Abstract
Glutamate transporter associated protein 3–18 (GTRAP3-18) is an endoplasmic reticulum (ER)-localized protein belonging to the prenylated rab-acceptor-family interacting with small Rab GTPases, which regulate intracellular trafficking events. Its impact on secretory trafficking has not been investigated. We report here that GTRAP3-18 has an inhibitory effect on Rab1, which is involved in ER-to-Golg trafficking. The effects on the early secretory pathway in HEK293 cells were: reduction of the rate of ER-to-Golgi transport of the vesicular stomatitis virus glycoprotein (VSVG), slowed accumulation of a Golgi marker plasmid in pre-Golgi structures after Brefeldin A treatment and inhibition of cargo concentration of the neuronal glutamate transporter excitatory amino-acid carrier 1 (EAAC1) into transpor complexes in HEK293 cells, an effect that could be completely reversed in the presence of an excess of Rab1. In accordance with the known role of Rab1 in neurite formation, overexpression of GTRAP3-18 significantly inhibited the length of outgrowing neurites in differentiated CAD cells. The inhibitory effect of GTRAP3-18 on neurite growth was rescued by co-expression with Rab1, supporting the conclusion that GTRAP 3-18 acted by inhibiting Rab1 action. Finally, we hypothesized that expression of GTRAP3-18 in the brain shoul be lower at stages of active synaptogenesis compared to early developmental stages. This was the case as expression of GTRAP3-18 declined from E17 to P0 and adult rat brains. Thus, we propose a model where protein trafficking and neuronal differentiation are directly linked by the interaction of Rab1 and its regulator GTRAP3-18.
Keywords: PRA1 family proteins, Rab GTPases, ER-Golgi trafficking, neuronal differentiation
Introduction
Newly translated proteins are transported from the endoplasmic reticulum (ER) to the Golgi via the intermediate compartment (IC) in either vesicles [1] or larger transport carriers [2, 3] depending on the travelling cargo. The coat protomer II complex (COPII) mediates cargo selection and concentration into so-called ER-exit sites (ERES) [1]. After fission of COPII vesicles from ERES, they homotypically fuse to generate vesicular-tubular clusters (VTC), which are alternatively termed the ER-Golgi intermediate compartment (ERGIC) [4]. Homotypic fusion of COPII vesicles is dependent on the tethering complex transport protein particle I (TRAPPI) [5, 6]. The latter serves as a guanine nucleotide exchange factor for the small GTPase Rab1. Therefore, VTC formation is dependent on Rab1. It is still heavily debated where Rab1 is activated, although there is evidence that it is already activated on ERES. This is supported by the fact that TRAPPI was shown to be located at ERES and not in VTC/ERGIC [5]. In addition, Golgi-specific Brefeldin A resistance factor 1 (GBF1), a newly identified Rab1 effector, is found on ERES after co-expression of constitutively active Rab1 [7]. Finally, in cells overexpressing a dominant negative Rab1 mutant, cargo, for example the temperature-sensitive ts045 mutant of VSVG, is not able to be concentrated into ERES [8, 9].
Interestingly, upon induction of differentiation of pheochromocytoma (PC12) cells, the IC expands and segregates into globular structures, which are involved in ER to Golgi transport and remain in the cell body, and tubular structures that are enriched in Rab1 and move directly to the outgrowing neurites [10]. These tubular domains are accompanied by ERES and are thought to play a role in membrane (and most likely also protein) delivery to the newly forming neurites. Despite this, it remains unclear what the role of Rab1 is in neuronal differentiation. This uncertainty stems from the fact that the precise role of Rab1 in the early secretory pathway remains not fully understood.
In the current study, we focused on the prenylated Rab acceptor 1 (PRA1) family member GTRAP3-18 (also called PRA2). GTRAP3-18 was initially identified as an interaction partner for the glutamate transporter EAAC1 [11]. Contradicting reports were published on its subcellular localization [11–13] and the question on its cellular function still begs an answer. We found that GTRAP3-18 is a protein that resides in the ER, binds Rab1 and thus inhibits its actions in the early secretory pathway. In addition, we show that GTRAP3-18, most likely due to its Rab1 inhibitory action, inhibits neurite growth in the neuronal-like CAD cells. Finally, we report that expression of GTRAP3-18 is higher in the embryonal rat brain compared with new born or adult rat.
Materials and methods
Reagents, plasmids and antibodies
Endoglycosidase H (EndoH) and Complete protease inhibitor cocktail were purchased from Roche Applied Biosciences (Indianapolis, IN, USA). Brefeldin A (BFA) was from Sigma Aldrich (Saint Louis, MO, USA). GTRAP siRNAs (SI00903602, SI00903609) and All Stars Negative siRNA (1027280) were from Qiagen (Valencia, CA, USA).
The yellow fluorescent protein (YFP)-Golgi and cyan fluorescent protein (CFP)-ER subcellular localization plasmids were from Clontech (Mountain View, CA, USA). EAAC1 from rat GFP-EAAC1 (Dr. M. B. Robinson; University of Pennsylvania, Philadelphia, PA), GTRAP3-18 from rat HA-GTRAP3-18 (Dr. J. D. Rothstein, Johns Hopkins University School of Medicine, Baltimore, MD, USA) and Rab1a from human Rab1a (Dr. W.E. Balch; The Scripps Research Institute, La Jolla, CA, USA) were cloned into pECFP/pEYFP-C1 (Clontech). The last 47 amino acids of GTRAP3-18 were cloned into pGEX-5x-1 vector (Amersham-Pharmacia, Buckinghamshire, UK) to produce GST-cGTRAP. CFP-Rab1a-N124I was generated using the QuickChange®II XL Site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). The plasmids encoding YFP-Sec24D, Sar1a, Sar1a-T39N, GST-cGAT1 [14] and CFP-GAT1d37 [15] have been described elsewhere.
Rabbit polyclonal antibodies used were anti-GFP (Clontech), anti-Rab1 and anti-HA (both Santa Cruz Biotechnology, Santa Cruz, CA, USA). Anti-HA Agarose was from Sigma Aldrich.
