Abstract
MicroRNAs (miRNA) are small RNAs that regulate the translation of thousands of message RNAs and play a profound role in mammalian biology. Over the past 5 years, significant advances have been made towards understanding the pathways that generate miRNAs and the mechanisms by which miRNAs exert their regulatory functions. An emerging theme is that miRNAs are both generated by and utilized by large and complex macromolecular assemblies. Here, we review the biology of mammalian miRNAs with a focus on the macromolecular complexes that generate and control the biogenesis of miRNAs.
Keywords: microRNA, drosha, dicer, Microprocessor, exportin-5
Introduction
MicroRNAs (miRNAs) are single-stranded RNAs of 22-nucleotides in length that play a critical role in regulating gene expression in multi-cellular organisms [1]. MiRNAs guide gene silencing by base pairing with the target mRNAs, and lead to translational repression and/or mRNA cleavage. MiRNAs direct diverse regulatory pathways, including developmental timing control, cell differentiation and proliferation, apoptosis and organ development [2]. Currently, over 600 miRNAs have been identified in human cells and it is estimated that thousands of genes in the human genome are regulated by miRNAs [3].
The human genome contains over 3 billion base pairs of DNA, about 15% of which is believed to be transcribed [4]. From within this vast sea of RNA, the miRNA biogenesis machinery must recognize and process the tiny RNA fragments destined to become miRNAs. Here, we review the current understanding of miRNA biogenesis with an emphasis on the molecular machines that mediate this essential cellular process. We also discuss recent findings that have begun to shed light on how miRNA recognition and maturation are controlled.
miRNAs originate from primary RNA transcripts
Most miRNAs arise from long RNA transcripts generated by RNA polymerase II [5, 6], although RNA polymerase III is also reported to generate a subset of miRNAs as well [7]. These primary transcripts, or pri-miRNA, often contain multiple miRNA sequences in tandem and can be up to several kilobases long. Pri-miRNA can be non-coding transcripts, bearing a 5′ 7-methyl guanylate (m7G) cap and a 3′ poly (A) tail or be encoded within introns or untranslated regions of coding RNAs [8–12]. About half of the known miRNAs in human beings are derived from the introns of messenger RNAs (mRNAs) [13, 14]. Regardless of the origin of the pri-miRNA, the consensus requirement for the recognition and subsequent processing of these molecules relies upon the ability of the sequence to form a stable hairpin structure of at least 30 base-pairs. This extended hairpin structure serves as signal for entry into miRNA maturation pathway (Fig. 1).
1.
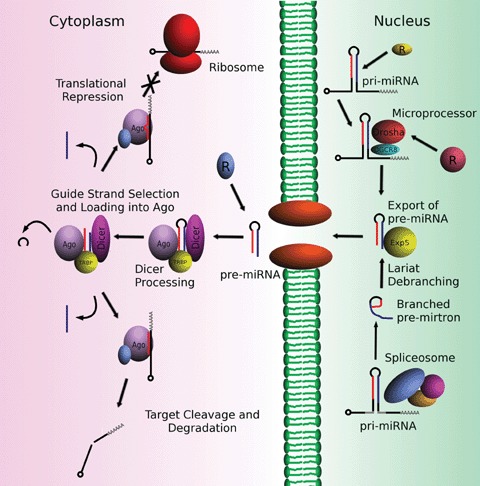
The miRNA biogenesis pathway. Primary miRNAs (pri-miRNAs) contain hairpin structures that are recognized by either the Microprocessor or splicing machinery in the nucleus. Pre-miRNAs are then exported to the cytoplasm where they are processed into mature single-stranded miRNAs and silence target genes. Regulatory proteins are generically labelled ‘R’ at known points of regulation in maturation of specific miRNAs.
