Abstract
Reactive oxygen species (ROS) play an important role in normal biological functions and pathological processes. ROS is one of the driving forces for oxidizing proteins, especially on cysteine thiols. The labile, transient, and dynamic nature of oxidative modifications poses enormous technical challenges for both accurate modification site determination and quantitation of cysteine thiols. The present study describes a mass spectrometry-based approach that allows effective discovery and quantification of irreversible cysteine modifications. The utilization of a long reverse phase column provides high-resolution chromatography to separate different forms of modified cysteine thiols from protein complexes or cell lysates. This FT-MS approach enabled detection and quantitation of ATM complex cysteine sulfoxidation states using Skyline MS1 filtering. When we applied the long column UPLC-MS/MS analysis, 61 and 44 peptides from cell lysates and cell were identified with cysteine modifications in response to in vitro and in vivo H2O2 oxidation, respectively. Long column Ultra High Pressure Liquid Chromatography -pseudo Selected Reaction Monitoring (UPLC-pSRM) was then developed to monitor the oxidative level of cysteine thiols in cell lysate under varying concentrations of H2O2 treatment. From UPLC-pSRM analysis, the dynamic conversion of sulfinic (S-O2H) and sulfonic acid (S-O3H) was observed within nucleoside diphosphate kinase (Nm23-H1) and heat shock 70 kDa protein 8 (Hsc70). These methods are suitable for proteome-wide studies, providing a highly sensitive, straight-forward approach to identify proteins containing redox-sensitive cysteine thiols in biological systems.
Keywords: cysteine oxidation, UPLC, pSRM, long column, label-free quantitation, Skyline, MS1 filtering
INTRODUCTION
Cysteine residues are major targets of reactive oxygen species (ROS), such as hydrogen peroxide (H2O2) and hydroxyl radical (OH.), as well as reactive nitrogen species. Control of the intracellular concentration of ROS is critical for cell proliferation and survival1. ROS can behave as effective second messengers that participate in many normal cellular functions such as transduction of cell surface receptor signaling. However, a substantial increase in the intracellular concentration of ROS can result in damage of cellular components, compromise cell viability and lead to mutations affecting tumor suppressor and oncogenic pathways2, 3. Many cancer cells generate more ROS than non-transformed cells in response to stimuli such as phorbol esters and hypoxia/reoxygenation4. Among the amino acids susceptible to ROS modification, cysteine is particularly reactive owing to its nucelophilic properties5, 6, 7.
Thiols of cysteine residues in proteins can form a wide variety of sulfoxidation products including reversible oxidation states S-glutathionylation (P-SS-G), disulfide bonds (P-SS-P), S-nitrosylation (P-SNO), sulfenic acid (P–SOH), and the irreversible higher oxidation states of sulfinic acid (P-SO2H)8 and sulfonic (P–SO3H) acid9. Modifications of specific thiols are known to play important roles in regulation of enzymatic activity. For example, nucleoside diphosphate kinase A (Nm23-H1) is involved in the ROS signaling pathway, and disulfide bond cross-linking of Nm23-H1 is formed in response to H2O2. Conformational changes in the oxidized Nm23-H1 cause the dissociation of the native hexameric structure to a dimeric form, resulting in loss of its kinase activity10.
Oxidative modifications of cysteinyl residues in proteins are difficult to identify and quantify in biological samples due to the low abundance of the modified proteins, the diversity of oxidation products, and the absence of distinguishing UV or visible spectrophotometric absorbance/fluorescence properties. Sample preparation steps routinely reduce and alkylate existing oxidized cysteines or alternatively introduce oxidation artifacts that complicate interpretation of the results. Comprehensive assessment of the dynamic changes in site-specific oxidation states would allow us to understand the distribution of redox status in disease pathology and to characterize the molecular mechanisms under different physiological states. Reversible oxidation states mediate fluctuations in cellular oxidation status, while the higher oxidation states serve as hallmarks of diseases associated with oxidative stress. Recent reviews have summarized the regulatory role and oxidative damage of cysteine oxidation products and described advances in proteomic methods for detection using chemical derivatization or antibodies combined with gel electrophoresis and/or LC-MS/MS approaches7, 11, 12, 13.
Mass spectrometry has recently evolved into a high-throughput methodology for detecting and quantifying proteins with post-translational modifications (PTMs) 14. Progress has been made in proteomic-scale studies of the reversible cysteine oxidation states by taking advantage of the high reactivity of cysteine sulfhydryls with alkylating agents. Approaches based on Isotope-Coded Affinity Tags (ICAT) have been widely used in redox proteomics because they label the cysteine sulfhydryl moiety and can be enriched through biotin affinity purification15. For redox studies, a three step approach uses differential alkylation to detect the oxidation status of cysteine residues indirectly16. The OxICAT method uses MS intensities to calculate the percentage of reversibly oxidized cysteine at the peptide level and then monitors changes due to oxidative stress17, 18. The peptide redox status is then normalized to the total peptide expression level, avoiding the pitfall of confounding changes in oxidation status with changes in protein expression level. This method has been used to detect and quantify 40 proteins whose thiol oxidation status changed at least 1.5-fold in Caenorhabditis elegans following in vivo treatment with H2O2 18. Together, these methods have advanced the field of quantitative proteomics for studying reversible oxidation states, but they are not applicable to sulfinic and sulfonic acid.
A recent extension of the cysteine-labeling approach is the oxidation multiple reaction monitoring (OxMRM) method. In this method, a stable isotope is used to label the cysteines via differential alkylation, followed by affinity purification of the protein of interest and quantitation with multiple reaction monitoring19. With this approach, it is possible to detect low abundance sulfinic and sulfonic acid modifications, as well as those reversibly oxidized, albeit for a limited number of enriched proteins. Hansen et al. quantified an increase in cellular global protein glutathionylation levels from steady state levels of <0.1% to >15% of protein cysteines upon oxidant exposure20. Sulfinic and sulfonic acid modifications are expected to be at similarly low levels in the steady state with OxMRM evidence of a 100-fold increase upon hydrogen peroxide treatment19.
An alternative to stable isotope labeling strategies is the use of label-free quantitation methods. The introduction of Fourier transform (FT) mass spectrometers such as the LTQ-Orbitrap with high mass accuracy, resolution, and sensitivity, in combination with the high reproducibility of nano-HPLC are crucial for the development of MS and MS/MS ion intensity measurements to quantify modified peptides. The open source software Skyline facilitates label-free quantitation via MS1 filtering21, 22.
Use of label-free MS1 intensity for quantitation in highly complex samples, such as cell lysates, can be problematic due to interference between co-eluting peptides with similar m/z values. High resolution MS1 reduces interference, but there may still be co-eluting species at the low abundance levels typical for PTMs. Hewel et al. have utilized the LTQ Orbitrap Velos to conduct high resolution MS and targeted MS/MS and to then quantitate samples using extracted product ion chromatograms with stable isotope labeled reference peptides, calling the technique targeted peptide monitoring23. An analogous experiment was performed by Sherrod et al. who used low resolution MS/MS and Skyline in an approach termed pseudo Selected Reaction Monitoring (pSRM) 24. pSRM uses multiple product ions per peptide for specificity. They compared the use of internal reference peptides with that of stable isotope dilution for quantitation and found somewhat higher coefficients of variance observed from internal reference peptides (22-31%) versus stable isotope dilution (10-20%), with the same trends observed in both methods and consistent with immunoblot results24. This experiment demonstrated the suitability of pSRM with internal reference peptides for the relative quantitation of PTMs in a protein complex. The advantage of this pipeline is the ease of moving from data-dependent acquisition results to lists of precursors for targeted MS/MS and then to quantitation of the extracted ion chromatograms. However, the technique has not been demonstrated as effective on PTMs in complex samples.
In this study we aim to develop a label-free, LC-MS/MS method to detect and quantify SO2H and SO3H modified cysteine residues without further enrichment suitable for analysis of endogenous levels of protein expression. Proteome-wide analysis of protein-thiol oxidation is achieved by using both a long reverse phase column nano-UPLC method that offers high-chromatographic resolution to separate different forms of modified cysteine residues and a high resolution, high mass accuracy, and high sensitivity hybrid LTQ-Orbitrap mass analyzer for confident identification. After discovering novel cysteine oxidation sites, the ion-intensity based, label-free quantitation method UPLC-pSRM is adapted from Skyline, a freely available, open-source software application, for quantitative data processing of the cysteine sulfoxide peptides. Our data demonstrates the proficiency of the long column-based method to identify and quantify site-specific changes in complex mixtures in response to hydrogen peroxide exposure.
EXPERIMENTAL PROCEDURES
Materials and reagents
All chemicals and HPLC grade solvents were purchased from Sigma except as listed below: sequencing grade trypsin was purchased from Promega (Madison, WI); 6 Bovine Protein Digest Exponential Molar Mix was purchased from Michrom (Auburn, CA).
Protein immunoaffinity purification
Recombinant ATM complex purification was performed as previously described25. Briefly, wild-type ATM expressed in HEK 293T cells was isolated using anti-Flag M2 antibody resin, washed extensively, and eluted with Flag peptide.
In vitro hydrogen peroxide (H2O2) treatment
Both BSA and ATM protein complexes were incubated with H2O2 (final concentrations 0, 267 μM and 900 μM) for 90 min at 30°C. Then β-mercaptoethanol was added to a final concentration of 50 mM to fully reduce any disulfides. Trypsin was added and incubated overnight at 37°C.
Jurkat cell lysate purchased from Novus Biologicals (Littleton, CO) was diluted into 5 mM Tris-HCl pH 7.4, 15 mM NaCl, 0.1 mM EDTA, 0.1% Triton X-100, 0.01% SDS, 0.1% Sodium deoxycholate, 0.1 mM PMSF. The detergent was removed from the cell lysate using HiPPRTM Detergent Removal Resin from Thermo (Rockford, IL) following the manufacturer's instructions before incubation with H2O2 and the H2O2 was diluted in 1XPBS to desired concentrations. 25 μg of detergent-free lysate was treated either with control (1XPBS) or H2O2 (final concentrations 80 μM, 440 μM, 2 mM and 50 mM) and incubated for 90 min at 37°C. The incubated mixture was reduced with DTT (final concentration 2 mM) and alkylated with iodoacetamide (final concentration 5.5 mM) before in-solution digest with trypsin overnight at 37°C, and proteolysis was stopped by addition of 5% acetic acid.