Cell culture and transfection
HEK293 and CAD cells were cultured as described before [15, 16]. All cell culture reagents were from PAA Laboratories (Pasching, Upper Austria) except fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA, USA). If not otherwise indicated, all transfections were performed 24 hrs after seeding either using the CaPO4 precipitation method for co-localization and BFA assays or Lipofectamine®-Plus (Invitrogen) for all other experiments. For microscopy cells were seeded on 15 mm or 24 mm poly-D-lysine (PDL)-coated coverslips (Marienfeld, Lauda-Königshofen) in 12 wells or 6 wells, respectively.
CAD differentiation and RNA interference experiments
CAD cells were transfected with pEYFP-C1 alone (as a control) or together with CFP-tagged GAT1d37, CFP- or HA-tagged GTRAP3-18 or CFP-tagged Rab1-wild type. Eight hours after transfection, the medium was changed to either complete medium (to keep cells non-differentiated) or medium lacking FBS (to induced differentiation) for 20 hrs before imaging. For RNAi treatment before differentiation, reverse transfection of a final concentration of 100 nM siRNA was done with Lipofectamine 2000 (Invitrogen), followed by pEYFP-C1 transfection after 48 hrs in the presence of FBS using Fugene 6 (Roche Applied Biosciences) and starvation for 20 hrs before imaging. Cells displaying outgrowing neurites were scored (300–500 cells per sample for CAD starvation experiments and 200–300 cells for siRNA experiments) and neurite length was measured.
Fluorescence microscopy
Epifluorescence imaging was performed on a Zeiss Axiovert 200M inverted microscope equipped with a CoolSNAP fx cooled CCD camera (Photometrics, Roper Scientific, Tucson, AZ, USA) and fluorescence filters (Chroma Technology Corp., Brattleboro, VT, USA). Images were acquired and analysed with the MetaMorph of the MetaSeries software package (Universal Imaging Corp., Downingtown, PA, USA). Confocal imaging was done on a Zeiss LSM510 confocal laser scanning microscope and analysed with the Zeiss LSM Image Browser software (Zeiss AG, Oberkochen).
Protein purification, GST-pulldown and co-immunoprecipitation
Protein purification of the GST-tagged GTRAP3-18 C-terminus and GST-pulldown were done as described before [15]. For co-immunoprecipitation cells were transfected with HA-GTRAP3-18 and lysed on ice in radio immuno precipitation assay (RIPA) buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid [EDTA]) with 1% NP40 and freshly added phenylmethanesulfonylfluoride (PMSF) and protease inhibitors. After rotation of lysates on 4°C for 1 hr and centrifugation at 13,000 rpm at 4°C for 10 min., the supernatant was collected and 3.5 mg of protein were incubated with 50 μl of pre-washed anti-HA-agarose (Sigma Aldrich) at 4°C overnight. After several washing steps complexes were eluted with sample buffer (2% SDS, 100 mM β-mercaptoethanol) and subjected to SDS-PAGE.
ER-to-Golgi transport assay
Cells were transfected with VSVG-ts045-YFP together with pECFP-C1 or CFP-GTRAP3-18 and kept at 40°C overnight, followed by incubation at 32°C for indicated time-points and harvesting. Microsomes were isolated in Hepes/Sorbitol buffer (0.375 M Sorbitol, 20 mM Hepes-KOH pH 7.2) with protease inhibitors and digested with EndoH at 37°C overnight. Protein fractions of 10 μg were loaded on a SDS-polyacrylamide gel. Band intensities were quantified with ScionImage™ 4.0.2 (Scion Corporation, Frederick, MD, USA) and the Golgi-accumulation over time was calculated as the ratio of the signal in the upper Golgi band normalized to the total signal per time point. Subsequently, each time point was normalized to the amount of protein in the Golgi at the time-point of 0 min., which was set to 1. For each time-point we measured six paired determinants (n= 6). The paired determinants of each group were pooled and tested in a two-tailed, paired t-test.
In-vitro concentration assay
Cells on glass coverslips were transfected with either CFP-EAAC1 and dominant-negative Sar1-T39N alone or together with YFP-GTRAP3-18. Twenty-four hours later, cells were rendered semi-intact and incubated with an ER budding reaction originally adapted by Xu and Hay [4]. The reaction mixture consisted of 25 μl 25/90 buffer (90 mM KOAc, 25 mM Hepes-KOH pH 7.2), 60 μl cytosol (3–4 μg/μl protein concentration), 20 μ,l assay buffer (18 mM CaOAc, 50 mM EGTA, 20 mM Hepes-KOH pH 7.2), 5 μl 0.11 M MgOAc, 22 μl H2O, 58 μl 25/125 buffer (125 mM KOAc, 25 mM Hepes-KOH pH 7.2) and 10 μl ATP-regeneration system (5 mM creatine phosphate, 4 ng/ml creatine kinase, 1 mM adenosine-5′-triphosphate (ATP), 0.5 mM GTP). Cytosol preparation and preparation of semi-intact cells were described previously [14]. Images were taken right after incubation with ice-cold budding reaction and after 15 minutes on 37° and cells with and without transport complexes (TC) were scored.
Brefeldin A Golgi-recovery assay
Cells on glass coverslips were transfected with either YFP-Golgi alone or together with CFP-GTRAP3-18. Cells were then treated with 5 μm BFA for 30 min. at 37°C, following thorough BFA washout. Images were acquired at indicated time-points after washout and cells with accumulation of the marker plasmid in pre-Golgi structures were counted.
Animal experiments, RNA extraction and semi-quantitative RT-PCR
Sprague-Dawley rats were from the Institut für Labortierkunde und Genetik (Himberg, Lower Austria). Timed pregnant rats (day E17) were sacrificed and embryonic brains were removed as described before [17]. This was also done with newborn and adult rat brains. RNA extraction of total brains and of CAD cells after RNAi-mediated knockdown were done as described previously [14]. 200 ng of total RNA were used for a duplex GTRAP3-18 and GAPDH RT-PCR with the Superscript® III Platinum One-Step RT-PCR kit (Invitrogen). Band intensities were quantified with ImageJ (U. S. National Institutes of Health, Bethesda, MD, USA). GTRAP3-18 expression levels were normalized to GAPDH levels and the resulting ratio was plotted as arbitrary units (A.U.).
