Abstract
The nuclear factor κB (NF-κB) signalling pathway controls important cellular events such as cell proliferation, differentiation, apoptosis and immune responses. Pathway activation occurs rapidly upon TNFα stimulation and is highly dependent on ubiquitination events. Using cytoplasmic to nuclear translocation of the NF-κB transcription factor family member p65 as a read-out, we screened a synthetic siRNA library targeting enzymes involved in ubiquitin conjugation and de-conjugation for modifiers of regulatory ubiquitination events in NF-κB signalling. We identified F-box protein only 7 (FBXO7), a component of Skp, Cullin, F-box (SCF)-ubiquitin ligase complexes, as a negative regulator of NF-κB signalling. F-box protein only 7 binds to, and mediates ubiquitin conjugation to cIAP1 and TRAF2, resulting in decreased RIP1 ubiquitination and lowered NF-κB signalling activity.
Keywords: FBXO7, NF-κB signalling, RNA interference, siRNA, regulatory ubiquitination, cIAP1
Introduction
The NF-κB signalling pathway controls important cellular events such as cell proliferation, differentiation, apoptosis and immune responses [1, 2]. Binding of TNFα to its cognate receptor, TNF-Receptor 1 (TNF-R1), promotes receptor trimerization and rapid recruitment of a multi-protein complex, also known as the TNF-R1 signalling complex (TNF-RSC). In addition to TNF-R1, this TNF-RSC consists of TRADD, TRAF2/5, cIAP1/2, the linear ubiquitin chain assembly complex (LUBAC) and RIP1, with TRADD functioning as an adaptor protein recruiting TRAF2, which through constitutive association co-recruits cIAP1 and 2 (cIAP1/2) (reviewed in Ref. [3, 4]). It is well established that activation of NF-κB signalling relies heavily on ubiquitination events. Both TRAF2 and cIAP1/2 are ubiquitin ligases that have been proposed to attach K63-linked ubiquitin chains to RIP1. However, because K63-ubiquitination of RIP1 and NF-κB signalling activity are restored upon reconstitution with an ubiquitin-ligase deficient TRAF2 mutant in TRAF2−/− MEFs [5], it is a likely scenario that the role of TRAF2 rather is to serve as a scaffold, bringing RIP1 and cIAP1/2 into the vicinity of each other and thus enabling cIAP1/2-mediated ubiquitination of RIP1 [6, 7]. The K63-ubiquitination of RIP1 leads to recruitment and activation of the TAK/TAB kinase complex and the inhibitor of ικB Kinase (IKK) complex, consisting of IKKα, β and NEMO, by binding to the K63-linked ubiquitin chains on RIP1 [8–10]. In addition, K63-linked ubiquitination of TRAF2 has been reported to aid in recruitment of the IKK complex [11]. Recently it was also found that linear ubiquitin chains formed by the LUBAC enhance recruitment of NEMO to RIP1, thus increasing NF-κB signalling activity [12, 13]. Once the IKK complex is active, it phosphorylates IκBα, targeting this inhibitor for ubiquitination by SCFβTRCP and proteasomal degradation [14]. Subsequently, the NF-κB transcription factor heterodimer (p50/p65) translocates from the cytoplasm to the nucleus and induces target gene transcription [15].
Apart from their likely role in K63-linked ubiquitination of RIP1, cIAPs have several other targets within the NF-κB signalling pathway. It is believed that K63-linked ubiquitination of NEMO is important for full IKK complex activation, with cIAP1 being the responsible ubiquitin ligase [16]. Furthermore, overexpression studies have shown that wild-type (WT) cIAP1 can ubiquitinate TRAF2, resulting in repression of NF-κB signalling. This indirectly suggests that cIAP1 may promote K48-linked ubiquitination of TRAF2 and proteasomal degradation of this scaffolding partner [17]. Finally the cIAPs can also auto-ubiquitinate themselves, resulting in their proteasomal degradation [18, 19]. Despite these observations little is known about how the ubiquitin ligase activity and substrate specificity of the cIAPs is regulated.
A number of de-ubiquitinating enzymes (DUBs) have been firmly placed as negative regulators of NF-κB signalling. The Cylindroma tumour suppressor protein (CYLD) de-ubiquitinates NEMO and TRAF2 [20–22], while USP15 reverses βTRCP-mediated ubiquitination of IκBα [23]. In addition, the dual-activity protein A20 removes K63-linked ubiquitin chains from RIP1, whereupon it targets this RIP1 for proteasomal degradation via its K48-ubiquitin ligase activity [24]. Finally a number of additional DUBs have been shown to influence NF-κB signalling upstream of, or at the level of IκBα [25–28].
Recruitment of the TNF-RSC and the ensuing ubiquitination events that trigger signalling activation and NF-κB target gene transcription occur quite rapidly, in fact, within minutes after TNFα stimulation. In the context of this short timeframe we set out to identify additional modulators of regulatory ubiquitination within NF-κB signalling. We performed an imaging-based siRNA screen, monitoring the nuclear accumulation of endogenous p65 upon TNFα stimulation, using a gene-family siRNA library targeting DUBs and other proteins involved in ubiquitin conjugation. We identified several negative regulators of NF-κB signalling and focussed our attention on the FBXO7, a member of the F-box protein family, which confers substrate specificity to SCF-ubiquitin ligase complexes. We show herein that FBXO7 is a negative regulator of NF-κB signalling, modulating ubiquitination of several components of the TNF-RSC and ultimately lowering NF-κB signalling activity.
Materials and methods
DUB siRNA screen and high-throughput immunofluorescence microscopy
The siGenome DUB siRNA library (Thermo Scientific, Lafayette, CO, USA) was aliquoted into black μClear 384 well plates (BD Biosciences, Franklin Lakes, NJ, USA). U2OS cells were transfected using a reverse transfection protocol and the Dharmafect 1 (Thermo Scientific) transfection reagent. Medium was refreshed 24 hrs after transfection and cells were left to propagate for an additional 48 hrs. Next, cells were treated with 10 ng/ml TNFα (Sigma-Aldrich, St. Louis, MO, USA) for 20 min. or left untreated, after which they were fixed using 4% formaldehyde PBS.