In vivo hydrogen peroxide (H2O2) treatment
HEK 293T cells were maintained in 5% CO2 at 37°C, humidified atmosphere. Cells were not serum starved. Once the cells reached to 80-90% confluence, and incubated with 440 μM H2O2 concentrations at 37°C for 12min. After the reaction, the medium was removed and quickly washed with 1XPBS three times. To prevent post-lytic oxidation, 20% (v/v) ice cold trichloroacetic acid (TCA) was immediately added to the cells and spread to cover the entire plate, followed by incubation at 4°C for 20min. Cells were scraped into an 1.5mL microcentrifuge tube, and washed once with 10% (v/v) ice cold TCA, twice with 5% (v/v) ice cold TCA. Samples were resuspended in lysis buffer (50 mM tri-ethylene ammonium bicarbonate pH 8.0, 5 M Urea, 10 mM EDTA, 1% SDS and 0.1% protease inhibitor cocktail followed by sonication on ice for 20 min. Samples were then reduced with 10mM TCEP (tris(2-carboxyethy)phosphine) at 50°C for 30min and alkylated with 100 mM iodoacetamide at 37°C for 60min. Samples were then proceed to in solution digest with trypsin/Lys-C mix (Promega) at 45°C for 2hr. , The digest was stopped by addition of 5% acetic acid. The digest was analyzed two time using 0.4 μg by LC-MS/MS.
Mass spectrometry and data analysis
Data-dependent acquisition and dose response UPLC-pSRM analyses were both performed on a Thermo Scientific LTQ-Orbitrap Elite (San Jose, CA) mass spectrometer equipped with an ultra-high pressure Dionex Ultimate 3000 RSLCnano system (Sunnyvale, CA) with buffer A (0.1% (v/v) formic acid in water) and buffer B (0.1% (v/v) formic acid in acetonitrile). Peptides were concentrated onto an in-house packed 100-μm ID × 2-cm C18 column (Magic C18, 3 Mm, 100 Å, Michrom Bioresources Inc.) then separated on a 75-μm ID × 50 cm C18 fused silica column. Liquid chromatography was performed using a gradient of 5-40% buffer B over 90 min. To identify peptides and post-translational modifications, the mass spectrometer was operated in positive mode using data-dependent acquisition of MS2. For FT MS1/ion trap MS2 experiments on both H2O2 treated cell and lysate samples, one scan cycle included an MS1 scan (300-1700) at a resolution of 120,000 followed by MS2 on the twenty most abundant precursors found in the MS1 spectrum. For FT MS1/MS2 experiments on 6 Bovine Protein Digest and ATM, FT MS1 were performed at resolution 120,000 with FT CID MS2 at resolution 15,000 on the 10 most abundant precursors or with FT CID and HCD at resolution 15,000 on the top 5 precursors. For UPLC-pSRM, the FT MS1 scan (300-1500) at a resolution of 30,000 was followed by MS2 for targeted peptides in the ion trap (Tables 1 and 2). Automatic gain control target values were set as follows: FT MS at 1e6, FT MS2 at 5e4, IT MS2 1e4. Maximum ion injection times were set as follows: FT MS and MS2 at 500 ms, IT MS2 at 150 ms. For IT CID, minimum ion count for MS2 was set to 5,000; isolation width 1.9 Da; collision energy 35; activation Q 0.25; and activation time 10 ms. For FT CID, minimum ion count for MS2 was set to 1,000; isolation width 3.0; other parameters set as for IT CID. For FT HCD, relevant parameters were set as for FT CID except for activation time of 0.100 ms. Charge state determination was enabled during FT MS prescan, determining scan range for MS2 scans.
Table 1.
ATM and PRMT5 MS1 ions selected for quantitation
| Protein | Peptides selected for MS1 filtering | Precursor m/z | Precursor [M+1] m/z | Precursor [M+2] m/z |
|---|---|---|---|---|
| ATM | ||||
| Internal reference | LLDPFPDHVVFK | 476.2625+++ | 476.5968+++ | 476.9311+++ |
| Internal reference | TILNHVLHVVK | 424.9311+++ | 425.2654+++ | 425.5996+++ |
| Cys633 (Sulfinic acid) | AAMNFFQSVPECEHHQK | 678.9647+++ | 679.2990+++ | 679.6328+++ |
| Cys1396 (Sulfinic acid) | ATFAYISNCHK | 643.7953++ | 644.2968++ | 644.7974++ |
| Cys2074 (Sulfinic acid) | QAGIIQALQNLGLCHILSVYLK | 809.7858+++ | 810.1201+++ | 810.4542+++ |
| Cys2074 (Sulfonic acid) | QAGIIQALQNLGLCHILSVYLK | 1222.1725++ | 1222.6740++ | 1223.1751++ |
| Cys2991 (Sulfinic acid) | ALYLQQRPEDETELHPTLNADDQECK | 1030.1437+++ | 1030.4779+++ | 1030.8121+++ |
| PRM T5 | ||||
| Internal reference | AAILPTSIFLTNKK | 758.9585++ | 759.4600++ | 759.9614++ |
| Internal reference | GPLVNASLR | 463.7745++ | 464.2759++ | 464.7772++ |
| Cys518 (Sulfinic acid) | LHNFHQLSAPQPCFTFSHPNR | 628.3000++++ | 628.5507++++ | 628.8013++++ |
| Cys22 (Sulfonic acid) | DLNCVPEIADTLGAVAK | 888.9378++ | 889.4393++ | 889.9402++ |
| Cys42 (Sulfinic acid) | QGFDFLCMPVFHPR | 863.3974++ | 863.8989++ | 864.3995++ |
Table 2.
Selected peptides and MS2 transitions for UPLC-pSRM quantitation
| Protein /modified cysteine residue | Peptide sequence | Precursor m/z | Product m/z |
|---|---|---|---|
| DJ-1 | |||
| Cys106 sulfonic acid | GLIAAICAGPTALLAHEIGFGSK | 1129.59 (2+) | 1497.8 (y15+), 1719.8 (y17+), 784.9 (y162+), 916.9 (y182+), 988.0 (y202+) |
| Internal reference | ALVILAK | 364.25 (2+) | 331.2 (y3+), 444.3 (y4+), 543.3 (y5+) |
| Internal reference | GPGTSFEFALAIVEALNGK | 961.00 (2+) | 730.4 (y7+), 843.5 (y8+), 914.5 (y9+), 1027.6 (y10+), 1245.7 (y12+) |
| Nm23-H1 | |||
| Cys109 sulfinic acid | GDFCIQVGR | 513.73 (2+) | 331.2 (y3+), 572.3 (y5+), 707.3 (y6+), 354.1 (y62+) |
| Cys109 sulfonic acid | GDFCIQVGR | 521.73 (2+) | 331.2 (y3+), 459.2 (y4+), 572.3 (y5+), 723.3 (y6+) |
| Internal reference | NIIHGSDSVESAEK | 495.91 (3+) | 434.2 (y4+), 563.2 (y5+), 662.3 (y6+), 331.6 (y62+), 629.7 (y122+) |
| Internal reference | TFIAIKPDGVQR | 448.92 (3+) | 671.3 (y6+), 336.1 (y62+), 456.7 (y82+), 492.2 (y92+), 548.8 (y102+) |
| Hsc70 | |||
| Cys17 sulfinic acid | GPAVGIDLGTTYSCVGVFQHGK | 755.04 (3+) | 871.4 (y8+), 872.4 (y162+), 928.9 (y172+), 957.4 (y182+) |
| Cys17 sulfonic acid | GPAVGIDLGTTYSCVGVFQHGK | 1127.54 (2+) | 616.3 (y5+), 772.4 (y7+), 1272.5 (y11+), 1373.6 (y12+), 1474.6 (y13+) |
| Internal reference | SQIHDIVLVGGSTR | 741.40 (2+) | 477.2 (y5+), 689.3 (y7+), 788.4 (y8+), 1016.5 (y10+), 577.3 (y112+) |
| Internal reference | SINPDEAVAYGAAVQAAILSGDK | 1130.57 (2+) | 1001.5 (y10+), 1072.5 (y11+), 1200.6 (y13+), 1363.7 (y14+), 1434.7 (y15+) |
Peptide/protein identification was performed with Proteome Discoverer 1.4 embedded with SEQUEST (Thermo Scientific), and MASCOT (Matrix Science) using the Uniprot Human reference proteome database including canonical and manually reviewed isoforms, version 2013_04, with 89,430 sequences or the Uniprot SwissProt database version 2011_11 with 448077sequences for 6 Bovine Protein Digest experiments. MSPepSearch spectral library search for was also used for the H2O2 treated cell (in vivo) and H2O2 treated whole lysate (in vitro) samples with the NIST human peptide tandem mass spectral library (06-05-2013 with 328,791 spectra). The search parameters used were as follows: two missed cleavages on a trypsin digest were permitted, no fixed modifications, variable modifications on oxidized methionine (monoxidation and dioxidation) and cysteine residues: sulfinic (SO2H, +31.990), and sulfonic (SO3H, +47.985) acid and carbamidomethylation; 10 ppm precursor tolerance, 0.02 Da MS/MS tolerance if acquired in the Orbitrap and 0.8 Da MS/MS tolerance if acquired in the LTQ. Variable modification of STY phosphorylation was used in ATM complex searches. MASCOT also includes protein N-terminal acetylation as a variable modification, with removal of the protein N-terminal Methionine, if present. The MASCOT modification search for sulfinic acid peptides included neutral loss of H2SO2 as an additional fragment ion. A reverse sequence database was generated from the Uniprot Human database and used to perform a decoy search for peptide validation. Peptide identifications were filtered using Percolator, where a false discovery rate (FDR) of 2% (q < 0.02) was applied and filtered applying a minimum of 4 high confidence peptides per protein.
