Abstract
MicroRNA (miRNA)s are a class of non-coding RNAs that regulate gene expression post-transcriptionally. Muscle-specific miRNA, miRNA (miR)-1, miR-133 and miR-206 play a crucial role in the regulation of muscle development and homeostasis. Muscle injuries are a common muscloskeletal disorder, and the most effective treatment has not been established yet. The purpose of this study was to demonstrate that a local injection of double-stranded (ds) miR-1, miR-133 and 206 can accelerate muscle regeneration in a rat skeletal muscle injury model. After the laceration of the rat tibialis anterior muscle, ds miR-1, 133 and 206 mixture mediated atelocollagen was injected into the injured site. The control group was injected with control siRNA. At 1 week after injury, an injection of miRNAs could enhance muscle regeneration morphologically and physiologically, and prevent fibrosis effectively compared to the control siRNA. Administration of exogenous miR-1, 133 and 206 can induce expression of myogenic markers, MyoD1, myogenin and Pax7 in mRNA and expression in the protein level at 3 and 7 days after injury. The combination of miR-1, 133 and 206 can promote myotube differentiation, and the expression of MyoD1, myogenin and Pax7 were up-regulated in C2C12 cells in vitro. Local injection of miR-1, 133 and 206 could be a novel therapeutic strategy in the treatment of skeletal muscle injury.
Keywords: microRNA, muscle, MyoD1, myogenin, Pax7
Introduction
Muscle injuries are common muscloskeletal disorders, especially in sports and traumatology medicine. They are routinely treated conservatively, but the most effective treatment has not been established yet. Complications such as muscle contractures, atrophy and residual pain are often encountered. Several studies have attempted to attain complete muscle recovery using several drugs, cytokines and cell therapy [1–3].
MicroRNAs (miRNA)s are small, ∼22-nucleotide, a class of non-coding RNAs that regulate gene expression post-transcriptionally [4, 5]. Many miRNAs are evolutionarily conserved across phyla, identified from nematodes to human beings [6]. MiRNAs regulate gene expression by binding the 3′-untranslated region of their target mRNAs leading to translational repression or mRNA degradation. Several miRNAs exhibit a tissue-specific or developmental stage-specific expression pattern and have been reported to be associated with the pathogenesis of human diseases such as cancer, leukaemia and rheumatoid arthritis [7–9]. MiRNA has been shown to be one of the important key players in a variety of biological processes including several diseases.
Muscle-specific miRNAs, miRNA (miR)-1, miR-133 and miR-206 have been well investigated in skeletal, smooth and cardiac muscles [10–14]. These three miRNAs play a crucial role in the regulation of muscle development. Several studies have clarified that miR-1 promotes the differentiation of skeletal muscle progenitors by post-trasnscriptional down-regulation of histone deacetylase 4 (HDAC4) and MEF2C, which inhibits muscle differentiation and skeletal muscle gene expression [10, 15, 16]. On the other hand, miR-133 inhibits myoblast differentiation and promotes its proliferation by repressing the expression of serum response factor, which plays a critical role in muscle proliferation and differentiation [10, 12, 17]. MiR-206 is only expressed in skeletal muscle while miR-1 and miR-133 are expressed in both skeletal and cardiac muscles. MiR-206 also promotes muscle differentiation by the repression of follistatin-like 1 and Utrophin genes expression, and its expression is induced by MyoD and myogenin, both of which are critical transcriptional factors for muscle differentiation [14, 18, 19]. These three miRNAs contribute to muscle development, differentiation and homeostasis in cooperation with each other. In addition, these miRNAs participate in the pathogenesis of several muscle diseases such as cardiac hypertrophy, arrhythmia and muscular dystrophy [20–22]. Yang et al. demonstrated that administration of an antisense inhibitor for miR-1 in infarcted rat hearts could relieve arrhythmogenesis, which strongly suggests that targeting miRNAs could be the novel therapeutic strategy [21].
Based on this evidence, we suggested that overexpression of these three miRNAs, miR-1, miR-133 and miR-206, in muscle injury model could accelerate muscle regeneration. All three muscle-specific miRNAs are important factors in muscle development, therefore we administered a local injection of double-stranded (ds) miR-1, miR-133 and miR-206 mediated atelocollagen to injured muscle. The purpose of this study was to demonstrate the acceleration of skeletal muscle regeneration by single local injection of muscle specific miRNAs in the rat tibialis anterior muscle laceration model. Skeletal muscle injury in animal models consists of three distinct phases: degeneration and inflammation, regeneration and fibrosis [23–26]. Since the initial 1 week after injury necrosis and inflammation phase is the most critical phase for enhancing muscle regeneration, we focused on this 7 days after injury phase to evaluate the skeletal muscle regeneration.
Materials and methods
C2C12 cell culture and transfection
C2C12 cells were seeded from a 1.0 × 104/well into a 12-well plate with Dulbecco’s modified Eagle’s medium (Invitrogen, Calsband, CA, USA) containing 10% foetal bovine serum (Invitrogen) and 1% penicillin/streptomycin (Invitrogen). Myogenic differentiation was induced by changing the medium to Dulbecco’s modified Eagle’s medium containing 2% horse serum and 1% penicillin/streptomycin. Transfections were carried out using Lipofectamine™ LTX transfection reagent (Invitrogen) according to the manufacturer’s instructions. A total of 20 nM of double-stranded miRNA or siRNA were used. The sequence of each mmu-miRNA is as follows: miR-1; 5′-ACA UAC UUC UUU AUA UGC CCA UA-3′, 3′-UGG AAU GUA AAG AAG UAU GUA U-5′ miR-133; 5′-GCU GGU AAA AUG GAA CCA AAU-3′, 3′-UUU GGU CCC UUC AAC CAG CUG-5′ miR-206; 5′-ACA UGC UUC UUU AUA UCC UCA U-3′, 3′-UGG AAU GUA AGG AAG UGU GUG G-5′. Control siRNAs with no specific function were also prepared for the control group (sequences; 5′-ATC CGC GCG ATA GTA CGT A-3′ and 3′-overhung dTdT/dTdT (sense/antisense); siRNA negative control, B-Bridge International, Inc., Mountain View, CA, USA). The medium was replaced with fresh medium every 2 days. The cells were incubated at 37°C in 5% CO2 for a total of 4 days.
