Abstract
MicroRNAs (miRNAs) are small non-coding RNAs, which regulate gene expression. Single nucleotide polymorphisms (SNPs) may occur in miRNA biogenesis pathway genes, primary miRNA, pre-miRNA or a mature miRNA sequence. Such polymorphisms may be functional with respect to biogenesis and actions of mature miRNA. Specific SNPs were identified in predicted miRNA target sites within 3′ untranslated regions of mRNAs. These SNPs have a potential to affect the efficiency of miRNA binding to the target sites or can create or disrupt binding sites. Resulting gene dysregulation may involve changes in phenotype and may eventually prove critical for the susceptibility to cancer and its onset as well as for estimates of prognosis and therapy response. In this review, we provide a comprehensive list of potentially functional miRNA-related SNPs and summarize their importance as candidate cancer biomarkers.
Keywords: MicroRNA, polymorphism, SNP, cancer, risk factor, diagnosis, prognosis, prediction
Introduction
miRNAs are short non-coding RNAs, 18–25 nucleotides in length, which regulate gene expression. In the view of their complementarity with 3′ untranslated region (3′-UTR) of target mRNA, functional effects of miRNAs are provided by two mechanisms: by post-transcriptional regulation of gene expression leading to target mRNA degradation or repression of its translation with consequent decrease of particular protein levels or on the contrary by up-regulation of their targets. Bioinformatic approach was used previously for prediction of capacity of miRNAs to regulate approximately half of the mammalian genes, with significant number of important oncogenes and tumour suppressor genes involved [1, 2]. miRNAs have been studied most intensively in the field of oncological research, and emerging evidence suggests that altered miRNA regulation is involved in the pathogenesis of cancer [3–6]. Changes in the expression of miRNAs have been observed in a variety of human solid cancers [7, 8]: breast [9, 10], colorectal [11, 12], lung [13, 14], kidney [15, 16], prostate [17], cervical [18], gastric [19], bladder [20], pancreatic [21], oesophageal [22], head and neck [23], thyroid [24] and ovarian cancer [25], hepatocellular carcinoma [26] and glioma [27, 28]. In addition, approximately 50% of all annotated human miRNA genes are located in fragile sites or areas of the genome that are frequently associated with cancer [29]. Up-regulation of mature miRNA may occur as a consequence of transcriptional activation or amplification of the miRNA encoding gene, whereas silencing or reduced expression may result from deletion of a particular chromosomal region, epigenetic silencing or defects in their biogenesis.
From the mechanistic point of view, miRNAs represent ideal candidates for cancer predisposition loci: limited variation in quantity may affect thousands of target mRNAs and can result in diverse functional consequences [30]. The initial demonstration that miRNA-related SNPs can affect phenotype was elegantly provided by Abelson et al. who found that a mutation in the miR-189–binding site of SLITRK1 was associated with Tourette's syndrome [31]. The first evidence that point mutations in miRNA genes may affect function and result in cancer susceptibility comes from a pioneering study conducted by Carlo Croce's group in which a germline mutation in pri-mir-16-1 resulted in low levels of miR-16-1 expression in familial chronic lymphocytic leukaemia [32, 33]. Since then, several studies have used systematic sequencing or in silico approaches to identify SNPs in miRNA-related genes, catalogues of which have been created and published [34–36]. Taken together, these facts provide sufficient theoretical basis for follow-up case-control studies to determine the association between such genetic markers and cancer risk. This study provides brief outlook on miRNA biogenesis, biology and the functional effects of miRNA-related SNPs, and—to the best of our knowledge—a complete summary of case-control studies performed in the field of solid cancer and miRNA SNPs.
Classification of miRNA-related SNPs
SNPs have been widely implicated in cancer development [37, 38]. However, polymorphisms in the miRNA regulatory pathway are a novel class of functional polymorphisms present in the human genome. In principle, SNPs in miRNA genes are thought to exert their effects by one of three mechanisms: first, by means of transcription of the primary transcript (pri-miRNA SNPs and SNPs in miRNA biogenesis genes); secondly, by means of primary miRNAs (pri-miRNA) and pre-miRNA processing (pri-, pre-miRNA SNPs and SNPs in miRNA biogenesis genes) and thirdly, by means of miRNA–mRNA interactions (SNPs in mature miRNA sequences and miRNA-binding sites) [39, 40]. SNPs summarized in Table 1 are arranged according to this classification.
Table 1.
List of miRNA SNPs evaluated in solid cancer epidemiologic studies
| Polymorphism | SNP ID | miRNA/gene | Allele | Description | Size of cohort cases/controls | Population | Risk (95% CI) | Reference |
|---|---|---|---|---|---|---|---|---|
| Breast cancer | ||||||||
| Pre-miRNA | rs11614913 | miR-196-a2 | T/C | Increased risk | 1009/1093 | Chinese | 1.23 (1.02–1.48) | 45 |
| C/T | Decreased risk | 475/502 | United States | 0.44 (0.28–0.70) | 46 | |||
| Lack of association | 1894/2760 | Italian, German | NS | 47 | ||||
| rs3746444 | miR-499 | A/G | Lack of association | 1894/2760 | Italian, German | NS | 47 | |
| Increased risk | 1009/1093 | Chinese | 1.25 (1.02–1.51) | 45 | ||||
| rs2910164 | miR-146a | G/C | Earlier age of onset | 42 case-only | United States | Not given | 48 | |
| Lack of association | 1894/2760 | Italian, German | NS | 47 | ||||
| Lack of association | 1894/2760 | Italian, German | NS | 47 | ||||
| rs895819 | miR-27a | A/G | Decreased risk | 1217/1422 | German | 0.88 (0.78–0.99) | 49 | |
| C/T | Decreased risk | 363BRCA1/125 BRCA2 | Jewish | 1.96 (1.16–3.33) for TC versus TT | 50 | |||
| rs6505162 | miR-423 | A/C | Increased risk | 363BRCA1/125 BRCA2 | Jewish | 2.77 (1.11–6.9) | 50 | |
| miRNA-binding site | rs743554 | ITGB4 (miR-34a) | G/A | Reduced survival | 749/1493 | Swedish | 2.11 (1.21–3.68) | 51 |
| rs2747648 | ESR1 (miR-453) | C/T | Increased risk | 1223/1495 | German | 0.60 (0.41–0.89) for C versus T allele | 52 | |
| rs16917496 | SET8 (miR-502) | T/C | Earlier age of onset | 1110/1097 | Chinese | 1.66 (1.06–2.61) for CC versus TT | 53 | |
| rs1434536 | BMPR1B (miR-125b) | C/T | Increased risk | 459+1145/1142 | US/ethnicity data not available | 1.94 (1.40–2.71) for TT versus CC | 54 | |
| Colorectal cancer | ||||||||
| miRNA biogenesis | frameshift mut. | Ago2 | Associated with MSI-H | 100 case-only | Korean | Not given | 55 | |
| frameshift mut. | TNRC6A | Associated with MSI-H | 100 case-only | Korean | Not given | 55 | ||