Skyline quantitation
A spectral library was created from the Proteome Discoverer protein identification search results for the 6 Bovine Protein Digest, ATM and Jurkat cell lysate samples treated with H2O2. Skyline v1.4 was applied for chromatogram extraction of MS1 precursor ions (Table 1) for peak area calculation in ATM and PRMT5 quantitation. The MS1 precursor isotopic import filter was set to a count of three, (M, M+1, and M+2) at a resolution of 120,000. For UPLC-pSRM experiments on cell lysate, Skyline used the spectral library to rank the y fragment ions from target peptides by intensity and selected transitions for calculating peak area automatically using the following criteria: y or precursor ion, precursor charge state 2, 3, 4; ion charge state 1, 2, 3; product ion m/z > precursor; always select fragment ion N-terminal to Proline; precursor m/z exclusion window of 3 Th. Targeted LC-MS/MS were acquired, and extracted ion count peak areas for MS2 fragment ions were summed for 3-5 transitions for each targeted peptide using Skyline (Table 2). Skyline calculates the area under the curve for each peak by interpolating peak height between the raw data points for the duration of the chromatographic peak elution. Savitzky-Golay smoothing was used for display purposes in the calculated peak area vs. retention time plots.
Each cysteine modified peptide was normalized by the average peak area of two peptides from the same protein that are distinct from the cysteine modified peptide, designated as internal reference peptides. The internal reference peptides are chosen based on observation in the data dependent acquisition mode and absence of cysteine or methionine residues, so that the peptides will not be oxidized in response to hydrogen peroxide treatment and thus the ion intensity is not expected to vary between samples. For MS1 quantitation, the precursor peak areas (M, M+1, M+2) of a modified peptide were divided by the average of precursor peak areas (M, M+1, M+2) of two non-modified peptides from the same protein. For pSRM quantitation normalization, the total peak area of 4-5 fragment ion transitions was divided by the average of fragment ion peak areas of two non-modified peptides from the same protein. This normalized the amount of oxidation relative to the protein expression in a given sample.
Statistical analysis
All data are reported as mean ± standard deviation (SD) unless otherwise stated.
RESULTS
Optimization of the nano-LC/MS approach for detecting and quantitating cysteine sulfoxide reaction products with standards
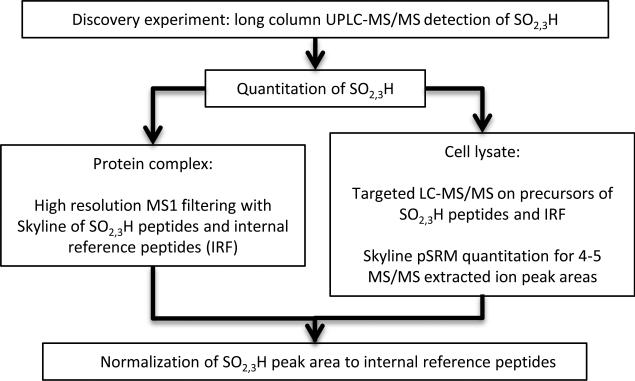
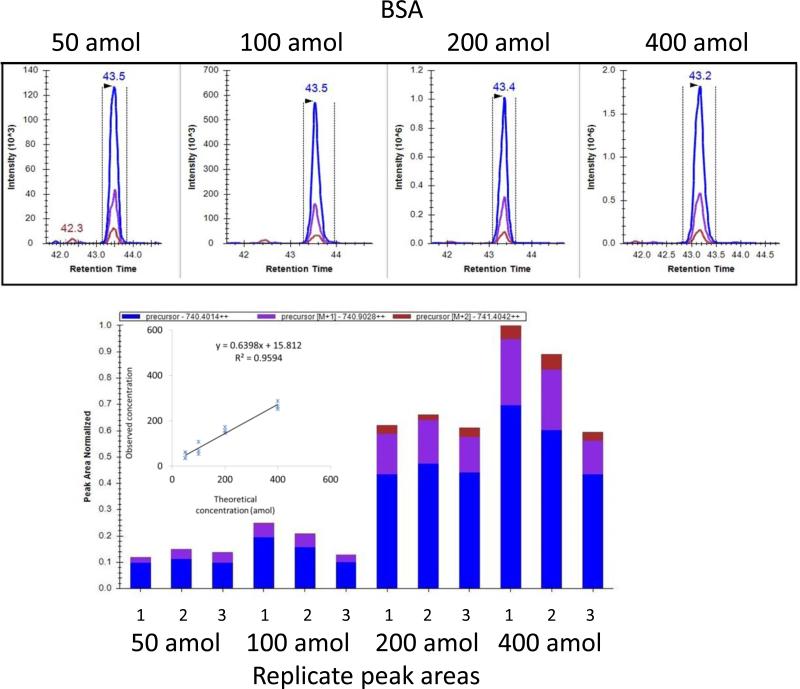
The experimental workflow is shown in Figure 1. The analytical procedure was first optimized using protein standards. A custom-packed long (50cm) reverse phase nano-column was used to separate peptide mixtures at room temperature. UPLC-MS/MS of tryptic peptide mixtures from hydrogen peroxide treated BSA identified sulfenic (−SOH), sulfinic (−SO2H), and sulfonic acid (−SO3H) reaction products (data not shown). For quantification, we adapted a recently developed platform-independent software approach termed Skyline MS1 filtering. The software extracts quantitative information from multiple LC-MS/MS experiments by integrating MS1 ion intensity across the peak elution profile22. To verify the applicability of this method for detecting low level species in a mixture of higher abundance peptides, we analyzed BSA and casein, ranging from 50 amol to 4 fmol by serial dilution of a standard 6 protein digest mixture where the BSA:casein molar ratio was 1:10. The response curve was generated by analyzing three technical replicates at each of four concentration points. Two peptides from BSA and two peptides from casein were selected for the calculation of peak area. High correlation (r2=0.95) was obtained between the experimentally-derived relative abundance measurements using both BSA and casein standards, and the theoretical ratios calculated from the known concentration specified by the manufacturer (Figures 2 and S1).
Figure 1.
Workflow for detection and quantitation of cysteine sulfoxide reaction products.
Figure 2.
Reproducible peak area and retention times and good correlation between peak area and concentration are observed for peptides from BSA. Tryptic digest peptide mixtures were analyzed by LC-MS/MS on the LTQ-Orbitrap Elite mass spectrometer where MS1 spectra were acquired in high-resolution FT mode. Skyline extracts and displays MS1 intensity values for M (blue), M+1(purple) and M+2 (red) precursor ions at varying concentrations from a single run in the upper panel and integrates peak area in the selected retention time window for triplicate runs at four concentrations (50-400 amol) in the lower panel for LGEYGFQNALIVR. The correlation coefficient (R2) is calculated based on the average of triplicate runs using the peptides HLVDEPQNLIK and LGEYGFQNALIVR. The regression line demonstrates that the linear range extends to the attomole level.
MS1 based quantification of sulfinic and sulfonic acid reaction products from affinity purified ATM protein complex
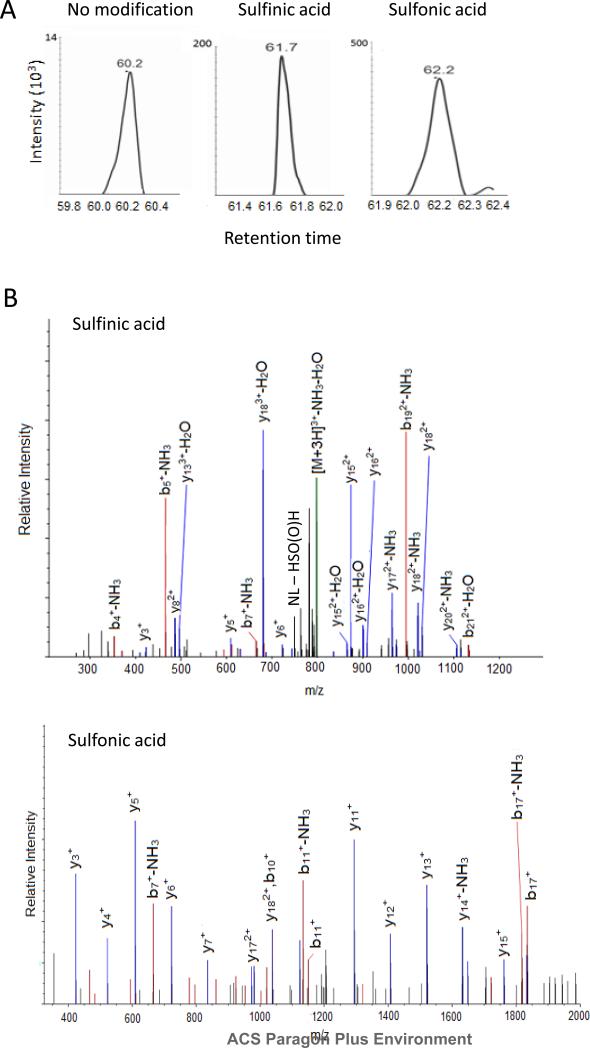
We then applied the same method to study affinity purified ATM treated with hydrogen peroxide from high resolution MS and MS/MS spectra acquired in the Orbitrap following the workflow outlined in Figure 1. ATM is a Serine/Threonine kinase present at low abundance in the human nucleus which may participate in regulating and maintaining redox balance in cells26. Treatment by thiol oxidizing agents, diamide at low μM levels or 100 μM to 20 mM H2O2 for 30 minutes, can activate ATM kinase activity by inducing disulfide formation27. The combined results of triplicate run from 0, 267 μM and 900 μM H2O2 treated ATM pull down complex allow us to detect 50 of 89 cysteines of ATM, 27 of which were oxidized to sulfinic and sulfonic acid forms (Table S1 shows all peptides identified for ATM and protein arginine N-methyltransferase 5 (PRMT5), Table S2 lists cysteine oxidized peptides, Figure S2 contains FT MS/MS for modified peptides), including Cys2991 which was previously identified in our labs as critical for ATM activation27. Similar results were found in the endogenous co-purified protein, PRMT5, where 9 of 12 cysteines were detected including 6 oxidized cysteine residues. Figure 3A shows the MS extracted ion chromatograms for the unmodified, sulfinic and sulfonic acid modified ATM QAGIIQALQNLGLCHILSVYLK peptide that contains Cys2074 modifications, demonstrating baseline chromatographic resolution of the peptide forms using the long column, with representative tandem mass spectra shown in Figure 3B. It is worth noting that we observed the characteristic sulfinic acid neutral loss of 66 Da from HS(O)OH found previously in the study by Hoffman et al., but the ion does not dominate the MS/MS28. In our data set, we observed strong neutral loss of 66 Da in 42% of the sulfinic acid MS/MS. MS3 experiments were also conducted to exploit the sulfinic acid neutral loss of 66 Da, but did not yield additional sites. Men and Wang previously reported facile cleavage of the amide bond C terminal to the sulfinic acid modified cysteine in a study of glycine rich model peptides on a quadrupole ion trap instrument29. In the MS/MS for ATM and PRMT5 peptides (Figure S2), we observed a strong y ion C terminal to the sulfinic acid modified cysteine 58% of the time. The neutral loss of HS(O)OH and y ion C terminal cleavages were rarely observed as strong fragment ions in sulfonic acid modified peptides, at frequencies of 6% and 3%, respectively, consistent with previous observation29.