Immunocytochemistry
To examine the effect of overexpression of miR-1, miR-133 and miR-206 on the myogenic differentiation of C2C12 cells, immunocytochemistry was conducted as previously described [27]. The cells were fixed in cold methanol for 2 min., and washed in phosphate-buffered saline (PBS) for 10 min. at room temperature (RT). Afterwards, the cells were washed three times in PBS, and incubated in blocking buffer for 30 min. at RT. After washing the cells in PBS, they were incubated overnight at 4°C with primary antibody using monoclonal antiskeletal myosin [fast] clone MY-32 (Sigma-Aldrich, St. Louis, MO, USA), MyoD1 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), myogenin (Chemicon, Temecula, CA, USA) and Pax7 (R&D Systems, Inc., Mineapolis, MN, USA). After washing the cells in PBS, they were incubated with secondary antibody using Alexa Fluor 488 or 568 -conjugated goat antimouse IgG (Molecular Probes; Invitrogen, Eugene, OR, USA) for 1 hr at RT. 4,6,-diamidino-2-phenylindole (DAPI) (Dojindo Laboratories, Kumamoto, Japan) solution was applied for 5 min. for nuclear staining. For each well, five microscopic areas at ×200 magnification were randomly chosen, and the fusion index (ratio of nuclei in myotubes to all nuclei) and total cell number were calculated to evaluate the myogenic differentiation and cell proliferation capacity of C2C12 cells. And MyoD, myogenin and Pax7+ cells were counted to evaluate the expression of myogenic marker genes.
Preparation of double-stranded RNA / atelocollagen complex
All animals were divided into two groups. One was used for the injection of ds miR-1, 133 and 206, and the other was used as a siRNA control. Atelocollagen is a highly purified type I collagen isolated from the calf dermis with pepsin treatment (Koken Co., Ltd., Tokyo, Japan). The ds miR-1 (the sequence of rno-miR-1 is as follows. miR-1; 5′-ACA UAC UUC UUU AUA UGC CCA UA-3′, 3′-UGG AAU GUA AAG AAG UGU GUA U-5′), miR-133 and miR-206 or control siRNA and atelocollagen complex was prepared as follows: An equal volume of atelocollagen (in PBS at pH 7.4) and miRNAs solution (30 μg/25 μl; the volume of each miRNA is 10 μg) was combined and mixed by rotation at 4°C for 20 min. The non-specific control siRNA and atelocollagen complex was also prepared in the same way.
Skeletal muscle injury model and local injection
All animals were housed individually and fed a commercial diet. This study was reviewed and approved by the Ethics Committee for Experimental Animals of Hiroshima University, and all animals were treated according to the guidelines of the Institutional Animal Care and Use Committee.
Twelve-week-old male adult SD rats were used in this study. All rats were obtained from CLEA Japan, Inc. (Meguro-ku, Tokyo, Japan). The rat skeletal muscle injury model was made according to a previous report [3]. Rats were anaesthetized with ketamine and xylazine (60 and 10 mg/kg), and an adequate depth of anaesthesia was maintained. An anterolateral skin incision was made in the right leg, and the tibialis anterior muscle was exposed. The fascia was incised longitudinally, and carefully released from the muscle belly. The muscle belly was lacerated transversely at the mid portion using scalpel. The defect shape was a wedge, and its size was approximately 6 mm length, 4 mm wide and 5 mm depth. A single local injection of atelocollagen mediated ds miRNA or control siRNA was administered into the surrounding area of the muscle defect after closure of the fascia with a suture. The skin was closed and the rats were placed in their cages, and were permitted unrestrained mobility.
Mechanical evaluation of the regenerated muscle
One week after the local injection, isometric tensile strength produced by stimulating the common peroneal nerve was measured with transducer load cells (LVS-1KA; Kyowa Electronic Instruments, Tokyo, Japan), and recorded with a sensor interface (PCD-300A; Kyowa Electronic Instruments) and software (PCD-30A; Kyowa Electronic Instruments) as previously described [3]. Under general anaesthesia by intraperitoneal injection of ketamine (60 mg/kg), all rats were set in a supine position and the tibialis anterior muscles and common peroneal nerves in both legs were exposed (n= 5 in each group). Both legs were fixed tightly to the leg holder, and the common peroneal nerves of both legs were stimulated with an electrostimulator (SEN-2201, Nihon Koden, Tokyo, Japan). Frequencies of stimulation were 1 Hz (fast twitch) and 50 Hz (tetanus). The minimum voltage that caused the tibialis anterior muscle to visibly contract was measured as a threshold to determine the voltage of stimulation. The nerve was stimulated by a voltage 10-fold higher than that of the threshold, and maximum isometric tensile strength produced by the tibialis anterior muscles of both legs was measured. To minimize inter-animal variation, the data were normalized to non-treated controls as follows: The strength of the experimental muscle was divided by that of the control non-injured muscle measured from the contralateral side and shown as a strength ratio. Strength ratios of the lacerated side to the contralateral side were also calculated in fast twitch and tetanus stimulation.
Histological evaluation
After electromechanical evaluation, all rats were killed with an overdose of sodium pentobarbital at the end of the electromechanical studies. The regenerated tibialis anterior muscles of each group were harvested and were quickly embedded in Tissue Freezing Medium (Triangle Biomedical Sciences, Durham, NC, USA), snap-frozen in liquid nitrogen and stored at –80°C.