| Pre-miRNA | rs2289030 | miR-492 | C/G | Reduced survival | 426 case-only | Korean | 1.19 (0.73–1.93) | 56 |
| miRNA-binding site | rs17281995 | CD86 (5 miRNAs) | G/C | Increased risk | 697/624 | Central-European | 2.93 (1.29–6.67) | 57 |
| rs1051690 | INSR (miR-612) | G/A | Increased risk | 697/624 | Central- | 1.86 (0.99–3.50) | 57 | |
| rs61764370 | KRAS (let-7) | T/G | EGFR therapy response | 130 case-only | European United States | Not given | 58 | |
| Lung cancer (NSCLC) | ||||||||
| Pre-miRNA | rs11614913 | mir-196-a2 | T/C | Increased risk | 1058/1035 | Chinese | 1.25 (1.01–1.54) | 59 |
| Reduced survival | 663 case-only | Chinese | 1.29 (1.01–1.65) | 60 | ||||
| miRNA-binding site | rs61764370 | KRAS (let-7) | T/G | Increased risk | 2433 -pooled | Worldwide | 2.39 (1.1–4.6) | 61 |
| Lack of association | 461 case-only | United States | NS | 62 | ||||
| Prostate cancer | ||||||||
| Pre-miRNA | rs2910164 | miR-146a | G/C | Decreased risk | 251/280 | Chinese | 0.65 (0.43–0.99) | 63 |
| Renal cell carcinoma | ||||||||
| miRNA biogenesis | rs2740348 | GEMIN4 | G/C | Decreased risk | 276/278 | US Caucasians | 0.67 (0.47–0.96) | 64 |
| rs7813 | GEMIN4 | T/C | Decreased risk | 277/278 | US Caucasians | 0.68 (0.47–0.96) | 64 | |
| rs197412 | GEMIN3 | T/C | Increased risk | 277/278 | US Caucasians | 1.31 (0.93–1.85) | 64 | |
| rs11077 | XPO5 | A/C | Increased risk | 276/277 | US Caucasians | 1.55 (0.98–2.44) | 64 | |
| Cervical cancer | ||||||||
| Pri-miRNA | rs11134527 | miR-218 | A/G | Decreased risk | 703/713 | Chinese | 0.72 (0.52–0.99) | 66 |
| miRNA-binding site | rs2566 | LAMB3 | C/T | Increased risk | 703/713 | Chinese | 1.57 (1.25–1.96) | 66 |
| Ovarian cancer | ||||||||
| Pre-miRNA | rs2910164 | miR-146a | G/C | Earlier age of onset | 82 case-only | United States | Not given | 48 |
| rs6505162 | miR-423 | A/C | Increased risk | 363BRCA1/125BRCA2 | Jewish | 2.77 (1.11–6.9) | 50 | |
| rs2740351 | GEMIN4 | G/C | Decreased risk | 339/349 | United States | 0.71 (0.57–0.87) | 67 | |
| rs7813 | GEMIN4 | T/C | Decreased risk | 339/349 | United States | 0.71 (0.57–0.88) | 67 | |
| miRNA biogenesis | rs12194974 | LIN28B | G/A | Decreased risk | 1815/1900 | United States | 0.90 (0.82–0.98) | 68 |
| Gastric cancer | ||||||||
| Pre-miRNA | rs895819 | miR-27a | A/G | Increased risk | 340/304 | Chinese | 1.48 (1.06–2.05) | 69 |
| rs11614913 | miR-196-a2 | T/C | Increased risk | 213/213 | Chinese | 1.57 (1.03–2.39) | 70 | |
| Bladder cancer | ||||||||
| miRNA biogenesis | rs7813 | GEMIN4 | T/C | Decreased risk | 736/736 | United States | 0.78 (0.60–1.02) | 71 |
| rs197414 | GEMIN3 | A/C | Increased risk | 740/742 | United States | 2.50 (1.08–5.78) | 71 | |
| rs784567 | TRBP | C/T | Decreased risk | 737/729 | United States | 0.80 (0.62–1.02) | 71 | |
| Pri-miRNA | rs7372209 | miR-26a-1 | C/T | Decreased risk | 728/728 | United States | 0.68 (0.45–1.04) | 71 |
| rs531564 | miR-124–1 | C/G | Increased risk | 739/740 | United States | 2.44 (0.96–6.20) | 71 | |
| Pre-miRNA | rs2289030 | miR-492 | C/G | Increased risk | 739/734 | United States | 1.35 (0.98–1.87) | 71 |
| rs6505162 | miR-423 | A/C | Increased risk | 723/733 | United States | 1.25 (0.96–1.61) | 71 | |
| Oesophageal cancer | ||||||||
| miRNA biogenesis | rs11077 | XPO5 | A/C | Increased risk | 346/346 | United States | 1.58(1.03–2.45) | 72 |
| rs14035 | RAN | C/T | Increased risk | 346/346 | United States | 1.99(1.17–3.38) | 72 | |
| Pri-miRNA | rs7372209 | miR-26a-1 | C/T | Increased risk | 346/346 | United States | 1.35(1.04–1.76) | 72 |
| rs213210 | miR-219–1 | T/C | Increased risk | 346/346 | United States | 1.75(1.10–2.80) | 72 | |
| Pre-miRNA | rs11614913 | miR-196-a2 | T/C | Increased risk | 346/346 | United States | 1.73(1.16–2.56) | 72 |
| rs2910164 | miR-146a | G/C | Increased risk | 444/468 | Chinese | 2.39 (1.36–4.20) | 73 | |
| rs6505162 | miR-423 | A/C | Increased risk | 346/346 | United States | 0.64(0.51–0.80) | 72 | |
| Hepatocellular carcinoma | ||||||||
| Pre-miRNA | rs2910164 | miR-146a | G/C | Increased risk | 479/504 | Chinese | 2.02 (1.06–3.85) for GG versus CC | 74 |
| rs11614913 | miR-196-a2 | T/C | Increased risk | 560/391 | Chinese | 1.18 (0.73–1.93) for CC versus TT | 75 | |
| miRNA-binding site | rs3783553 | IL1A (miR-122, miR-378) | Del/Ins | Increased risk | 1477/1673 | Chinese | 0.62 (0.49–0.78) for ++ versus (+− and –) | 76 |
| Head and neck cancer | ||||||||
| Pre-miRNA | rs11614913 | miR-196-a2 | T/C | Increased risk | 1039 | United States | 7.4 (1.9–28.2) | 77 |
| Thyroid cancer | ||||||||
| Pre-miRNA | rs2910164 | miR-146a | G/C | Decreased risk | 206/274 | Finnish | 0.42 (0.24–0.73) for CC against GC_GG | 78 |
| Glioma | ||||||||
| Pre-miRNA | rs11614913 | miR-196-a2 | T/C | Decreased risk | 670/680 | Chinese | 0.74 (0.56–0.98) | 80 |
SNPs in miRNA processing machinery
miRNA genes are primarily transcribed by RNA polymerase II/III into pri-miRNAs that have several hundred nucleotides. The processing of pri-miRNA by the nuclear RNase DROSHA within the microprocessor complex, which also includes DGCR8/Pasha, produces the 70- to 100-nt pre-miRNAs. The pre-miRNAs are then exported into the cytoplasm by the Exportin-5 (XPO5)/Ran-GTP complex [41]. The pre-miRNA is further cleaved in the cytoplasm by an RNase III endonuclease, Dicer, to release two complementary short RNA molecules. The argonaut proteins (Ago1–4) form complex together with GEMIN3 and GEMIN4 and selectively bind to the guide strand and facilitates the formation of a miRNA–RNA-induced silencing complex (RISC) [41]. Upon miRNA binding, the RISC complex is activated—through a mechanism which remains to be elucidated—and it recognizes the binding site in 3′-UTR of the target mRNA and contributes to regulation of the gene expression (Fig. 1).
Fig 1.
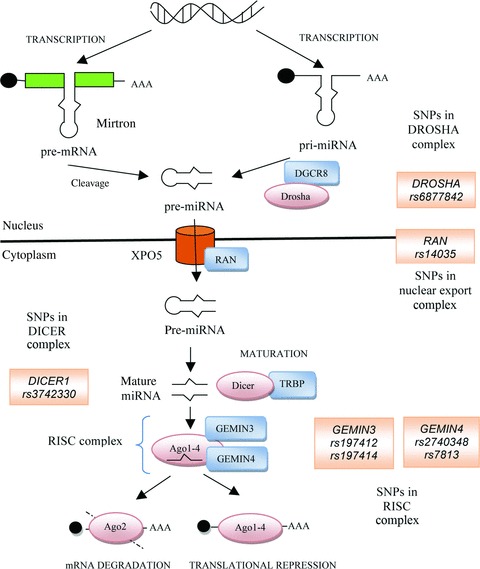
Biogenesis and SNPs in miRNA processing machinery.
Polymorphisms which affect expression of proteins in miRNA biogenesis pathway may affect miRNA-mediated regulation within the cell. SNPs in core components of the silencing machinery may affect overall silencing efficiency. In view of their predictably highly deleterious consequences, mutations which drastically perturb RNA silencing are naturally rare. Nevertheless, SNPs with subtle effects on gene function may occur. As distinct targets may be more or less sensitive to variations in miRNA concentration or in silencing efficiency, such SNPs may affect certain pathways to a greater degree than the others. Thus, SNPs affecting the silencing machinery may contribute to the genetic variation observed in specific phenotypes [39]. Recently, a few reports focused on the SNPs in silencing machinery and their association with cancer. Many SNPs in these genes, including GEMIN4, GEMIN3, XPO5, AGO1, AGO2, TRBP and RAN play a role in cancer risk [39, 40].