Results
The resident ER protein GTRAP3-18 does not get exported from the ER
Because of the contradicting reports on the subcellular localization of GTRAP3-18 [11–13], we started by determining it ourselves. When we expressed YFP-tagged GTRAP3-18 in HEK293 cells it co-localized extensively with the ER marker construct CFP-ER (Fig. 1A). Others have shown that the carboxyl-terminus of PRA1 family proteins is responsible for their subcellular localization [12]. They postulated that PRA2/GTRAP3-18 leaves the ER and is subsequently retrieved from the ERGIC/VTC by means of its last four amino-acids – KARE, which mimics the di-basic ER-retrieval motif-KKXX. In our opinion -KARE should not function as a retrieval motif. To prove this we deleted the terminal 4 amino acids of GTRAP3-18 (GTRAPd4). We stress that the sequence KARE does not harbour any known ER export motif. If GTRAP3-18 is retrieved by means of the KARE motif, its deletion should release the protein to post-ER compartments. This was not the case. GTRAPd4 showed the same distribution as wild-type GTRAP3-18 and was located in the ER (Fig. 1B). It may be argued here that GTRAPd4 is misfolded. Therefore, we wanted to reach a higher level of confidence. We incubated HEK293 cells expressing YFP-GTRAP3-18 at 15°C for 2 hrs. This treatment blocks protein secretion at the level of the ERGIC/VTC [18]. No change in the subcellular distribution of GTRAP3-18 could be detected (Fig. 1C). Finally, if GTRAP3-18 escapes the ER, it ought to have an ER export motif that mediates an interaction with Sec24, the cargo receptor of the COPII coat. We incubated the GST-tagged C-terminus of GTRAP3-18 with cytosol prepared from HEK293 cells overexpressing YFP-tagged Sec24D and performed a GST-pulldown. As a positive control we used the GST-tagged C-terminus of the GABA transporter 1 (GAT1), which we previously found to contain an ER export motif that interacts with COPII [14]. We could not demonstrate an interaction of GTRAP3-18 with Sec24D (Fig. 1D). Taken together, we demonstrated unequivocally that GTRAP3-18 is an ER resident protein that does not leave the ER.
1.
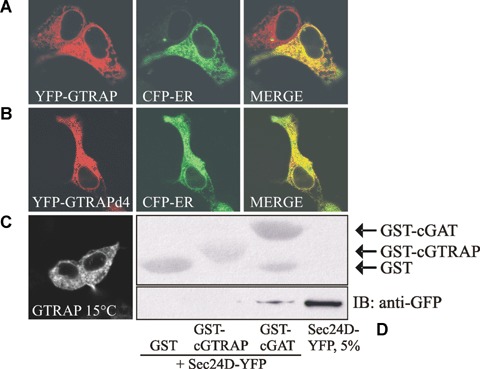
GTRAP3-18 is a resident endoplasmic reticulum (ER) protein and does not interact with Sec24D. (A, B) HEK293 were transfected with plasmids encoding either YFP-tagged wild-type GTRAP3-18 or a mutant with a deletion of the last four amino acids (GTRAPd4) together with a plasmid encoding CFP-ER. Images were acquired 24 hrs after transfection and co-localized using the Zeiss LSM Image Browser software. (C) HEK293 cells expressing YFP-GTRAP3-18 were kept at 15°C for 2 hrs prior to image acquisition. (D) Cytosol (100 μg) prepared from cells overexpressing YFP-Sec24D was co-incubated either with GST, GST-tagged GTRAP3-18 C-terminus or GST-tagged GAT1 C-terminus. GST-pulldown was performed as indicated in Materials and methods. The top panel represents the Ponceau-stained membrane to visualize the loading of the various GST-constructs.
GTRAP3-18 slows ER to Golgi transport of VSVG-ts045
Co-expression with GTRAP3-18 reduces the plasma membrane expression of several membrane proteins [19]. To clarify the influence of GTRAP3-18 on trafficking in the early secretory pathway we employed an ER-to-Golgi transport assay. We used the YFP-tagged version of a temperature sensitive mutant of VSVG (VSVG-ts045) as cargo protein. This protein is misfolded at 40°C and retained in the ER. After a temperature shift to 32°C, VSVG-ts045 rapidly folds and enters the secretory pathway. Its presence in the ER or the Golgi can easily be assessed by testing for its sensitivity to EndoH. When the protein reaches the Golgi it becomes resistant to Endo H. We transfected HEK293 cells with a plasmidencoding YFP-tagged VSVG-ts045 and either a plasmid encoding for CFP or a plasmid encoding for CFP-tagged GTRAP3-18. Cells were maintained over night at 40°C to synchronize VSVG-ts045 in the ER. As shown in Fig. 2A, co-expression of CFP-GTRAP3-18 noticeably slowed the transport kinetics. A densitometric quantification of six independent experiments revealed a statistically significant difference between the pooled paired determinants of the control and the GTRAP3-18 group (Fig. 2B, t= 4.057; P< 0.001 in a two-tailed, paired t-test).
2.
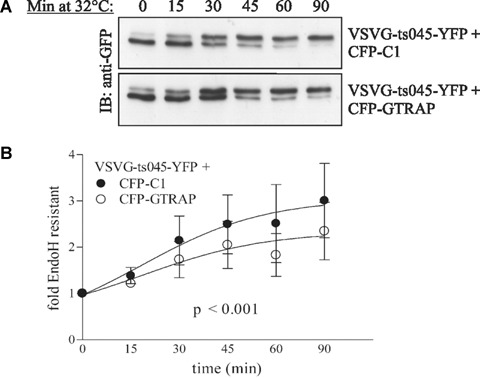
GTRAP3-18 slows ER to Golgi transport of VSVG-ts045. (A) A representative example of a single Western blot of VSVG-ts045-YFP and CFP (as a control) or CFP-GTRAP3-18 after synchronization at 40°C overnight, incubation at 32°C for indicated time-points and Endoglycosidase H digest. Lower gel bands represent EndoH sensitive ER-localized fractions and higher bands represent EndoH resistant Golgi-localized fractions. (B) Band intensities of six independent experiments were quantified and the Golgi-accumulation over time was calculated as explained in Materials and methods. Statistical significance was obtained in a paired, two-tailed t-test. Symbols show mean ± S.E.M.