Fixed cells were washed with PBS, permeabilized with 0.2% Triton-X100 PBS for 10 min., blocked with 5% BSA PBS for 1 hr, and incubated with rabbit-anti-p65 (C-20; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) antibody at 1:10 dilution for 1 hr. After four wash steps with 0.5% Tween-20 PBS (PBST), cells were incubated with a fluorescent conjugated goat-anti-rabbit Alexa488 (Invitrogen Life Technologies, Grand Island, NY, USA) antibody at 1:75 dilution for 1 hr. After another four wash steps with PBST, 4′,6-diamidino-2-phenylindole (DAPI) was used to stain the DNA content.
Representative images were acquired at 20× with the BD Pathway 855 High Content Imager (BD Biosciences) and analysed by Cell Profiler image analysis software (http://www.cellprofiler.org/) [29, 30]. The DAPI channel was used for nuclei segmentation allowing for quantification of the Alexa488 signal in both the nuclei as well as the cytoplasmic compartment of every cell. The ratios of nuclei over cytoplasmic (Nuc/Cyt) p65 intensities were calculated, normalized to the negative controls and the three technical replicates were summarized by mean ± standard deviation to allow for hit selection. Wells that contained <30% of cells compared to the negative control were excluded from further analysis, and siRNA pools that displayed a Nuc/Cyt p65 ratio beyond negative control ± 3× standard deviation were considered for validation.
siRNA transfections and qRT PCR
The U2OS cells and immortalized BJ primary human fibroblasts (BJET) were transfected using a reverse or a double transfection protocol and Dharmafect 1 or Dharmafect 3 (Thermo Scientific) transfection reagents, respectively. Medium was refreshed 24 hrs after transfection and cells were left to propagate for an additional 48 hrs. Next, cells were treated with 10 ng/ml TNFα (Sigma-Aldrich Corp.) for indicated times, after which RNA was isolated using either Trizol (Invitrogen Life Technologies) or the RNAeasy mini kit (Qiagen, Valencia, CA, USA). Subsequently, 1 µg RNA was used for generating cDNA with the SuperScript II kit (Invitrogen Life Technologies). Using qRT primers listed below, and the FastStart MasterPLUS SYBR Green kit (Roche Diagnostics Limited, Burgess Hill, UK), relative mRNA levels for genes of interest were measured. The relative levels were first normalized to the levels of RPL13 or RPL4 control gene mRNA, and then normalized to the negative control. The mean ± standard deviation of technical replicates is either shown directly (representative figures), or used to summarize three independent experiments.
siRNA sequences
The following siRNAs gave reproducible phenotypes and knockdown:
| FBXO7 no. 1 (D-013606-02, Thermo Scientific) | GGAAUGACGAUCGUAUGUU |
| FBXO7 no. 2 (J-013606-06, Thermo Scientific) | CUGAGUCAAUUCAAGAUAA |
| OTUB2 no. 1 (D-010983-02, Thermo Scientific) | CCGUUUACCUGCUCUAUAA |
| OTUB2 no. 4 (D-010983-04, Thermo Scientific) | AAAGAACGCGUACUGCAGA |
| STAMBP no. 2 (D-012202-02, Thermo Scientific) | GAGAAGCCCUCCUUAGAUG |
| STAMBP no. 4 (D-012202-04, Thermo Scientific) | GCAAGGAUCCACCUCUGUU |
qRT primer sequences
The following TaqMan#x00AE; Gene Expression Assays (Applied Biosystems, Inchinnan Business Park, UK) were used for knockdown validation experiments:
| FBXO7 | Hs00201825_m1 |
| OTUB2 | Hs01027047_m1 |
| STAMBP | Hs00197726_m1 |
The following primers were used for the NF-κB target gene activation assays:
| IL8_Fw | AGCACTCCTTGGCAAAACTG |
| IL8_Rv | CGGAAGGAACCATCTCACTG |
| A20_Fw | TCTTCTGGAGTTCTCTCCCGT |
| A20_Rv | TGACCAGGACTTGGGACTTT |
| IRF1_Fw | GACCCTGGCTAGAGATGCAG |
| IRF1_Rv | ATCCTTGTTGATGTCCCAGC |
| STX11_Fw | GCTTCTCGGTTCGCACTCT |
| STX11_Rv | TGCTGGTCATATTGCTTGGA |
Materials, antibodies and plasmids
The Luciferase plasmid NF-κB-Luc was obtained from Clontech (Mountain View, CA, USA). SV40-Renilla was obtained from Promega (Madison, WI, USA). pEGFP-FBXO7 and pVlag-FBXO7 were cloned by PCR amplification of IMAGE clone 3611049. Sal I site containing PCR primers (GATC GTC GAC CAA CCC AAA TAC ATC TGG/GATC GTC GAC CCA CTC CTG TGG AGG TT) were used to construct the ΔF-box mutant. pEGFP-cIAP1 was cloned by PCR amplification of IMAGE clone 3908352.
Antibodies used were anti-GFP (FL) and (B-2), anti-p65 (C-20), anti-CDK4 (C-22), anti-Ubiquitin (P4D1) from Santa Cruz Biotechnology, Inc., anti-flag (M2) from Sigma-Aldrich Corp., anti-pIkBα (5α5) and anti-IκBα (L35A5) from Cell Signaling Technology, Inc. (Danvers, MA, USA), anti-cIAP1 (AF8181) from R&D Systems Inc. (Minneapolis, MN, USA), and anti-HA (12CA5) from a hybridoma culture supernatant grown in our laboratory. F-box protein only 7 rabbit antibody is described in Ref. [31]. Normal mouse IgG from Santa Cruz Biotechnology, Inc. was used as non-immune control in immuno-precipitation experiments.