The Microprocessor initiates miRNA processing
Within the nucleus, the hairpin structures formed in pri-miRNAs are recognized and cropped out of primary transcripts by a ∼650 kD protein complex called the Microprocessor [15, 16]. The catalytic subunit of the Microprocessor is a protein in the RNase III family of enzymes named Drosha (also known as RNASEN) [9, 17, 18]. Drosha cleaves the pri-miRNA near the base of the hairpin to liberate a 65–100 nucleotide RNA product. Recognition of hairpin structure and cleavage site selection are mediated by another subunit in the Microprocessor named DiGeorge syndrome Critical Region gene 8 (DGCR8), which is also called Pasha (partner of Drosha) [16, 19, 20]. DGCR8 acts by recognizing the junction between single-stranded and duplex RNA at the base of the pri-miRNA hairpin thereby anchoring the Microprocessor to the bottom of the hairpin structure [21]. Such anchoring of the complex by DGCR8 positions Drosha for cleavage of the pri-miRNA by placing the Drosha active site about 11 base-pairs (one dsRNA helical turn) from the hairpin base. Drosha makes a double-stranded cleavage, generating a pre-miRNA hairpin with the 3′ 2-nucleotide overhang and 5′ terminal phosphate characteristic of RNase III products [9].
Recombinant Drosha in complex with DGCR8 is sufficient to recognize and properly cleave synthetic pri-miRNA substrates in vitro[15]. Therefore, the Drosha-DGCR8 complex is thought to form the functional core of the Microprocessor. However, the endogenous Microprocessor, when isolated from human or Drosophila cells, contains multiple additional protein subunits [15, 22, 23]. Precise roles for these additional factors in the Microprocessor function have not yet been established. However recent biochemical studies have shown that the DGCR8-associated RNA helicases, p68 (also called DDX5) and p72 (also called DDX17), are required for the recognition and processing of some of pri-miRNAs in vivo[23]. Steady state levels of a subset of pre-miRNAs (94 out of 266 examined) were diminished in p72−/− mouse embryos [23]. Intriguingly, p72 point mutants lacking only putative ATPase activity do not rescue pri-miRNA processing, suggesting that ATP-dependant alteration of RNA structure or removal of RNA binding proteins may be an important feature in the recognition and cleavage of many pri-miRNAs by the Microprocessor.
Nuclear proteins regulate activity of the Microprocessor
The Microprocessor carries out the first step in miRNA biogenesis pathway (after transcription) and hence also poses as a potentially powerful step for regulation of miRNA silencing. An emerging theme for the regulation of miRNA biogenesis is the interaction of various RNA-binding proteins with specific pri-miRNA transcripts. For example, two groups recently found that the human protein Lin-28 specifically binds to the terminal loop in let-7 pri-miRNA hairpin and prevents cleavage by the Microprocessor [24, 25]. The binding of site of Lin28 on pri-let7 shares considerable overlap with the binding site of the Microprocessor [26]. In this case, it is therefore likely that inhibition of the Microprocessor is the result of direct competitive binding.
RNA-binding proteins can also influence Microprocessor function by altering pri-miRNA structure. A recent study showed that the ubiquitous RNA-binding protein hnRNP A1 binds to the hairpin of pri-miR-18a and confers greater processing efficiency for the pri-miRNA via facilitating substrate recognition by the Microprocessor [27]. Intriguingly, the requirement for hnRNP A1 is context-dependent. That is, moving the pri-miRNA18a hairpin structure to other positions in the primary transcript obviated the requirement for hnRNP A1 binding. This shows that sequences outside of the hairpin structure can contribute to pri-miRNA recognition and suggests a mechanism in which hnRNP A1 binding stabilizes the hairpin structure relative to other RNA structures that may be formed with regions flanking the miRNA in the primary transcript.
Proteins that directly interact with the Microprocessor can also influence pri-miRNA recognition and processing. It was recently shown in vascular smooth muscle cells that activation of TGF-β and BMP signalling pathways recruits SMAD signal transducers to the Microprocessor through interactions with its p68 subunit [28]. Association with SMAD proteins in turn stimulates Micro processor activity on pri-miR-21, which leads to induction of a contractile phenotype in vascular smooth muscle cells. Intriguingly, activation of the Microprocessor by TGF-β and BMP was shown to be specific for only a subset of pri-miRNAs. Understanding the mechanism by which SMAD proteins activate the Microprocessor towards a handful of specific pri-miRNA targets is an important challenge for the future.
