 peptide fused to eGFP (EC-eGFP) [24]. When the biotinylated cells were treated with 5 μM EC-Fc, the internalized fraction of ErbB2 was increased up to three folds compared to that of the untreated cells at 37°C (Fig. 2C). We previously described that the internalization of ErbB2 was only observed in SK-OV-3 cells, when SK-BR-3 cells and SK-OV-3 cells were treated with EC-eGFP [24]. Taking the internalization demonstrated previously into consideration, this level of internalization was conceivable in SK-OV-3 cells, so that 5 μM EC-Fc should correspond to 10 μM EC-eGFP. The dimeric form of EC-Fc induced ErbB2 internalization whereas with EC-eGFP treatment ErbB2 was retained on the surface of the SK-BR-3 cells without internalization.
Fig 2.
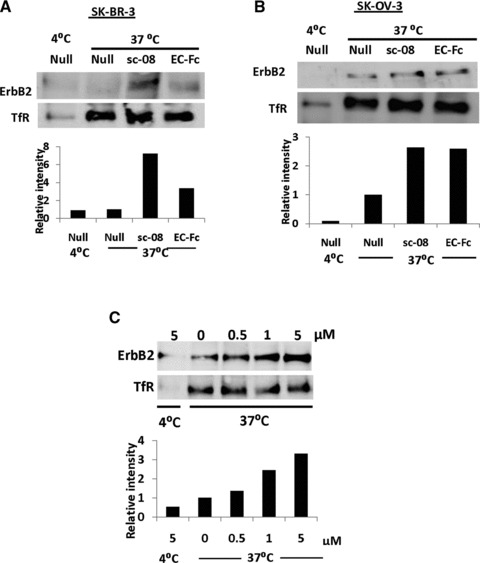
Uptake of ErbB2 in SK-BR-3 and SK-OV-3 cells. The surface of SK-BR-3 cells (A) and SK-OV-3 cells (B, C) were labelled with biotin and were stimulated with EC-Fc. After treatment with trypsin internalized ErbB2 was immunoprecipitated with Avidin agarose and detected with HRP-labelled Avidin. (A, B) Cells were incubated at 4°C and 37°C without stimulation (Null), stimulated with antibody sc-08 for 1 hr and incubated with anti-mouse IgG for another 1 hr at 37°C (sc-08) and stimulated with EC-Fc 2 hrs at 37°C (EC-Fc). (C) SK-OV-3 cells were treated with various concentrations of the EC-Fc starting from 0 to 5 μM at 37°C and incubated at 4°C without stimulation for 2 hrs. Transferrin receptor (TfR) was monitored simultaneously as the control for the internalization experiments. This data set represents the representative of two independent experiments. The intensity of ErbB2 was densitometrically analysed by ImageJ and plotted into each graph to evaluate internalization. The density from the cells left untreated at 37°C was normalized as 1 in each graph.
Internalization of ErbB2 was enhanced by multivalent form of EC-Fc in SK-BR-3 cells
The multivalent form of EC-1 peptide was prepared exploiting the affinity of ZZ-BNC for IgG Fc region (Fig. 3A). When ZZ-BNC was mixed with EC-Fc, the ligand EC-1 was multivalently displayed on the surface of ZZ-BNC (EC-Fc/BNC). On the other hand, ZZ-BNC displaying null ligand was just prepared by mixing Fc protein with ZZ- BNC (Fc/BNC). First of all, binding capacity and binding efficiency of the ZZ-BNC with ligand was optimized and characterized. To optimize the ratio of Fc fusion molecule to BNC, FITC-labelled ZZ-BNC was mixed with Fc protein at variable molar ratios of 1:10, 1:20, 1:40, 1:60, 1:80 and 1:100, respectively, and the residual fluorescent intensity in the supernatant was measured. As shown in Figure 3B, the soluble Fc/BNC was reduced to 30% at a molar ratio of ZZ-BNC to Fc protein at 1:100 when judged from the fluorescence in the supernatant. Similarly, when human IgG was used, the soluble IgG/BNC was found reduced to 10% at a molar ratio of ZZ-BNC to IgG protein at 1:100. The amount of Fc protein bound to ZZ-BNC in the supernatant was further estimated from intensity of the band detected by Western blotting (Fig. 3C). As the result, the amount of Fc protein bound to ZZ-BNC was determined maximum when the molar ratio of ZZ-BNC to Fc protein was at 1:20. Thus, EC-Fc/ZZ-BNC was prepared by mixing ZZ-BNC and EC-Fc at the molar ratio of 1:20 for further experiments.
Fig 3.
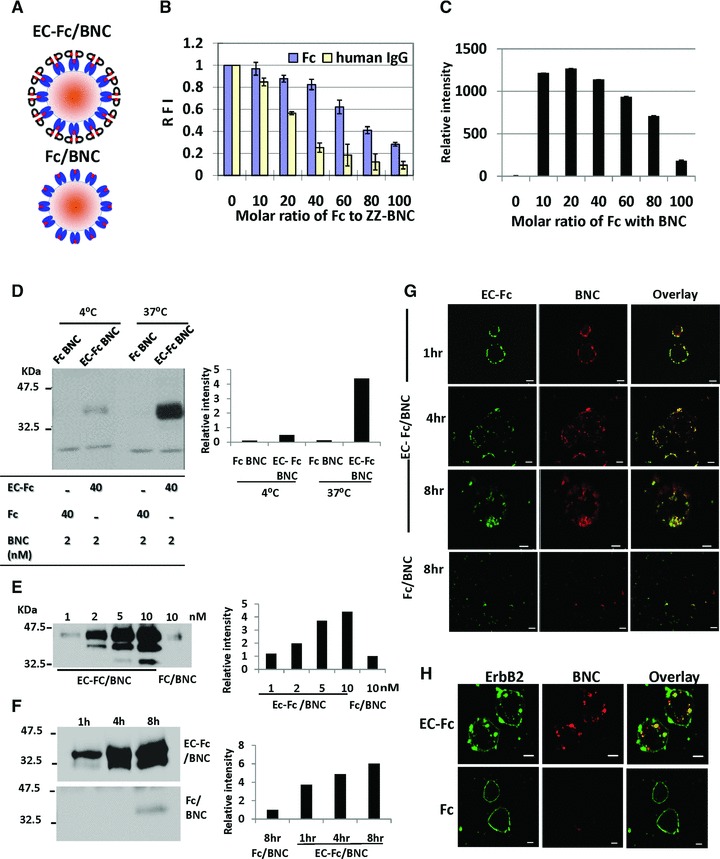
Characterization of BNC displaying EC-Fc. (A) Schematic representation of the multivalent display of EC-Fc on ZZ-BNC (EC-Fc/BNC) and Fc on ZZ-BNC (Fc/BNC). (B) The solubility of BNC-displaying Fc/BNC and IgG/BNC was evaluated with FITC-labelled BNC. Residual fluorescence in supernatant was measured in varying molecular ratio of Fc or human IgG to ZZ-BNC. The intensity from the FITC-labelled ZZ-BNC without Fc was calculated as 1 in each graph. (C) Western blot analysis of Fc in the supernatant obtained in (B). Fc/BNC in the supernatant was immunoprecipitated with anti-HBsAg antibody conjugated to micro beads and was subjected to Western blotting. The Fc on the blot was detected with anti-human IgG. The bands were densitometrically analysed with ImageJ and relative intensity of each lane was plotted. (D–F) Assessment of internalization of EC-Fc/BNC in SK-BR-3 cells through Western blot. (D) EC-Fc/BNC (40 nM/2 nM) or Fc/BNC (40 nM/2 nM) was incubated with SK-BR-3 cells for 5 hrs at 4°C and 37°C. (E) SK-BR-3 cells were treated