For the evaluation of the fibrotic area, the myofibre diameter, and the number of the centronucleated myofibre, Masson trichrome staining was carried out on each sample. The average area of fibrosis in the sagittal sections and the average diameter of myofibres in the axial sections were analysed using a digital microscope (BZ-9000, Keyence, Japan). At 200× magnification in the axial sections, the myofibre diameters of five randomly selected fields per muscle were evaluated as previously described [3]. The total number of centronucleated myofibres was measured within the injured site in five random fields of each sample as previously described [3].
Synthesis of complementary DNA (cDNA)
Three and 7 days after local injection, total RNA was isolated from the regenerated muscle (n= 5) that had been homogenized on ice with Trizol reagent (Invitrogen) for PCR analysis. Total RNA yields were calculated and quality was determined using absorption spectrochemical analysis. One microgram of total RNA was reverse-transcribed using the QuantiTect® Reverse Transcription Kit (Qiagen, Chatsworth, CA, USA) according to the manufacturer’s protocol. The genomic DNA elimination reaction was carried out using 2 μl of gDNA wipeout buffer, 1 μg (1 μl) template RNA and 11 μl RNase-free water at 42°C for 2 min. Reverse transcription was performed in 1 μl quantiscript reverse transcriptase, 4 μl quantiscript RT buffer, 1 μl RT primer mix and 14 μl template RNA (the entire genomic DNA elimination reaction) at 42°C for 15 min. and 95°C for 3 min. and then the cDNA product was maintained at 4°C.
Quantitative (real time) PCR
Quantitative RT-PCR assays were performed with a TaqMan miRNA assay kit (Applied Biosystems, Mountain View, CA, USA) for the expression of rno-miR-1, 133 and 206 and SYBR Green (Invitrogen) for the MyoD, myogenin, Pax7 and transforming growth factor (TGF)-β1. Reverse transcriptase reactions of mature miRNAs contained a sample of total RNA, 50 nM stem-loop RT primer, 10× RT buffer, 100 mM each dNTPs, 50 U/μl MultiScribe reverse transcriptase and 20 U/μl RNase inhibitor. Fifteen microlitre reactions were incubated in a thermocycler (BioRad, Hercules, CA, USA) for 30 min. at 16°C, 30 min. at 42°C, 5 min. at 85°C, and held at 4°C. Real-time PCR was performed with a Mini Opticon Real-time PCR System (BioRad) in a 10 μl PCR mixture containing 1.33 μl RT product, 21× TaqMan Universal PCR Master Mix, 0.2 μM TaqMan probe, 15 μM forward primer and 0.7 μM reverse primer. Each SYBR Green reaction was performed with 1.0 μl template cDNA, 10 μl SYBR Green mix, 1.5 μM primer and water to adjust the final volume to 20 μl. Primer sequences were as follows: mouse MyoD1, 5′-ACA TAG ACT TGA CAG GCC CCG A-3′ (forward), 5′-AGA CCT TCG ATG TAG CGG ATG G-3′ (reverse); mouse myogenin, 5′-TAC GTC CAT CGT GGA CAG CAT-3′ (forward), 5′-TCA GCT AAA TTC CCT CGC TGG-3′ (reverse); mouse Pax7, 5′-CTG GAT GAG GGC TCA GAT GT-3′ (forward), 5′-GGT TAG CTC CTG CCT GCT TA-3′ (reverse); mouse ACTB, 5′-AGC TGC CTG ACG GCC A-3′ (forward), 5′-GAT TCC ATA CCC AAG AAG GAA GG-3′ (reverse); rat myostatin, 5′-TGC TGT AAC CTT CCC AGG ACC A-3′ (forward), 5′-GTG AGG GGG TAG CGA CAG CAC-3′ (reverse); rat TGF-β1, 5′-CCA CGT GGA AAT CAA TGG GA-3′ (forward), 5′-GGC CAT GAG GAG CAG GAA G-3′ (reverse); rat MyoD1, 5′-GCG ACA AGC CGA TGA CTT CTA T-3′ (forward), 5′-GGT CCA GGT CCT CAA AAA AGC-3′ (reverse); rat myogenin, 5′-GAC CCT ACA GGT GCC CAC AA-3′ (forward), 5′-ACA TAT CCT CCA CCG TGA TGC T-3′ (reverse); rat Pax7, 5′-GCC CTC AGT GAG TTC GAT TAG C-3′ (forward), 5′-TCC TTC CTC ATC GTC CTC TTT C-3′ (reverse); and rat ACTB, 5′-GAT CAT TGC TCC TCC TGA GCG-3′ (forward), 5′-TGC TGA TCC ACA TCT GCT GGA-3′ (reverse). All reactions were incubated in a 48 well plate at 95°C for 10 min., followed by 40 cycles of 95°C for 15 sec. and 60°C for 1 min. and performed in triplicate. The snoRNA-135 or ACTB gene was used as a control to normalize differences in total RNA levels in each sample. A threshold cycle (CT) was observed in the exponential phase of amplification, and quantification of relative expression levels was performed with standard curves for target genes and the endogenous control. Geometric means were used to calculate the ΔΔCT (delta-delta CT) values and expressed as 2−ΔΔCT. The value of each control sample was set at 1 and was used to calculate the fold-change of target genes.