Cell cultures, transient transfections and reporter assays
The U2OS, BJET and HEK293 cells were cultured in DMEM, supplemented with 10% foetal calf serum. All cell lines were acquired from the American Type Culture Collection. DNA transfections of U2OS and HEK293 were done with the calcium phosphate method. For luciferase reporter assays 0.125 μg NF-kB-Luc, 0.25 ng SV40-renilla and 0.625 μg pcDNA3.1 or pVlag-FBXO7 plasmids were transfected per 24-well. Forty-eight hours after transfection, cells were stimulated with 10 ng/ml TNF-α and luciferase activity was measured 72 hrs after transfection.
Immunoblotting and immunoprecipitation
Western blots were performed by using whole cell extracts, separated on 8–10% SDS-PAGE gels or pre-cast gradient gels (Invitrogen Life Technologies) and transferred to polyvinylidine difluoride membranes (Millipore, Billerica, MA, USA). Western blots were probed with the indicated antibodies. Co-immunoprecipitation experiments were essentially done as in Ref. [32]. U2OS cells were transfected by calcium phosphate precipitation with the indicated plasmids. Seventy-two hours after transfection cells were lysed in either ELB or ELB Plus buffer, supplemented with ‘complete’ protease inhibitors (Roche Diagnostics Limited), and proteins were immunoprecipitated with 2 μg of the indicated antibody conjugated to protein G sepharose beads. To detect flag-TRAF2 ubiquitination, HEK293 cells were transfected by calcium phosphate precipitation with the indicated plasmids, and immunoprecipitations were performed in the more denaturing radioimmunoprecipitation assay (RIPA) buffer, with vortexing during cellular lysis.
Results
A siRNA screen for novel regulators of NF-κB signalling
To identify novel modulators of regulatory ubiquitination in NF-κB signalling, we performed an imaging-based screen measuring sub-cellular redistribution of NF-κB family member p65, using a commercially available siRNA library targeting DUBs and other proteins involved in ubiquitination. U2OS cells were transfected with the siRNA library consisting of pools of four unique siRNAs targeting a specific gene, and left to propagate for 72 hrs, after which they were stimulated with TNFα for 20 min., or left untreated prior to fixation (Fig. 1A). Subsequently, endogenous p65 was visualized by immunofluorescence and DNA was stained with DAPI. Images were acquired by using an automated microscope and analysed with CellProfiler software [29, 30]. The nuclear over cytoplasmic (Nuc/Cyt) ratio of the p65 signal was calculated and used to select candidate modulators of NF-κB signalling (Fig. 1B).
Fig 1.
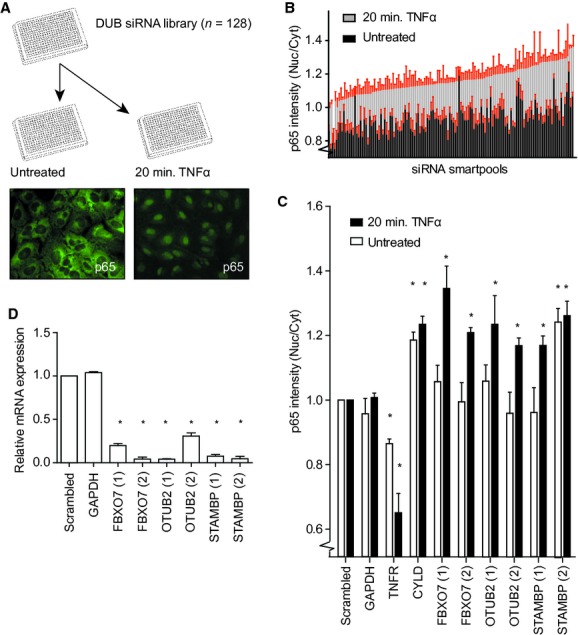
Screen for modulators of NF-κB signalling. (A) Screen setup: U2OS cells were transfected in a single well format with the DUB siRNA library (Thermo Scientific), stimulated with TNFα or left untreated, and analysed for cellular redistribution of NF-κB p65. Representative photos of unstimulated and TNFα-stimulated cells showing endogenous p65 staining. (B) Control normalized ratios of nuclear over cytoplasmic p65 intensity for the siRNA smartpools included in the screen. Error bars shown in red. (C) Summary of three independent experiments for cellular redistribution of p65 upon knockdown of FBXO7, OTUB2, STAMBP, and control genes, GAPDH, TNF-R1 and CYLD. A non-targeting (scrambled) siRNA smartpool was used as negative control. (D) Summary of three independent experiments showing the knockdown efficiency determined by qRT PCR for mRNA levels of the identified genes. (C and D) *P < 0.05, values are mean ± standard deviation. P-value computed from unpaired two-tailed t-test.
Wells that contained <30% of cells compared to the negative control were excluded from further analysis, and siRNA pools that displayed a Nuc/Cyt p65 ratio beyond negative control ± 3× standard deviation were considered for validation. Of the 128 screened siRNA pools, 15 matched these criteria including those targeting the known negative regulators of NF-κB signalling CYLD and A20. Further validation led to the selection of six hits that showed reproducible phenotypes, and three of these could be validated with two unique siRNAs, decreasing the likelihood of potential off-target effects (Fig. 1C). These validated and novel modulators of NF-κB signalling include FBXO7, OTU domain-containing ubiquitin aldehyde-binding protein 2 (OTUB2), and signal transducing adaptor molecule binding protein (STAMBP). The siRNA-induced suppression of each of these genes results in increased nuclear accumulation of p65 upon TNFα stimulation compared to a pool of non-targeting siRNAs (Scrambled), and a pool of siRNAs targeting GAPDH (Fig. 1C). This is in contrast to inhibition of CYLD, which even in the absence of TNFα stimulation promotes an increase in nuclear accumulation of p65. As expected, knockdown of the TNF-R1 abrogates the cellular redistribution of p65. We also tested the knock-down efficiency of the individual siRNAs that affected p65 nuclear accumulation. For each individual siRNA we observe a strong reduction in the mRNA levels of their intended targets (Fig. 1D).