with various concentration of EC-Fc/BNC from 1 to 10 nM. Fc/BNC in 10 nM was taken as control. (F) SK-BR-3 cells were treated with 2 nM EC-Fc/BNC at various time periods. Simultaneously 2 nM Fc/BNC was taken as control. (D–E) After the incubation the cells were trypsinised and lysed followed by immunoprecipitation with anti-HBsAg antibody conjugated to micro beads. The precipitates were immunoblotted and were detected with anti–pre-S1 antibody. The bands of BNC were densitometrically analysed by ImageJ and plotted into each graph to evaluate amount endocytosed. (G, H) Confocal microscopic observation of SK-BR-3 cells treated with EC-Fc/BNC or Fc/BNC. Cells were incubated for various time periods (G) and for 4 hrs (H). The RITC-labelled ZZ-BNC was used and the cells were fixed and permeabilized. EC-Fc or Fc were detected with anti-human IgG labelled with FITC (G) and ErbB2 was detected with sc-08 antibody followed by rabbit anti-mouse IgG Alexa 488 (H). Bars, 10 μm.
ErbB2 internalization mediated by EC-Fc/BNC was evaluated by the cellular uptake of EC-Fc/BNC (Fig. 3D–F). The SK-BR-3 cells were incubated with either EC-Fc/BNC or Fc/BNC for 5 hrs at 37°C or 4°C (Fig. 3D). As a result, EC-Fc/BNC showed significant cellular uptake at 37°C in SK-BR-3 cells. The EC-Fc/BNC cellular uptake into SK-BR-3 cells was found not only dose dependent in the range from 1 to 10 nM (Fig. 3E) and time course dependent up to 8 hrs at 2 nM (Fig. 3F). These results suggest that multivalent EC-1 peptide displayed on BNC might make its internalization into the cells efficient. The time course changes in the internalization of BNC in SK-BR-3 cells were evaluated by incubating cells with EC-Fc/BNC for various time periods under a confocal microscope (Fig. 3G). To demonstrate that observed localization or internalization of the receptor is not an artefact of fixation and cell permeabilization, we assessed the internalization of the EC-Fc/BNC with live cell imaging (Fig. S1). The colocalization of EC-Fc/BNC was confirmed by merging the location of BNC and EC-Fc, whereas Fc/BNC did not show any internalization. The internalization of ErbB2 induced by EC-Fc/BNC was further confirmed under confocal microscope (Fig. 3H). SK-BR-3 cells were incubated with RITC-labelled BNC displaying EC-Fc at 2 nM for 4 hrs. Then the cells were further probed for the ErbB2 by anti-ErbB2 monoclonal antibody sc-08 followed by Alexa 488–labelled secondary antibody. The internalized ErbB2 was observed to be colocalized together with BNC in SK-BR-3 whereas the Fc/BNC did not show any detectable internalization of ErbB2 as well as for BNC. As the result, the internalization of ErbB2 is concluded to be dependent on the uptake of BNC displaying EC-1 peptide multivalently. Internalization of EC-Fc/BNC was found to be dependent on both time course and ErbB2 in SK-BR-3 cells. Simultaneously, the internalization of ErbB2 was confirmed to be induced when EC-Fc/BNC was added. This observation implies that the cellular uptake of EC-Fc/BNC should be associated with the receptor-mediated endocytosis.
Comparative study of the effect by EC-Fc and EC-Fc/BNC
The specific binding of EC-1 ligand to cells overexpressing ErbB2 was assessed by immunostaining (Fig. 4A). Three cell lines MCF-7, SK-BR-3 and SK-OV-3 cells were selected and the binding of EC-Fc protein and EC-Fc/BNC was evaluated. The expression level ErbB2 was approximately 104 sites/cell in MCF-7 [30], 106 sites/cell in SK-BR-3 and SK-OV-3, respectively [31, 32]. The cells were incubated with EC-Fc or Fc or EC-Fc/BNC or Fc/BNC for 1 hr at 37°C followed by staining with anti-human IgG Fc-labelled with FITC. The fluorescence was observed only at the linage of SK-BR-3 and SK-OV-3 cells but not with MCF-7 whereas Fc and Fc/BNC did not show any positive staining in all cell lines. No detectable fluorescence at the confocal microscopy to MCF-7 attributes to the low ErbB2 expression in the cells. The surface binding of the EC-Fc/BNC as well as EC-Fc was then qualitatively analysed by flow cytometry. As shown in Figure 4A in SK-BR-3 and SK-OV-3 cells, EC-Fc (green line) as well as Ec-Fc/BNC (blue line) exhibited to bind to cell surface specifically. The binding efficiency of EC-Fc was found to be better than EC-Fc/BNC for SK-BR-3 cells. As the results, the specific binding of EC-Fc and EC-Fc/BNC to ErbB2 and overexpressing cells was successfully demonstrated.
Fig 4.