GTRAP3-18 binds Rab1a and reduces cargo concentration at transport complexes
GTRAP3-18 has been shown to bind to recombinant Rab1 [12]. To show that this interaction occurs in vivo, we expressed HA-tagged GTRAP3-18 in HEK293 cells and performed a co-immunoprecipitation experiment with endogenous Rab1. As shown in Fig. 3A, endogenous Rab1 could only be co-immunoprecipitated by an anti-HA antibody when HA-GTRAP3-18 was expressed. Therefore we conclude that this interaction is of relevance in living cells.
3.
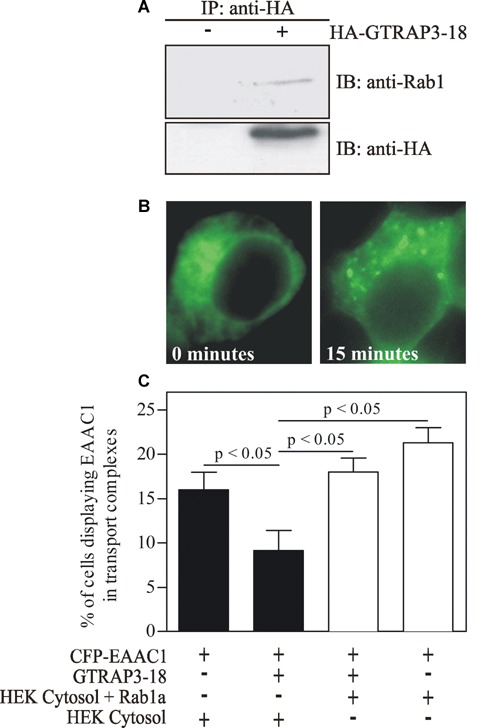
GTRAP3-18 binds Rab1a in vivo and thus significantly reduces cargo concentration of the EAAC1 glutamate transporter at transport complexes, an effect that can be completely overcome by an excess of Rab1a. (A) Equal amounts of lysates of untransfected or HA-GTRAP3-18 trans-fected HEK293 cells were immunoprecipitated using HA-agarose. Immune complexes were subjected to SDS-PAGE and co-immunoprecipitated, endogenous Rab1a was visualized with an anti-Rab1 antibody. (C) HEK293 cells on coverslips were transfected with CFP-EAAC1 as cargo, Sar1-T39N to synchronize the cargo in the ER and GTRAP3-18 if indicated. Cells were permeabilized after 24 hrs and a concentration assay was performed introducing fresh cytosol (either untransfected or Rab1a enriched cytosol as indicated) and therefore also wild type Sar1 to start ER export. Directly after adding of the ER budding reaction (0 min.; see a in B displaying diffuse staining of EAAC1) as well as after incubation at 37° for 15 min. (see b in B displaying EAAC1 in concentrated complexes) pictures were taken and cells with diffuse staining and with formed transport complexes (TC) were counted. P-values were obtained with an anova followed by Scheffe post-hoc comparisons test). Bars show mean ± S.E.M.
Transmembrane cargo is concentrated into ERES by the COPII complex [20]. COPII vesicles undergo homotypic fusion to form the ERGIC/VTC [4–6] or they may fuse with the pre-existing ERGIC/VTC [21]. It has been shown that further maturation steps of ERES may exist, which are dependent on Rab1 and its effector GBF1 [7, 22, 23]. Because GTRAP3-18 binds Rab1, we wanted to determine the impact of this interaction on Rab1-controlled trafficking events. First, we wanted to clarify the effect on cargo concentration at/from the ER using a method that we have previously established [14]. This method is based on synchronization of membrane cargo in the ER by co-expression of the dominant negative Sar1-T39N which blocks COPII assembly. Preparation of semi-intact cells enables the introduction of fresh cytosol from non-transfected cells together with an ATP-regeneration system and GTP. After 15 min., punctuated structures can be observed which we term TC (see Fig. 3B for representative images). We have previously shown that their formation is dependent on COPII [14]. We stress that our assay, unlike other's, does not use a GTP-clamped Sar1 mutant, which would inhibit COPII vesicle uncoating and VTC formation. Therefore, TC represent ERES and VTC (which are connected to the ER) as well as ERGIC (which is discontinuous from the ER). As a cargo we used the CFP-tagged glutamate transporter EAAC1. We performed the assay in presence or absence of GTRAP3-18. Co-expression of GTRAP3-18 reduced the number of cells displaying CFP-EAAC1 in TC by nearly 50% (Fig. 3C). We like to pose that this effect is due to Rab1 binding by GTRAP3-18, thereby limiting its accessibility for TC formation. If this is the case, an excess of Rab1 should saturate these ‘binding sites’ and overcome the inhibitory effect. When we performed the assay with cytosol prepared from HEK293 cells overexpressing Rab1, the inhibitory effect of GTRAP3-18 could be completely overcome. This result tempts to the conclusion that GTRAP3-18 blocks Rab1-mediated maturation of ERES and thus cargo concentration.