SNPs in pri-, pre-, mat-miRNAs
In general, sequence variations in miRNA genes, including pri-miRNAs, pre-miRNAs and mature miRNAs, have the potential of influencing the processing and/or target selection of miRNAs. As a consequence of SNPs present in pri-, pre- and mature-miRNA, aberrant expression of hundreds of genes and pathways, greatly affecting miRNA function, may occur [42]. There are three options of interfering with miRNA function on the basis of SNPs in miRNA sequences: (1) mutations in the pri- or pre-miRNA may affect their stability or processing efficiency; (2) mutations acting in cis or trans on the pri-miRNA promoter may influence the transcription rate and (3) a sequence of the mat-miRNA might be altered, thereby stabilizing or destabilizing its interaction with mRNA targets; SNPs in mat-miRNA sequences may be further sub-classified into the following two sub-categories: (i) SNPs within the miRNA 5′-seed region, from positions 2—to 7, which are responsible for specificity of target recognition and (ii) SNPs within miRNA 3′-mismatch tolerant region that is capable of tolerating mismatches to a certain extent.
In a study focusing on the occurrence of SNPs within pre-miRNAs sequences, Saunders et al. identified 65 SNPs in 474 pre-miRNAs using publicly available SNP database (dbSNP). Pre-miRNAs were found with the SNP density of 1.3 SNPs per kb. As the regions flanking miRNAs are the most frequent intergenic regions, most likely with either weak or no functional constraint whatsoever, these regions show a higher SNP density of three SNPs per kb. Consequently, a low level of variation in functional regions of miRNAs was subsequently established [35].
SNPs in miRNA-binding sites
In contrast to the SNPs in pri-, pre- and mat-miRNA, sequence variations within the 3′-UTR of a target (coding) gene are more abundant in the human genome and express a more defined and limited range of effects. SNPs within miRNA target sites influence only their encoded target-mRNAs and their downstream effectors, and therefore are more specific. A recent genome-wide association study has suggested that a gene with more than two miRNA target sites will have increased expression variability compared to a gene which is not regulated by a miRNA. The variability is further increased by SNPs in the miRNA target sites [43].
There are two possible mechanistic consequences of presence of SNPs within miRNA-binding region [32]: (1) SNPs may affect functional target sites, thereby destabilizing or stabilizing the interaction with the miRNA; (2) mutations may create illegitimate miRNA target sites (in the 3′-UTR or even in other segments of the transcript) which will be particularly relevant if occurring in anti-targets (Fig. 2).
Fig 2.

SNPs in miRNA-binding sites.
MiRNA-related SNPs and solid cancer
Both the central role of miRNAs as regulators of translation and their functional significance in cancer pathogenesis have been established some time ago, whereas the relationships between genetic variation in genes of the miRNA regulatory pathway and cancer risk have only begun to be explored recently [39]. As of today, over 30 epidemiologic studies were carried out that focus on the importance of miRNA-related SNPs in cancer susceptibility and therapeutic and clinical outcome in patients with a wide range of solid cancers (summarized in Table 1).
Breast cancer
Breast cancer is the leading cause of cancer-related death in women and the second most common cancer in the world after lung cancer. As up to 10% of the total number of women diagnosed with breast cancer report a family history, miRNAs [9, 10] could represent a promising new class of potentially susceptibility genes.
The most frequently studied SNP is rs11614913 in the pre-miRNA region of miR-196-a2 which was first identified by Hu et al. (2008) in a case-control study of 1009 breast cancer cases and 1093 cancer-free controls in a population of Chinese women. Although most miRNAs repress target translation, hsa-mir-196 was found recently to direct mRNA cleavage of its target, Hoxb8 [44]. The hsa-mir-196a2 rs11614913:T>C variant genotype is associated with significantly increased risk of breast cancer [45]. These observations were further validated by Hoffman et al. (2010) who performed a screening of genetic variants in 15 miRNA genes in a study of 441 cases and 479 controls and detected that a common sequence variant in hsa-miR-196a-2 (rs11614913:C>T) is significantly associated with decreased breast cancer risk. Moreover, by delivering expression vectors containing either wild-type or mutant precursors of miR-196a-2 into breast cancer cells, they have showed that this variant leads to the less efficient processing of the miRNA precursor to its mature form as well as to its diminished capacity to regulate target genes [46]. On the contrary, the study conducted by Catucci et al. (2010) on 1894 German and Italian familial breast cancer cases suggested a lack of association between SNPs rs11614913 and both breast cancer risk and age at breast cancer onset [47].
A G to C polymorphism (rs2910164) located within the sequence of a miR-146a precursor, which leads to a change from a G:U pair to a C:U mismatch in its stem region was primarily studied by Shen et al. (2008) [48]. The predicted miR-146a target genes include BRCA1 and BRCA2, that is the key breast and ovarian cancer susceptibility genes. To examine whether rs2910164 plays any role in breast and ovarian cancer, the associations between this polymorphism and the age at diagnosis were studied in 42 patients with familial breast cancer and 82 patients with familial ovarian cancer. Breast cancer patients who had at least one miR-146a variant allele were diagnosed at an earlier age than women with no variant alleles (median age 45 versus 56 years, P = 0.029) and ovarian cancer patients who had at least one miR-146a variant allele were diagnosed at younger age than women with no variant allele (median age 45 versus 50 years, P = 0.014) [48]. Unfortunately, the association of rs2910164 and breast cancer was not confirmed in further larger studies by Hu et al. (2008) and Catucci et al. (2010).
An SNP in the pre-miRNA region of hsa-mir-499 (rs3746444:A>G) was detected in a cell line of invasive breast cancer. Therefore, it is biologically plausible that these polymorphisms of hsa-mir-499 may play a role in the development of breast cancer. Hu et al. (2008) reported that the hsa-mir-499 variant genotype is associated with significantly increased risk of breast cancer; on the contrary, Catucci et al. (2010) [47] reports lack of association among their total of 1894 breast cancer cases who were negative for disease-causing mutations or unclassified variants in BRCA1 and BRCA2, and 2760 controls from Germany and Italy. The obvious inconsistency of these observations could be explained by different ethnicity of investigated populations.
SNP rs895819, located in pre-miR-27a, was selected for an association study by systematic approach based on the scanning for SNPs located in the recently identified breast cancer-relevant miRNA genes (including pre-miRNAs and about ±200 bp flanking regions) by sequencing of these regions. In a group of 1217 German familial breast cancer patients and 1422 unrelated healthy German women, Yang et al. found that the G allele of rs895819, located in the terminal loop of a pre-miR-27a oncogene, was associated with the reduced familial breast cancer risk (P = 0.0215) [49]. Significance of the rs895819 was further confirmed in a most interesting study by Kontorovich et al., who hypothesized that aberrant gene silencing by miRNA may affect mutant BRCA penetrance: to test this presumption, the frequency of 42 SNPs within predicted miRNA-binding sites or miRNA precursors was determined and compared in 363 BRCA1 and 125 BRCA2 mutation carriers in high-risk Jewish women. In addition to other important outcomes of this study, the authors report that BRCA2 mutation carriers had a significantly higher risk of developing breast cancer when being the TT homozygotes compared to T/C heterozygotes of pre-miR-27a (rs895819; P = 0.013) and when carrying AC alleles compared to the AA carriers of miR-423 (rs6505162; P = 0.021). This study provides preliminary evidence for another regulatory level of deleterious mutations penetrance in cancer predisposition genes [50].
In addition to the polymorphisms within miRNA and pre-miRNA sequences, several SNPs in miRNAs binding regions 3′-UTR of coding genes were also identified as novel biomarkers of breast cancer susceptibility.
Brendle et al. [51] examined the effect of SNPs in predicted miRNA target sites of six integrin genes (ITGA3, ITGA6, ITGAv, ITGB3, ITGB4 and ITGB5) on breast cancer risk and clinical outcome. Six SNPs were genotyped in 749 Swedish breast cancer cases with detailed clinical data and the follow-up included up to 15 years of study with 1493 matched controls. The strongest association was observed between the A allele of the SNP rs743554 in the ITGB4 gene and oestrogen receptor-negative tumours (OR 2.09; 95% CI 1.19–3.67). The A allele carriers had a worse survival rate compared to the carriers of the wild-type genotypes. None of the SNPs were significantly associated with breast cancer risk [51].