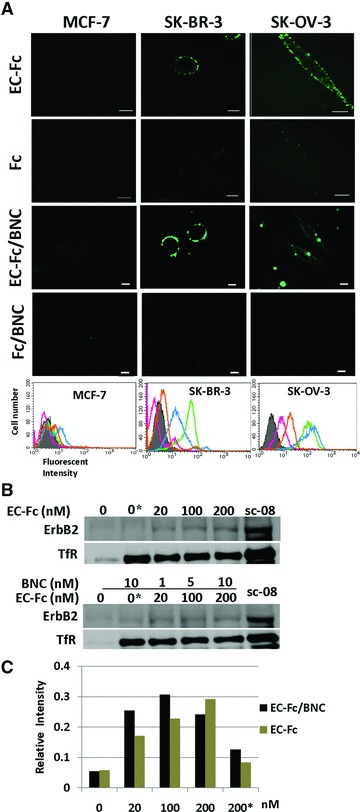
Evaluation of ErbB2 internalization in SK-BR-3 cells treated with EC-Fc/BNC. (A) Comparison of EC-Fc/BNC bound to the surface of the cells over expressing ErbB2. Cells were incubated with EC-Fc (1 μM), Fc (1 μM), EC-Fc/BNC (40 nM/2 nM), and Fc/BNC (40 nM/2 nM) for 1 hr at 37°C in MCF-7 cells, SK-BR-3 cells, and SK-OV-3 cells. MCF-7 cells were used as the control for low ErbB2 expression. The cells were stained with anti-human IgG labelled with FITC. Bars depict 10 μm. The treated cells were also subjected to flow cytometoric analysis for the surface bound fraction of EC-Fc and Fc ligand with and without BNC in MCF-7, SK-BR-3 and SK-OV-3, respectively. Untreated cells were shown in shadow (grey). Treatment with EC-Fc ligand Fc ligand, EC-Fc/BNC and Fc/BNC was marked as green, magenta, blue and orange, respectively. (B) SK-BR-3 cells were treated with different concentration of EC-Fc or Ec-Fc/BNC. The cell surface receptors were reversibly biotinylated with NHS-SS-Biotin and were incubated with EC-Fc, Fc, EC-Fc/BNC or Fc/BNC for 2 hrs at 37°C. SK-BR-3 cells left untreated at 4°C for 2 hrs were shown as lane ‘0’. ‘0*’ corresponds to the cells treated with Fc ligand (top) and Fc/BNC (lower) at a concentration of 200 nM, respectively. After the treatments, cells were lysed, immunoprecipitated with avidin agarose and were subjected to Western blot to detect ErbB2 with anti-ErbB2 antibody. sc-08 antibody treatment was taken as positive control for endocytosis of ErbB2. Transferrin receptor (TfR) was monitored as an internal control for the experiments. (C) The bands from the Western blots were densitometrically analysed with ImageJ and the intensity each band was normalized by the intensity when treated with antibody sc-08.
ErbB2 internalization depending on the doses of EC-Fc and EC-Fc/BNC was assessed in the range of 20–200 nM in SK-BR-3 cells (Fig. 4B). ErbB2 internalization was observed with both EC-Fc and EC-Fc/BNC. When the intensity of the band analysed densitometrically showed the internalization of ErbB2 was more efficient by the stimulation for 2 hrs up to 100 nM of the ligand multivalently displayed on BNC than by EC-Fc ligand alone (Fig. 4C).
EC-Fc/BNC internalized through GEEC pathway in SK-BR-3 cells
Cells adopt divergent pathways for the endocytosis of the cargos and receptors. The key pathways were categorized into clathrin-dependent and clathrin-independent mechanisms [33]. The clathrin-independent pathway is further classified into caveolar and GPI-anchored early endocytic compartments (GEEC) pathways [33, 34]. ErbB2 is thought to be internalizing in SK-OV-3 through clathrin-mediated internalization [24]. In this study we found that the mechanism of internalization of ErbB2 present in SK-OV-3 cells is deficient in SK-BR-3 cells. We tried to identify the difference between the two cell lines using DNA microarray (Fig. S2). As a result the expression of claudin 16, caveolin-1 and caveolin-2 were found to be extensively down-regulated in SK-BR-3 cells when compared to SK-OV-3 cells. This absence of caveolin-1 in SK-BR-3 cells was further confirmed by immunostaining using anti-Cav1 antibody (Fig. 5A). SK-BR-3 did not show the presence of caveolin-1 suggesting that the impaired caveolar mechanism prevailing in the cell, which was consistent with the finding by the previous reports [13, 35]. Because caveolin-1 was detected in SK-OV-3 cells the internalization pathway deficient in SK-BR-3 cells might be attributed to caveolae.
Fig 5.
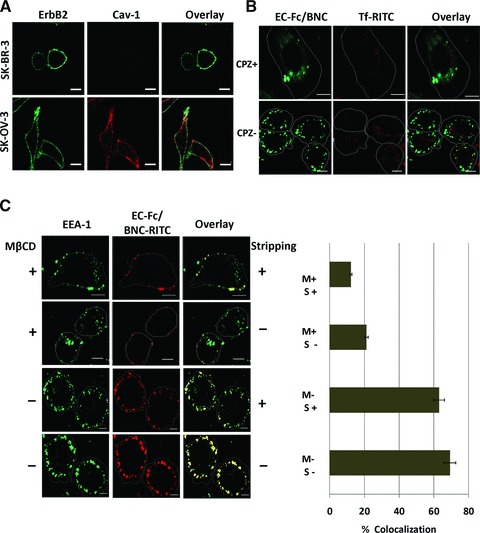
Assessment of the mechanism of internalization of EC-Fc/BNC in SK-BR-3 cells. (A) SK-BR-3 cells and SK-OV-3 cells were stained with antibodies against ErbB2 (green) and Cav-1 (red). (B) SK-BR-3 cells were treated with EC-Fc/BNC in the presence or absence of 100 nM of CPZ and stained with anti-human IgG antibody labelled with FITC. Transferrin-RITC was used as a control for the internalization. (C) SK-BR-3 cells were treated with EC-Fc/BNC labelled with RITC in the presence or absence of mβCD. The cells were stained with anti–EEA-1 antibody followed by secondary antibody against mouse IgG labelled with AlexaFlour-488. The cells were then stripped with and without acid treatment to remove the surface bound fraction and to visualize the internalized fraction. Bars, 10 μm.
The mechanistic approach for the uptake of the EC-Fc/BNC was assessed in SK-BR-3 cells using inhibitors for other pathways. SK-BR-3 cells were treated with EC-Fc/BNC in the presence or absence of inhibitors for clathrin, and GEEC pathway and the internalization was assessed. When the cells were treated with 100 nM CPZ, an amphiphilic drug, which inhibits the clathrin-mediated pathway, the internalization of the EC-Fc/BNC was unaffected (Fig. 5B).