Immunofluorescent analysis of the regenerated muscle
Tissues embedded in compound were sectioned into 6 μm sections serially in the sagittal or axial planes. For immunofluorescent staining of desmin, isolectin B4, Pax7, MyoD, vimentin and TGF-β1, 6-μm serial sections were mounted on saline-coated glass slides and air-dried before being fixed with 4.0% paraformaldehyde at 4°C for 5 min. and stained immediately. Immunohistochemistry was performed with the following antibodies: desmin (Santa Cruz Biotechnology, Inc.), isolectin B4 (Vector Laboratories, Burlingame, CA, USA), Pax7 (R&D Systems, Inc.), MyoD1 (Santa Cruz Biotechnology, Inc.) and Vimentin (Santa Cruz Biotechnology, Inc.). The secondary antibodies for each immunostaining were as follows: Alexa Fluor 488 or 568-conjugated goat antimouse IgG (Molecular Probes; Invitrogen) for Pax7 and TGF-β1, Alexa Flour 488 or 568-conjugated goat anti-rabbit IgG (Molecular Probes; Invitrogen) for desmin, MyoD1 and vimentin. DAPI solution was applied for 5 min. for nuclear staining. For the evaluation of capillary density, five randomly selected areas around the fibrosis area in axial section at 200× magnification were evaluated. Capillaries were recognized as tubular structures positive isolectin B4, the marker for rat endothelial cell.
Statistical analyses
All results in this study were expressed as the mean ± S.D. Comparison among six groups was done using the Tukey–Kramer’s post hoc test, and the Mann Whitney U-test was used for the detection of the differences between control siRNA group and miRNA group. P-values of less than 0.05 were considered to be statistically significant.
Results
A combination of miR-1, miR-133 and miR-206 enhances the differentiation and proliferation of C2C12 cells
To determine which is the most effective for the rat injured skeletal muscle in vivo, each muscle-specific miRNA or the combination of all muscle-specific miRNAs were administered, after which we measured the fusion index and cell number in C2C12 cells in vitro. The fusion index of C2C12 cells in the combination of miR-1, 133 and 206 was significantly higher in comparison with other groups (Fig. 1A, B). MiR-1 and miR-206, which play a role in muscle differentiation, also significantly enhanced muscle differentiation, but the combination of miR-1, 133 and 206 was the most effective for muscle differentiation. The cell number in the miR-133, which plays a role in proliferation, was significantly higher than in other groups (Fig. 1B). The combination of miR-1, 133 and 206 also could increase cell numbers, but there was no significant difference between miR-133 and the combination of miR-1, 133 and 206. According to these results, we determined that the most appropriate administration for muscle regeneration is the combination of miR-1, 133 and 206.
Fig 1.
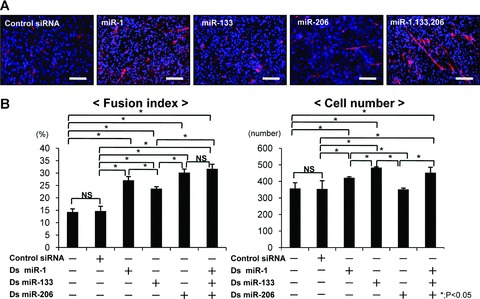
The effect of the combination of miR-1, 133 and 206 for the myogenic differentiation in C2C12 cells. (A) Immunocytochemistry for fast myosin heavy chain at 4 days after transfection. Myotube was shown as red, and nuclei as blue. Original magnification ×200. Bar; 100 μm. (B) Fusion index and cell number to evaluate for myoblast differentiation and cell proliferation in C2C12 cells. Data were calculated as means ± S.D. Statistical analysis was performed with the Tukey– Kramer’s post hoc test. *; P < 0.05. NS; no significance.
The expression pattern of miR-1, miR-133 and miR-206 after skeletal muscle injury
To confirm the temporal expression pattern of each muscle-specific miRNA after muscle laceration, rats were killed at days 0, 1, 2, 3, 5, 7 by an overdose of pentobarbital (n= 3 at each time-point). RNA was isolated from the harvested tibialis anterior muscle, and real-time PCR for mature miR-1, miR-133 and miR-206 was conducted. The expression level of each miRNA on the first day after injury decreased less than 0.5-fold of the pre-injured level. As each day passed, the expression level of each miRNA gradually elevated towards 7 days after injury, with miR-1 and miR-206 increasing 1.2-fold, and miR-133 returning to approximately the same level as that of pre-injury (Fig. 2).
Fig 2.
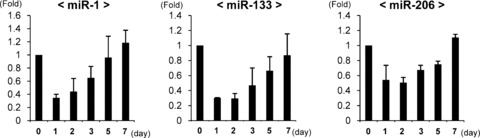
Temporal expression changes of mature rno-miR-1, 133 and 206 after rat tibialis anterior muscle injury by real-time PCR analysis (each time-point; n= 3). snoRNA-135 was used as an internal control. Data were calculated as means ± S.D.
A local injection of miR-1, miR-133 and miR-206 accelerates the muscle morphology and functional regeneration
To evaluate the muscle regeneration morphologically after local injection, Masson Tricrome staining and immunohistochemistry of desmin, which is expressed during skeletal muscle development and myotube formation, were performed. Many more regenerated centronucleated myofibres were observed near the perifibrosis site in the miRNA group compared to the control siRNA group (Fig. 3A). Desmin+ myofibres increased in the miRNA group compared to the control siRNA group (Fig. 3B). The diameter of myofibres at 1 week after injection in the miRNA group (60.3 ± 2.4 μm) was significantly greater than that in the control siRNA group (44.7 ± 3.5 μm) (Fig. 3C). The number of centronucleated myofibres in the miRNA group (27.4 ± 3.4) was higher than that in the control siRNA group (11.6 ± 1.7) (Fig. 3C).
Fig 3.