Both OTUB2 and STAMBP are members of the family of DUB enzymes [33]. On the other hand, FBXO7 belongs to the F-box protein family, which in the context of SCF ubiquitin ligase complexes, confers substrate specificity in the ubiquitination process [34, 35]. Interestingly, depending on the context, FBXO7 is able to both stabilize and destabilize its interaction partners [31, 36], and certain recessive mutants of FBXO7 have been linked to early-onset Parkinson-pyramidal syndromes [37, 38]. Furthermore, a study by Chang et al. suggests that cIAP1, a well established positive regulator of NF-κB signalling, is ubiquitinated by the SCFFBXO7 complex [39]. However, the influence of this modification on NF-κB signalling was not examined [39]. We therefore set out to investigate how FBXO7-mediated ubiquitination might influence NF-κB signalling events.
F-box only protein 7 is a negative regulator of NF-κB signalling
Having established that FBXO7 inhibition leads to increased nuclear accumulation of p65, we next examined whether or not NF-κB target gene activation is also affected. The NF-κB target genes IL8, A20, IRF1 and STX11 were selected for these experiments based on micro-array studies in U2OS cells treated with TNFα for 1–2 hrs (data not shown). We measured the TNFα-induced activation of these NF-κB target genes by qRT PCR on RNA isolates of U2OS cells transfected with siRNAs targeting TNF-R1, CYLD or FBXO7 (Fig. 2A and S1). As expected, knockdown of TNF-R1 impairs the activation of these target genes. In contrast, knockdown of CYLD results in an increase of transcriptional activation of IL8, A20 and IRF1. Similarly to CYLD, knockdown of FBXO7, using two unique siRNAs, results in hyper-activation of IL8, A20, IRF1 and STX11. Importantly, we observed similar effects on target gene activation in an independent cell line, BJ primary fibroblasts immortalized by stable expression of human telomerase (hTERT). In these cells, suppression of FBXO7 expression also leads to increased transcriptional activation of IL8 and A20 upon TNFα stimulation (Fig. 2B).
Fig 2.
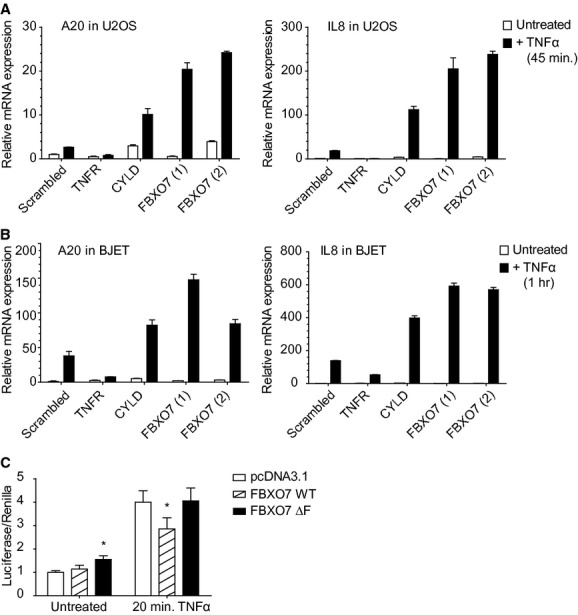
FBXO7 regulates NF-κB signalling. (A and B) NF-κB target gene, A20 and IL8, activation determined by qRT PCR upon stimulation with TNFα in (A) U2OS and (B) immortalized primary human fibroblasts (BJET) transfected with siRNAs targeting FBXO7, TNF-R1, CYLD or a non-targeting (scrambled) control. Values are mean ± standard deviation of replicate measurements from a representative experiment. (C) NF-κB luciferase reporter assay in U2OS cells expressing the NF-κB luciferase reporter construct, a SV40-Renilla construct, and either empty vector (pcDNA3.1), Flag-tagged wild-type FBXO7 (WT), or Flag-tagged F-box deletion mutant FBXO7 (ΔF). Values represent ratio of Luciferase activity over Renilla control activity in the absence or presence of TNFα. *P < 0.05, values are mean ± standard deviation of four independent experiments. P-value computed from unpaired two-tailed t-test.
Next, we tested whether or not overexpression of FBXO7 would suppress NF-κB signalling. A NF-κB luciferase reporter was co-transfected with plasmids encoding either WT FBXO7, or a FBXO7 F-box deletion mutant (ΔF), which no longer forms a SCF complex. We found that overexpression of WT FBXO7 but not the ΔF mutant suppresses the activity of the NF-κB luciferase reporter upon stimulation with TNFα (Fig. 2C). We conclude that the regulation of NF-κB signalling by FBXO7 depends on SCFFBXO7 complex formation, and that SCFFBXO7 negatively regulates NF-κB signalling.
FBXO7 acts upstream of IκBα degradation
To gain more insight into the function of FBXO7 as a negative regulator of NF-κB signalling, we investigated the influence of FBXO7 on the rate of IκBα protein degradation upon TNFα stimulation. This is a key regulatory step in the NF-κB pathway, separating effectors directly acting on NF-κB transcription factors and transcriptional activation of downstream targets, from upstream signalling events at the level of the TNF-RSC. We transfected U2OS cells with control siRNAs as well as siRNAs targeting CYLD and FBXO7, performed TNFα time-course experiments and used Western blot analysis to visualize IκBα phosphorylation and degradation. As expected, knockdown of CYLD increases the rate of IkBα protein degradation independent of TNFα stimulation (Fig. 3A). Suppression of FBXO7 expression results in a similar phenotype, however, degradation of IκBα is primarily affected upon treatment with TNFα, in agreement with the results of the nuclear translocation assays (Fig. 1C). Conversely, overexpression of WT but not ΔF FBXO7 results in stabilization of IκBα, explaining the diminished signalling observed upon overexpression in the NF-κB reporter experiments (Figs 2C and 3B). These observations point to the TNF-RSC as the site of action for FBXO7, which is supported by the previously reported interaction between FBXO7 and cIAP1 [39].
Fig 3.