The above examples show that the Microprocessor's selectivity for specific pri-miRNA substrates can depend on factors beyond Drosha-DGCR8 alone. Although Drosha-DGCR8 is sufficient to process simple synthetic pri-miRNAs in vitro, pri-miRNAs generated in vivo are apt to have greater structural features allowing an additional layer of selectivity imposed by other RNA-binding factors in the cell. Moreover, recent studies have found that pri-miRNA transcripts are processed co-transcriptionally [29], suggesting that kinetics of RNA folding may also be a factor in pri-miRNA recognition. Morlando and co workers also found that in the case where the target lies within the intron of a gene, Drosha-mediated cleavage occurs prior to splicing, and that this promotes intron degradation by exonucleases [29]. This finding suggests that in some cases miRNA generation and mRNA maturation could be tightly coupled processes. Like splicing and alternative splicing of mRNAs, recognition of pri-miRNAs by the Microprocessor is likely to be highly dependent on the notoriously dynamic property of RNA folding. It is therefore reasonable to predict that pri-miRNA processing is integrated with nuclear RNA metabolism in general and that a host of RNA-interacting factors that regulate biogenesis of specific miRNAs remain to be discovered.
Intronic pre-miRNAs can bypass the Microprocessor
Originally identified in Drosophila and C. elegans, there is also a subset of miRNAs that originate from short intronic hairpins termed ‘mirtrons’[30, 31]. These miRNA precursors draw distinction from the conventional pri-miRNA in that they bypass the need for cleavage by Drosha. Instead, mirtrons are generated through action of the splicing machinery and lariat-debranching enzyme. The mirtrons rejoin the canonical miRNA biosynthesis pathway preceding cytoplasmic export.
The independence of mirtrons from Drosha/DGCR8 processing provides the possibility of having pre-miRNA originating from different pathways. Recently, it was found that both plants and mammals generate mirtrons as well [32]. The presence of mirtrons could be a strategy of evolution in diversifying miRNA-based gene silencing.
The Exportin-5 Complex transports pre-miRNA to the cytoplasm
Upon being generated by the Microprocessor or through the mirtron pathway, pre-miRNA hairpins are exported to the cytoplasm of the cell [8]. The nuclear export process is mediated by a ∼230 kD protein complex containing the nucleoplasmic transport factor Exportin-5 (Exp5) [33–37]. Exp5 is a member of the karyo-pherin family of transport proteins that interact directly with the small GTPase Ran (RAs-related Nuclear protein). Exp5 recognizes the characteristic 3′-overhang terminal end of the pre-miRNA and part of its duplex structure [38, 39]. Pre-miRNA binding to Exp5-Ran requires Ran to be in the GTP bound state (RanGTP). The pre-miRNA bound complex then moves to the cytoplasm through the nuclear pore complex. In the cytoplasm, the Exp5 complex interacts with Ran GTPase activating protein (RanGAP), which stimulates the GTPase activity of Ran [40]. GTP hydrolysis then induces Exp5 to release its cargo into the cytoplasmic milieu.
Exp5 has also been shown to interact with the protein inter-leukin enhancer-binding factor (ILF3, also called NF90 and NFAR1) [35]. Intriguingly, ILF3 also interacts with the Microprocessor protein DGCR8 [22]. Because ILF3 binds to both DGCR8 and Exp5, it has been proposed that ILF3 functions as a shuttling factor that delivers pri-miRNAs formed by the Microprocessor to Exp5 for export out of the nucleus. Important biochemical studies for the future include determining the effect of ILF3 binding on Microprocessor activity, particularly with respect to how ILF3 influences release of the pre-miRNA product.
The RISC-loading complex completes miRNA maturation
In the cytoplasm, pre-miRNAs are processed into mature single-stranded miRNAs by a ∼500 kD protein assembly called the RISC-loading complex (RLC) [41–43]. The RLC cleaves the pre-miRNA to remove its hairpin loop, resulting in a miRNA duplex (also called miRNA:miRNA* duplex, where miRNA* denotes the passenger RNA strand that will be discarded in the final step of miRNA maturation, see below). After the duplex is formed, one of the RNA strands is selected and loaded into a member of the Argonaute (Ago) family of proteins. Once loaded with a single-stranded miRNA Ago proteins form the core subunit of the RNA-induced silencing complex (RISC, also called miRNP when loaded with a miRNA), where all mature and functional miRNAs are thought to reside [44–46].