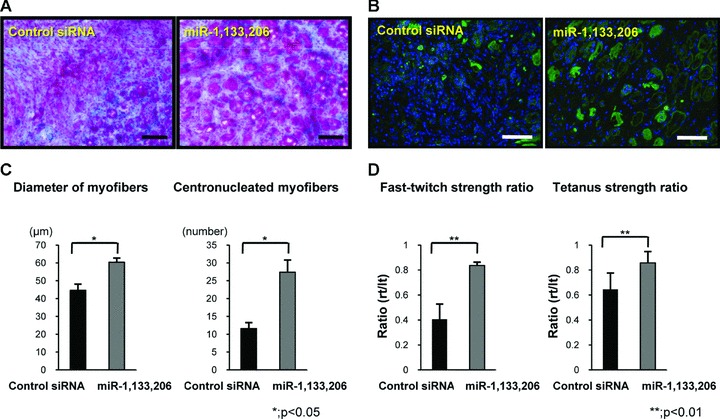
Histological and functional analysis of the acceleration of muscle regeneration by miR-1, 133 and 206. (A) Masson Trichrome staining in the axial section of each group. (B) Axial section of immunohistochemistry of desmin (lower). The bar indicates 100 μm. (C) Diameter of regenerated muscle fibres and the number of the centronucleated myofibres in each group. There was a significant difference between both groups. (D) Fast twitch and tetanus strength ratios in both groups. Fast twitch and tetanus strength ratios in the miRNAs group were significantly higher than in the control siRNA group. Data were calculated as means ± S.D. *; P < 0.05, **; P < 0.01. The P-value was determined by the Mann-Whitney U-test.
To evaluate the functional recovery at 1 week after injection, we conducted physiological measurements of strength, the fast-twitch and tetanus strength ratio. The fast-twitch strength ratio in the miRNA group (0.84 ± 0.03 fold) was significantly higher than in the control siRNA group (0.4 ± 0.1 fold; P < 0.01) (Fig. 3D). The tetanus strength ratio in the miRNA group (0.84 ± 0.1 fold) was also significantly higher than in the control siRNA group (0.64 ± 0.1 fold; P < 0.01). The mean twitch strength of injured side in miRNA group (0.65 ± 0.08 N) was significantly higher than in control siRNA group (0.46 ± 0.11 N) (P < 0.01). There was no significant difference of mean twitch strength of uninjured side in both groups (miRNA group; 0.82 ± 0.03 N, control siRNA group; 0.82 ± 0.05 N). The mean tetanus strength of injured side in miRNA group (1.17 ± 0.19 N) was significantly higher than in control siRNA group (0.92 ± 0.17 N) (P < 0.01). There was also no significant difference of mean tetanus strength of uninjured side in both groups (miRNA group; 1.32 ± 0.11 N, control siRNA group; 1.36 ± 0.12 N). Injected miR-1, miR-133 and miR-206 could accelerate morphological and functional muscle regeneration.
Enhancement of angiogenesis by miR-1, 133 and 206 injection
Angiogenesis plays a crucial role in muscle repair, therefore, we evaluated angiogenesis during muscle regeneration in both group by capillary density at 1 week. Vascular staining of isolectine B4 demonstrated that neovascularization around fibrosis area in miRNA group was enhanced compared to control siRNA group (Fig. 4A). Capillary density in the miRNA group (59.6 ± 7.1 per mm2) was significantly increased than that in the control siRNA group (45.2 ± 5.7 per square mm2) (P < 0.05) (Fig. 4B).
Fig 4.
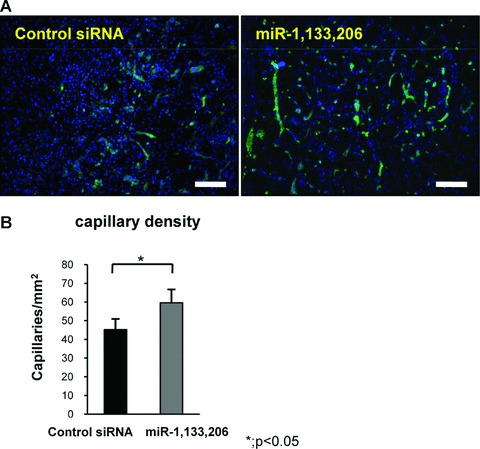
Evaluation of angiogenesis. (A) Immunofluorescent analysis of isolectin B4 in the axial section of both groups. The bar indicates 100 μm. (B) Capillary density in both groups. The capillary density in the miRNAs group was higher compared to the control siRNA group. There was a significant difference between both groups. Data were calculated as means ± S.D. *; P < 0.05. The P-value was determined by the Mann-Whitney U-test.
Local injection of miR-1, 133 and 206 prevented fibrosis during muscle regeneration
At 1 week after local injection, Masson Tricrome staining on the sagittal section revealed that muscle-specific miRNAs could replace the muscle fibres at the injured site, while retaining the large defect and inflammatory cell accumulation at the injured site in the control siRNA group (Fig. 5A). To evaluate fibrosis at 1 week after the injection, an immunofluorescent assessment of vimentin, a corrective indicator of fibrosis scar formation, was performed. MiR-1, 133 and 206 could effectively prevent fibrosis compared to the control siRNA group (Fig. 5A). An immunofluorescent assessment of TGF-β1, a key factor of fibrosis, revealed that miR-1, 133 and 206 also effectively prevented TGF-β1 expression in the protein level (Fig. 5A). The area of fibrosis in the miRNA group (837.848 ± 15.848 μm2) was significantly lower than that in the control siRNA (4424.044 ± 813.163 μm2) (Fig. 5B). mRNA of TGF-β1 was significantly down-regulated in the miRNA group compared to the siRNA group on days 3 and 7 (Fig. 5C). Administration of miR-1, 133 and 206 could prevent fibrosis during muscle regeneration through down-regulation of TGF-β1.
Fig 5.