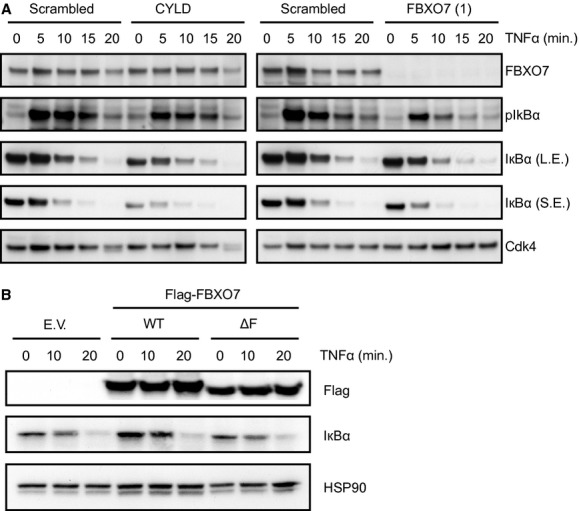
FBXO7 regulates the NF-κB signalling upstream of IκBα degradation. (A) Western blot analysis of U2OS cells transfected with non-targeting (scrambled) control, CYLD or FBXO7 siRNAs stimulated with TNFα for 5, 10, 15, 20 min., or left untreated. Western blot analysis was performed for FBXO7, pIκBα, IκBα and Cdk4 (loading control). L.E.: long exposure; S.E.: short exposure. (B) Western blot analysis of U2OS cells expressing empty vector (EV), Flag-FBXO7 WT, or ΔF. Western blot analysis was performed for the Flag epitope, IκBα and HSP90 (loading control).
FBXO7 promotes ubiquitination of bound cIAP1
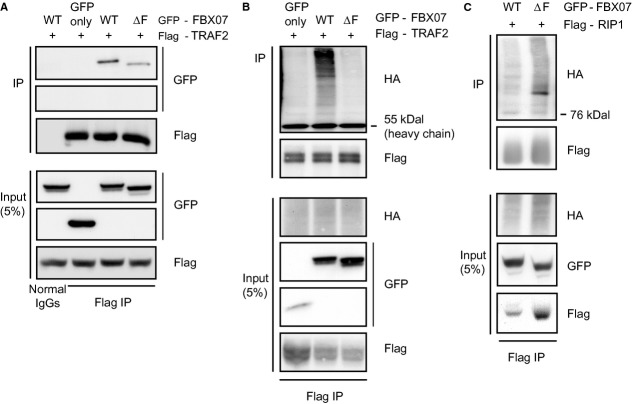
To gain further insight into possible sites of action of FBXO7 in the context of the TNF-RSC, we examined whether or not FBXO7 interacts with components of the TNF-RSC that are known to be ubiquitinated. First, in co-transfection experiments with tagged versions of either WT or ΔF FBXO7 and cIAP1, we confirmed that cIAP1 not only binds to WT, but also ΔF FBXO7 (Fig. 4A). These interactions were not influenced by TNFα stimulation (data not shown). Next, we investigated whether or not cIAP1 is ubiquitinated by SCFFBXO7. We observed no change in the ubiquitination levels of total cellular cIAP1 in the presence of exogenous FBXO7 (data not shown). However, when scaling up FBXO7-cIAP1 co-immunoprecipitation experiments, we found that cIAP1, associated with WT but not FBXO7 ΔF, is highly subject to ubiquitination (Fig. 4B).
Fig 4.
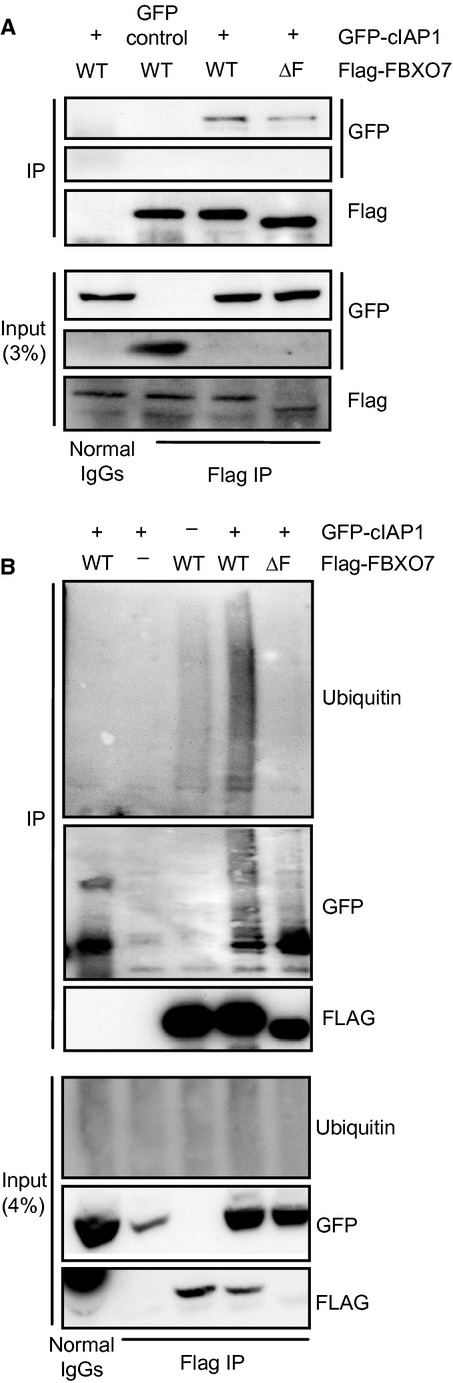
FBXO7 interacts with and mediates ubiquitination of cIAP1. (A) Western blot analysis of a co-immunoprecipitation experiment in U2OS cells expressing GFP-cIAP1 and Flag-FBXO7 WT or ΔF. (B) Western blot analysis of in vivo ubiquitinated GFP-cIAP1 co-immunoprecipitated with Flag-FBXO7 WT or ΔF.