Most of the cargos irrespective of their route merges with Rab5 and early endosome antigen-1 (EEA-1) enriched in early endosomes, which is further sorted into various intracellular destinations [36, 37]. The colocalization of EC-Fc/BNC with EEA-1 was assessed with anti–EEA-1 antibody under confocal microscope at various time periods starting from 5, 30 and 90 min and the colocalization was found to be maximum at 30 min (Fig. S3).
When the cells were treated with 5 mM mβCD, which dislodges the cholesterol from the surface and inhibit the cdc42 activity, for 30 min the internalization was blocked and the cells untreated showed considerable uptake of EC-Fc/BNC. The bound fractions of EC-Fc/BNC were treated with and without acid stripping to ensure the colocalized fraction inside the cells (Fig. 5C). The colocalization was calculated to more than 60%, which was reduced in the internalization of EC-Fc/BNC with the mβCD treatment. To assess the surface binding of the EC-Fc/BNC after the inhibitor treatment the cells were stained for the ErbB2 and EC-Fc/BNC at various concentration of the mβCD (Fig. S4). The inhibitor treatment did not deteriorate the binding affinity of the EC-Fc/BNC to ErbB2. These results were further confirmed with flow cytometric studies (Fig. S5).
Discussion
The EC-Fc was designed as dimer due to the hinge region in IgG Fc domain (Fig. 1A). However, we found higher oligomeric forms such as tetramer and hexamer. These higher order forms were supposed to be divalent as described in ‘Results’ and Figure 1C. Further multivalent form of EC-1 peptide was prepared by displaying EC-Fc on BNC which was optimized to a ratio of EC-Fc to BNC = 1:20. These divalent and multivalent forms of EC-Fc induced internalization of ErbB2 into SK-BR-3 cells whereas monovalent form of EC-1 peptide did not induce internalization [24]. The internalization of ErbB2 was found dependent on the concentration in the range of 20–200 nM for EC-Fc and appeared to be enhanced when 20–100 nM EC-Fc was displayed on BNC (Fig. 4B). EC-Fc/BNC appeared more efficient in inducing the internalization of ErbB2 when compared to EC-Fc ligand alone. The time course dependence of the BNC internalization was found to be also in the same range of EC-Fc/BNC concentration. The binding affinity of the EC-Fc was estimated to be about 26 nM whereas monovalent EC-1 peptide as EC-eGFP fusion showed 1 μM (Table 1). The affinity of the multivalent display for ErbB2 was 33 nM, which did not ameliorate the affinity of EC-Fc for the ErbB2. Therefore, the affinity should elucidate the enhanced internalization of ErbB2 induced by EC-Fc/BNC in SK-BR-3 cells. It is believed that the heterodimerization between ErbB2 and ErbB3 is one of the reasons for retention of ErbB2 on the cell surface, so that monovalent form of the EC-1 peptide could not induce internalization of ErbB2 in SK-BR-3 cells. Shuttling between homodimers and heterodimers should increase depending on the distance of the dimers. EC-Fc supposedly keeps the receptors close enough to facilitate homodimer formation, which will allow the internalization of ErbB2. Therefore, the cells treated with EC-Fc/BNC showed considerable increment in the internalization of the BNC, although the effect of EC-Fc moiety appeared conceivable because Fc/BNC did not show significant internalization of ErbB2 as the background (Fig. 3D–F).
Table 1.
Affinity for ErbB2 of EC-1 peptide in various forms and internalization of ErbB2 in SK-OV-3 and SK-BR-3 cells
| Forms of EC-1 peptide | EC-eGFP | EC-Fc | EC-Fc/BNC |
|---|---|---|---|
| Kd (nM) | 1.0 × 103* | 2.6 × 10 | 3.3 × 10 |
| Internalization of ErbB2 observed in | |||
| SK-OV-3 cells | +† | ++‡ | ++ |
| SK-BR-3 cells | –§ | + | ++ |
[24].
Internalization was observed in the range of μM orders of ligands.
Internalization was observed in the range of nM orders of ligands.
Internalization was not observed.
It is thought that the processing and degradation of the ErbB2 is different in various cell lines [24, 34, 35]. The CPZ at 100 nM, which inhibits the formation of clathrin-coated pits, inhibited the internalization of ErbB2 by EC-1 peptide fused to eGFP in SK-OV-3 [24]. On the contrary, the same concentration of CPZ did not block the internalization of ErbB2 in SK-BR-3 cells. In this study, we confirmed the absence of caveolin-1 in SK-BR-3 cells using microarray and immunostaining where it showed around 300-fold increment in the expression of Cav-1 in SK-OV-3 cells (Fig. S1). Recent studies support our observation that the SK-BR-3 cells lack caveolae [13, 35]. ErbB2 internalization after geldanamycin treatment showed GEEC or clathrin-independent compartments (CLIC) pathway, which ultimately fuses with the classical pathway for the degradation in lysosomes, is independent of caveolar and clathrin pathway [35].
In this study we employed mβCD, a cholesterol dislodging oligosaccharide, for deciphering the mechanism behind the internalization of the EC-Fc/BNC in SK-BR-3 cells, as an inhibitor for the blockade of GEEC pathway. Cholesterol depletion with mβCD inhibits both caveolar [38] and GEEC pathway [39, 40]. Earlier it was shown that cholesterol level determines the internalization of GPI-anchored proteins and with clostridium difficile toxin B [34, 39]. Because SK-BR-3 cells lack caveolar machinery, the blockade with mβCD will only correspond its effect in blocking the internalization of GEEC-mediated mechanism. The internalization of ErbB2 is sensitive to the depletion of cholesterol, which is essential for most of the clathrin-independent pathways. GEECs are formed as compartments free of clathrin, caveolin and dynamin, which are short lived (2–5 min) and ultimately fuses with EEA-1 enriched early endosome [33, 38]. It was shown that cdc42 is responsible for the dynamin-dependent and clathrin-independent internalization of GPI-anchored proteins. It was also reported earlier that cdc42 activation recruits actin polymerization machinery rendering this pathway sensitive to inhibitors of cholesterol depletion and actin polymerization [39]. This pathway specifies the visible difference in the uptake of the EC-Fc/BNC between SK-OV-3 and SK-BR-3 cells.
Another important molecule, which plays a key role in the GEEC pathway, is EEA-1 that contains an FYVE finger, which interacts with PI3K [41, 42]. PI3K phosphorylates Rab 5 that helps the EEA-1 to localize to early endocytic compartments [36]. The colocalization of EC-Fc/BNC with EEA-1, which corresponds the preliminary step in the endosomal pathways before transferring it to the sorting endosomes, was observed (Fig. S2). Because both caveolar and clathrin pathways are impaired in SK-BR-3 cells as described earlier, the colocalization should correspond to GEEC pathway.