Tchatchou et al. [52] reported significant association of a variant (rs2747648: C>T) affecting a miRNA target site in the oestrogen receptor 1 (ESR1). Age stratification of the cases in this study revealed that the association was stronger in pre-menopausal women (OR 0.60; 95% CI 0.41–0.89; P = 0.010). Furthermore, the effect was stronger in high-risk familial cases (OR 0.42; 95% CI 0.25–0.71; P = 0.0009). According to in silico analysis, rs2747648 affects the binding capacity of miR-453, which is increased in the presence of the C allele. By contrast, the T allele attenuates the binding of miR-453, which may lead to a reduced miRNA-mediated ESR1 repression and consequently to higher ESR1 protein levels and an increased breast cancer risk [52].
SET8 methylates TP53 and regulates genome stability. Song et al. [53] evaluated the SNP (rs16917496) within the miR-502 seed binding region in the 3′-UTR of the SET8 gene in a case-control study on 1110 breast cancer cases and 1097 controls. The SET8 CC genotype was independently associated with an earlier age of breast cancer onset in an allele-dose-dependent manner (52.2 years for TT, 51.4 for TC and 49.5 for CC) [53].
Bone morphogenetic protein receptor type-1 (BMPR1B) binds bone morphogenetic proteins (BMP) that are multifunctional signalling molecules that belong to the TGF-β superfamily and were first identified based on their ability to form bone tissue the extraskeletal sites. miR-125b negatively regulates BMPR1B and C/T allelic variation (rs1434536) within the target site, disrupting the regulation by miR-125b, increasing BMPR1B expression and ultimately elevating disease risk [54]. Findings of Saetrom et al. [54] indicated in a cohort of 428 breast cancer patients and 1064 controls indicate that carriers of the TT genotype in the BMPR1B gene are at higher breast cancer risk [54].
There are also several experimentally evaluated miRNA-related SNPs lacking association with breast cancer risk or clinical outcome rs12983273 in miR-373 [49], rs3807348 in miR-335 [49], rs731085 in let-7a-3 [49] and rs2292832 in miR-149 [45].
Colorectal cancer
Several genome-wide profiling studies described dysregulation of miRNAs in colorectal cancer (CRC) tissue and blood serum [11, 12]. A number of experimental studies on these miRNAs revealed insight into miRNA-mediated regulatory links to well-known oncogenic and tumour suppressor signalling pathways. The polymorphisms within the miRNA sequences and binding regions within 3′-UTRs of their mRNA targets were reported to be novel CRC risk factors [12].
By analysing human gene sequences obtained from the public database (http://genome.cse.ucsc.edu/), Kim et al. [55] found that six miRNA regulation genes (AGO1, AGO2, TARBP2, TNRC6A, TNRC6C and EXPORTIN5) have mononucleotide repeats with seven or more nucleotides in the coding sequences which could be potential targets for frameshift mutation in cancers with microsatellite instability (MSI) [55]. Mutations in analysed genes except the AGO1 were detected in 27% of CRC samples. Significant differences in mutation frequency between the cancers with MSI-H (19/58), MSI-L (0/32) and MSS (0/90) indicated that association of the mutations with CRC could be MSI-H specific. It seems that frameshift mutations of these genes could cause alterations of miRNA regulation and could contribute to the development of CRC indicating MSI-H phenotype [55].
A study of Lee et al. [56] included 426 consecutive Korean patients with surgically treated CRC: 40 polymorphisms in miRNA-related genes were determined. In a univariate analysis, the progression-free survival of the patients with the combined miR-492 C/G and G/G genotype was significantly worse than that of the patients with the mir-492 C/C genotype (rs2289030; P = 0.0426), however there was no difference in the overall survival [56].
Pioneering design in the field of research of miRNAs-associated SNPs was presented in the study by Landi et al. [57], who selected 3′-UTRs of 104 genes as candidates for CRC and identified putative miRNA-binding sites by specialized algorithms (PicTar, DianaMicroT, miRBase, miRanda, TargetScan and microInspector). Fifty-seven SNPs were identified in miRNA-binding sites and evaluated for their ability to affect binding of miRNA to its target, by assessing the variation of Gibbs free energy between the two alleles of each SNP. A total of eight common SNPs were identified and further investigated in a case-control association study. The study was carried out on a series of 968 cases and 697 controls from the Czech Republic, a population with the highest worldwide incidence of CRC. Statistically significant associations were found between CRC risk and variant alleles of CD86 and INSR genes. These observations are the first to report positive associations between SNPs in miRNA-binding regions and cancer risk [57].
Recent studies have found that KRAS mutations predict resistance to monoclonal antibodies targeting the epidermal growth factor receptor in metastatic colorectal cancer (mCRC). Zhang et al. [58] have tested the hypothesis whether SNP in a let-7 microRNA complementary site (lcs6) in the KRAS 3′-UTR may be associated with a clinical outcome in 130 KRAS wild-type (KRASwt) mCRC patients enrolled in a phase II study of cetuximab monotherapy (IMCL-0144). KRAS let-7 lcs6 SNP was found to be related to object response rate (ORR) in mCRC patients whose tumours had KRASwt. The 12 KRASwt patients harbouring at least a variant G allele (TG or GG) had a 42% ORR compared to a 9% ORR in 55 KRASwt patients with let-7 lcs6 TT genotype (P = 0.02). KRASwt patients with TG/GG genotypes also presented with a trend of longer median progression-free survival (3.9 versus 1.3 months) and OS (10.7 versus 6.4 months) compared to the individuals with TT genotypes [58].
Lung cancer
The discovery of biomarkers and their application in conjunction with traditional cancer diagnosis, staging and prognosis could substantially improve timely diagnosis and patient care. However, despite the efforts made to date, reliable markers are still missing and prognosis of the disease remains poor. In case of lung cancer, miRNA expression profiles and specific miRNAs have been shown to correlate with tumoural tissue and survival of lung adenocarcinomas [13, 14].
Tian et al. [59] conducted a study focusing on significance of four miRNA SNPs (miR-146a rs2910164, miR-499 rs3746444, miR-149 rs2292832, miR-196-a2 rs11614913) as potential risk factors of non–small cell lung cancer (NSCLC) in a case-control study of 1058 incident lung cancer patients and 1035 cancer-free controls from a Chinese population. The miR-196a2 rs11614913 variant homozygote CC was associated with a 25% increase in lung cancer risk compared to their wild-type homozygote TT and heterozygote TC. However, no significant effects were observed in case of the association between the three remaining SNPs and lung cancer risk. These findings suggest that functional SNP rs11614913 in miR-196a2 could also contribute to susceptibility to lung cancer [59].
The same research group evaluated in detail the association between the same SNPs and the survival of patients with NSCLC [60]. As they assumed that disease susceptibility was inherited as a recessive phenotype, they found that the rs11614913 SNP in hsa-mir-196a2 was associated with survival in 556 individuals with NSCLC (107 patients in the validation set). Specifically, the survival rate was significantly lower in case of CC homozygotes. Binding assays revealed that rs11614913 can affect binding of mature hsa-mir-196a2 to its target mRNA. This is the first study to describe miRNA SNPs and NSCLC outcome on a large study population size with high statistical power (if α = 0.05, based on the data set for hsa-mir-196a2 rs11614913, an 80% power to detect an HR of 1.41 was reached) [60].
The let-7 family of miRNAs seems to play a key role in lung cancer: their levels are decreased in NSCLC; the lower levels are biomarkers of poor outcome; these miRNAs regulate multiple important oncogenes involved in lung cancer, including KRAS, and they inhibit growth of lung cancer cell lines both in vitro and in vivo [61]. The aim of the seminal study by Franck J. Slack's group from Yale was to identify SNPs, which could potentially modify let-7 binding as well as to assess the effects of these SNPs on target gene regulation and the risk of NSCLC. let-7 complementary sites (lcs) were sequenced in the KRAS 3′ untranslated region in 74 NSCLC cases to identify mutations and SNPs correlating with NSCLC. The allele frequency of a previously unidentified SNP at lcs6 was characterized in 2433 people (representing 46 human populations). The frequency of the variant allele was 18.1–20.3% in NSCLC patients and 5.8% in worldwide populations. The association between this SNP and the risk of NSCLC was also investigated in a case-control study of lung cancer cases from the New Mexico that showed a 2.4-fold increased risk (95% CI 1.1–4.6; P = 0.02) for NSCLC cancer. Functionally, the variant allele resulted in KRAS overexpression in vitro. The lcs6 variant allele in a KRAS miRNA complementary site was significantly associated with increased risk of NSCLC among moderate smokers and represents a new paradigm for let-7 miRNAs in lung cancer susceptibility [61].