Figure 3.
MS/MS spectra and extracted ion chromatogram (XIC) of the unmodified and oxidized ATM Cys2074 peptides. A, The XIC plot vs. retention time of unmodified and modified cysteine variants of ATM Cys 2074 peptide QAGIIQALQNLGLCHILSVYLK demonstrates chromatographic separation, as follows: unmodified (m/z 799.1,+3) sulfinic acid (m/z 809.78,+3) and sulfonic acid (m/z 1222.17,+2) B, The MS/MS spectra of the QAGIIQALQNLGLCHILSVYLK precursor ions for sulfinic acid (m/z 809.78,+3) and sulfonic acid (m/z 1222.17,+2) modified peptides. The sulfinic acid containing peptide fragmented to produce the characteristic 66 Da neutral loss of HS(O)OH.
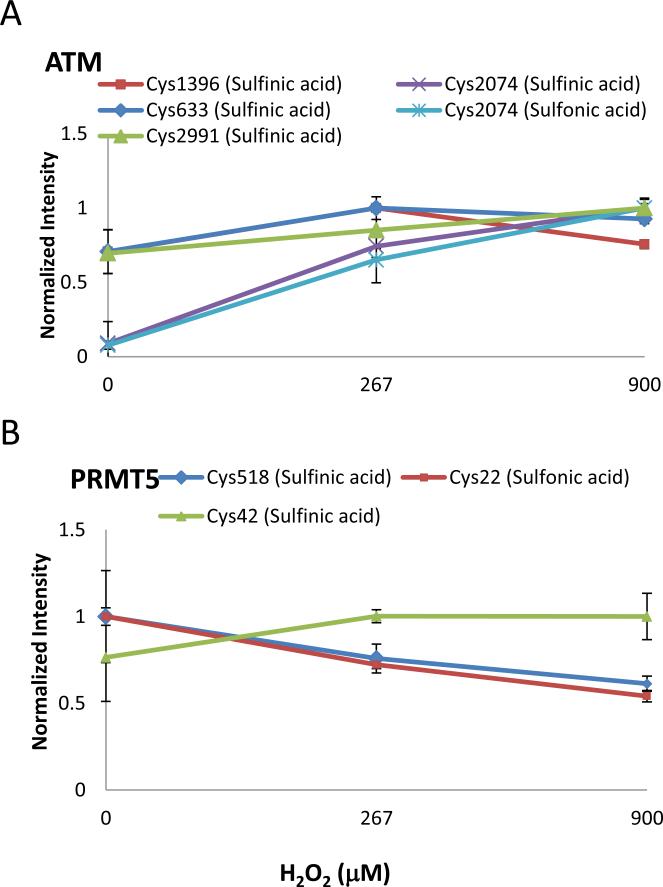
MS1 relative quantitation of ATM and PRMT5 was used to measure dose response under varying concentrations of H2O2. The MS1 peak areas from cysteine containing peptides were calculated to compare relative abundance between samples using Skyline. The peptide ratios were normalized using two non-cysteine/methionine containing, unmodified peptides from each protein as listed in Table 1, following the internal reference peptide method to control for protein abundance24. We chose not to correct the variability based on the same, but unmodified peptide because the cysteine residues are converted to sulfinic or sulfonic acid at higher H2O2 concentrations, and thus the amount of unmodified peptide is expected to decrease correspondingly. In addition, modification near a protease cleavage site may alter the efficiency of the cleavage and cause changes in ratios of modified:unmodified peptide. The use of distinct unmodified peptides from the protein of interest reduces these sources of variability. Due to the low stoichiometry of modified cysteine, not all detected cysteine oxidation sites could be quantitated. The Skyline results of ATM and PRMT5 cysteine oxidation levels are shown in Figure 4A and B. Among all the monitored cysteine residues, only ATM Cys2074 sulfinic and sulfonic acid levels showed a marked increase relative to control (Figure 4A). The induction levels of other examined cysteine sulfinic or sulfonic acid-modified peptides, including ATM Cys633, Cys1396, Cys2991 and PRMT5 Cys22, Cys42, and Cys518, showed no significant change in response to H2O2 oxidation.
Figure 4.
Quantitation of irreversible oxidation of ATM and endogenous PRMT5 by H2O2. The oxidation level of selected A, ATM and B, PRMT5 cysteines were calculated based on Skyline MS1 filtering results. The raw peak area data were corrected using peak areas from two unmodified peptides from PRMT5 or ATM and normalized to the most intense value of each dose response. The level of ATM Cys2074 sulfinic and sulfonic acid increases in a dose-dependent manner. ATM Cys633, Cys1396, and PRMT5 Cys22, Cys42, Cys518 show no change in response to H2O2 oxidation. In all cases, error bars represent the standard error of three biological replicates.
Global analysis for discovery of cysteine oxidation from cell lysate
After demonstrating that the use of the long reverse phase column provided a high-resolution chromatographic separation that enabled detection of low abundance modified peptides in protein mixtures, we next assessed its suitability for measuring the oxidation status of endogenous proteins. Jurkat cell lysate was treated with 440 μM H2O2 followed by alkylation with iodoacetamide, and then 0.4 μg was analyzed by LC MS/MS on the LTQ Orbitrap Elite, with MS/MS spectra acquired in the ion trap. Four replicate injections were analyzed, where modified peptides were identified from the database search using a 2% false discovery rate. The list of modified peptides was reduced further by utilizing a protein filter requiring 4 high confidence peptides per protein. The use of the stringent protein filter resulted in similar levels of modified and unmodified peptide filtering. When a single peptide is required for protein identification, changing the FDR from 5% to 2% results in 90% of bulk peptides remaining, while the number of oxidized peptide is down to 67%, and 37% of sulfonic acid modified peptides pass the 2% FDR filter. When 4 peptides/protein filter is applied the ratio of peptides at 2% FDR/5%FDR is 95%, 93%, and 90% for all peptides, oxidized peptides, and sulfonic acid modified peptides, respectively. This resulted in the detection of 61 cysteine oxidized peptides identified from 43 proteins in the H2O2 oxidized lysate. The 61 modified peptides contain 10 sulfinic acid cysteines and 51 sulfonic acid cysteines, respectively (Table S3, S4, and Figure S3 and S4).
Global analysis for discovery of cysteine oxidation from cells
HEK 293T cells were incubated with 440YM H2O2 for 12 min, and then lysed in TCA to minimize artifacts during sample preparation. Samples were reduced and alkylated before mass spectrometry analysis. The data was interrogated and filtered using the same criteria as described above, a total of 44 modified peptides were identified from 40 proteins. The 44 modified peptides contain 7 sulfinic acid cysteines and 38 sulfonic acid cysteines, respectively (Table S5, S6, and Figure S5).
Quantitative analysis of proteins susceptible to hydrogen peroxide oxidation
Our data set identified several known oxidation sites including Cys247 of Glyceraldehyde 3-phosphate dehydrogenase (GAPDH)28, Cys111 of Superoxide dismutase [Cu-Zn] (SOD1)30, Cys109 of Nm23-H110, Cys106 of High mobility group box (HMGB) 131 , Cys106 of DJ-132, and other proteins33,34 (Table 3 and 4). Based on these findings, Nm23-H1, DJ-1, and Hsc70 were further examined to determine the H2O2 dose dependence of the cysteine sulfoxide levels using long column UPLC-pSRM transitions (Table 2). Doses included 0, 80 μM, 440 μM, 2 mM, and 50 mM H2O2. Use of a long reversed phase column improved peptide separation of modified species. Targeted UPLC-MS/MS data were acquired on both the selected precursor ions for sulfinic and sulfonic acid modified peptides and the separate, non-cysteine/methionine containing peptides from the same protein that served as internal reference peptides. pSRM quantitation occurred post-acquisition, through extraction of product ion chromatograms using Skyline24. Table 2 lists peptide sequences and precursor ions selected for targeted MS/MS and product ions used for quantitation, while MS/MS are shown in Figure S4. For a given peptide, the peak areas of the product ions listed in Table 2 are summed and then normalized to the internal reference peptides to determine the modified peptide quantitation relative to the protein of interest. Since the oxidation treatments were performed on cell lysates, protein abundance is not expected to vary between experiments.
Table 3.