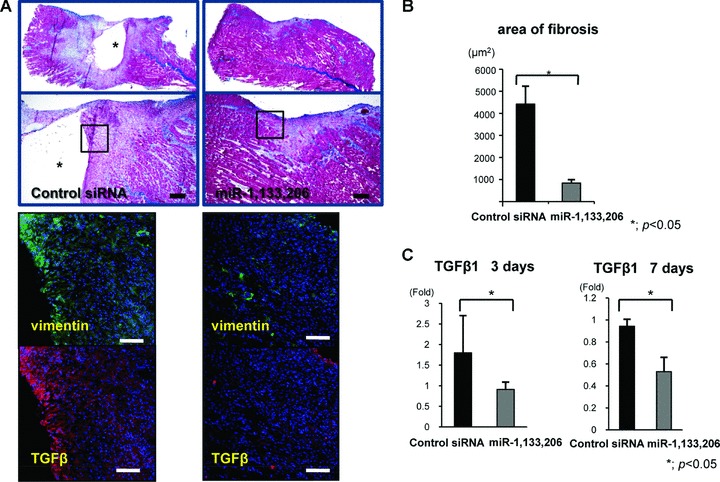
(A) Masson Trichrome staining in the sagittal section of each group (upper). * shows the defect at the laceration site. The bar indicates 300 μm. Immunofluorescent analysis of vimentin and TGF-β in the axial section of both groups (lower). These figures show the square part of the upper figures. The bar indicates 100 μm. (B) Area of fibrosis. The area in the miRNAs group was low compared to the control siRNA group. There was a significant difference between both groups. *; P < 0.05. (C) Real-time PCR analysis of TGF-β1 at days and 7 days after injury. The expression level of TGF-β1 was significantly down-regulated in the miRNAs group. Data were calculated as means ± S.D. *; P < 0.05. The P-value was determined by the Mann-Whitney U-test.
Injected muscle-specific miRNAs can up-regulate the expression of the muscle-specific genes, and down-regulate myostatin
To further examine muscle regeneration, we performed real-time PCR for MyoD1, myogenin and Pax7, markers for myogenesis. The expression level of MyoD, myogenin and Pax7 in the miRNA group was significantly higher than in the control siRNA group at 3 and 7 days (Fig. 6A). The expression of these genes increased in the period from 3 to 7 days. Furthermore, we investigated the expression of myostatin (GDF8), one of the validated target genes of miR-1 and miR-206, at 3 and 7 days. The expression level of myostatin in the miRNA group was significantly lower than in the control siRNA group at 3 and 7 days (Fig. 6A).
Fig 6.
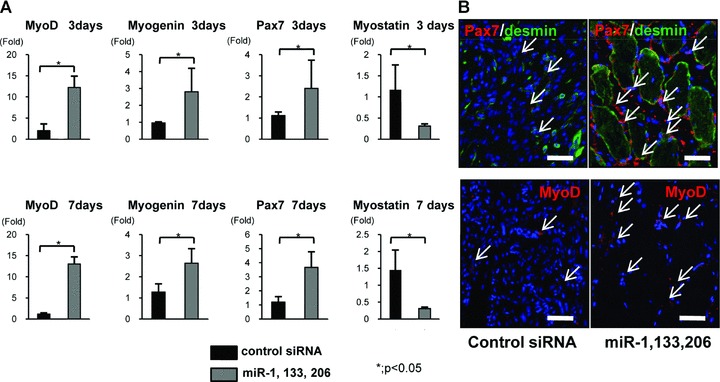
(A) Real-time PCR analysis of the marker genes of myogenesis, MyoD1, myogenin and Pax7, and myostatin at 3 and 7 days after injury. MyoD1, myogenin and Pax7 were significantly up-regulated in the miRNA group at 3 and 7 days. Myostatin was significantly down-regulated in the miRNA group at 3 and 7 days. Data were calculated as means ± S.D. *; P < 0.05. The P-value was determined by the Mann-Whitney U-test. (B) Immunofluorescent analysis of Pax7, desmin and MyoD1 in the axial section of both groups. An abundant number of Pax7+ cells around desmin+ myofibres were observed in the miRNA group. MyoD1+ cells were also abundant in the miRNA group compared to the control siRNA group. Bar: 50 μm.
To investigate the expression pattern of these myogenic markers, we performed an immunofluorescent assessment of Pax7 and MyoD1 (Fig. 6B). In the miRNA group, a significant number of Pax7+ cells were observed around the desmin+ regenerative muscle fibres at 1 week. A much greater number of MyoD1+ cells were also observed around peri-injured sites compared to the control siRNA group. Both early markers of skeletal muscle progenitor cells and myogenic committed markers were significantly up-regulated in the miRNA group compared to the control siRNA group.
miR-1, 133 and 206 can up-regulate MyoD1, myogenin and Pax7 in C2C12 cells
To confirm the effect of miR-1, 133 and 206 to induce the myogenic marker genes, MyoD1, myogenin and Pax7 in vitro, we performed real-time PCR using the mouse-specific primer of MyoD1, myogenin and Pax7. The expression of MyoD1, myogenin and Pax7 was significantly up-regulated in the combination of miR-1, 133 and 206 (Fig. 7A). Immunofluorescent analyses revealed that the number of MyoD1, myogenin and Pax7+ cells was significantly higher in the combination of miR-1, 133 and 206 compared to other groups (Fig. 7B, C).
Fig 7.
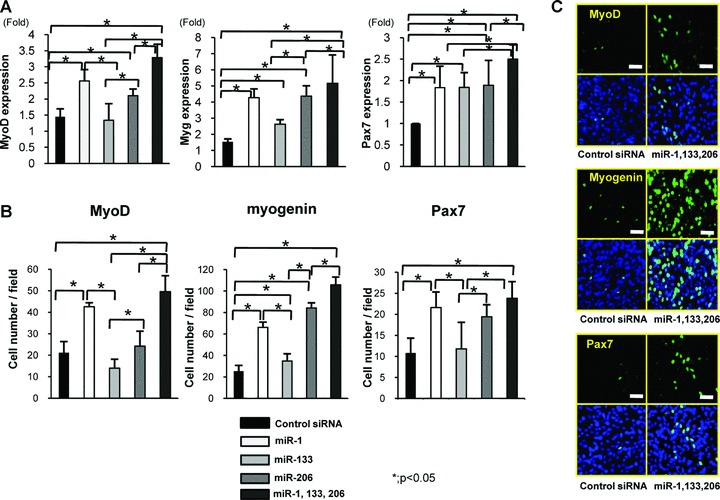
(A) Real-time PCR analysis of the expression of MyoD1, myogenin and Pax7 in C2C12 cells. Overexpression of miR-1, 133 and 206 could significantly induce the expression of MyoD1, myogenin and Pax7. (B) The number of MyoD, myogenin and Pax7+ cells in each group. The number of MyoD1, myogenin and Pax7+ cells was significantly higher in the combination of miR-1, 133 and 206 compared to other groups. Data were calculated as means ± S.D. *; P < 0.05. Statistical analysis was performed with the Tukey–Kramer’s post hoc test. (C) Immunocytochemistry of MyoD1, myogenin and Pax7. Abundant MyoD1, myogenin Pax7+ cells after differentiation of C2C12 cells were observed in the combination of miR-1, 133 and 206. Bar; 50 μm.