FBXO7 binds to and ubiquitinates TRAF2
The TRAF2 and cIAP1/2 are constitutively bound to each other and co-recruited to the TNF-RSC upon TNFα stimulation [3, 4]. We therefore posed the question whether or not FBXO7 also interacts with TRAF2 and represents a constitutive member of the complex. To answer this question, we immuno-precipitated Flag-TRAF2 from cells that had been co-transfected with GFP-FBXO7 WT or ΔF constructs. We found that FBXO7 binds to TRAF2 (Fig. 5A), independently of TNFα (data not shown). We next examined whether or not FBXO7 also affects TRAF2 ubiquitination. HEK293 cells were transfected with constructs expressing Flag-TRAF2, HA-ubiquitin and GFP-FBXO7 WT or ΔF. To avoid possible ubiquitin contamination from co-immunoprecipitated proteins, we lysed cells in denaturing RIPA buffer. We then performed Flag-immunoprecipitations and immuno-blots for HA-ubiquitin. We found that there is a significant increase in TRAF2 ubiquitination upon overexpression of WT but not ΔF FBXO7 (Fig. 5B). In summary FBXO7 binds to cIAP1 and TRAF2, and promotes ubiquitination of both proteins, the consequence being decreased NF-κB signalling activity.
Fig 5.
FBXO7 interacts with TRAF2, and alters ubiquitination of TRAF2 and RIP1. (A) Western blot analysis of a co-immunoprecipitation experiment in U2OS cells expressing Flag-TRAF2 and GFP-tagged wild-type FBXO7 WT or ΔF. (B and C) Western blot analysis of an in vivo ubiquitination assays in HEK293 cells expressing HA-ubiquitin, Flag-TRAF2 or Flag-RIP1, GFP only, GFP-FBXO7 WT or ΔF.
FBXO7 lowers RIP1 ubiquitination
As TRAF2 and cIAP1 are both highly important for ubiquitin chain formation on RIP1, we next investigated whether or not FBXO7-induced changes in TRAF2 and cIAP1 ubiquitination levels affects RIP1 ubiquitination. First, in co-immunoprecipitation experiments we tried to determine whether or not FBXO7 and RIP1 interact. However, we were unable to detect this interaction (data not shown). We then measured RIP1 ubiquitination levels in HEK293 cells transfected with constructs expressing Flag-RIP1, HA-ubiquitin and GFP-FBXO7 WT or ΔF. We found that overexpression of GFP-FBXO7 WT diminishes RIP1 ubiquitination compared to ΔF FBXO7 (Fig. 5C). We suggest that this decrease reduces the recruitment and activation of kinase complexes, preventing IκBα degradation, ultimately resulting in reduced NF-κB signalling.
Discussion
Here, we screened a synthetic siRNA library for modulators of regulatory ubiquitination in NF-κB signalling. Employing cytoplasmic to nuclear redistribution of the endogenous NF-κB transcription factor subunit p65 as a read-out, we identified and verified three novel regulators. We focused our attention on FBXO7, and showed that suppression of FBXO7 expression leads to increased nuclear accumulation of p65 upon TNFα stimulation, and hyper-activation of NF-κB transcriptional targets. By performing TNFα time-course experiments we placed FBXO7 upstream of IκBα degradation. We found that FBXO7 interacts with cIAP1 and TRAF2 and promotes ubiquitination of both proteins, which in turn results in decreased RIP1 ubiquitination, and lower levels of NF-κB signalling.
The FBXO7 has been reported to have different functions. In the context of SCFFBXO7 proteasomal degradation of Hepatoma Up-Regulated Protein is promoted [36]. Other functions, such as stabilization of Cdk6/CyclinD complexes, and pro-B cell differentiation have recently been linked to FBXO7 nuclear localization [40, 41]. Contrarily, our data show that in the context of NF-κB signalling, FBXO7 exerts its function in the cytoplasm.
The RIP1 is a key protein within the NF-κB signalling pathway with K63-linked ubiquitin chains serving as recruitment platforms for the two kinase complexes TAK/TAB and IKK [8–10]. This K63-linked ubiquitin conjugation depends on recruitment of constitutively interacting TRAF2-cIAP1/2 complexes to the TNF-RSC [6, 7]. We showed that FBXO7 binds to both cIAP1 and TRAF2 independently of TNFα, suggesting constitutive binding and co-recruitment to the TNF-RSC upon stimulation. We found that FBXO7 promotes ubiquitination of both cIAP1 and TRAF2, leading to a decrease in RIP1 ubiquitination and lowered NF-κB signalling activity. Different scenarios can be envisioned to account for the lowered levels of RIP1 ubiquitination. Within the context of NF-κB signalling, in addition to RIP1, the cIAP1/2 proteins can promote ubiquitination of NEMO, TRAF2 as well as auto-ubiquitination [16–19]. However, at present it remains unclear how cIAP1/2 substrate selectivity is determined. We observed that cIAP1 specifically bound by FBXO7 is ubiquitinated. Perhaps this post-translational modification could switch cIAP1 substrate selectivity away from RIP1, thus decreasing kinase complex recruitment. In a second negative regulatory step one could even speculate that the ubiquitin ligase activity of cIAP1 could be turned towards other targets within the signalling pathway, possibly triggering their proteasomal degradation. A change from K63- to K48-linked ubiquitin conjugation likely requires the exchange of ubiquitin conjugating enzyme (E2) partner (as discussed in Ref. [42]), however, whether or not SCFFBXO7 mediated ubiquitination of cIAP1 triggers such a change remains to be investigated. Interestingly, binding of two E2s, UbcH5, which is primarily responsible for K48 linked ubiquitination and Ubc13 (K63-specific), has been observed for the C-terminus of Hsp70 interacting protein (CHIP) [43–45]. This ubiquitin ligase plays an important role in protein quality control through binding to the chaperone proteins Hsp70 and Hsp90, targeting their client proteins for proteasomal degradation (reviewed in Ref. [46]). However, at present there are no known targets for CHIP mediated K63-linked ubiquitination, and how E2 choice is determined also remains unknown. Specific cellular localization of certain E2s has been proposed to control some interactions [44]. In addition, we speculate that post-translational modifications of ubiquitin ligases, causing conformational changes could also promote a switch in E2 interactions.