GTRAP3-18 slows accumulation of a Golgi marker into pre-Golgi intermediates after Golgi disruption by Brefeldin A
The Golgi apparatus is a highly dynamic organelle that can be disrupted by treatment with BFA [24]. Upon BFA washout, the Golgi recovers rapidly and completely [24]. Reformation of the Golgi after BFA treatment was shown to be Rab1-dependent [25]. We, therefore, reasoned that overexpression of GTRAP3-18 ought to inhibit Golgi reformation upon BFA washout. The Golgi was visualized by using YFP-Golgi, which represents the YFP-tagged 84 amino acids of galactosyl-transferase that carry a Golgi targeting motif. Directly after BFA washout, YFP-Golgi showed a reticular ER-like staining (Fig. 4A, panel a) but afterwards accumulated rapidly in pre-Golgi intermediates (Fig. 4A, panel b). After 15 min. at 37°C, about 55% of cells displayed YFP-Golgi in transport intermediates (Fig. 4B). In contrast, only 30% of cells expressing GTRAP3-18 exhibited the same distribution. After 45 min., the difference was even more pronounced and statistically significant (82% without versus 55% with GTRAP3-18 co-transfection).
4.
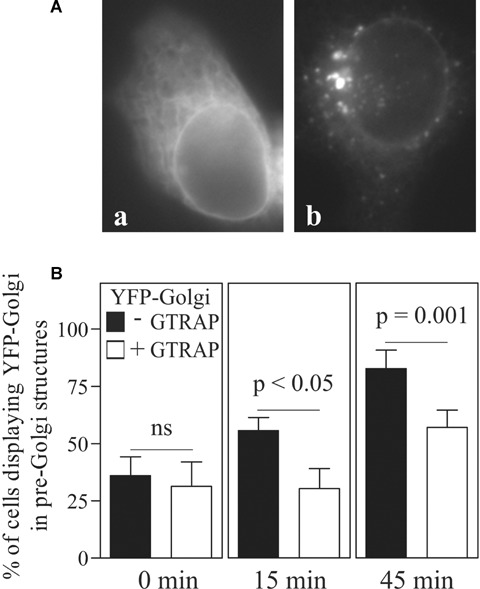
GTRAP3-18 slows recovery of YFP-Golgi into pre-Golgi structures after treatment with Brefeldin A. HEK293 cells were transfected with YFP-Golgi alone or together with CFP-tagged GTRAP3-18. After Brefeldin A washout, cells were kept at 37°C and images were acquired at indicated time-points. Cells with diffuse distribution of the marker plasmid (A, a) and with the marker in pre-Golgi intermediates (A, b) were scored. The graph in (B) represents the percentage of marker in pre-Golgi intermediates at 0 min. (a), 15 min. (b) and 45 min. (c) after Brefeldin A washout. P-values were calculated in a paired t-test. Bars show mean ± S.E.M.
Neurite outgrowth in CAD cells is regulated by opposing effects of GTRAP3-18 and Rab1a
It has been shown recently in PC12 cells that Rab1a is involved in a Golgi-independent transport pathway [10] that might be necessary for delivery of lipids to the surfaces of newly outgrowing neurites.
We wanted to determine whether the negative effect of GTRAP3-18 on Rab1 function has an influence on neurite outgrowth. Therefore, we used CAD cells, a central nervous system (CNS) catecholaminergic cell line. The rationale for using this cell line was that they are easier to maintain than PC12 cells, differentiate by starvation without the need for extrinsic factors and can be transfected with a higher efficiency than PC12 cells. CAD cells express a number of neuron-specific proteins, among them β3-tubulin, GAP-43, SNAP-25 and synaptotagmin [16].
We hypothesized that if Rab1 is important for neurite outgrowth in CAD cells, expression of GTRAP3-18 ought to inhibit this process, without affecting the number of cells that principally undergo differentiation. In order to better visualize CAD cell neurites, we expressed YFP which ‘fills’ the cells and makes the neurites easy to recognize and measure. As shown in Fig. 5B (P-values in black), starvation significantly induced neurite outgrowth. Expression of CFP-GTRAP3-18 significantly reduced the length of outgrowing neurites compared to control conditions (Fig. 5B, P-values in blue). To exclude that filling the ER with a membrane protein alters cellular function and trafficking events, we expressed the CFP-tagged mutant of GAT1 lacking the last 37 amino acids (GAT1d37). We showed previously that this mutant is retained in the ER [15]. Expression of GAT1d37 did not affect neurite growth (Fig. 5A and B). Interestingly, over-expression of wild type Rab1 did not further increase neurite length compared to control cells. Fig. 5A shows the distribution for every single-measured neurite and their means depicted as a red line. To have a closer look on the population of longer neurites, we chose a cut-off value that excluded 95% of each sample based on the undifferentiated control groups. The graph in Fig. 5C illustrates the percentage of neurites per sample that lies above this cut-off value. The differentiated CAD cells containing GTRAP3-18 have the smallest population of longer neurites compared to the other ones. Notably, GTRAP3-18 had no significant influence on the number of cells that displayed outgrowing neurites (data not shown).
5.
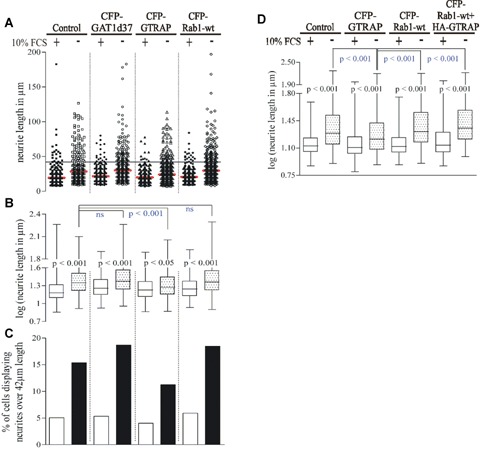
GTRAP3-18 hinders and Rab1 facilitates neurite outgrowth in differentiated CAD cells. CAD cells were transfected with pEYFP-C1 alone (control) or together with indicated plasmids in a 1: 1 ratio. Cells were then either kept in complete medium or in starving medium lacking FBS to induce differentiation for 20 hrs. Images were acquired and neurite length was measured. A Shows a scatter plot of all measured values per sample (red line represents the mean of all values). The black line represents a cut-off value of 42 μm that was chosen because it accounts for 95% of values in the undifferentiated control groups of the same experiment (YFP alone as control and YFP with CFP-GAT1d37). The box plot in (B) visualizes the data from (A) after log-transformation as median with 25% and 75% percentile as box and minimum and maximum as error bars. P-values were produced with an anova followed by Bonferroni's multiple comparison test. Ns indicates not significant. P-values in black show statistical significance between control and differentiated conditions within the same group, P-values in blue show statistical significance between the differentiated control sample and differentiated samples of all other groups. The percentage of cells with neurites over 42 μm length is shown in (C). Data for the box plot in (D) is obtained as in (B) and includes a subset that is triple-transfected with pEYFP-C1: CFP-Rab1-wt: HA-GTRAP in a 1: 3: 2 ratio. Only cells displaying a CFP- and YFP-signal were included.