Discussion
The current study demonstrated that the administration of miRNA provides a novel therapeutic strategy for muscle injury. MiRNAs have been shed a light, because they have been found to play a crucial role in the pathogenesis of human diseases. Many investigators have attempted to elucidate the precise role of individual miRNAs in human disease to develop a new strategy to target miRNAs. Tazawa et al. demonstrated that miR-34a is down-regulated in human colon cancer, and that tumour growth in mice is significantly inhibited by the administration of ds miR-34a/atelocollagen complex [28]. Several therapeutic trials to regulate the endogenous miRNAs related to various diseases have been conducted [21, 28–30].
Skeletal muscle injury in animal models consists of three distinct phases: degeneration and inflammation, regeneration and fibrosis [23–26]. The first phase in the first few days after injury is characterized by necrosis of muscle tissue, degeneration and an inflammatory response in the injured site. In the regeneration phase, which occurs at 5 to 10 days after injury, several growth factors that regulate myoblast proliferation and differentiation are promoted, to secrete and produce connective tissue and scar tissue. The fibrosis phase, the formation of the scar tissue, usually occurs between the second and third week after injury. It is important to accelerate muscle regeneration and prevent fibrosis within the initial 2 weeks of the muscle repair process, leading to successful muscle repair. From the view point of miRNA, the first week after injury is also important during skeletal muscle regeneration. Yuasa et al. showed the temporal expression profiles of miR-1, miR-133 and miR-206 after the cardiotoxin (CTX) injection into TA muscle in mice [31]. They demonstrated that the expression level of miR-1, miR-133 and miR-206 decreased after the CTX injection, after which its expression level gradually increased during the regeneration process. Its expression pattern was similar to our results. In the current study, the expression level of miR-1, miR-133 and miR-206 was decreased at day 1 after muscle injury, therefore, exogeneous miR-1, 133 and 206 were administered at day 0, which can promote muscle regeneration instead of deteriorated endogeneous miRNA-1, 133 and 206.
The target genes for miRNAs are estimated to range between one and several hundred, based on target predictions using the bioinformatics approach [32]. Therefore, many molecular networks via miR-1, 133 and 206 might interact to promote muscle regeneration after a local injection. Several validated target genes of miR-1, 133 and 206, which relate to muscle regeneration have been reported, such as HDAC4, GDF8, fibronectin and nPTB as miR-1 target genes, serum response factor, nPTB and RhoA as miR-133 target genes, Cx43, GDF8 and nPTB as miR-206 [10, 12, 33–37]. In the current study, injected miR-1, 133 and 206 might regulate these target genes to enhance muscle regeneration. The real-time PCR analyses revealed that the expression of MyoD1, myogenin and Pax7 was up-regulated at 3 and 7 days after injection. MyoD and myogenin were induced during muscle differentiation, and miR-1, 133 and 206 are up-regulated by the MyoD and myogenin [11]. Interestingly, Pax7 was significantly up-regulated by the administration of miR-1, 133 and 206. Pax7 has been implicated in satellite cell specification, the regulation of satellite cell self-renewal. We confirmed these results in C2C12 cells by the overexpression of miR-1, 133 and 206 in vitro. McFarlane et al. demonstrated that myostatin (GDF8) inhibits Pax7 expression during myoblast proliferation, implicating it in the negative regulation of satellite cell self-renewal. Myostatin is one of the validated target genes of miR-1 and 206. In the present study, the expression level of myostatin was down-regulated at 3 and 7 days after miR-1, 133 and 206 injections. This might be one of the mechanisms of up-regulation of Pax7. Our results suggest that the combination of miR-1, 133 and 206 can enhance not only myogenic differentiation but also myoblast proliferation and self-renewal. During muscle regeneration, angiogenesis was enhanced by miR-1, 133 and 206 injections. Administration of miR-1, 133 and 206 lead to enhancement of muscle regeneration by angiogenesis, however, direct mechanism of angiogenesis by miR-1, 133 and 206 is still unclear. Furthermore, injection of miR-1, 133 and 206 can significantly prevent fibrosis during muscle regeneration through the down-regulation of TGF-β, which is a major trigger of the fibrotic cascade. Undetermined target genes of these muscle-specific miRNAs might participate in antifibrotic event.
miR-1, miR-133 and miR-206 cooperate to maintain muscle homeostasis, balancing between cell proliferation and differentiation. We suggest that administration of all three of these miRNAs can enhance muscle regeneration in vivo. We also administered a local injection of the other miRNA, cartilage-specific miR-140, and atelocollagen only as a control to prove the specific therapeutic efficacy of miR-1, 133 and 206. The same results as for the control siRNA were obtained (data is not shown). Atelocollagen has been reported as a biomaterial carrier for gene delivery in vitro and in vivo. SiRNA or miRNA combined with atelocollagen is expected to stay stable for 1 week in this system [24]. Therefore, it is reasonable to expect that the effect of these miRNAs in the muscle-injured site will be long-lasting using this system. The current study showed the possibility of a new strategy for skeletal muscle repair regulating miRNAs in vivo by a local injection of ds miRNAs into the injured site. Further studies, including elucidation of the detailed mechanism with the determination of new target genes, are needed before a clinical trial is carried out.