The RISC-loading process has been best studied in Drosophila embryo extracts. Flies contain two distinct RLCs: RLC-1 contains the proteins Dicer-1 and Loquacious (also called R3D1) [47]; RLC-2 is composed of the proteins Dicer-2 and R2D2 [42, 48]. The function of the Dicer subunit in the RLC is to process dsRNA into small RNA duplexes of uniform length, typically about 20 base pairs [49]. This is accomplished by making a double-stranded cleavage roughly 20 nucleotides from the free open end of the pre-miRNA hairpin or long dsRNA [50, 51]. Like Drosha, Dicer is a member of the RNase III family of enzymes and thus also generates RNA products with 3′ 2-nucleotide overhangs and 5′ terminal phosphates [49, 52]. In Drosophila, Dicer-1 is responsible for miRNA production and is an essential protein that is required for fly development [53]. Dicer-2, on the other hand, is not required for development but is involved in the production of small interfering RNAs (siRNA), which are generated from long dsRNA fragments and mediate silencing of retrotransposons and RNA viruses [54, 55].
After cleavage by Dicer, the small RNA duplex is thought to dissociate from the RLC and then rebind [56]. The release and rebinding of mi- or siRNA duplexes to the RLC has two functions in flies. First, rebinding allows small RNA sorting so that duplexes generated by RLC-1 can be transferred to RLC-2 and vice versa[57, 58]. Second, rebinding allows the duplex to orient itself on the RLC in such a way that the correct ‘guide’ strand of RNA is loaded into Ago.
Selection of the guide strand is one of the final steps in miRNA biogenesis. The rule for choosing which RNA strand in the miRNA duplex is to be retained in Ago and which is to be discarded as the ‘passenger’ is well established: the RNA strand with its 5′ end on the less thermodynamically stable end of the duplex is designated as the guide strand [59, 60]. The mechanism by which thermodynamic asymmetry is measured is best understood for siRNAs in the Drosophila RLC-2. In this complex, the protein R2D2, which contains two double-stranded RNA binding domains (dsRBD), selectively binds to the end of the siRNA duplex that possesses the greatest double-stranded character [42]. This binding orients the siRNA duplex on the RLC for the next step in the process, which is the recruitment and passing of the duplex to the protein Argonaute-2 (Ago2). Once the duplex has been passed to Ago2 one siRNA strand (the passenger) is cleaved and discarded, whereas the other strand (the guide) is retained as a mature siRNA and used in subsequent gene silencing. The orientation of the duplex as it is passed from Dicer-2/R2D2 to Ago2 determines which strand will be retained. MiRNAs are thought to be loaded into Ago1 through RLC-1 using a similar mechanism, except that the passenger strands in miRNA duplexes are not cleaved, but instead removed by an uncharacterized bypass mechanism [61].
In human cells, Dicer is associated with Ago2 prior to miRNA duplex binding [41, 43]. The human RLC is believed to be composed of the proteins Dicer, Ago2 and TRBP. TRBP contains two functional dsRBDs similar to Drosophila R2D2 and, although it has not yet been demonstrated, likely plays a similar role in guide strand selection. Human Dicer also interacts with an RNA-binding protein called PACT, which is a paralog of TRBP [62]. The precise mechanistic function of PACT in pre-miRNA processing has not yet been determined. However, recombinant human RLC assembled in vitro can process pre-miRNA substrates and load them into Ago2 in the absence of PACT [63].
There are some distinct biochemical differences the RLCs found Drosophila and in mammalian systems. Although Drosophila has two Dicers that exist in distinct molecular complexes, the human genome encodes only a single Dicer gene. The human Dicer is likely the ortholog of Drosophila Dicer-1 because both process pre-miRNA substrates and neither require ATP hydrolysis for catalysis [64–69]. However, in contrast to the Drosophila system, human Ago2 has been shown to associate with the RLC even prior to pre-miRNA binding [41, 43]. Furthermore, after human Ago2 is loaded with the guide miRNA, it dissociates from the RLC [41, 63]. In contrast, Drosophila Ago2, Dicer-2 and R2D2 stay associated with each other and recruit additional protein components to form an 80S super assembly termed ‘Holo-RISC’[70].