In this study, we revealed the presence of GEEC pathway as ErbB2 internalization, which critically differs between SK-BR-3 cells and SK-OV-3 cells using EC-1 peptide in various forms. As for the nanoparticle internalization, the pathways of clathrin and caveolar- independent endocytosis have not been described until now [43]. The internalization of the carrier particle was described repeatedly on folate receptor (FR) tagged nanoparticles [44]. In these reports, mechanism of the carrier internalization emphasizes the FR internalization through GEEC pathway. Similarly, EC-Fc should utilize the mechanism of internalization of ErbB2 in this study even when they are displayed on BNC (Fig. 4B).
It is considered that ZZ-BNC is an efficient nano-machine for molecular targeting in drug delivery system displaying antibodies on the surface [25, 29, 45]. Multivalency of EC-Fc was successfully demonstrated to target ErbB2 enhancing the internalization of ErbB2, in this report. In this context, delivery of therapeutic substances into specific cells of various diseases including cancer can be feasible if EC-Fc/BNC is used as a drug delivery system. Multivalent display strategy proposed here should be useful for molecular targeting to introduce therapeutic drug into the cells followed by the internalization of receptors.
Acknowledgments
The authors thank Prof. N. Kanayama for his valuable help and suggestions in flow cytometric analysis. The authors also thank Prof. H. Matsui for helpful discussion throughout the work. This work was partly supported by Grant-in-Aid for Scientific Research (B) from the Ministry of Education, Culture, Sports, Science and Technology (No. 21300179) in Japan, Health and Labor Sciences Research Grants, Research on Nanotechnical Medical, H21-nano-general-004 and National Natural Science Foundation of China (Key Program, Grant No. 30930038).
Conflict of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Supporting Information
Live cell imaging. For live cell imaging, theSK-BR-3 cells were cultured in 18 mm coverslips and washed twicewith phenol red free RPMI (Sigma) containing 1 % glucose, 25 mMHEPES and 1 % BSA supplemented with 100 U/ml pencillin and 100μg/ml streptomycin. The EC-Fc BNC tagged with RITC was incubatedon ice for 30 min prior to the visualization in order to enable thebinding of the multivalent forms of BNC to the cell surface. Thecells were then washed thrice with warm media and were visualizedin 100X 1.3 N.A. oil immersion objectiveunder Olympus IX81 invertedmicroscope (Olympus) with temperature control followed by dataacquisition with Metamorph software (Molecular Devices). Theimaging was carried out with DP71 cooled CCD camera attached to themicroscope and the fluorescence of RITC was acquired along with theDIC field image. The time-lapse imaging was carried out for onehour with time intervals of 5 minutes each. In Metamorph software,each channel was split and overlayed separately. The acquiredstacks were analyzed with MBF Image J.
Fig. S2 Microarray analysis. Expression of genes inSK-BR-3 cells was analyzed by microarray procedure. Total RNApreparation and analysis was performed as described previously [46,47]. DNA microarray was carrying 1,795-oligonucleotide probes forhuman cell surface proteins. cDNAs were synthesized withSuperescript II reverse transcriptase (Invitrogen) with oligo dTprimers. Amino-allyl-dUTP was incorporated into cDNAs followed bycoupling with Cy-3 dye (Ambion, TX, USA) and were processed forhybridization at 55°C for 15 hours. The fluorescent images forthe hybridization were captured using FLA8000 scanner (Fuji Film,Japan) and analyzed with GenePix Pro5.1 software (Axon Instruments,CA). We found caveolins, Cav-1 and 2, and claudin 16 (CLDN-16) weredownregulated in SK-BR-3 cells when compared to SK-OV-3 cells whileErbB2 and GAPDH expression were equally expressed in both the celllines. RT-PCR (Fig. S1A) and qRT-PCR (Fig. S1B) further confirmedthese results. The conditions for the RT-PCR are as follows:94°C for 5min; followed by 30 cycles of 94°C for 30 sec,55°C for 30 sec, 72°C for 30 sec and 72°C for 7minutes. Quantitative real time PCR was performed with SYBR GreenRealtime Master Mix (Toyobo) in triplicates containing 5ng of cDNAalong with 400 nM primers using LightCycler™ (Roche). Thethermal cycling condition was as follows: 95°C for 1 minfollowed by 40 cycles of 95°C -10 sec, 55°C for 10 sec,72°C for 25 sec and 60°C for 1 min. The following sets ofthe primers were used for the PCR reaction. Claudin-16, (Forward)5′- GCTTGCCACAATGAGGGATCT-3′, (Reverse)5′-TGACTTGGCCATGGAAACACC-3′; Cav-1 (Forward)5′-GACTCGGAGGGACATCTCTAC-3′, (Reverse)5′-GTTGATGCGGACATTGCTGA-3′; Cav-2, (Forward)5′-ACGTACAGCTCTTCATGGAC-3′ (Reverse)5′-CAGTTGCAGGCTGACAGAAG-3′.
Fig. S3 Time dependent colocalization of EC-Fc BNC in earlyendosomes. Colocalization of EC-Fc/BNC-RITC with EEA-1 was found in the early endosomes. SK-BR-3 cells were treated with EC-Fc/BNC-RITC for 5, 30, 90 min, and the colocalization of the EC-Fc/BNC-RITC with the early endosomal marker, EEA-1 was assessed with anti EEA-1 antibody (BD Biosciences) and the secondary antibody labeled with AlexaFlour-488 (Molecular Probes). Error bars indicate standard error. JACoP plugin in Image J was employed for percentage colocalization assessment. Percentage colocalization was found to be maximum up to approximately 70% by 30 min. The data represented here is the representative of two independent experiments.
Fig. S4 Binding of EC-Fc BNC to the cell surface aftertreatment with varying concentration of mβCD. The bindingof EC-Fc/BNC’s to ErbB2, in the presence of 2 to 10 mM ofcholesterol dislodging oligosaccharide-mβCD, was not affected.SK-BR-3 cells were treated with 5 and 10 mM of mβCD for 30minutes followed by treatment with EC-Fc/BNC labeled with RITC areshown in Figure S2. The cells were stained with anti ErbB2 antibody(green) and red spots indicate EC-Fc/BNC labeled with RITC. Barsdepict 10 μm.