List of proteins of known cysteine oxidation sites detected with long column UPLC-MS/MS in Jurkat cell lysate
| Accession # | Protein Name | Sequence | Modification |
|---|---|---|---|
| P10809 | 60 kDa heat shock protein, mitochondrial | AAVEEGIVLGGGcALLR33,34 | Trioxidation |
| P10809 | 60 kDa heat shock protein, mitochondrial | GQKcEFQDAYVLLSEK33 | Trioxidation |
| P10809 | 60 kDa heat shock protein, mitochondrial | GQKcEFQDAYVLLSEKK33 | Trioxidation |
| P10809 | 60 kDa heat shock protein, mitochondrial | GQKcEFQDAYVLLSEK34 | Dioxidation |
| P63261 | Actin, gamma | EKLcYVALDFEQEMATAASSSSLEK23,32,a | Trioxidation |
| B4DVE7 | Annexin A11 | GVGTDEAcLIEILASR16,a | Trioxidation |
| B4DVE7 | Annexin A11 | BKDAISGIGTDEKcLIEILASR16 | Trioxidation |
| Q05639 | Elongation factor 1-alpha 2,EEF1A | SGDAAIVEMVPGKPMcVESFSQYPPLGR16 | Trioxidation |
| P04406 | Glyceraldehyde-3-phosphate dehydrogenase | VPTANVSVVDLTcRLEKRAK28 | Trioxidation |
| P04406 | Glyceraldehyde-3-phosphate dehydrogenase | VPTANVSVVDLTcR28 | Dioxidation |
| P04406 | Glyceraldehyde-3-phosphate dehydrogenase | VPTANVSVVDLTcR28 | Trioxidation |
| O43390 | Heterogeneous nuclear ribonucleoprotein R | GFcFLEYEDHKa | Trioxidation |
| P09429 | High mobility group box B1 | RPPSAFFLFcSEYRPK25 | Trioxidation |
| Q04760-2 | Lactoylglutathione lyase | GFGHIGIAVPDVYSAcKRa | Trioxidation |
| P24666 | LMW phosphotyrosine protein phosphatase | SVLFVcLGNIcRa | Trioxidation, |
| P15531 | Nucleoside diphosphate kinase, Nm23-H1 | GDFcIQVGR10,16,a | Dioxidation |
| P15531 | Nucleoside diphosphate kinase, Nm23-H1 | GDFcIQVGR10,16,a | Trioxidation |
| M(Oxidation), | |||
| P62937 | Peptidyl-prolyl cis-trans isomerase A | IIPGFmcQGGDFTR28,a | C(Trioxidation) |
| P62937 | Peptidyl-prolyl cis-trans isomerase A | KITIADcGQLE28 | Dioxidation |
| P62937 | Peptidyl-prolyl cis-trans isomerase A | KITIADcGQLE28 | Trioxidation |
| C(Trioxidation), | |||
| P30101 | Protein disulfide-isomerase A3 | FIQENIFGIcPHmTEDNKDLIQGKa | M(Oxidation) |
| Q99497 | Protein DJ-1, PARK7 | GLIAAIcAGFTALLAHEIGFGSKa,32 | Trioxidation |
| P00441 | Superoxide dismutase [Cu-Zn] , SOD1 | AVcVLKGDGPVQGIINFEQK30 | Dioxidation |
| P00441 | Superoxide dismutase [Cu-Zn], SOD1 | AVcVLKGDGPVQGIINFEQK30 | Trioxidation |
| P00441 | Superoxide dismutase [Cu-Zn], SOD1 | HVGDLGNVTADKDGVADVSIEDSVISLSGDHcIIGRa,30,48 | Trioxidation |
Human cysteine oxidation sites reported in RedoxDB (http://biocomputer.bio.cuhk.edu.hk/RedoxDB/)
Table 4.
List of proteins of known cysteine oxidation sites detected with long column UPLC-MS/MS in HEK 293T cells
| Accession # | Protein Name | Sequence | Modification |
|---|---|---|---|
| P68104 | Eongation factor 1-alpha 1 | KDGNASGTTLLEALDcILPPTRPTDKPLRLPLQDVYKa | Trioxidation |
| P04406 | Glyceraldehyde-3-phosphate dehydrogenase | IISNAScTTNCLAPLAKa,28 | Trioxidation |
| Q71D3 | Histone H3.2 | FQSSAVMALQEASEAYLVGLFEDTNLcAIHAK49 | Trioxidation |
| P30044-2 | Peroxiredoxin-5 | KGVLFGVPGAFTPGcSKa,48 | Trioxidation |
| P62937 | Peptidyl-prolyl cis-trans isomerase A | ALSTGEKGFGY KGScFHRa,34 | Trioxidation |
| P62937 | Peptidyl-prolyl cis-trans isomerase A | GFGYKGScFHRa,34 | Trioxidation |
| P30041 | Peroxiredoxin-6 | DFTPVcTTELGRa,48 | Trioxidation |
| Q99497 | Protein DJ-1, PARK7 | GLAAIcAGPTALLAHEIGFGSKa,32 | Trioxidation |
| Q99497 | Protein DJ-1, PARK7 | KGLIAAIcAGPTALLAHEIGFGSKa,32 | Trioxidation |
| P00441 | Superoxide dismutase [Cu-Zn], SOD1 | HVGDLGNVTADKDGVADVSIEDSVISLSGDHcIIGRa,30,48 | Trioxidation |
| M(Oxidation) | |||
| P07437 | Tubulin beta chain | mREIVHIQAGQcGNQIGAKa | C(Trioxidation) |
Human cysteine oxidation sites reported in RedoxDB (http://biocomputer.bio.cuhk.edu.hk/RedoxDB/)
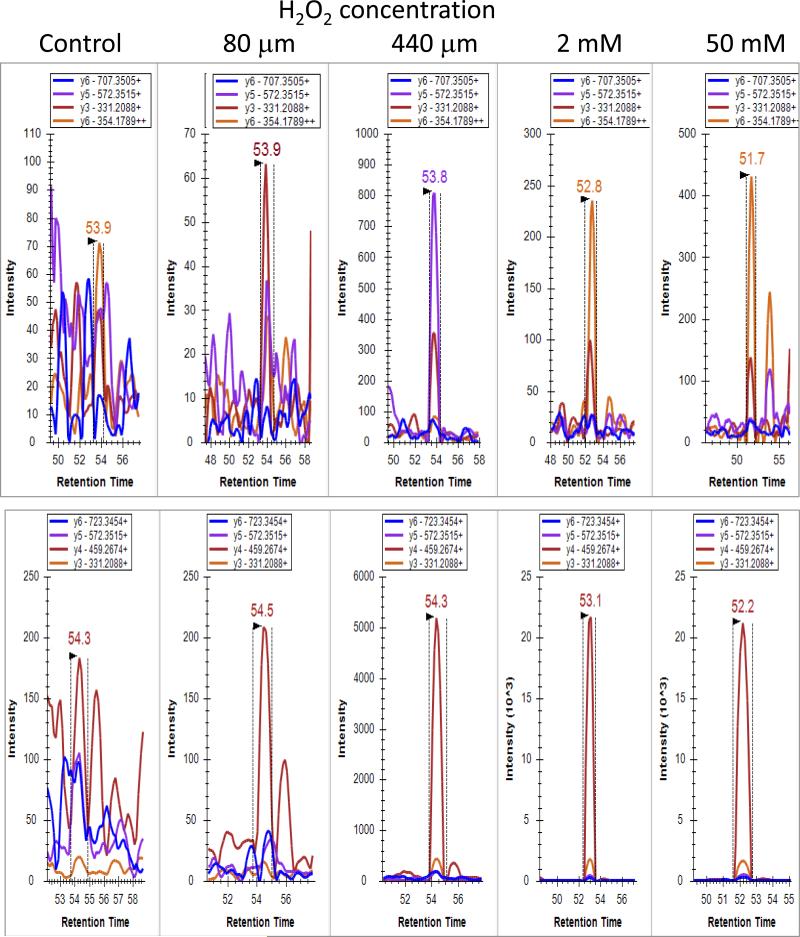
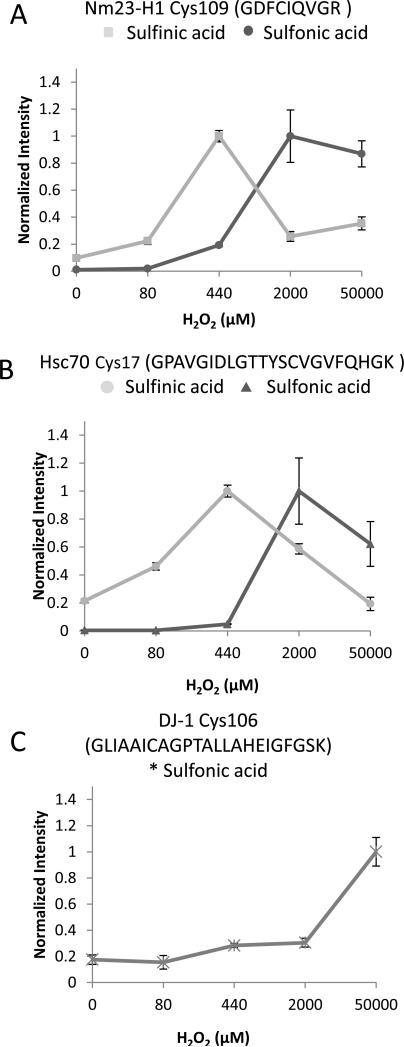
Fragment ion peak intensities as a function of retention time for Nm23-H1 Cys109 at each hydrogen peroxide dose are displayed in Figure 5 for sulfinic acid (upper panel) and sulfonic acid (lower panel). In the control, no cysteine sulfoxide was detectable above noise level counts in the UPLC-pSRM experiments, while intensity changes of 10 and 100 fold were observed for the sulfinic and sulfonic acid modifications, respectively, after treatment. Similar plots for DJ-1 Cys106 and Hsc70 Cys17 are shown in Figure S4. All 3 proteins containing modified cysteine peptides showed a dose-dependent increase in the level of sulfonic acid modification with increasing H2O2 concentration as measured by summing the peak area of y fragment ions from the pSRM transitions (Figure 6). Sulfonic acid modification on DJ-1 Cys106 showed a progressive increase starting at the 440 μM H2O2 dose and increasing up to the final 50 mM dose; whereas Nm23-H1 Cys109 and Hsc70 Cys17 showed dramatic responses and reached a plateau at 2 mM H2O2. The dose response curves of sulfinic acid modifications on both Nm23-H1 Cys109 and Hsc70 Cys17 climbed steeply at the 440 μM H2O2 dose and decreased at higher doses as the cysteine sulfinic acid converted to sulfonic acid. Consequently, the sulfonic acid modification at both sites displays a dramatic increase in the dose response between the 440 μM H2O2 dose and the 2 mM dose (Figure 6A, 6B, and Figure S4).
Figure 5.
Skyline displays of MS2 ion intensity as a function of retention time for Nm23-H1 Cys109. Fragment ion intensities are displayed at varying concentrations of H2O2 for sulfinic acid (upper panel) and sulfonic acid (lower panel). Differences in vertical axis for each panel show the change in ion intensity as a function of hydrogen peroxide concentration. The retention time indicates Cys109 sulfinic and sulfonic acid modified peptides elute at different time points. Savitzky-Golay smoothing used in display.