Mature miRNAs silence target genes from within RISC
Argonaute bound to a single-stranded guide miRNA forms the core subunit RISC (also called miRISC and miRNP), the effector complex of miRNA-mediated gene silencing [45, 46]. A ‘minimal RISC’, which is sufficient to recognize and cleave a target RNA, can be generated in vitro by simply incubating recombinant human Ago2 with a single-stranded guide RNA [71]. However, in vivo RISC is a much larger and possibly dynamic molecular machine. Over 50 proteins have been identified in association with Ago1/Ago2 [72] and reported sizes of RISC range from 100 kD [73] to over 2.5 MD [70, 74].
RISC uses the mature miRNA bound to Ago to guide the silencing of genes bearing full or partial sequence complementarity to the miRNA. In mammals, most miRNA-mediated gene silencing is thought to function through translational repression of targeted message RNAs. However, cases of Ago2-catalysed target cleavage have been reported as well [75]. The mechanism of translation repression by miRNAs is the subject of many reviews and to date remains somewhat controversial [76–82].
Discussion and future directions
The coordinated action of RNA polymerase II, the Microprocessor, Exp5 and the RLC results in the step-wise generation of mature miRNAs in human beings. Although the basic pathways for generating miRNAs are now understood, many mechanistic questions remain. For example, there are four Ago proteins in human cells and thus four possible destinations for any given miRNA. Is there a miRNA sorting mechanism in mammals and if so are all Ago proteins loaded by the same RLC? Recent results have shown that human Ago2 is post-translationally modified [83, 84]. Does modification alter the loading mechanism or contribute to miRNA sorting? Recent results show that cytoplasmic Lin-28 can bind pre-miRNAs and inhibit cleavage by Dicer [85]. Are there other regulators of RISC-loading remaining to be discovered? Is there a cytoplasmic counterpart to ILF3 that shuttles pre-miRNAs from Exp5 to the RLC? And finally, how are miRNAs turned over in the cell? Recent experiments in plants have identified a family of miRNA degrading exonucleases termed Small RNA degrading nuclease (SDN), which belong to the 3′-5′ Exoc family of nucle-ases [86]. These RNases were shown to degrade single-stranded mature miRNAs down to 7–8 nucleotide fragments in vitro. However, if mature miRNAs are tightly bound to Ago proteins, it is not clear how the SDNs will access their substrates in vivo. Clearly, many exciting discoveries in the field of miRNA biology are on the near horizon.
Acknowledgments
We are grateful to Ashley J. Pratt for stimulating conversations and critical reading of the manuscript. P.W.L. is a pre-doctoral fellow of the American Heart Association. I.J.M. is a Pew Scholar in the Biomedical Sciences.
References
- 1.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–6. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 4.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engström PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C. RIKEN Genome Exploration Research Group; Genome Science Group (Genome Network Project Core Group); FANTOM Consortium. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–6. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 5.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–66. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 8.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–70. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 10.Basyuk E, Suavet F, Doglio A, Bordonne R, Bertrand E. Human let-7 stem-loop precursors harbor features of RNase III cleavage products. Nucleic Acids Res. 2003;31:6593–7. doi: 10.1093/nar/gkg855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci USA. 2007;104:17719–24. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–9. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–83. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–10. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–40. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 16.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–5. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 17.Wu H, Xu H, Miraglia LJ, Crooke ST. Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J Biol Chem. 2000;275:36957–65. doi: 10.1074/jbc.M005494200. [DOI] [PubMed] [Google Scholar]
- 18.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–48. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–7. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Han J, Lee γ, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 22.Shiohama A, Sasaki T, Noda S, Minoshima S, Shimizu N. Nucleolar localization of DGCR8 and identification of eleven DGCR8-associated proteins. Exp Cell Res. 2007;313:4196–207. doi: 10.1016/j.yexcr.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 23.Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, Mihara M, Naitou M, Endoh H, Nakamura T, Akimoto C, Yamamoto Y, Katagiri T, Foulds C, Takezawa S, Kitagawa H, Takeyama K, O'Malley BW, Kato S. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol. 2007;9:604–11. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 24.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–49. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, Sliz P, Gregory RI. Determinants of microRNA processing inhibition by the developmen-tally regulated RNA-binding protein Lin28. J Biol Chem. 2008;283:21310–4. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 27.Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–6. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 28.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, Proudfoot NJ. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol. 2008 doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–6. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–36. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim VN. MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends Cell Biol. 2004;14:156–9. doi: 10.1016/j.tcb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 34.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 35.Brownawell AM, Macara IG. Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins. J Cell Biol. 2002;156:53–64. doi: 10.1083/jcb.200110082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–91. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeng Y, Cullen BR. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32:4776–85. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lund E, Dahlberg JE. Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs. Cold Spring Harb Symp Quant Biol. 2006;71:59–66. doi: 10.1101/sqb.2006.71.050. [DOI] [PubMed] [Google Scholar]