Fig. S5 FACS analysis for the surface binding andinternalization of EC-Fc BNC with inhibitor treatment. Surfacebinding and internalization of EC-Fc/BNC labeled with FITC inSK-BR-3 cells was quantified using FACS analysis. First, thebinding of EC-Fc/BNC on SK-BR-3 cells was treated with or without 5mM of mβCD on ice for 30 min. The cells were then fixed andanalyzed with FACS to determine the surface bound fraction (Fig.S5A). The histograms showed overlapping of the peak tops indicatingthat the surface binding is majorly unaffected by the mβCDtreatment. The peak (blue), which corresponds to approximately 20%of the total cell counts, orienting towards the untreated cells(grey peak) indicates the presence of a different population ofcells that are incapable of binding to EC-Fc/BNC after mβCDtreatment. Simultaneously, another set cells were treated withEC-Fc/BNC labeled with FITC in the presence or absence of 5 mM ofmβCD at 37°C for 30 minutes. Cells were then trypsinizedto remove the surface fraction, fixed and permeabilized, and theinternalized fraction was analyzed adopting the same procedure withFACS (Fig. S5B, C). mβCD treated SK-BR-3 cells showed a peakpoint in the histogram which overlaid the peak area of untreatedcells indicating the mβCD inhibits the internalization, whilethe cells without mβCD treatment drifted from the untreatedcells towards the right. Total intensity of the internalizedfraction was assessed from the FACS data were plotted on to a graph(Fig. S5). mβCD suppressed the internalization toapproximately 50% when compared to the absence of mβCDtreatment. M: mβCD; T: trypsin. Error bars indicate standarderror.
References
- 1.Yarden Y, Sliwkowski MX. Untangling the ErbB signaling network. Nature Rev Mol Cell Biol. 2001;2:127–37. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 2.Olayioye MA, Neve RM, Lane HA, et al. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J. 2000;9:3159–67. doi: 10.1093/emboj/19.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klapper LN, Waterman H, Sela M, et al. Tumor-inhibitory antibodies to HER-2/ErbB-2 may act by recruiting c-Cbl and enhancing ubiquitination of HER-2. Cancer Res. 2000;60:3384–8. [PubMed] [Google Scholar]
- 4.Slamon DJ, Clark GM, Wong SG, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–82. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 5.Hynes NE, Stern DF. The biology of ErbB2/neu/HER-2 and its role in Cancer. Biochem Biophys Acta. 1994;1198:164–84. doi: 10.1016/0304-419x(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 6.De Placido S, Carlamangno C, De Laurentiis M, et al. C-erbB2 expression predicts tamoxifen efficacy in breast cancer patients. Breast Cancer Res Treat. 1998;52:55–64. doi: 10.1023/a:1006159001039. [DOI] [PubMed] [Google Scholar]
- 7.Pinkas-Kramarski R, Soussan L, Waterman H, et al. Diversification of Neu differentiation factor and epidermal growth factor signaling by combinatorial receptor interactions. EMBO J. 1996;15:2452–67. [PMC free article] [PubMed] [Google Scholar]
- 8.Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009;9:463–75. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- 9.Salomon DS, Brandt R, Ciardiello F, et al. Epidermal growth factor-related peptide and their receptors in human malignancies. Crit Rev Oncol Ematol. 1995;19:183–232. doi: 10.1016/1040-8428(94)00144-i. [DOI] [PubMed] [Google Scholar]
- 10.Slamon DJ, Godolphin W, Jones LA, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–12. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Silva E, Gershenson D, et al. Amplification and rearrangement of c-erb B proto-oncogenes in cancer of human female genital tract. Oncogene. 1989;4:985–9. [PubMed] [Google Scholar]
- 12.Austin CD, De Maziere AM, Pisacane PI, et al. Endocytosis and sorting of ErbB2 and the site of action of cancer therapeutics trastuzumab and geldanamycin. Mol Biol Cell. 2004;15:5268–82. doi: 10.1091/mbc.E04-07-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hommelgaard AM, Lerdrup M, van Deurs B. Association with membrane protrusions makes ErbB2 and internalization-resistant receptor. Mol Biol Cell. 2004;15:1557–67. doi: 10.1091/mbc.E03-08-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lerdrup M, Brunn S, Grandal MV, et al. Endocytic downregulation of ErbB2 is stimulated by cleavage of its C-terminus. Mol Biol Cell. 2007;18:3656–66. doi: 10.1091/mbc.E07-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lerdrup M, Hommelgaard AM, Grandal MV, et al. Geldanamycin stimulates internalization of ErbB2 in a proteosome-dependent pathway. J Cell Sci. 2006;119:85–95. doi: 10.1242/jcs.02707. [DOI] [PubMed] [Google Scholar]
- 16.Hudziak RM, Lewis GD, Winget M, et al. P185HER2 monoclonal antibody has antiproliferative effects in vitro and sensitizes human breast tumor cells to tumor necrosis factor. Mol. Cell. Biol. 1989;9:1165–72. doi: 10.1128/mcb.9.3.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis GD, Lofgren JA, McMurtey AE, et al. Growth regulation of human breast and ovarian tumor cells by heregulin: evidence for the requirement of ErbB2 as a critical component in mediating heregulin responsiveness. Cancer Res. 1996;56:1457–65. [PubMed] [Google Scholar]
- 18.Cuello M, Ettenberg SA, Clark AS, et al. Downregulation of the erbB2 receptor by trastuzumab (herceptin) enhances tumor necrosis factor related apoptosis inducing ligand mediated apoptosis in breast and ovarian cancer cell lines that overexpress erbB2. Cancer Res. 2001;61:4892–900. [PubMed] [Google Scholar]
- 19.Baselga J, Albanell J, Molina MA, et al. Mechanism of action of trastuzumab and scientific update. Semin Oncol. 2001;28:4–11. doi: 10.1016/s0093-7754(01)90276-3. [DOI] [PubMed] [Google Scholar]
- 20.Urbanelli L, Ronchini C, Fontana L, et al. Targeted gene transduction of mammalian cells expressing the HER2/neu receptor by filamentous phage. J Mol Biol. 2001;313:965–76. doi: 10.1006/jmbi.2001.5111. [DOI] [PubMed] [Google Scholar]
- 21.Karasseva NG, Glinsky VV, Chen NX, et al. Identification and characterization of peptides that bind human ErbB2 selected from a bacteriophage display library. J Protein Chem. 2002;21:287–96. doi: 10.1023/a:1019749504418. [DOI] [PubMed] [Google Scholar]
- 22.Houimel M, Schneider P, Terskikh A, et al. Selection of peptides and synthesis of pentameric peptabody molecules reacting specifically with ErbB-2 receptor. Int J Cancer. 2001;92:748–55. doi: 10.1002/1097-0215(20010601)92:5<748::aid-ijc1258>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 23.Pero SC, Shukla GS, Armstrong AL, et al. Identification of a small peptide that inhibits the phosphorylation of ErbB2 and proliferation of ErbB2 overexpressing breast cancer cells. Int J Cancer. 2004;111:951–60. doi: 10.1002/ijc.20306. [DOI] [PubMed] [Google Scholar]