Figure 6.
UPLC-pSRM of irreversible oxidation of endogenous proteins by H2O2. A, Relative quantitation of the pSRM transitions of the modified peptide Nm23-H1 Cys109; B, Hsc70 Cys17; and C, DJ-1 Cys106, was using doses of H2O2 from 0 to 50 mM. The intensity of both Nm23-H1 Cys109 (■) and Hsc70 Cys17 (•) sulfinic acid reaches the highest level at 440μM and decreases at higher doses. H2O2 increases the level of sulfonic acid in all three proteins in a dose dependent manner and the intensity reaches a plateau at 2 mM in both Nm23-H1 and Hsc70. Data were corrected for internal standard intensities and normalized to the most intense value of each dose response. Graphs show means and error bars show ± standard error of the means from n = 3 separate experiments.
DISCUSSION
Protein oxidation is one of the most challenging post-translational modifications to study, mainly due to the low abundance and stoichiometry of this event, the diversity of reaction products, and the likelihood of altering native oxidation states during sample handling. However, protein oxidation is critical for many cellular processes, especially in ROS signal transduction. This study demonstrates a mass spectrometric approach for identification and relative quantitation of H2O2 induced, irreversible cysteine oxidation products. The utilization of a long column to separate different thiol oxidation products using purified recombinant protein matrix metalloproteinase-9 (MMP-9) has been demonstrated by Krishnatry et al35. We adapted and further improved this analytical strategy to create a high-resolution chromatographic separation for identifying cysteine sulfinic and sulfonic acid peptides in pull-down protein complexes, and whole cell. In order to confidently assign the modifications, we used a 2% peptide FDR and further filtered the list for protein identifications with at least 4 peptide identifications. Incorrect peptide identifications often result from “one hit wonder” protein identifications. Modified peptides, present at lower abundance than unmodified peptides, usually have higher FDR rates. Thompson et al. calculated 2% peptide FDR for modified peptides from 4 peptide/protein filtering using the ion score cutoff values that produce 2% FDR with 2 peptides/protein for searches on all peptides36. Using the stringent filter of 4 high confidence peptides per protein, we attain equal levels of filtering for modified and unmodified peptides at the level of all monitored oxidative modifications and specifically for cysteine sulfonic acid. The second concern for PTM searches is matching of peptides containing multiple modifications resulting from the expansion of the search space due to variable modifications. Our search was conducted with methionine oxidation and dioxidation along with cysteine sulfination, sulfonation, and carbamidomethylation as variable modifications. The 2% peptide FDR combined with 4 peptides/protein disproportionately removed peptides with multiple modifications, resulting in a list of high confidence modified peptides. A caveat in studying oxidant treated samples is that extensive methionine oxidation is observed and should be included in the search, while other oxidative modifications are less abundant where inclusion in the search results in more false positive matches.
Label-free Skyline MS1 and long column UPLC-pSRM were applied to quantify oxidation levels from H2O2 treated samples. Approaches such as OxICAT and OxMRM have been applied to quantify numerous oxidized cysteines15, 17,19, 37. However, ICAT suffers limited dynamic range and OxMRM relies on affinity purification or immunoprecipitation of target protein complexes. Other approaches such as O18 labeling and Isobaric Tagging for Relative Quantitation (iTRAQ) have been applied to study oxidative modifications from purified recombinant proteins38,39. However, most approaches require labor-intensive preparations and may not recapitulate the protein oxidation status under physiological conditions. To our knowledge, the long column combined with label-free-pSRM quantitation approach has not been applied previously to quantitative analysis of oxidative protein modifications from complex mixtures. In this approach, the long column is helpful in separating the unmodified, sulfinic and sulfonic acid versions of each peptide sequence. The ability to identify related peptides in a narrow retention time window increases confidence that the modified peptide identifications are correct. Additionally, pSRM, using high resolution MS1 and low resolution, targeted MS/MS ensures selective quantitation of the correctly identified peptide. An advantage of this approach is that UPLC-pSRM measures endogenous protein oxidation levels without further affinity purification and thus minimizes potential oxidation artifacts. It also prevents the potential drawback of labels such as iTRAQ which can produce unpredictable fragmentation patterns that interfere with MS data interpretation40. The long column UPLC-pSRM strategy is easily implemented utilizing the tools in Skyline for generating transitions for targeted LC-MS/MS and for quantitating data of extracted MS/MS ion chromatograms.
In the affinity purified ATM protein, 27 cysteine sulfoxidation sites were identified. Previously, we used mass spectrometry methods to identify ATM cysteine residues Cys819, Cys1045, Cys1177, Cys1396, Cys2021, Cys2801, and Cys2991 as forming disulfide bridges upon oxidation with 0.5 mM H2O2 for 30 minutes27. These sites were identified as forming sulfinic or sulfonic acid modifications in the current study. In addition, the evolutionarily conserved sites Cys1266, Cys1948, Cys2704 and the Ataxia-Telangiectasia mutation site Cys2824 were also detected as susceptible to sulfinic and/or sulfonic acid modification. Sites involved in disulfide bridge formation are able to be further oxidized into sulfinic and sulfonic acid, consistent with models of redox regulation that can move to higher oxidation states under conditions of oxidative stress. We previously determined that mutation of most of the disulfide bridge forming cysteines did not impair activation of ATM27. Only the mutation of the Cys2991 residue generated an ATM protein that is deficient in activation by oxidative stress. The observation that other cysteines are oxidized form to sulfinic and sulfonic acid is consistent with the idea that ATM is generally susceptible to oxidative stress but Cys2991 is critical to redox signaling. Current studies are exploring the role of the newly identified sites of cysteine oxidation on ATM. In addition, we also discovered novel modification sites from endogenous PRMT5 (Tables 1 and S2). Quantitation of oxidized cysteine levels was measured using Skyline MS1 filtering. The results showed both Cys2074 sulfinic and sulfonic acid of ATM increased in response to increasing H2O2 dose. Other measured modified cysteines did not show similar trends, presumably due to oxidation arising during the affinity purification process (Figure 4).
A recent review by Ratnayake et al. covering experimental approaches to stably trap reduced and oxidized cysteines noted the lack of probes available for enrichment of sulfinic and sulfonic acid modifications41. In contrast, our approach can be applied for global identification of sulfinic and sulfonic acid. The in vitro experiments identified 61 cysteine sulfinic and sulfonic acid oxidized peptides (Tables 3, S3 and S4) and 44 modified peptides found in vivo (Tables 4, S5 and S6). When comparing the results to previously published datasets obtained from recombinant proteins or protein over-expression studies, more than 30 cysteine-oxidized peptides had overlap as shown in Tables 3 and 4. Sites were identified which covered a wide range of functions and abundances, including chaperone proteins (e.g. Peptidyl-prolyl cis-trans isomerase A (PPIA), Hsc70,and 60 kDa heat shock protein), metabolic proteins (e.g. GAPDH and Nm23-H1), and signaling proteins (Receptor-type tyrosine-protein phosphatase C, and low molecular weight phosphotyrosine protein phosphatase). Among the 61 oxidized peptides identified from the in vitro global analysis, five were chosen for label-free quantitation of the hydrogen peroxide dose-dependence of the sulfinic and sulfonic acid modifications. Oxidative modifications of a multifunctional protein, DJ-1, have been characterized by mass spectrometry previously in human brains following Parkinson's disease42. Both of our in vivo and in vitro studies identified oxidation of Cys106 of DJ-1 to sulfonic acid with 440 μM H2O2, a concentration of hydrogen peroxide within the range of doses where sulfinic or sulfonic acid has been detected in previous findings. Kinumi et al. detected the presence of Cys106 sulfonic acid at 100 μM H2O2 treatment of human umbilical vein epithelial cells while in vitro studies used H2O2 concentrations ranging from 250 μM to 50 mM to detect the sulfinic acid modified Cys106 32,43,44. This range of concentrations and variation in level of oxidation may be attributed to differences in exposure time, cell type, method of detection and the difference between exposures of native protein in a cell versus purified recombinant protein. Although we were able to identify the sulfonic acid modification, there were no confident spectra that could be assigned to Cys106 sulfinic acid under the same oxidizing conditions.
Nm23-H1 plays a key role in tumor metastasis, oncogenesis, proliferation, and development. Studies have shown that the kinase activity is regulated by oxreduction of Cys109 in response to ROS formation10, 45. We identified both Cys109 sulfinic and sulfonic acid modifications of Nm23-H1 with 440 μM H2O2. A dramatic increase in Cys109 sulfinic acid modification was observed with 440 μM H2O2, but then decreased at higher mM concentrations. Interestingly, we found the level of Cys109 sulfonic acid was steadily elevated between 80-440 μM H2O2 before it sharply increased with 2 mM and reached a plateau at 50 mM (Figure 6A). Similar dynamic sulfinic to sulfonic acid conversion response trends were also observed in Hsc70 Cys17. Ten to 100 fold increases in peak areas for cysteine oxidation were observed in these proteins, showing significant effects from millimolar levels of hydrogen peroxide.
Cysteine sulfinic acid and sulfonic acid modifications are proposed to be the products of oxidative stress that tips the balance from the reversible oxidation processes used for redox sensing in cellular signaling to an irreversible state leading to senescence or cell death7. However, repair of sulfinic acid modification of peroxiredoxin has been observed in eukaryotes, suggesting apparently irreversible modifications can function as redox switches13. From the in vivo experiment, we identified sulfonic acid modifications on the active site Cys48 from peroxiredoxin (Prx) 5 and on the active site Cys47 of Prx6, both of which proceed through sulfenic acid (−SOH) intermediates46,47. Similarly, the Cys111 of SOD1 found in both in vitro and in vivo experiments, is known to be oxidatively modified and this is thought to play a role in aggregation30, 48. The active site Cys152 of GAPDH was also detected with the sulfonic acid modification. We observe oxidation of the 2 cysteine residues in Histone H3.1, Cys96 and Cys110, and Cys110 sulfonic acid modification in Histone H3.2. A recent report documents Histone H3 could function as a redox sensor through Cys110 glutathionylation which increases during cell proliferation and decreases in aged cells49. The novel sites may also contain important redox sensitive cysteine residues.