Our observation that GTRAP3-18 inhibits neurite growth raises the question whether this effect is due to inhibition of Rab1 function. If this is true, neurite growth in GTRAP3-18 expressing cells ought to be rescued by an excess of Rab1, which can be achieved by overexpression of Rab1. Therefore, we transfected CAD cells either with a plasmid encoding for YFP alone or together with CFP-GTRAP3-18 or CFP-Rab1. In one experimental group this transfection mixture further included HA-tagged GTRAP3-18. As shown in Fig. 5D, the inhibitory effect of GTRAP3-18 on neurite growth was reversed by co-expression of Rab1. Taken together, these results unambiguously show that GTRAP3-18 inhibits neurite growth in CAD cells viablocking Rab1 function.
One major concern in this study is that all the results are based on overexpression of GTRAP3-18. We do not know whether GTRAP3-18 operates as a part of a large multi-protein complex. If so, its overexpression would perturb the action of this complex, as has been shown for other multi-protein complexes [26, 27]. In this case lack of GTRAP3-18 ought to have the same effect as the overexpression. If not, its knockdown would either have the opposite effect or it would more likely be without effect because the physiologic level of GTRAP3-18 is very low. To address this concern we tested CAD cell neurite growth in cells where GTRAP3-18 has been knocked down by siRNA. We used two working siRNAs which achieved a high knockdown efficiency after 48 hrs (Fig. 6A) and had no negative influence on cell viability (data not shown). None of the siRNAs did elicit any inhibitory effect on average neurite length (Fig. 6B). Like in the case of overexpression, GTRAP3-18 knockdown did not change the percentage of cells that showed outgrowing neurites (data not shown).
6.
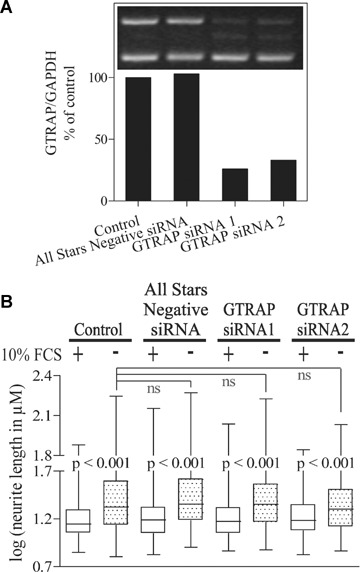
RNAi-mediated knockdown of GTRAP3-18 has no effect on neurite outgrowth of CAD cells. (A) Knockdown efficiency of two siRNAs against GTRAP3-18 after 48 hrs as determined by semi-quantitative RT-PCR (Inset: Representative agarose gel detail). GTRAP expression levels were corrected for GAPDH levels of the same PCR and the value of the control PCR was set to 100% of expression. A shows a representative level of knockdown for conditions used in all RNAi experiments. B measured neurite length in CAD cells after siRNA-mediated knockdown of GTRAP3-18 and differentiation. For detailed explanation of (B) see corresponding graph in Fig. 5.
GTRAP3-18 is differentially expressed during development
It has been shown for addicsin (mouse GTRAP3-18) that the protein follows a distinct expression pattern during development with low levels in early stages, a peak at day P5 and a subsequent decline until P42 [28]. Synaptogenesis is higher in the postnatal period compared with the embryonic phase of development [29].
In order to confirm that the expression of GTRAP3-18 shows a drop also in our case we tested the expression levels of GTRAP3-18 (by semi-quantitative RT-PCR) at specific developmental stages (embryonic day 17 [E17], the first post-natal day [P0] and in adult rats). We found that the expression of GTRAP3-18 showed a statistically significant decline from E17 to P0 and adult rat (Fig. 7).
7.
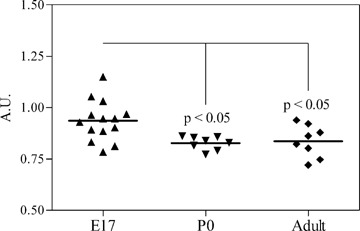
Expression pattern of GTRAP3-18 during developmental stages that are relevant for neurite outgrowth and synaptogenesis. Total brain RNA of Sprague-Dawley rats of different developmental stages was isolated and subjected to GTRAP and GAPDH double semi-quantitative RT-PCR. A scatter plot shows all measured values for GTRAP3-18 expression after correction for GAPDH levels as arbitrary units (A.U.). The black line represents the mean per sample. P-values were calculated with an anova followed by Bonferroni's multiple comparison test.
Discussion
GTRAP3-18 has originally been found in a yeast two-hybrid screen as a modulator of the neuronal glutamate transporter EAAC1 [11]. The protein is expressed in the brain and in several non-neuronal tissues, for example, mainly in the kidney but also in heart and in muscle [30]. Co-expression with GTRAP3-18 reduces the surface expression of EAAC1. Interestingly, JM4, which shares 62% sequence homology with human JWA and rat GTRAP3-18, is also an ER-localized four transmembrane domain protein that binds to the CC chemokine receptor 5 and interferes negatively with its ER to Golgi trafficking [31]. While this would suggest that the inhibitory effect of GTRAP3-18 is due to a direct interaction with the protein, it turned out that GTRAP3-18 co-expression perturbs trafficking of many other proteins [19]. This implies that the effect of GTRAP3-18 is rather general and thus tempts to the conclusion that it may affect one or more regulator(s) of membrane trafficking. Here we show clearly that the effect is due to binding of Rab1. This is supported by the observation that all Rab1-controlled trafficking events are inhibited.