- 24.Hashizume T, Fukuda T, Nagaoka T, et al. Cell type dependent endocytic internalization of ErbB2 with an artificial peptide ligand that binds to ErbB2. Cell Biol Int. 2008;32:814–26. doi: 10.1016/j.cellbi.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 25.Tsutsui Y, Tomizawa K, Nagita M, et al. Development of bionanocapsules targeting brain tumors. J Control Release. 2007;122:159–64. doi: 10.1016/j.jconrel.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 26.Tada H, Kurokawa T, Seita T, et al. Expression and characterization of a chimeric bispecific antibody against fibrin and against urokinase-type plasminogen activator. J Biotech. 1994;33:157–74. doi: 10.1016/0168-1656(94)90108-2. [DOI] [PubMed] [Google Scholar]
- 27.Seno M, Futami J, Kosaka M, et al. Nucleotide sequence encoding human pancreatic ribonuclease. Biochim Biophys Acta. 1994;1218:466–8. doi: 10.1016/0167-4781(94)90208-9. [DOI] [PubMed] [Google Scholar]
- 28.Nagaoka T, Fukuda T, Hashizume T, et al. A betacellulin mutant promotes differentiation of pancreatic acinar AR42J cells into insulin-producing cells with low affinity of binding to ErbB1. J Mol Biol. 2008;380:83–94. doi: 10.1016/j.jmb.2008.03.054. [DOI] [PubMed] [Google Scholar]
- 29.Yamada T, Iwasaki Y, Tada H, et al. Nanoparticles for the delivery of genes and drugs to human hepatocytes. Nat Biotechnol. 2003;21:885–90. doi: 10.1038/nbt843. [DOI] [PubMed] [Google Scholar]
- 30.Dini M, Jafari K, Faiferman I. Cell-mediated cytotoxicity in pre invasive and invasive squamous cell carcinoma of the cervix. Obstet Gynecol. 1980;55:728–31. [PubMed] [Google Scholar]
- 31.Hughes DPM, Thomas DG, Giordano TJ, et al. Cell surface expression of epidermal growth factor receptor and Her-2 with nuclear expression of Her-4 in primary osteosarcoma. Cancer Res. 2004;64:2047–53. doi: 10.1158/0008-5472.can-03-3096. [DOI] [PubMed] [Google Scholar]
- 32.Friedman LM, Rinon A, Schechter B, et al. Synergistic down-regulation of receptor tyrosine kinases by combinations of mAbs: implications for cancer immunotherapy. Proc Natl Acad Sci. 2005;102:1915–20. doi: 10.1073/pnas.0409610102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayor S, Pagano RE. Pathways of clathrin-independent endocytosis. Nat. Rev. Mol. Cell. Biol. 2007;8:603–12. doi: 10.1038/nrm2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirkham M, Parton RG. Clathrin-independent endocytosis: new insights into caveolae and non-caveolar lipid raft carriers. Biochim Biophys Acta. 2005;1745:272–86. doi: 10.1016/j.bbamcr.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 35.Barr DJ, Ostermeyer-Fay AG, Matundan RA, et al. Clathrin-independent endocytosis of ErbB2 in geldanamycin-treated human breast cancer cells. J Cell Sci. 2008;121:3155–66. doi: 10.1242/jcs.020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sönnichsen B, De Renzis S, Neilsen E, et al. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab 11. J Cell Biol. 2000;149:901–14. doi: 10.1083/jcb.149.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Falcone S, Cocucci E, Podini P, et al. Macropinocytosis: regulated coordination of endocytic and exocytic membrane traffic events. J Cell Sci. 2006;119:4758–69. doi: 10.1242/jcs.03238. [DOI] [PubMed] [Google Scholar]
- 38.Sabharanjhak S, Sharma P, Parton RG, et al. GPI-anchored proteins are delivered to recycling endosomems via a distinct cdc42-regulated, clathrin-independent pinocytic pathway. Dev Cell. 2002;2:411–23. doi: 10.1016/s1534-5807(02)00145-4. [DOI] [PubMed] [Google Scholar]
- 39.Chadda R, Howes MT, Plowman SJ, et al. Cholesterol-sensitive Cdc42 activation regulates actin polymerization for endocytosis via the GEEC pathway. Traffic. 2007;8:702–17. doi: 10.1111/j.1600-0854.2007.00565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown FD, Rozelle AL, Yin HL, et al. Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. J Cell Biol. 2001;154:1007–18. doi: 10.1083/jcb.200103107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mu F-T, Callaghan JM, Steele-Mortimer O, et al. EEA1, an early endosome-associated protein. EEA 1 is a conserved alpha-helical peripheral membrane protein flanked by systeine “fingers” and contains a calmodulin-binding IQ motif. J Biol Chem. 1995;270:13503–11. doi: 10.1074/jbc.270.22.13503. [DOI] [PubMed] [Google Scholar]
- 42.Mills I, Jones A, Clague M. Involvement of the endosomal autoantigen EEA1 in homotypic fusion of early endosomes. Curr Biol. 1998;8:881–4. doi: 10.1016/s0960-9822(07)00351-x. [DOI] [PubMed] [Google Scholar]
- 43.Sahay G, Alakhova YD, Kabanov VA. Endocytosis of nanomedicines. J. Con. Rel. 2010;145:182–95. doi: 10.1016/j.jconrel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu Y, Low PS. Folate mediated delivery of macromolecular anticancer therapeutic agents. Adv Drug Deliv Rev. 2002;54:675–93. doi: 10.1016/s0169-409x(02)00042-x. [DOI] [PubMed] [Google Scholar]
- 45.Yu D, Fukuda T, Kuroda S, et al. Engineered bio-nanocapsules, the selective vector for drug delivery system. IUBMB Life. 2006;58:1–6. doi: 10.1080/15216540500484368. [DOI] [PubMed] [Google Scholar]
- 46.Tuoya, Hirayama K, Nagaoka T, et al. Identification of cell surface marker candidates on SV-T2 cells using DNA microarray on DLC-coated glass. Biochem Biophys Res Commun. 2005;334:263–8. doi: 10.1016/j.bbrc.2005.06.083. [DOI] [PubMed] [Google Scholar]
- 47.Samah AS, Yuh Sugii Tuoya, et al. Identification of TM9SF2 as a candidate of the cell surface marker common to breast carcinoma cells. Clin Oncol Cancer Res. 2009;6:1–9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Live cell imaging. For live cell imaging, theSK-BR-3 cells were cultured in 18 mm coverslips and washed twicewith phenol red free RPMI (Sigma) containing 1 % glucose, 25 mMHEPES and 1 % BSA supplemented with 100 U/ml pencillin and 100μg/ml streptomycin. The EC-Fc BNC tagged with RITC was incubatedon ice for 30 min prior to the visualization in order to enable thebinding of the multivalent forms of BNC to the cell surface. Thecells were then washed thrice with warm media and were visualizedin 100X 1.3 N.A. oil immersion objectiveunder Olympus IX81 invertedmicroscope (Olympus) with temperature control followed by dataacquisition with Metamorph software (Molecular Devices). Theimaging was carried out with DP71 cooled CCD camera attached to themicroscope and the fluorescence of RITC was acquired along with theDIC field image. The time-lapse imaging was carried out for onehour with time intervals of 5 minutes each. In Metamorph software,each channel was split and overlayed separately. The acquiredstacks were analyzed with MBF Image J.