Several oxidized proteins such as SOD1, DJ-1, GAPDH, PPIA, and Elongation factor 1-alpha 1 (EEF1A) were both found in in vivo and in vitro datasets; however, only two peroxide-sensitive thiols overlap. This could due to the differences between cell lines and treatments. Undersampling also contributes to this observation. Indeed, the in vitro oxidized thiols of EEF1A and Nm23-H1 were also observed in 2DLC-MS/MS experiments from in vivo H2O2 treated sample (data not shown). We observed the oxidized forms of Prx5 and Prx6 from in vivo experiments. Prx family proteins are antioxidant enzymes which function as key regulators of the cellular redox balance. All Prx enzymes contain a conserved Cys residue that is peroxide-sensitive50. A recent study shows Prx6 may participate in different stages of skin carcinogenesis51. The targeted pSRM analysis was performed on H2O2 treated cell lysate and both oxidized cysteines of Prx5and Prx6 were below detection levels (data not shown). This finding suggests that the thiols of Prx5 and Prx6 may only highly reactive when cells expose to H2O2.
There are several considerations in experimental design unique to the study of sulfinic and sulfonic acid modifications. Since these oxidative modifications cannot be ‘locked in’, more sample handling and steps that involve exposure to air such as bead beating will induce higher levels of oxidation, confounding dose dependence measurements. The samples should be kept at low pH prior to digest to reduce cysteine disulfide bond formation. Other techniques to displace oxygen such as bubbling nitrogen through the liquid are not applicable in microliter volume samples. Thus, minimizing sample handling is the simplest practice to limit environmental oxidation. Performing dose dependence experiments ensures that artifact oxidation is contributing less effect than the hydrogen peroxide treatment. In our lysate experiments, we observed dose dependent increases in oxidation level at all sites quantitated, with sulfinic acid oxidation resulting in conversion to sulfonic acid (Figure 6). A baseline measurement for sample oxidation can be made by monitoring the ratio of oxidized peptide/all peptides. For in vitro and in vivo studies, 8% of high confidence peptides had methionine and cysteine oxidative modifications, while 35% of peptides in the pulldown ATM complex sample had oxidative modifications.
The use of a long column UPLC enabled us to identify and quantify different cysteine modifications from complex mixtures. The study using pull-down protein complexes showed 26 cysteines of ATM and 2 cysteines of PRMT5 were not covered (data not shown), due to the lack of appropriate trypsin cleavage sites. Other proteases with well-defined cleavage specifications such as Glu-C and Asp-N could be used in future studies. Alternative approaches with different complementary activation methods (UV photodissociation, electron-induced dissociation or high collision induced dissociation) could also be applied to obtain better protein coverage. While there are advantages in the simplicity of experimental design and execution of our workflow, there are several limitations. The primary drawback is the sensitivity limitations inherent in running lysate samples without enrichment for the modification of interest. We are able to detect the relatively abundant modifications and/or modified peptides on relatively abundant proteins in the lysate, but cannot access the low level modifications. The complexity of the sample also limits our ability to target proteins of interest and look for their modifications. We envision that in future experiments, the combination of two or three dimensional of sample fractionation such as strong cation exchange, size exclusion chromatography, gas phase fractionation and organelle purification would greatly enhance our ability to accurately quantitate oxidation on low level proteins.
In the current study, we developed mass spectrometric approaches to study irreversible cysteine oxidation products. MS1 quantitation is effective for samples such as the ATM protein complex because the quantitation can be performed on data dependent acquisition LC-MS/MS without necessitating further method development. Long column UPLC and high resolution MS are needed to separate co-eluting peptides by retention time and accurate mass, and the MS provides higher ion counts than MS2 quantitation methods. However, for lysate samples containing thousands of proteins, the resolving power of UPLC and high resolution MS is not enough to obtain accurate quantitation of modified peptides. In this case, the second mass filter provided pSRM method gives more reliable quantitation, at the cost of additional method development and instrument run time. The long column LC approach provides a highly sensitive, proteome-wide, unbiased screen designed to uncover redox sensitive cysteines. UPLC-pSRM delivers robust quantitation with wide dynamic range to explore endogenous regulation of redox-regulated cysteines which were undetectable by other less sensitive methods. Another advantage is that the methods described do not rely on immunoprecipitation thus avoiding the limitations of the availability and specificity of antibodies. Furthermore, the technology required for implementing long column UPLC-pSRM is readily available, as UPLC is becoming more widely adopted. The quantitation can be performed with the open source Skyline program using either MS1 filtering or fragment ions. Method development can be streamlined by using the initial data dependent LC-MS/MS runs to generate the transitions lists for targeted LC-MS/MS with quantitation of pSRM fragment ions. Long column UPLC-pSRM can be particularly useful in studying cysteine oxidation from precious clinical specimens that aid our understanding of cysteine oxidation in human health and disease.
Supplementary Material
ACKNOWLEDGEMENT
We are thankful for Marvin Mercado, Michelle Gadush, and Jacqulyn Tolson for assistance in the preparation of the manuscript. This work is supported by the Cancer Prevention Research Institute of Texas Grant RP110782 to M.P. and RP100670 to T.T.P, the NIEHS Center for Research on Environmental Disease Grant 5P30ES007784 sub-project 9010 to M.P. and NCI/NIH grant CA132813 to T.T.P.
ABBREVIATIONS
- Ox
oxidation
- pSRM
pseudo selected reaction monitoring
- UPLC
ultra-high pressure liquid chromatography
- MS
mass spectrometry
- PTM
post-translational modification
- ROS
reactive oxygen species
- ICAT
isotope-coded affinity tags
- MRM
multiple reaction monitoring
- BSA
bovine serum albumin
- ATM
ataxia telangiectasia mutated
- PRMT5
protein arginine N-methyltransferase 5
- Nm23-H1
nucleoside diphosphate kinase A
- Hsc70
heat shock 70 kDa protein 8
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
- SOD1
Superoxide dismutase [Cu-Zn]
- HMGB
High mobility group box
- EEF1A
Elongation factor 1-alpha 1
- PPIA
Peptidyl-prolyl cis-trans isomerase A
- Prx
Peroxiredoxin
- FT
Fourier-Transform
- TCA
trichloroacetic acid
REFERENCES
- 1.Tonks NK. Redox redux: Revisiting PTPs and the control of cell signaling. Cell. 2005;121:667–670. doi: 10.1016/j.cell.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 2.Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, Wahl GM. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol Cell. 2002;9:1031–1044. doi: 10.1016/s1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- 3.Wellen KE, Thompson CB. Cellular metabolic stress: considering how cells respond to nutrient excess. Mol. Cell. 2011;40:323–332. doi: 10.1016/j.molcel.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szatrowski TP, Nathan CF. Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res. 1991;51:794–798. [PubMed] [Google Scholar]
- 5.Bielski BH, Shiue GG. Reaction rates of superoxide radicals with the essential amino acids. Ciba Found Symp. 1978;65:43–56. doi: 10.1002/9780470715413.ch4. [DOI] [PubMed] [Google Scholar]
- 6.Di Simplicio P, Franconi F, Frosali S, Di Giuseppe D. Thiolation and nitrosation o1f cysteines in biological fluids and cells. Amino Acids. 2003;25:3–4. 323–39. doi: 10.1007/s00726-003-0020-1. [DOI] [PubMed] [Google Scholar]
- 7.Held JM, Gibson BW. Regulatory control or oxidative damage? Proteomic approaches to interrogate the role of cysteine oxidation status in biological processes. Mol. Cell. Proteomics. 2012;11:1–14. doi: 10.1074/mcp.R111.013037. R111.013037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woo HA, Chae HZ, Hwang SC, Yang KS, Kang SW, Kim K, Rhee SG. Reversing the inactivation of peroxiredoxins caused by cysteine sulfinic acid formation. Science. 2003;300:653–656. doi: 10.1126/science.1080273. [DOI] [PubMed] [Google Scholar]
- 9.Lindahl M, Mata-Cabana A, Kieselbach T. The disulfide proteome and other reactive cysteine proteomes: analysis and functional significance. Antioxid. Redox. Signal. 2011;14:2581–2642. doi: 10.1089/ars.2010.3551. [DOI] [PubMed] [Google Scholar]