One problem at the beginning of this study was to clarify the subcellular localization of GTRAP3-18. We unequivocally show that GTRAP3-18 is resident in the ER and does not leave it. This is supported by the following observations: (i) GTRAP3-18 co-labels with an ER marker (ii) deletion of the reputed dilysine (dibasic) motif (KARE) did not change its distribution and (iii) GTRAP3-18 did not bind COPII, which would be expected if it ought to leave the ER. Another problem that arose from this finding is the fact that the presence of Rab1 on the ER and therefore its role in ER exit are very heavily debated. The role of Rab1 in ER export is supported by the following findings: (i) the TRAPPI complex, which is the activator of Rab1 is found at ERES [6], (ii) the Rab1 deactivator TBC1D20 is found at the ER [8] and (iii) the Rab1 effector GBF1 is also found at ERES [7]. This implies that Rab1 can be activated at the ER, finds its effector at the ER and can be deactivated at the ER. Therefore, we are fully convinced that Rab1 plays an essential role in ER export.
We showed that GTRAP3-18 co-immunoprecipitates with endogenous Rab1. Indeed, an interaction between Rab proteins and PRA1 family proteins has been shown in the literature [12]. GTRAP3-18 inhibited the formation of TC when we monitored EAAC1 export in vitro. Importantly, exogenous excess of Rab1 rescued the concentration of EAAC1 into TC, implying that the action of GTRAP3-18 is most likely due to interaction with Rab1. In addition, GTRAP3-18 overexpression delayed reformation of the Golgi after BFA-induced breakdown. We attribute this effect to the block of the action of Rab1a on GBF1. Therefore, less ARF1 is active, which is needed for Golgi formation.
During neuronal development, vesicles are targeted to the growth cone to promote neurite outgrowth and synaptogenesis. Therefore it is not surprising that a number of membrane traffic regulators were found to be involved in this process, like the exocyst complex [27], Arf6 [32] and many others. Here, we clearly show that GTRAP3-18, while it does not affect the induction of differentiation in CAD cells, clearly affects growth of the neurite. We also found that expression levels of GTRAP3-18 were declining from E17 rats to P0 or adult rats. We conclude from this result that a drop of GTRAP3-18 allows stronger neurite outgrowth and hence active synaptogenesis.
In conclusion, our study (like others) links protein secretion to neuronal differentiation. However, we establish for the first time a role for Rab1a and its regulator GTRAP3-18 in this process, which provide a direct link between ER-to-Golgi trafficking and neuronal differentiation.
Acknowledgments
We gratefully acknowledge the skilled technical assistance of Marion Holy and the insightful discussions with Drs. Vladimir M. Korkhov, Michael Freissmuth and Michael Kiebler. This work was supported by the FWF (P17076, P18076 to H.H.S., SFB F35 to H.H.S. and Michael Freissmuth, and P18072 to H.F.) and the Schram foundation (to Michael Kiebler).
References
- 1.Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–66. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 2.Presley JF, Cole NB, Schroer TA, Hirschberg K, Zaal KJ, Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–5. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- 3.Stephens DJ, Pepperkok R. Imaging of procollagen transport reveals COPI-dependent cargo sorting during ER-to-Golgi transport in mammalian cells. J Cell Sci. 2002;115:1149–60. doi: 10.1242/jcs.115.6.1149. [DOI] [PubMed] [Google Scholar]
- 4.Xu D, Hay JC. Reconstitution of COPII vesicle fusion to generate a pre-Golgi intermediate compartment. J Cell Biol. 2004;167:997–1003. doi: 10.1083/jcb.200408135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu S, Satoh A, Pypaert M, Mullen K, Hay JC, Ferro-Novick S. mBet3p is required for homotypic COPII vesicle tethering in mammalian cells. J Cell Biol. 2006;174:359–68. doi: 10.1083/jcb.200603044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai H, Yu S, Menon S, Cai Y, Lazarova D, Fu C, Reinisch K, Hay JC, Ferro-Novick S. TRAPPI tethers COPII vesicles by binding the coat subunit Sec23. Nature. 2007;445:941–4. doi: 10.1038/nature05527. [DOI] [PubMed] [Google Scholar]
- 7.Monetta P, Slavin I, Romero N, Alvarez C. Rab1b interacts with GBF1 and modulates both ARF1 dynamics and COPI association. Mol Biol Cell. 2007;18:2400–10. doi: 10.1091/mbc.E06-11-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haas AK, Yoshimura S, Stephens DJ, Preisinger C, Fuchs E, Barr FA. Analysis of GTPase-activating proteins: Rab1 and Rab43 are key Rabs required to maintain a functional Golgi complex in human cells. J Cell Sci. 2007;120:2997–3010. doi: 10.1242/jcs.014225. [DOI] [PubMed] [Google Scholar]
- 9.Nuoffer C, Davidson HW, Matteson J, Meinkoth J, Balch WE. A GDP-bound of rab1 inhibits protein export from the endo-plasmic reticulum and transport between Golgi compartments. J Cell Biol. 1994;125:225–37. doi: 10.1083/jcb.125.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sannerud R, Marie M, Nizak C, Dale HA, Pernet-Gallay K, Perez F, Goud B, Saraste J. Rab1 defines a novel pathway connecting the pre-Golgi intermediate compartment with the cell periphery. Mol Biol Cell. 2006;17:1514–26. doi: 10.1091/mbc.E05-08-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin CI, Orlov I, Ruggiero AM, Dykes-Hoberg M, Lee A, Jackson M, Rothstein JD. Modulation of the neuronal glutamate transporter EAAC1 by the interacting protein GTRAP3-18. Nature. 2001;410:84–8. doi: 10.1038/35065084. [DOI] [PubMed] [Google Scholar]