Some evidence exists that NF-κB induced inflammation may be involved in development of Parkinson's disease (reviewed in Ref. [47]). As mentioned previously, certain recessive mutants of FBXO7 have been linked to early-onset Parkinson-pyramidal syndromes [37, 38]. We investigated whether or not such mutations might influence NF-κB signalling, and thereby promote disease phenotypes using two different FBXO7 patient mutants (R378G and R498X). However, neither binding to, nor ubiquitination of cIAP1 or TRAF2 was affected significantly by these mutations, and in NF-κB luciferase experiments patient mutants behaved like WT FBXO7 (data not shown). It remains possible that the investigated patient mutations give highly subtle phenotypes, or that defects are only noticeable in neuronal cell types. Alternatively, development of Parkinson-pyramidal syndrome may be a consequence of yet to be determined functions of FBXO7.
In conclusion, this study strongly implicates a novel inhibitory role of FBXO7 at the TNF-RSC in modulating regulatory ubiquitin events in NF-κB signalling. Future experiments are required to elucidate the precise mode of action further, expanding our knowledge of this complexly regulated signalling pathway.
Acknowledgments
The Netherlands Genomics Initiative/Netherlands Organization for Scientific Research. A.M.D. was supported by grants from the Dutch Cancer Society and the EU Network of Excellence ‘RUBICON’. H.J.K. was supported by a grant from Top Institute Pharma, project T3-105.
Conflict of interest
The authors confirm that there are no conflicts of interest.
Supporting information
Additional Supporting Information may be found in the online version of this article:
Fig. S1 FBXO7 regulates NF-κB signalling. NF-κB target gene, IRF1 and STX11, activation determined by qRT PCR upon stimulation with TNFα in U2OS cells transfected with siRNAs targeting FBXO7, TNF-R1, CYLD, or non-targeting (scrambled) control. Values are mean ± standard deviation of replicate measurements from a representative experiment.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Chen G, Goeddel DV. TNF-R1 signaling: a beautiful pathway. Science. 2002;296:1634–5. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- 2.Karin M, Cao Y, Greten FR, et al. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2:301–10. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 3.Chan FK. Three is better than one: pre-ligand receptor assembly in the regulation of TNF receptor signaling. Cytokine. 2007;37:101–7. doi: 10.1016/j.cyto.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wajant H, Scheurich P. TNFR1-induced activation of the classical NF-kappaB pathway. FEBS J. 2011;278:862–76. doi: 10.1111/j.1742-4658.2011.08015.x. [DOI] [PubMed] [Google Scholar]
- 5.Vince JE, Pantaki D, Feltham R, et al. TRAF2 must bind to cellular inhibitors of apoptosis for tumour necrosis factor (tnf) to efficiently activate nf-{kappa}b and to prevent tnf-induced apoptosis. J Biol Chem. 2009;284:35906–15. doi: 10.1074/jbc.M109.072256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertrand MJ, Milutinovic S, Dickson KM, et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 7.Mahoney DJ, Cheung HH, Mrad RL, et al. Both cIAP1 and cIAP2 regulate TNFalpha-mediated NF-kappaB activation. Proc Natl Acad Sci USA. 2008;105:11778–83. doi: 10.1073/pnas.0711122105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanayama A, Seth RB, Sun L, et al. TAB 2 and TAB 3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15:535–48. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 9.Ea CK, Deng L, Xia ZP, et al. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–57. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 10.Wu CJ, Conze DB, Li T, et al. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation [corrected] Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 11.Li S, Wang L, Dorf ME. PKC phosphorylation of TRAF2 mediates IKKalpha/beta recruitment and K63-linked polyubiquitination. Mol Cell. 2009;33:30–42. doi: 10.1016/j.molcel.2008.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haas TL, Emmerich CH, Gerlach B, et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 complex and is required for TNF-mediated gene induction. Mol Cell. 2009;36:831–44. doi: 10.1016/j.molcel.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 13.Tokunaga F, Sakata S, Saeki Y, et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11:123–32. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 14.Yaron A, Hatzubai A, Davis M, et al. Identification of the receptor component of the IkappaBalpha-ubiquitin ligase. Nature. 1998;396:590–4. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- 15.Baldwin AS., Jr The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–83. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 16.Tang ED, Wang CY, Xiong Y, et al. A role for NF-kappaB essential modifier/IkappaB kinase-gamma (NEMO/IKKgamma) ubiquitination in the activation of the IkappaB kinase complex by tumour necrosis factor-alpha. J Biol Chem. 2003;278:37297–305. doi: 10.1074/jbc.M303389200. [DOI] [PubMed] [Google Scholar]
- 17.Samuel T, Welsh K, Lober T, et al. Distinct BIR domains of cIAP1 mediate binding to and ubiquitination of tumour necrosis factor receptor-associated factor 2 and second mitochondrial activator of caspases. J Biol Chem. 2006;281:1080–90. doi: 10.1074/jbc.M509381200. [DOI] [PubMed] [Google Scholar]
- 18.Varfolomeev E, Blankenship JW, Wayson SM, et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell. 2007;131:669–81. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 19.Vince JE, Wong WW, Khan N, et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell. 2007;131:682–93. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