- 10.Lee E, Jeong J, Kim SE, Song EJ, Kang SW, Lee KJ. Multiple functions of Nm23-H1 are regulated by oxido-reduction system. PLoS ONE. 2009;4:e7949. doi: 10.1371/journal.pone.0007949. doi:10.1371/journal.pone.0007949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leonard SE, Carroll KS. Chemical ‘omics’ approaches for understanding protein cysteine oxidation in biology. Curr. Opin. Chem. Biol. 2011;15:88–102. doi: 10.1016/j.cbpa.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 12.Seo Y,H, Carroll KS. Profiling protein thiol oxidation in tumor cells using sulfenic acid-specific antibodies. Proc. Natl. Acad. Sci. U S A. 2009;106:16163–16168. doi: 10.1073/pnas.0903015106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paulsen CE, Carroll KS. Orchestrating redox signaling networks through regulatory cysteine switches. ACS Chemical Biology. 2009;5:47–62. doi: 10.1021/cb900258z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 2007;389:1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- 15.Sethuraman M, McComb ME, Heibeck T, Costello CE, Cohen RA. Isotope-coded affinity tag approach to identify and quantify oxidant-sensitive protein thiols. Mol. Cell. Proteomics. 2004;3:273–278. doi: 10.1074/mcp.T300011-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci. STKE. 2001:l1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 17.Sethuraman M, Clavreul N, Huang H, McComb ME, Costello CE, Cohen RA. Quantification of oxidative posttranslational modifications of cysteine thiols of p21ras associated with redox modulation of activity using isotope-coded affinity tags and mass spectrometry. Free Radic. Biol. Med. 2007;42:823–829. doi: 10.1016/j.freeradbiomed.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumsta C, Thamsen M, Jakob U. Effects of oxidative stress on behavior, physiology, and the redox thiol proteome of Caenorhabditis elegans. Antioxid. Redox. Signal. 2011;14:1023–1037. doi: 10.1089/ars.2010.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Held JM, Danielson SR, Behring JB, Atsriku C, Britton DJ, Puckett RL, Schilling B, Campisi J, Benz CC, Gibson BW. Targeted quantitation of site-specific cysteine oxidation in endogenous proteins using a differential alkylation and multiple reaction monitoring mass spectrometry approach. Mol. Cell. Proteomics. 2010;9:1400–1410. doi: 10.1074/mcp.M900643-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen R,E, Roth D, Winther JR. Quantifying the global cellular thiol-disulfide status. Proc. Nat.l Acad. Sci. USA. 2009;106:422–427. doi: 10.1073/pnas.0812149106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schilling B, Rardin MJ, Maclean BX, Zawadzka AM, Frewen BE, Cusack MP, Sorensen DJ, Bereman MS, Jing E, Wu CC, Verdin E, Kahn CR, MacCoss MJ, Gibson BW. Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in Skyline: application to protein acetylation and phosphorylation. Mol. Cell. Proteomics. 2012;11:202–214. doi: 10.1074/mcp.M112.017707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hewel JA, Phanse S, Liu J, Bousette N, Gramolini A, Emili A. Targeted protein identification, quantification and reporting for high-resolution nanoflow targeted peptide monitoring. Journal of Proteomics. 2012 doi: 10.1016/j.jprot.2012.10.020. 10.1016/j.jprot.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 24.Sherrod SD, Myers MV, Li M, Myers JS, Carpenter KL, MacLean B, MacCoss MJ, Liebler DC, Ham AL. Label-free quantitation of protein modifications by pseudo selected reaction monitoring with internal reference peptides. Journal of Proteome Research. 2012;11:3467–3479. doi: 10.1021/pr201240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JH, Paull TT. Purification and biochemical characterization of ataxia-telangiectasia mutated and Mre11/Rad50/Nbs1. Methods Enzymol. 2006;408:529–39. doi: 10.1016/S0076-6879(06)08033-5. [DOI] [PubMed] [Google Scholar]
- 26.Ditch S, Paull TT. The ATM protein kinase and cellular redox signaling: beyond the DNA damage response. Trends Biochem. Sci. 2012;37:15–22. doi: 10.1016/j.tibs.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science. 2010;330:517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 28.Hoffman M, Walsh GM, Rogalski JC, Kast J. Identification of Nitroxyl-induced modifications in human platelet proteins using a novel mass spectrometric detection method. Mol. Cell. Proteomics. 2009;8:887–903. doi: 10.1074/mcp.M800230-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Men L, Wang Y. Further studies on the fragmentation of protonated ions of peptides containing aspartic acid, glutamic acid, cysteine sulfinic acid, and cysteine sulfonic acid. Rapid Commun. Mass Spectrom. 2005;19:23–30. doi: 10.1002/rcm.1748. [DOI] [PubMed] [Google Scholar]
- 30.Fujiwara N, Nakano M, Kato S, Yoshihara D, Ookawara T, Eguchi H, Taniguchi N, Suzuki K. Oxidative modification to cysteine sulfonic acid of Cys111 in human Copper-Zinc superoxide dismutase. J. Biol. Chem. 2007;282:35933–35944. doi: 10.1074/jbc.M702941200. [DOI] [PubMed] [Google Scholar]
- 31.Yang H, Lundbäck P, Ottosson L, Erlandsson-Harris H, Venereau E, Bianchi ME, Al-Abed Y, Andersson U, Tracey KJ, Antoine DJ. Redox modification of cysteine residues regulate the cytokine activity high motility group box-1(HMGB1) Molecular Medicine. 2012;30:250. doi: 10.2119/molmed.2011.00389. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Kinumi T, Kimata J, Taira T, Ariga H, Niki E. Cysteine-106 of DJ-1 is the most sensitive cysteine residue to hydrogen peroxide-mediated oxidation in vivo in human umbilical vein endothelial cells. Biochem. Biophys. Res. Commun. 2004;317:722–728. doi: 10.1016/j.bbrc.2004.03.110. [DOI] [PubMed] [Google Scholar]
- 33.Jeong J, Jung Y, Na S, Jeong J, Lee E, Kim MS, Choi S, Shin DH, Paek E, Lee H,Y, Lee KJ. Novel oxidative modifications in redox-active cysteine residues. Mol Cell Proteomics. 2011;10:1–13. doi: 10.1074/mcp.M110.000513. M110.000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gevaert K, Ghesquière B, Staes A, Martens L, Van Damme J, Thomas GR, Vandekerckhove J. Reversible labeling of cysteine-containing peptides allows their specific chromatographic isolation for non-gel proteome studies. Proteomics. 2004;4:897–908. doi: 10.1002/pmic.200300641. [DOI] [PubMed] [Google Scholar]
- 35.Krishnatry AS, Kamei T, Wang H, Qu J, Fung HL. Identification of nitroglycerin-induced cysteine modifications of pro-matrix metalloproteinase-9. Rapid Commun. Mass Spectrom. 2011;30:2291–2298. doi: 10.1002/rcm.5118. [DOI] [PubMed] [Google Scholar]
- 36.Thompson MR, Thompson DK, Hettich RL. Systematic assessment of the benefits and caveats in mining microbial post-translational modifications from shotgun proteomic data: the response of Shewanella oneidensis to chromate exposure. J. Proteome Res. 2008;7:648–658. doi: 10.1021/pr070531n. [DOI] [PubMed] [Google Scholar]
- 37.Brandes N, Tienson H, Lindemann A, Vitvitsky V, Reichmann D, Banerjee R, Jakob U. A sudden fall in the levels of the coenzyme NADPH within cells could have a central role in the process of aging. eLife. 2013:e00306. doi: 10.7554/eLife.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madian AG, Hindupur J, Hulleman JD, Diaz-Maldonado N, Mishra VR, Guigard E, Kay CM, Rochet J, Regnier FE. Effect of single amino acid substitution on oxidative modifications of the Parkinson's disease-related protein, DJ 1. Mol. Cell. Proteomics. 2012;11:1–15. doi: 10.1074/mcp.M111.010892. M111.010892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pimenova T, Pereira CP, Gehrig P, Buehler PW, Schaer DJ, Zenobi R. Quantitative mass spectrometry defines an oxidative hotspot in hemoglobin that is specifically protected by haptoglobin. J. Proteome Res. 2010;9:4061–4070. doi: 10.1021/pr100252e. [DOI] [PubMed] [Google Scholar]
- 40.Aggarwal K, Choe LH, Lee KH. Shotgun proteomics using the iTRAQ isobaric tags. Brief Funct. Genomic Proteomic. 2006;5:112–120. doi: 10.1093/bfgp/ell018. [DOI] [PubMed] [Google Scholar]
- 41.Ratnayake S, Dias IH, Lattman E, Griffiths HR. Stabilising cysteinyl thiol oxidation and nitrosation for proteomic analysis. J. Proteomics. 2013 doi: 10.1016/j.jprot.2013.06.019. http://dx.doi.org/10.1016/j.jprot.2013.06.019. [DOI] [PubMed]
- 42.Choi J, Sullards MC, Olzmann JA, Rees HD, Weintraub ST, Bostwick DE, Gearing M, Levey AI, Chin LS, Li L. Oxidative damage of DJ-1 is linked to sporadic Parkinson and Alzheimer diseases. J. Biol. Chem. 2006;281:10816–10824. doi: 10.1074/jbc.M509079200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andres-Mateos E, Perier C, Zhang L, Blanchard-Fillion B, Greco TM, Thomas B, Ko HS, Sasaki M, Ischiropoulos H, Przedborski S, Dawson TM, Dawson VL. DJ-1 gene deletion reveals that DJ-1 is an atypical peroxiredoxin-like peroxidase. Proc. Natl. Acad. Sci. U.S.A. 2007;104:14807–14812. doi: 10.1073/pnas.0703219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou W, Zhu M, Wilson MA, Petsko GA, Fink AL. The oxidation state of DJ 1 regulates its chaperone activity toward alpha-synuclein. J. Mol. Biol. 2006;356:1036–1048. doi: 10.1016/j.jmb.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 45.Song EJ, Kim YS, Chung JY, Kim E, Chae SK. Oxidative modification of nucleoside diphosphate kinase and its identification by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Biochemistry. 2000;39:10090–10097. doi: 10.1021/bi000267a. [DOI] [PubMed] [Google Scholar]
- 46.Knoops B, Goemaere J, Van der Eecken V, Declercq JP. Peroxiredoxin 5: structure, mechanism, and function of the mammalian atypical 2-Cys peroxiredoxin. Antioxid. Redox Signal. 2011;15:817–829. doi: 10.1089/ars.2010.3584. [DOI] [PubMed] [Google Scholar]
- 47.Fisher AB. Peroxiredoxin 6: a bifunctional enzyme with glutathione peroxidase and phospholipase A2 activities. Antioxid. Redox Signal. 2011;15:831–844. doi: 10.1089/ars.2010.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilcox KC, Zhou L, Jordon JK, Huang Y, Yu Y, Redler RL, Chen X, Caplow M, Dokholyan NV. Modifications of superoxide dismutase (SOD1) in human erythrocytes: a possible role in amyotrophic lateral sclerosis. J. Biol. Chem. 2009;284:13940–13947. doi: 10.1074/jbc.M809687200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.García-Giménez JL, Olaso G, Hake SB, Bönisch C, Wiedemann SM, Markovic J, Dasí F, Gimeno A, Pérez-Quilis C, Palacio,s O, Capdevila M, Viña J, Pallardó FV. Histone H3 glutathionylation in proliferating mammalian cells destabilizes nucleosomal structure. Antioxid. Redox Signal. 2013 May 21; doi: 10.1089/ars.2012.5021. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rhee SG, Chae HZ, Kim K. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic. Biol. Med. 2005;38:1543–52. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 51.Rolfs F, Huber Marcel., Gruber F, Böhm F, Pfister HJ, Bochkov VN, Tschachler E, Dummer R, Hohl D, Schäfer M, Werner S. Dual Role of the antioxidant enzyme Peroxiredoxin 6 in skin carcinogenesis. Cancer Res. 2013;73:3460–9. doi: 10.1158/0008-5472.CAN-12-4369. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.