- 12.Abdul-Ghani M, Gougeon PY, Prosser DC, Da Silva LF, Ngsee JK. PRA isoforms are targeted to distinct membrane compartments. J Biol Chem. 2001;276:6225–33. doi: 10.1074/jbc.M009073200. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Vidensky S, Ruggiero AM, Maier S, Sitte HH, Rothstein JD. Reticulon RTN2B regulates trafficking and function of neuronal glutamate transporter EAAC1. J Biol Chem. 2007 doi: 10.1074/jbc.M708096200. doi: DOI: 10.1074/jbc.M708096200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farhan H, Reiterer V, Korkhov VM, Schmid JA, Freissmuth M, Sitte HH. Concentrative export from the endoplasmic reticulum of the gamma-aminobutyric acid transporter 1 requires binding to SEC24D. J Biol Chem. 2007;282:7679–89. doi: 10.1074/jbc.M609720200. [DOI] [PubMed] [Google Scholar]
- 15.Farhan H, Korkhov VM, Paulitschke V, Dorostkar MM, Scholze P, Kudlacek O, Freissmuth M, Sitte HH. Two discontinuous segments in the carboxyl terminus are required for membrane targeting of the rat gamma-aminobutyric acid transporter-1 (GAT1) J Biol Chem. 2004;279:28553–63. doi: 10.1074/jbc.M307325200. [DOI] [PubMed] [Google Scholar]
- 16.Qi Y, Wang JK, McMillian M, Chikaraishi DM. Characterization of a CNS cell line, CAD, in which morphological differentiation is initiated by serum deprivation. J Neurosci. 1997;17:1217–25. doi: 10.1523/JNEUROSCI.17-04-01217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeitelhofer M, Vessey JP, Xie Y, Tubing F, Thomas S, Kiebler M, Dahm R. High-efficiency transfection of mammalian neurons via nucleofection. Nat Protoc. 2007;2:1692–704. doi: 10.1038/nprot.2007.226. [DOI] [PubMed] [Google Scholar]
- 18.Saraste J, Palade GE, Farquhar MG. Temperature-sensitive steps in the transport of secretory proteins through the Golgi complex in exocrine pancreatic cells. Proc Natl Acad Sci USA. 1986;83:6425–9. doi: 10.1073/pnas.83.17.6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruggiero AM, Liu Y, Vidensky S, Maier S, Jung E, Farhan H, Robinson MB, Sitte H, Rothstein JD. The ER exit of glutamate transporter is regulated by the inducible mammalian Yip6b/GTRAP3-18 protein. J Biol Chem. 2007 doi: 10.1074/jbc.M701008200. doi: DOI: 10.1074/jbc.M706499200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aridor M, Weissman J, Bannykh S, Nuoffer C, Balch WE. Cargo selection by the COPII budding machinery during export from the ER. J Cell Biol. 1998;141:61–70. doi: 10.1083/jcb.141.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aridor M, Bannykh SI, Rowe T, Balch WE. Sequential coupling between COPII and COPI vesicle coats in endoplasmic reticulum to Golgi transport. J Cell Biol. 1995;131:875–93. doi: 10.1083/jcb.131.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alvarez C, Garcia-Mata R, Brandon E, Sztul E. COPI recruitment is modulated by a Rab1b-dependent mechanism. Mol Biol Cell. 2003;14:2116–27. doi: 10.1091/mbc.E02-09-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Mata R, Szul T, Alvarez C, Sztul E. ADP-ribosylation factor/COPI-dependent events at the endoplasmic reticulum-Golgi interface are regulated by the guanine nucleotide exchange factor GBF1. Mol Biol Cell. 2003;14:2250–61. doi: 10.1091/mbc.E02-11-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lippincott-Schwartz J, Yuan LC, Bonifacino JS, Klausner RD. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 1989;56:801–13. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bannykh SI, Plutner H, Matteson J, Balch WE. The role of ARF1 and rab GTPases in polarization of the Golgi stack. Traffic. 2005;6:803–19. doi: 10.1111/j.1600-0854.2005.00319.x. [DOI] [PubMed] [Google Scholar]
- 26.Gommel D, Orci L, Emig EM, Hannah MJ, Ravazzola M, Nickel W, Helms JB, Wieland FT, Sohn K. p24 and p23, the major transmembrane proteins of COPI-coated transport vesicles, form hetero-oligomeric complexes and cycle between the organelles of the early secretory pathway. FEBS Lett. 1999;447:179–85. doi: 10.1016/s0014-5793(99)00246-x. [DOI] [PubMed] [Google Scholar]
- 27.Vega IE, Hsu SC. The exocyst complex associates with microtubules to mediate vesicle targeting and neurite outgrowth. J Neurosci. 2001;21:3839–48. doi: 10.1523/JNEUROSCI.21-11-03839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inoue K, Akiduki S, Ikemoto MJ. Expression profile of addicsin/GTRAP3-18 mRNA in mouse brain. Neurosci Lett. 2005;386:184–8. doi: 10.1016/j.neulet.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 29.Ledda F, Paratcha G, Sandoval-Guzman T, Ibanez CF. GDNF and GFRalpha1 promote formation of neuronal synapses by ligand-induced cell adhesion. Nat Neurosci. 2007;10:293–300. doi: 10.1038/nn1855. [DOI] [PubMed] [Google Scholar]
- 30.Butchbach ME, Lai L, Lin CL. Molecular cloning, gene structure, expression profile and functional characterization of the mouse glutamate transporter (EAAT3) interacting protein GTRAP3-18. Gene. 2002;292:81–90. doi: 10.1016/s0378-1119(02)00669-8. [DOI] [PubMed] [Google Scholar]
- 31.Schweneker M, Bachmann AS, Moelling K. JM4 is a four-transmembrane protein binding to the CCR5 receptor. FEBS Lett. 2005;579:1751–8. doi: 10.1016/j.febslet.2005.02.037. [DOI] [PubMed] [Google Scholar]
- 32.Albertinazzi C, Za L, Paris S, De Curtis I. ADP-ribosylation factor 6 and a functional PIX/p95-APP1 complex are required for Rac1B-mediated neurite outgrowth. Mol Biol Cell. 2003;14:1295–307. doi: 10.1091/mbc.E02-07-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]