Fig. S2 Microarray analysis. Expression of genes inSK-BR-3 cells was analyzed by microarray procedure. Total RNApreparation and analysis was performed as described previously [46,47]. DNA microarray was carrying 1,795-oligonucleotide probes forhuman cell surface proteins. cDNAs were synthesized withSuperescript II reverse transcriptase (Invitrogen) with oligo dTprimers. Amino-allyl-dUTP was incorporated into cDNAs followed bycoupling with Cy-3 dye (Ambion, TX, USA) and were processed forhybridization at 55°C for 15 hours. The fluorescent images forthe hybridization were captured using FLA8000 scanner (Fuji Film,Japan) and analyzed with GenePix Pro5.1 software (Axon Instruments,CA). We found caveolins, Cav-1 and 2, and claudin 16 (CLDN-16) weredownregulated in SK-BR-3 cells when compared to SK-OV-3 cells whileErbB2 and GAPDH expression were equally expressed in both the celllines. RT-PCR (Fig. S1A) and qRT-PCR (Fig. S1B) further confirmedthese results. The conditions for the RT-PCR are as follows:94°C for 5min; followed by 30 cycles of 94°C for 30 sec,55°C for 30 sec, 72°C for 30 sec and 72°C for 7minutes. Quantitative real time PCR was performed with SYBR GreenRealtime Master Mix (Toyobo) in triplicates containing 5ng of cDNAalong with 400 nM primers using LightCycler™ (Roche). Thethermal cycling condition was as follows: 95°C for 1 minfollowed by 40 cycles of 95°C -10 sec, 55°C for 10 sec,72°C for 25 sec and 60°C for 1 min. The following sets ofthe primers were used for the PCR reaction. Claudin-16, (Forward)5′- GCTTGCCACAATGAGGGATCT-3′, (Reverse)5′-TGACTTGGCCATGGAAACACC-3′; Cav-1 (Forward)5′-GACTCGGAGGGACATCTCTAC-3′, (Reverse)5′-GTTGATGCGGACATTGCTGA-3′; Cav-2, (Forward)5′-ACGTACAGCTCTTCATGGAC-3′ (Reverse)5′-CAGTTGCAGGCTGACAGAAG-3′.
Fig. S3 Time dependent colocalization of EC-Fc BNC in earlyendosomes. Colocalization of EC-Fc/BNC-RITC with EEA-1 was found in the early endosomes. SK-BR-3 cells were treated with EC-Fc/BNC-RITC for 5, 30, 90 min, and the colocalization of the EC-Fc/BNC-RITC with the early endosomal marker, EEA-1 was assessed with anti EEA-1 antibody (BD Biosciences) and the secondary antibody labeled with AlexaFlour-488 (Molecular Probes). Error bars indicate standard error. JACoP plugin in Image J was employed for percentage colocalization assessment. Percentage colocalization was found to be maximum up to approximately 70% by 30 min. The data represented here is the representative of two independent experiments.
Fig. S4 Binding of EC-Fc BNC to the cell surface aftertreatment with varying concentration of mβCD. The bindingof EC-Fc/BNC’s to ErbB2, in the presence of 2 to 10 mM ofcholesterol dislodging oligosaccharide-mβCD, was not affected.SK-BR-3 cells were treated with 5 and 10 mM of mβCD for 30minutes followed by treatment with EC-Fc/BNC labeled with RITC areshown in Figure S2. The cells were stained with anti ErbB2 antibody(green) and red spots indicate EC-Fc/BNC labeled with RITC. Barsdepict 10 μm.
Fig. S5 FACS analysis for the surface binding andinternalization of EC-Fc BNC with inhibitor treatment. Surfacebinding and internalization of EC-Fc/BNC labeled with FITC inSK-BR-3 cells was quantified using FACS analysis. First, thebinding of EC-Fc/BNC on SK-BR-3 cells was treated with or without 5mM of mβCD on ice for 30 min. The cells were then fixed andanalyzed with FACS to determine the surface bound fraction (Fig.S5A). The histograms showed overlapping of the peak tops indicatingthat the surface binding is majorly unaffected by the mβCDtreatment. The peak (blue), which corresponds to approximately 20%of the total cell counts, orienting towards the untreated cells(grey peak) indicates the presence of a different population ofcells that are incapable of binding to EC-Fc/BNC after mβCDtreatment. Simultaneously, another set cells were treated withEC-Fc/BNC labeled with FITC in the presence or absence of 5 mM ofmβCD at 37°C for 30 minutes. Cells were then trypsinizedto remove the surface fraction, fixed and permeabilized, and theinternalized fraction was analyzed adopting the same procedure withFACS (Fig. S5B, C). mβCD treated SK-BR-3 cells showed a peakpoint in the histogram which overlaid the peak area of untreatedcells indicating the mβCD inhibits the internalization, whilethe cells without mβCD treatment drifted from the untreatedcells towards the right. Total intensity of the internalizedfraction was assessed from the FACS data were plotted on to a graph(Fig. S5). mβCD suppressed the internalization toapproximately 50% when compared to the absence of mβCDtreatment. M: mβCD; T: trypsin. Error bars indicate standarderror.