Given the functionality of lcs6, Nelson et al. [62] evaluated the hypothesis that this SNP is associated with the occurrence of KRAS mutation as well as survival in a cohort of 218 NSCLC patients. No association was reported between the lcs6 KRAS polymorphism and KRAS mutational status (codon 12). Furthermore, no association between lcs6 SNP and NSCLC patients survival rate was observed [62].
Prostate cancer
Recently, many studies suggested that miRNAs are involved in prostate cancer carcinogenesis [7, 8, 17]. To explore whether the intensively studied polymorphism rs2910164 (G>C) in miR-146a plays any role in prostate cancer, Xu et al. [63] analysed the association between miR-146a polymorphism and risk of prostate cancer in 251 patients and 280 controls from a southern Han Chinese population. The patients carrying CC homozygote genotype had a 0.65-fold reduced risk of the disease (95% CI 0.43–0.99) in comparison with those carrying GG/GC genotypes (P = 0.03), and the C allele displayed a lower prevalence of prostate cancer compared to the G allele (OR 0.73; 95% CI 0.57–0.94, P = 0.01). Moreover, miR-146a quantification showed that homozygous carriers of the C-variant had significantly decreased miRNA level compared to the carriers of the GG/GC genotype. Based on these data, it seems that rs2910164 in miR-146a contributes to the genetic predisposition to prostate cancer [63].
Renal cell carcinoma
Horikawa et al. [64] hypothesized that genetic variations in miRNA genes and miRNA machinery genes could be associated with the risk of renal cell carcinoma (RCC). Forty SNPs in 11 miRNA processing genes (DROSHA, DGCR8, XPO5, RAN, DICER1, TARBP2, AGO1, AGO2, GEMIN3, GEMIN4, HIWI) and 15 miRNA genes were genotyped in 279 Caucasian patients with RCC and 278 matched controls. Two SNPs in the GEMIN4 gene were significantly associated with altered RCC risk. The variant-containing genotypes of Asn929Asp (rs2740348) and Cys1033Arg (rs7813) exhibited significantly reduced risks. A combined unfavourable genotype (UG) analysis was conducted including five promising SNPs (DICER1 [rs3742330], AA; AGO1 [rs595961], AA; GEMIN4 [rs2740330], GG; GEMIN4 [rs7813], TT; GEMIN3 [rs197412], TC+CC; GEMIN3 [rs197388], TA+AA) showing at least a borderline significant association with the risk of the disease. Compared to the low-risk reference group with one UG, the median-risk (with two UGs) and high-risk (with three to five UGs) groups exhibited a 1.55-fold (95% CI 0.96–.50) and 2.49-fold (95% CI 1.58–3.91) increased risk of RCC, respectively (P for trend < 0.001). This data suggest that SNPs in miRNA-machinery genes may affect RCC susceptibility [64].
The same research team has evaluated the effects of the similar SNPs on survival and recurrence among renal cell RCC patients. When the SNPs were analysed separately, seven SNPs were found to be significantly associated with RCC survival and five with recurrence [65]. The most significant associations were SNPs in GEMIN4 with the variant alleles of both rs7813 and rs910925 associated with 1.74-fold (95% CI 1.15–2.62) increased risk of death, whereas the variant allele of rs3744741 conferred a decreased risk of death (HR 0.39; 95% CI 0.19–0.77). Several SNPs belonging to the pre-miRNA were identified to be significantly associated with RCC recurrence. Haplotypes of DICER and DROSHA were also associated with altered survival of the patients and recurrence of the disease. Compared to the patients carrying zero to two UGs, those individuals carrying three to five and six and more UGs had an increased risk of death with a HR of 2.49 (95% CI 1.24–5.00) and 6.66 (95% CI 2.49–17.86), respectively, with a significant dose–response trend (P for trend < 0.001). This data strongly suggest that miRNA-related SNPs may impact the recurrence and survival in RCC patients [65].
Cervical cancer
In cervical cancer, miR-218 can target laminin-5 β3 (LAMB3), but it is suppressed by HPV-16 E6 protein. Because laminin-5 is required in RAS and NF-κB blockade induced tumourigenesis of human squamous cell carcinoma and because it is a marker of invasiveness in cervical lesions, Zhou et al. [66] hypothesized that SNPs in pri-miR-218 rs11134527 and LAMB3 rs2566 may be individually and/or jointly involved in pathogenesis of cervical. Zhou et al. genotyped these two SNPs in a case-control study of 703 cervical cancer cases and 713 cancer-free controls of Chinese origin. In logistic regression analyses, the variant GG homozygote of pri-miR-218 rs11134527 was associated with the decreased risk of cervical cancer in comparison to the AA genotype, whereas the LAMB3 rs2566 variant CT/TT genotypes were associated with a significantly increased risk of cervical cancer, compared to the wild-type CC genotype. A significant dose–response effect was observed between the number of risk alleles, rs11134527A and rs2566 T, and the risk of cervical cancer (P for trend = 0.0006) [66].
Ovarian cancer
In a study by Liang et al. [67], the total of 26 SNPs in miRNA processing genes and miRNA-binding sites were analysed in 339 ovarian cancer cases and 349 healthy controls to assess association with cancer risk, overall survival and treatment response. Thirteen polymorphisms were found to have significant association with risk, whereas the most significant were two linked SNPs (r(2) = 0.99)), rs2740351 and rs7813 in GEMIN4.
In a large investigation by Permuth-Way et al. [68], a total of 318 SNPs in 18 genes were evaluated among 1815 epithelial ovarian cancer cases and 1900 controls, followed up by a replicative joint meta-analysis of data from an additional 2172 cases and 3052 controls. Of 23 SNPs from nine genes associated with risk (empirical P < 0.05) in the initial investigation, the meta-analysis replicated six SNPs from the DROSHA, FMR1, LIN28 and LIN28B genes, including rs12194974 (G>A), an SNP in a putative transcription factor binding site in the LIN28B promoter region (P = 0.015) and the authors conclude that based on their observations, variants in LIN28B and possibly other miRNA biogenesis genes may influence susceptibility to epithelial ovarian cancer.
Gastric cancer
To expand knowledge regarding the new SNP and biological function of miR-27a, Sun et al. [69] designed a study to determine whether A/G polymorphism (rs895819) within mir-27a is associated with a risk of gastric cancer. Authors also investigated miR-27a and its target gene Zinc finger and BTB domain containing 10 (ZBTB1) expression in consideration of the genotype [69]. In a case-control study on 304 gastric cancer cases and 304 cancer-free controls, they observed that the patients with variant genotypes (AG + GG) showed a significantly increased risk of gastric cancer compared to AA carriers (adjusted OR 1.48; 95% CI 1.06–2.05; P = 0.019). A significant association of hsa-mir-27a variant genotypes with lymph node metastasis was also observed. Further functional analyses indicated that variant genotypes might also be responsible for elevated miR-27a levels and reduced ZBTB10 mRNA [69].
The most frequently studied SNP in miRNAs, rs11614913 in miR-196a-2, was also studied in the gastric cancer. Peng et al. [70] conducted a hospital-based case-control study on 213 gastric cancer patients and 213 age- and sex-matched controls of Chinese origin. They found a significantly increased risk of gastric cancer in patients with the variant homozygote CC of miR-196a-2 in comparison with the wild-type homozygotes TT and CT heterozygote (adjusted OR 1.57; 95% CI 1.03–2.39; P = 0.038). Stratified analyses indicated that the variant homozygote CC genotype had a strong association with the lymph node metastasis of gastric cancer (adjusted OR 2.25, 95% CI 1.21–4.18, P = 0.011). These findings suggest that genetic variants within both miR-196a-2 and miR-27a could play an important role in the development and progression of gastric cancer [70].