- 40.Coutavas E, Ren M, Oppenheim JD, D'Eustachio P, Rush MG. Characterization of proteins that interact with the cell-cycle regulatory protein Ran/TC4. Nature. 1993;366:585–7. doi: 10.1038/366585a0. [DOI] [PubMed] [Google Scholar]
- 41.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–90. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–80. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 43.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscrip-tional gene silencing. Cell. 2005;123:631–40. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 44.Gu SG, Pak J, Barberan-Soler S, Ali M, Fire A, Zahler AM. Distinct ribonucleoprotein reservoirs for microRNA and siRNA populations in C. elegans. RNA. 2007;13:1492–504. doi: 10.1261/rna.581907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–41. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 46.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–50. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 47.Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 2005;19:1674–9. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–5. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 49.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonucle-ase in the initiation step of RNA interference. Nature. 2001;409:363–6. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 50.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 51.Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–8. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 52.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 54.Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–7. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- 55.Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, Hannon GJ, Brennecke J. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Preall JB, He Z, Gorra JM, Sontheimer EJ. Short interfering RNA strand selection is independent of dsRNA processing polarity during RNAi in Drosophila. Curr Biol. 2006;16:530–5. doi: 10.1016/j.cub.2006.01.061. [DOI] [PubMed] [Google Scholar]
- 57.Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130:287–97. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 60.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–16. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 61.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–20. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 62.Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–32. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci USA. 2008;105:512–7. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu X, Park JK, Jiang F, Liu Y, McKearin D, Liu Q. Dicer-1, but not Loquacious, is critical for assembly of miRNA-induced silencing complexes. RNA. 2007;13:2324–9. doi: 10.1261/rna.723707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ye X, Paroo Z, Liu Q. Functional anatomy of the Drosophila microRNA-generating enzyme. J Biol Chem. 2007;282:28373–8. doi: 10.1074/jbc.M705208200. [DOI] [PubMed] [Google Scholar]
- 66.Förstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–8. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 69.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 70.Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell. 2004;117:83–94. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- 71.Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombi-nant human RISC. Nat Struct Mol Biol. 2005;12:340–9. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 72.Höck J, Weinmann L, Ender C, Rüdel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–60. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–74. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 74.Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–8. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- 75.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–6. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 76.Xie W, Ted Brown W, Denman RB. Translational regulation by non-protein-coding RNAs: different targets, common themes. Biochem Biophys Res Commun. 2008;373:462–6. doi: 10.1016/j.bbrc.2008.06.075. [DOI] [PubMed] [Google Scholar]
- 77.Liu J. Control of protein synthesis and mRNA degradation by microRNAs. Curr Opin Cell Biol. 2008;20:214–21. doi: 10.1016/j.ceb.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 78.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 79.Richter JD. Think you know how miRNAs work? Think again. Nat Struct Mol Biol. 2008;15:334–6. doi: 10.1038/nsmb0408-334. [DOI] [PubMed] [Google Scholar]
- 80.Behm-Ansmant I, Rehwinkel J, Izaurralde E. MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay. Cold Spring Harb Symp Quant Biol. 2006;71:523–30. doi: 10.1101/sqb.2006.71.013. [DOI] [PubMed] [Google Scholar]
- 81.Kozak M. Faulty old ideas about translational regulation paved the way for current confusion about how microRNAs function. Gene. 2008;423:108–15. doi: 10.1016/j.gene.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 82.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–26. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 83.Qi HH, Ongusaha PP, Myllyharju J, Cheng D, Pakkanen O, Shi Y, Lee SW, Peng J, Shi Y. Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature. 2008;455:421–4. doi: 10.1038/nature07186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zeng Y, Sankala H, Zhang X, Graves PR. Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem J. 2008;413:429–36. doi: 10.1042/BJ20080599. [DOI] [PubMed] [Google Scholar]
- 85.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–93. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 86.Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321:1490–2. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]