- 20.Brummelkamp TR, Nijman SM, Dirac AM, et al. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- 21.Kovalenko A, Chable-Bessia C, Cantarella G, et al. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424:801–5. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- 22.Trompouli E, Hatzivassiliou E, Tsichritzis T, et al. The familial cyclindromatosis gene product is a deubiquitinating enzyme that negatively regulates NF-kB activation by members of the TNF receptor family. Nature. 2003;424:793–6. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- 23.Schweitzer K, Bozko PM, Dubiel W, et al. CSN controls NF-kappaB by deubiquitinylation of IkappaBalpha. EMBO J. 2007;26:1532–41. doi: 10.1038/sj.emboj.7601600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wertz IE, O'Rourke KM, Zhou H, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–9. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 25.Metzig M, Nickles D, Falschlehner C, et al. An RNAi screen identifies USP2 as a factor required for TNF-alpha-induced NF-kappaB signaling. Int J Cancer. 2011;129:607–18. doi: 10.1002/ijc.26124. [DOI] [PubMed] [Google Scholar]
- 26.Xu G, Tan X, Wang H, et al. Ubiquitin-specific peptidase 21 inhibits tumour necrosis factor alpha-induced nuclear factor kappaB activation via binding to and deubiquitinating receptor-interacting protein 1. J Biol Chem. 2010;285:969–78. doi: 10.1074/jbc.M109.042689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tzimas C, Michailidou G, Arsenakis M, et al. Human ubiquitin specific protease 31 is a deubiquitinating enzyme implicated in activation of nuclear factor-kappaB. Cell Signal. 2006;18:83–92. doi: 10.1016/j.cellsig.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 28.Enesa K, Zakkar M, Chaudhury H, et al. NF-kappaB suppression by the deubiquitinating enzyme Cezanne: a novel negative feedback loop in pro-inflammatory signaling. J Biol Chem. 2008;283:7036–45. doi: 10.1074/jbc.M708690200. [DOI] [PubMed] [Google Scholar]
- 29.Jones TR, Kang IH, Wheeler DB, et al. CellProfiler Analyst: data exploration and analysis software for complex image-based screens. BMC Bioinformatics. 2008;9:482. doi: 10.1186/1471-2105-9-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carpenter AE, Jones TR, Lamprecht MR, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laman H, Funes JM, Ye H, et al. Transforming activity of Fbxo7 is mediated specifically through regulation of cyclin D/cdk6. EMBO J. 2005;24:3104–16. doi: 10.1038/sj.emboj.7600775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dirac AM, Bernards R. The deubiquitinating enzyme USP26 is a regulator of androgen receptor signaling. Mol Cancer Res. 2010;8:844–54. doi: 10.1158/1541-7786.MCR-09-0424. [DOI] [PubMed] [Google Scholar]
- 33.Nijman SM, Luna-Vargas MP, Velds A, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–86. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 34.Winston JT, Koepp DM, Zhu C, et al. A family of mammalian F-box proteins. Curr Biol. 1999;9:1180–2. doi: 10.1016/S0960-9822(00)80021-4. [DOI] [PubMed] [Google Scholar]
- 35.Cenciarelli C, Chiaur DS, Guardavaccaro D, et al. Identification of a family of human F-box proteins. Curr Biol. 1999;9:1177–9. doi: 10.1016/S0960-9822(00)80020-2. [DOI] [PubMed] [Google Scholar]
- 36.Hsu JM, Lee YC, Yu CT, et al. Fbx7 functions in the SCF complex regulating Cdk1-cyclin B-phosphorylated hepatoma up-regulated protein (HURP) proteolysis by a proline-rich region. J Biol Chem. 2004;279:32592–602. doi: 10.1074/jbc.M404950200. [DOI] [PubMed] [Google Scholar]
- 37.Shojaee S, Sina F, Banihosseini SS, et al. Genome-wide linkage analysis of a Parkinsonian-pyramidal syndrome pedigree by 500 K SNP arrays. Am J Hum Genet. 2008;82:1375–84. doi: 10.1016/j.ajhg.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Fonzo A, Dekker MC, Montagna P, et al. FBXO7 mutations cause autosomal recessive, early-onset parkinsonian-pyramidal syndrome. Neurology. 2009;72:240–5. doi: 10.1212/01.wnl.0000338144.10967.2b. [DOI] [PubMed] [Google Scholar]
- 39.Chang YF, Cheng CM, Chang LK, et al. The F-box protein Fbxo7 interacts with human inhibitor of apoptosis protein cIAP1 and promotes cIAP1 ubiquitination. Biochem Biophys Res Commun. 2006;342:1022–6. doi: 10.1016/j.bbrc.2006.02.061. [DOI] [PubMed] [Google Scholar]
- 40.Nelson DE, Laman H. A competitive binding mechanism between Skp1 and Exportin 1 (CRM1) controls the localization of a subset of F-box proteins. J Biol Chem. 2011;286:19804–15. doi: 10.1074/jbc.M111.220079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meziane el K, Randle SJ, Nelson DE, et al. Knockdown of Fbxo7 reveals its regulatory role in proliferation and differentiation of haematopoietic precursor cells. J Cell Sci. 2011;124:2175–86. doi: 10.1242/jcs.080465. [DOI] [PubMed] [Google Scholar]
- 42.Nagy V, Dikic I. Ubiquitin ligase complexes: from substrate selectivity to conjugational specificity. Biol Chem. 2010;391:163–9. doi: 10.1515/bc.2010.021. [DOI] [PubMed] [Google Scholar]
- 43.Soss SE, Yue Y, Dhe-Paganon S, et al. E2 conjugating enzyme selectivity and requirements for function of the E3 ubiquitin ligase CHIP. J Biol Chem. 2011;286:21277–86. doi: 10.1074/jbc.M111.224006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu Z, Kohli E, Devlin KI, et al. Interactions between the quality control ubiquitin ligase CHIP and ubiquitin conjugating enzymes. BMC Struct Biol. 2008;8:26. doi: 10.1186/1472-6807-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graf C, Stankiewicz M, Nikolay R, et al. Insights into the conformational dynamics of the E3 ubiquitin ligase CHIP in complex with chaperones and E2 enzymes. Biochemistry. 2010;49:2121–9. doi: 10.1021/bi901829f. [DOI] [PubMed] [Google Scholar]
- 46.McDonough H, Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones. 2003;8:303–8. doi: 10.1379/1466-1268(2003)008<0303:calbtc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Flood PM, Qian L, Peterson LJ, et al. Transcriptional factor NF-kappaB as a target for therapy in Parkinson's disease. Parkinsons Dis. 2011:216298–305. doi: 10.4061/2011/216298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.