Bladder cancer
The first study to evaluate the significance of SNPs in miRNA processing pathway genes in bladder cancer predisposition was published by Yang et al. [71]. In this case-control study, the authors tested the hypothesis that common sequence variants in genes of miRNA and miRNA biogenesis pathway affect susceptibility to bladder cancer. To better understand this effect, the total of 41 SNPs from 24 miRNA genes were genotyped in a study conducted on 746 Caucasian patients with bladder cancer and 746 matched controls. The homozygous variant genotype of a non-synonymous SNP in the GEMIN3 gene (rs197414) was associated with significantly increased risk of bladder cancer. Several additional miRNA-related SNPs, which showed a borderline significant association with risk of bladder cancer were also identified. To assess the cumulative effects of the promising SNPs, we performed a combined UG analysis that included all SNPs showing at least a borderline statistical significance. Compared to the low-risk reference group with less than two UGs, the medium-risk group with two UGs exhibited a 1.29-fold (0.92–1.81) increased risk, whereas the high-risk group with more than two UGs exhibited a 1.92-fold (1.36–2.71) increased risk (P < 0.0001) [71].
Oesophageal cancer
The associations between oesophageal cancer risk and 41 potentially functional SNPs in 26 miRNA-related genes in a case-control study on 346 Caucasian oesophageal cancer patients (85.5% with oesophageal adenocarcinoma) and 346 frequency-matched (age, gender and ethnicity) controls were explored in a study by Ye et al. [72]. Seven SNPs were significantly associated with oesophageal cancer risk. The most notable finding was that the rs6505162, which is located in the pre-mir423 region, was associated with a per-allele odds ratio of 0.64 (95% CI 0.51–0.80; P for trend < 0.0001). A common haplotype of the GEMIN4 gene was associated with a significantly reduced risk of oesophageal cancer (OR 0.65; 95% CI 0.42–0.99). An analysis to further evaluate the cumulative effects of the promising (risk associated) combined UG SNPs was also performed. In comparison with the low-risk group (fewer than three UGs), the medium-risk group (three UGs) had a 2.00-fold (95% CI 1.31–3.08) increased risk and the high-risk group (three UGs and more) had a 3.14-fold (95% CI 2.03–4.85) increased risk (P for trend < 0.0001) [72].
Promising findings suggesting that functional rs2910164 in pre-miR-146a could contribute to susceptibility to oesophageal squamous cell carcinoma (ESCC) and clinical outcome were reported by the group of Guo et al. [73] in their case-control study on 444 sporadic ESCC patients and 468 matched cancer-free controls from a Han Chinese population. Compared to CC variant genotype of rs2910164, the GG genotype was associated with increased risk of ESCC (OR 2.39; 95% CI 1.36–4.20). Among smokers, the risk of GG genotype of rs2910164 was more pronounced (OR 3.17; 95% CI 1.71–4.46). In the stratification analyses, a strong correlation was identified between rs2910164 C/G variant and the clinical TNM stage (P < 0.01) [73].
Hepatocellular carcinoma
The first association study of miRNA polymorphisms and hepatocellular cancer (HCC) involved 479 HCC and 504 control patients. Xu et al. [74] found that the genotype distribution of miR-146a polymorphism rs2910164 in HCC cases was significantly different from that in control patients (P = 0.026). The results revealed that male individuals with GG genotype were twice as susceptible to HCC (P = 0.034) compared to those with CC genotype. When investigating the influence of this SNP on production of mature miR-146a, G-allelic miR-146a precursor displayed increased production of mature miR-146a compared to the C-allelic one [74].
Recent studies have implied that the rs11614913 SNP in miR-196-a2 is associated with susceptibility to a broad spectrum of solid tumours. To assess whether this SNP is associated with susceptibility to and clinicopathologic characteristics of the hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC), a total of 560 patients with chronic HBV infection and 391 healthy volunteers were studied, and a miR-196-a2 polymorphism was genotyped [75]. In this study, there was no significant association between miR-196-a2 SNP and the risk of HBV-related HCC in all patients; however, the risk of HCC was significantly higher with miR-196-a2 rs11614913 CC genotype or C allele compared to those with the TT genotype or T allele in male patients (P = 0.031 and 0.046, respectively). Moreover, the T allele was significantly more frequent in male HCC patients with lymphatic metastasis (P = 0.03) [75].
Gao et al. [76] predicted polymorphisms falling in miRNA-binding regions of HCC genes. Considering the effects of IL-1α, not only in carcinogenesis, tumour growth and invasiveness, but also in terms of interaction patterns between malignant cells and the host's immune system, the authors selected an insertion/deletion (Indel) polymorphism (rs3783553) in the 3′-UTR of interleukin-1α (IL-1α) for a case-control study in a Chinese population. With samples from 403 HCC patients and 434 healthy control individuals, strong evidence of association was observed for the variant homozygote. This association was validated in a second independent case-control study with 1074 HCC patients and 1239 healthy control individuals. ‘TTCA’ insertion allele for rs3783553 disrupts a binding site for miR-122 and miR-378, thereby increasing transcription of IL-1α in vitro and in vivo. These findings suggest that functional polymorphism rs3783553 in IL-1α could contribute to HCC susceptibility [76].
Head and neck cancer
A polymorphism in the frequently studied miR-196-a2 locus which was reported to have associations with a wide range of solid cancers was also examined in head and neck cancer by Christensen et al. [77]. SNP in miR-196-a2 (rs11614913, C/T) was evaluated to determine whether the miR-196-a2 genotype is associated with altered risk of disease and patient survival rate in a population-based case-control study (n = 1039) of head and neck squamous cell carcinoma cases (HNSCC). The presence of any variant allele was associated with a significantly reduced risk of HNSCC (OR 0.8; 95% CI 0.56–0.99). Homozygous variant allele carriers with pharyngeal tumours had a significantly reduced survival rate compared to the wild-type and heterozygous cases. This data successfully demonstrated an important role of miR-196-a2 genetic variability not only in HNSCC susceptibility, but also in prognosis of HNSCC patients [77].
Thyroid cancer
Although papillary thyroid carcinoma (PTC) displays strong heritability, no predisposing germ-line mutations have been found so far. In an association study of 608 PTC patients and 901 controls, marked differences in genotype distribution of rs2910164 in miR-146a (P < 0.001) were found, the GC heterozygotes being associated with an increased risk of developing PTC (OR 1.62, P = 0.001), and both homozygous states being protective with an odds ratio = 0.42 for the CC genotype (P = 0.003) and odds ratio = 0.69 for the GG genotype (P < 0.001) [78]. Jazdzewski et al. [79] explain this interesting observation in another study of theirs by pointing out a mechanism based on the fact that GC heterozygotes differ from both GG and CC homozygotes by producing three mature microRNAs: one from the leading strand (miR-146a) and two from the passenger strand (miR-146a*G and miR-146a*C), each with a distinct set of target genes [79].
Moreover, the authors showed that a common G/C polymorphism (rs2910164) within the pre-miR-146a sequence reduced the amount of pre and mature miR-146a from the C allele 1.9- and 1.8-fold, respectively, compared to the G allele. This is followed by a similar decrease in the amount of each pre-miRNAs generated from the corresponding pri-miR-146a in an in vitro processing reaction. The reduction in miR-146a led to less efficient inhibition of target genes involved in the Toll-like receptor and cytokine signalling pathway (TRAF6, IRAK1), and PTC1 (also known as CCDC6 or H4), a gene frequently rearranged with RET proto-oncogene in PTC [79].
Glioma
In gliomas, only one polymorphism within a mature miRNA sequence, specific rs11614913 gene variant of miR-196a, has ever been studied so far [80]. The obtained data suggest that the CC genotype of miR-196a SNP is associated with decreased risk of glioma in the Chinese population (OR 0.74; 95% CI 0.56–0.98). Significant association was similarly observed between these genotype and risk of particular glioma subgroups: patients over 18 years (OR 0.73; 95% CI 0.55–0.98), male glioma patients (OR 0.69; 95% CI 0.48–0.99) and patients with high-grade glioma–glioblastoma (OR 0.58; 95% CI 0.37–0.91). In contrast to other solid cancers, such as lung cancer [59] and breast cancer [45, 46], data from glioblastoma cases showed an opposite association between miR-196a genotype and cancer risk, which could be explained by the diversity of tissue origin and subsequent characteristic molecular alterations in different types of cancer [80].
Conclusions and future directions
It is obvious that miRNA-associated SNPs represent extremely interesting variations that have a huge potential to interfere with the function of miRNAs, which regulate genes involved in crucial signalling pathways. The effects of miRNA polymorphisms on their carrier may be either beneficial or adverse; for example a loss of function of a miRNA polymorphism may abolish or weaken binding of the miRNA to the target 3′-UTR and as a result, it may lead to the less potent regulation of the target. If the down-regulated target were a tumour suppressor or the up-regulated target were an oncogene, this may lead both to initiation or suppression of tumourigenesis.