Acknowledgments
This work was supported in part by a grant-in-aid for the General Insurance Association of Japan.
References
- 1.Chan YS, Li Y, Foster W, et al. The use of suramin, an antifibrotic agent, to improve muscle recovery after strain injury. Am J Sports Med. 2005;33:43–51. doi: 10.1177/0363546504265190. [DOI] [PubMed] [Google Scholar]
- 2.Bernasconi P, Di Blasi C, Mora M, et al. Transforming growth factor beta1 and fibrosis in congenital muscular dystrophies. Neuromuscul Disord. 1999;9:28–33. doi: 10.1016/s0960-8966(98)00093-5. [DOI] [PubMed] [Google Scholar]
- 3.Shi M, Ishikawa M, Kamei N, et al. Acceleration of skeletal muscle regeneration in a rat skeletal muscle injury model by local injection of human peripheral blood-derived CD133-positive cells. Stem Cells. 2009;27:949–60. doi: 10.1002/stem.4. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Farh KK, Grimson A, Jan C, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–21. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 7.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcucci G, Radmacher MD, Maharry K, et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. Engl J Med. 2008;358:1919–28. doi: 10.1056/NEJMoa074256. [DOI] [PubMed] [Google Scholar]
- 9.Nakasa T, Miyaki S, Okubo A, et al. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum. 2008;58:1284–92. doi: 10.1002/art.23429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen JF, Mandel EM, Thomson JM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–33. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rao PK, Kumar RM, Farkhondeh M, et al. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci USA. 2006;103:8721–6. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y, Samal E, Strivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–20. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 13.Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by twist and is required in muscles during larval growth. Genes Dev. 2005;19:2343–54. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenberg MI, Georges SA, Asawachaicharn A, et al. MyoD inhibits Fstl1 and Utrn expression by including transcription of miR-206. J Cell Biol. 2006;175:77–85. doi: 10.1083/jcb.200603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu J, McKinsey TA, Zhang CL, et al. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol Cell. 2000;6:233–44. doi: 10.1016/s1097-2765(00)00025-3. [DOI] [PubMed] [Google Scholar]
- 16.Lu J, McKinsey TA, Nicol RL, et al. Signal- dependent activation of the MEF2 trasncription factor by dissociation from histone deacetylase. Proc Natl Acad Sci USA. 2000;97:4070–5. doi: 10.1073/pnas.080064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miano JM. Serum response factor: toggling between disparate programs of gene expression. J Mol Cell Cardiol. 2003;35:577–93. doi: 10.1016/s0022-2828(03)00110-x. [DOI] [PubMed] [Google Scholar]
- 18.Kim HK, Lee YS, Sivaprasad U, et al. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174:677–87. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taniguchi M, Seok HY, Callis TE, et al. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2007;42:1137–41. doi: 10.1016/j.yjmcc.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Rooij E, Sutherland LB, Liu N, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA. 2006;103:18255–60. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang B, Lin H, Xiao J, et al. The muscle-specific microRNA-1 regulates cardiac arrythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13:486–91. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 22.Eisenberg I, Eran A, Nishio I, et al. Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad Sci USA. 2007;104:17016–21. doi: 10.1073/pnas.0708115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukushima K, Badlani N, Usas A, et al. The use of an antifibrosis agent to improve muscle recovery after laceration. Am J Sports Med. 2001;29:394–402. doi: 10.1177/03635465010290040201. [DOI] [PubMed] [Google Scholar]
- 24.Garrett WE, Jr, Safran MR, Seaber AV, et al. Biomechanical comparison of stimulated and nonstimulated skeletal muscle pulled to failure. Am J Sports Med. 1987;15:448–54. doi: 10.1177/036354658701500504. [DOI] [PubMed] [Google Scholar]
- 25.Garrett WE, Jr, Seaber AV, Boswick J, et al. Recovery of skeletal muscle after laceration and repair. J Hand Surg. 1984;9:683–92. doi: 10.1016/s0363-5023(84)80014-3. [DOI] [PubMed] [Google Scholar]
- 26.Kasemkijwattana C, Menetrey J, Somogyl G, et al. Development of approaches to improve the healing following muscle contusion. Cell Transplant. 1998;7:585–98. doi: 10.1177/096368979800700609. [DOI] [PubMed] [Google Scholar]
- 27.Nozaki M, Li Y, Zhu J, et al. Improved muscle healing after contusion injury by the inhibitory effect of suramin on myostatin, a negative regulater of muscle growth. Am J Sports Med. 2008;36:2354–62. doi: 10.1177/0363546508322886. [DOI] [PubMed] [Google Scholar]
- 28.Tazawa H, Tsuchiya N, Izumiya M, et al. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA. 2007;104:15472–7. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krützfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 30.Krützfeldt J, Kuwajima S, Braich R, et al. Specificity, duplex degradation and subcellular localization of antagomir. Nucleic Acids Res. 2007;39:2885–92. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuasa K, Hagiwara Y, Ando M, et al. MicroRNA-206 is highly expressed in newly formed muscle fibers: implications regarding potential for muscle regeneration and maturation in muscular dystrophy. Cell Structure and fuction. 2008;33:163–9. doi: 10.1247/csf.08022. [DOI] [PubMed] [Google Scholar]
- 32.Krek A, Grun D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 33.Sayed D, Hong C, Chen IY, et al. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100:416–24. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 34.Clop A, Marcq F, Takeda H, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813–8. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 35.Boutz PL, Chawla G, Stoilov P, et al. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev. 2007;21:71–84. doi: 10.1101/gad.1500707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson C, Catoe H, Werner R. MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Res. 2006;34:5863–71. doi: 10.1093/nar/gkl743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carè A, Catalucci D, Felicetti F, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–8. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]