Not only have the miRNA polymorphisms effect on biological functions of the miRNAs, thus resulting in different regulation of genes involved in apoptosis or proliferation; they might also substantially affect therapy response or may lead to therapy failure, which makes the understanding of the underlying processes a necessity. Characterization of the miRNA polymorphisms and identification of their functional impact may provide a good basis for miRNA-based therapeutic approaches in the future. The miRNA-based SNPs could provide the clinician with a good prediction data on drug adverse reactions or toxicity as well as with information on susceptibility to solid tumours and its related prognosis; thus, it may be a good starting point for the development of individually tailored miRNA-based therapy.
In our review, over 30 epidemiologic evidence have been summarized, most of them supporting the involvement of genetic polymorphisms in miRNA genes and miRNA biogenesis pathway genes in solid cancer. The results of these studies suggest that individual as well as combined genotypes of miRNA-related variants may be used to predict not only the risk of cancer, but in some cases also therapeutic and clinical outcome. Because miRNA polymorphisms were first noted in molecular epidemiology only in 2006, it is likely that some associations presented here are the accidental findings, but variants of several miRNAs (miR-196-a2, miR-146a, miR-27a) were identified repeatedly with a high level of statistical significance, and we suggest that these could be considered as new important candidate cancer genes. However, the analysis of large study populations of different ethnic groups in multicentric design is necessary to verify the associations and answer questions regarding the possible impact of this new direction of molecular epidemiology in cancer.
Acknowledgments
This work is supported by grants from Internal Grant Agency of Czech Ministry of Health (IGA MZ CR) NS 10352-3/2009 and NS/9814, project No. MZ0MOU2005 of the Czech Ministry of Health and by the project ‘CEITEC – Central European Institute of Technology’(CZ.1.05/1.1.00/02.0068).
Conflict of interest
The authors confirm that there are no conflicts of interest.
References
- 1.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 2.Fabbri M, Croce CM, Calin GA. MicroRNAs. Cancer J. 2008;14:1–6. doi: 10.1097/PPO.0b013e318164145e. [DOI] [PubMed] [Google Scholar]
- 3.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27:5848–56. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–89. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Novakova J, Slaby O, Vyzula R, Michalek J. MicroRNA involvement in glioblastoma pathogenesis. Biochem Biophys Res Commun. 2009;386:1–5. doi: 10.1016/j.bbrc.2009.06.034. [DOI] [PubMed] [Google Scholar]
- 7.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 8.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumours defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 10.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–8. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 11.Slaby O, Svoboda M, Fabian P, et al. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- 12.Slaby O, Svoboda M, Michalek J, Vyzula R. MicroRNAs in colorectal cancer: translation of molecular biology into clinical application. Mol Cancer. 2009;8:102. doi: 10.1186/1476-4598-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 14.Yu SL, Chen HY, Chang GC, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Jung M, Mollenkopf HJ, Grimm C, et al. MicroRNA profiling of clear cell renal cell cancer identifies a robust signature to define renal malignancy. J Cell Mol Med. 2009;13:3918–28. doi: 10.1111/j.1582-4934.2009.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slaby O, Jancovicova J, Lakomy R, et al. Expression of miRNA-106b in conventional renal cell carcinoma is a potential marker for prediction of early metastasis after nephrectomy. J Exp Clin Cancer Res. 2010;29:90. doi: 10.1186/1756-9966-29-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porkka KP, Pfeiffer MJ, Waltering KK, et al. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–5. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 18.Lee JW, Choi CH, Choi JJ, et al. Altered microRNA expression in cervical carcinomas. Clin Cancer Res. 2008;14:2535–42. doi: 10.1158/1078-0432.CCR-07-1231. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Zhang Y, Ding J, et al. Survival prediction of gastric cancer by a seven-microRNA signature. Gut. 2010;59:579–85. doi: 10.1136/gut.2008.175497. [DOI] [PubMed] [Google Scholar]
- 20.Catto JW, Miah S, Owen HC, et al. Distinct microRNA alterations characterize high- and low-grade bladder cancer. Cancer Res. 2009;69:8472–81. doi: 10.1158/0008-5472.CAN-09-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee EJ, Gusev Y, Jiang J, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–54. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feber A, Xi L, Luketich JD, et al. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg. 2008;135:255–60. doi: 10.1016/j.jtcvs.2007.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X, Chen Z, Yu J, et al. MicroRNA profiling and head and neck cancer. Comp Funct Genom. 2009:837514. doi: 10.1155/2009/837514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikiforova MN, Chiosea SI, Nikiforov YE. MicroRNA expression profiles in thyroid tumours. Endocr Pathol. 2009;20:85–91. doi: 10.1007/s12022-009-9069-z. [DOI] [PubMed] [Google Scholar]
- 25.Iorio MV, Visone R, Di Leva G, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 26.Jiang J, Gusev Y, Aderca I, et al. Association of microRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419–27. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong H, Siu H, Luo L, et al. Investigation gene and microRNA expression in glioblastoma. BMC Genom. 2010;11:S16. doi: 10.1186/1471-2164-11-S3-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slaby O, Lakomy R, Fadrus P, et al. MicroRNA-181 family predicts response to concomitant chemoradiotherapy with temozolomide in glioblastoma patients. Neoplasma. 2010;57:264–9. doi: 10.4149/neo_2010_03_264. [DOI] [PubMed] [Google Scholar]
- 29.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu W, Zhao W, Qi C, Qi X. MicroRNA polymorphisms, MicroRNA pharmacogenomics and cancer susceptibility. Curr Pharmacogenom Personal Med. 2010;8:289–305. [Google Scholar]
- 31.Abelson JF, Kwan KY, O’Roak BJ, et al. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science. 2005;310:317–20. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- 32.Calin GA, Ferracin M, Cimmino A, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 33.Wojcik SE, Rossi S, Shimizu M, et al. Non-coding RNA sequence variations in human chronic lymphocytic leukemia and colorectal cancer. Carcinogenesis. 2010;31:208–15. doi: 10.1093/carcin/bgp209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bao L, Zhou M, Wu L, et al. PolymiRTS database: linking polymorphisms in microRNA target sites with complex traits. Nucleic Acids Res. 2007;35:D51–4. doi: 10.1093/nar/gkl797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci USA. 2007;104:3300–5. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glinsky GV. An SNP-guided microRNA map of fifteen common human disorders identifies a consensus disease phenocode aiming at principal components of the nuclear import pathway. Cell Cycle. 2008;7:2570–83. doi: 10.4161/cc.7.16.6524. [DOI] [PubMed] [Google Scholar]
- 37.McLeod HL, Yu J. Cancer pharmacogenomics: SNPs, chips, and the individual patient. Cancer Invest. 2003;21:630–40. doi: 10.1081/cnv-120022384. [DOI] [PubMed] [Google Scholar]
- 38.Erichsen HC, Chanock SJ. SNPs in cancer research and treatment. Br J Cancer. 2004;90:747–51. doi: 10.1038/sj.bjc.6601574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mishra PJ, Bertino JR. MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics. 2009;10:399–416. doi: 10.2217/14622416.10.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 42.Georges M, Coppieters W, Charlier C. Polymorphic miRNA-mediated gene regulation: contribution to phenotypic variation and disease. Curr Opin Genet Dev. 2007;17:166–76. doi: 10.1016/j.gde.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Liu J, Rivas FV, Wohlschlegel J, et al. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2006;7:1261–6. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yekta S, Shih I, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–6. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 45.Hu Z, Liang J, Wang Z, et al. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum Mutat. 2009;30:79–84. doi: 10.1002/humu.20837. [DOI] [PubMed] [Google Scholar]
- 46.Hoffman AE, Zheng T, Yi C, et al. microRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer Res. 2009;69:5970–7. doi: 10.1158/0008-5472.CAN-09-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Catucci I, Yang R, Verderio P, et al. Evaluation of SNPs in miR-146a, miR196a2 and miR-499 as low-penetrance alleles in German and Italian familial breast cancer cases. Hum Mutat. 2010;31:E1052–7. doi: 10.1002/humu.21141. [DOI] [PubMed] [Google Scholar]
- 48.Shen J, Ambrosone CB, RA DiCioccio, et al. A functional polymorphism in the miR-146a gene and age of familial breast/ovarian cancer diagnosis. Carcinogenesis. 2008;29:1963–6. doi: 10.1093/carcin/bgn172. [DOI] [PubMed] [Google Scholar]
- 49.Yang R, Schlehe B, Hemminki K, et al. A genetic variant in the pre-miR-27a oncogene is associated with a reduced familial breast cancer risk. Breast Cancer Res Treat. 2010;121:693–702. doi: 10.1007/s10549-009-0633-5. [DOI] [PubMed] [Google Scholar]
- 50.Kontorovich T, Levy A, Korostishevsky M, et al. Single nucleotide polymorphisms in miRNA binding sites and miRNA genes as breast/ovarian cancer risk modifiers in Jewish high-risk women. Int J Cancer. 2010;127:589–97. doi: 10.1002/ijc.25065. [DOI] [PubMed] [Google Scholar]
- 51.Brendle A, Lei H, Brandt A, et al. Polymorphisms in predicted microRNA-binding sites in integrin genes and breast cancer: ITGB4 as prognostic marker. Carcinogenesis. 2008;29:1394–9. doi: 10.1093/carcin/bgn126. [DOI] [PubMed] [Google Scholar]
- 52.Tchatchou S, Jung A, Hemminki K, et al. A variant affecting a putative miRNA target site in estrogen receptor (ESR) 1 is associated with breast cancer risk in premenopausal women. Carcinogenesis. 2009;30:59–64. doi: 10.1093/carcin/bgn253. [DOI] [PubMed] [Google Scholar]
- 53.Song F, Zheng H, Liu B, et al. An miR-502-binding site single-nucleotide polymorphism in the 3′-untranslated region of the SET8 gene is associated with early age of breast cancer onset. Clin Cancer Res. 2009;15:6292–300. doi: 10.1158/1078-0432.CCR-09-0826. [DOI] [PubMed] [Google Scholar]
- 54.Saetrom P, Biesinger J, Li SM, et al. A risk variant in an miR-125b binding site in BMPR1B is associated with breast cancer pathogenesis. Cancer Res. 2009;69:7459–65. doi: 10.1158/0008-5472.CAN-09-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim MS, Oh JE, Kim YR, et al. Somatic mutations and losses of expression of microRNA regulation-related genes AGO2 and TNRC6A in gastric and colorectal cancers. J Pathol. 2010;221:139–46. doi: 10.1002/path.2683. [DOI] [PubMed] [Google Scholar]
- 56.Lee HC, Kim JG, Chae YS, et al. Prognostic impact of microRNA-related gene polymorphisms on survival of patients with colorectal cancer. J Cancer Res Clin Oncol. 2010;136:1073–8. doi: 10.1007/s00432-009-0754-6. [DOI] [PubMed] [Google Scholar]
- 57.Landi D, Gemignani F, Naccarati A, et al. Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis. 2008;29:579–84. doi: 10.1093/carcin/bgm304. [DOI] [PubMed] [Google Scholar]
- 58.Zhang W, Winder T, Ning Y, et al. A let-7 microRNA-binding site polymorphism in 3′-untranslated region of KRAS gene predicts response in wild-type KRAS patients with metastatic colorectal cancer treated with cetuximab monotherapy. Ann Oncol. 2011;22:104–9. doi: 10.1093/annonc/mdq315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tian T, Shu Y, Chen J, et al. A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol Biomarkers Prev. 2009;18:1183–7. doi: 10.1158/1055-9965.EPI-08-0814. [DOI] [PubMed] [Google Scholar]
- 60.Hu Z, Chen J, Tian T, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118:2600–8. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chin LJ, Ratner E, Leng S, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–40. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nelson HH, Christensen BC, Plaza SL, et al. KRAS mutation, KRAS-LCS6 polymorphism, and non-small cell lung cancer. Lung Cancer. 2010;69:51–3. doi: 10.1016/j.lungcan.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu B, Feng NH, Li PC, et al. A functional polymorphism in Pre-miR-146a gene is associated with prostate cancer risk and mature miR-146a expression in vivo. Prostate. 2010;70:467–72. doi: 10.1002/pros.21080. [DOI] [PubMed] [Google Scholar]
- 64.Horikawa Y, Wood CG, Yang H, et al. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res. 2008;14:7956–62. doi: 10.1158/1078-0432.CCR-08-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin J, Horikawa Y, Tamboli P, et al. Genetic variations in microRNA-related genes are associated with survival and recurrence in patients with renal cell carcinoma. Carcinogenesis. 2010;31:1805–12. doi: 10.1093/carcin/bgq168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou X, Chen X, Hu L, et al. Polymorphisms involved in the miR-218-LAMB3 pathway and susceptibility of cervical cancer, a case-control study in Chinese women. Gynecol Oncol. 2010;117:287–90. doi: 10.1016/j.ygyno.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 67.Liang D, Meyer L, Chang DW, et al. Genetic variants in microRNA biosynthesis pathways and binding sites modify ovarian cancer risk, survival, and treatment response. Cancer Res. 2010;70:9765–76. doi: 10.1158/0008-5472.CAN-10-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Permuth-Wey J, Kim D, Tsai YY, et al. LIN28B polymorphisms influence susceptibility to epithelial ovarian cancer. Cancer Res. 2011;71:3896–903. doi: 10.1158/0008-5472.CAN-10-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun Q, Gu H, Zeng Y, et al. Hsa-mir-27a genetic variant contributes to gastric cancer susceptibility through affecting miR-27a and target gene expression. Cancer Sci. 2010;101:2241–7. doi: 10.1111/j.1349-7006.2010.01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peng S, Kuang Z, Sheng C, et al. Association of microRNA-196a-2 gene polymorphism with gastric cancer risk in a Chinese population. Dig Dis Sci. 2010;55:2288–93. doi: 10.1007/s10620-009-1007-x. [DOI] [PubMed] [Google Scholar]
- 71.Yang H, Dinney CP, Ye Y, et al. Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. Cancer Res. 2008;68:2530–7. doi: 10.1158/0008-5472.CAN-07-5991. [DOI] [PubMed] [Google Scholar]
- 72.Ye Y, Wang KK, Gu J, et al. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev Res. 2008;1:460–9. doi: 10.1158/1940-6207.CAPR-08-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo H, Wang K, Xiong G, et al. A functional varient in microRNA-146a is associated with risk of esophageal squamous cell carcinoma in Chinese Han. Fam Cancer. 2010;9:599–603. doi: 10.1007/s10689-010-9370-5. [DOI] [PubMed] [Google Scholar]
- 74.Xu T, Zhu Y, Wei QK, et al. A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis. 2008;29:2126–31. doi: 10.1093/carcin/bgn195. [DOI] [PubMed] [Google Scholar]
- 75.Qi P, Dou TH, Geng L, et al. Association of a variant in MIR 196A2 with susceptibility to hepatocellular carcinoma in male Chinese patients with chronic hepatitis B virus infection. Hum Immunol. 2010;71:621–6. doi: 10.1016/j.humimm.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 76.Gao Y, He Y, Ding J, et al. An insertion/ deletion polymorphism at miRNA-122-binding site in the interleukin-1alpha 3′ untranslated region confers risk for hepatocellular carcinoma. Carcinogenesis. 2009;30:2064–9. doi: 10.1093/carcin/bgp283. [DOI] [PubMed] [Google Scholar]
- 77.Christensen BC, Avissar-Whiting M, Ouellet LG, et al. Mature microRNA sequence polymorphism in MIR196A2 is associated with risk and prognosis of head and neck cancer. Clin Cancer Res. 2010;16:3713–20. doi: 10.1158/1078-0432.CCR-10-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jazdzewski K, Murray EL, Franssila K, et al. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2008;105:7269–74. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jazdzewski K, Liyanarachchi S, Swierniak M, et al. Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer. Proc Natl Acad Sci USA. 2009;106:1502–5. doi: 10.1073/pnas.0812591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dou T, Wu Q, Chen X, et al. A polymorphism of microRNA196a genome region was associated with decreased risk of glioma in Chinese population. J Cancer Res Clin Oncol. 2010;136:1853–9. doi: 10.1007/s00432-010-0844-5. [DOI] [PubMed] [Google Scholar]


