Abstract
Kringle 5(K5) is the fifth kringle domain of human plasminogen and its anti-angiogenic activity is more potent than angiostatin that includes the first four kringle fragment of plasminogen. Our recent study demonstrated that K5 suppressed hepatocarcinoma growth by anti-angiogenesis. To find high efficacy and minimal peptide sequence required for the anti-angiogenic and anti-tumour activities of K5, two deletion mutants of K5 were generated. The amino acid residues outside kringle domain of intact K5 (Pro452-Ala542) were deleted to form K5mut1(Cys462-Cys541). The residue Cys462 was deleted again to form K5mut2(Met463-Cys541). K5mut1 specifically inhibited proliferation, migration and induced apoptosis of endothelial cells, with an apparent two-fold enhanced activity than K5. Intraperitoneal injection of K5mut1 resulted in more potent tumour growth inhibition and microvessel density reduction than K5 both in HepA-grafted and Bel7402-xenografted hepatocarcinoma mouse models. These results suggested that K5mut1 has more potent anti-angiogenic activity than intact K5. K5mut2, which lacks only the amino terminal cysteine of K5mut1, completely lost the activity, suggesting that the kringle domain is essential for the activity of K5. The activity was enhanced to K5mut1 level when five acidic amino acids of K5 in NH2 terminal outside kringle domain were replaced by five serine residues (K5mut3). The shielding effect of acidic amino acids may explain why K5mut1 has higher activity. K5, K5mut1 and K5mut3 held characteristic β-sheet spectrum while K5mut2 adopted random coil structure. These results suggest that K5mut1 with high efficacy is the minimal active peptide sequence of K5 and may have therapeutic potential in liver cancer.
Keywords: plasminogen kringle 5, structure, function, anti-angiogenesis, anti-tumour
Introduction
Angiogenesis, the growth of new blood vessels from preexisting capillaries, is necessary for solid tumour growth and metastasis [1, 2]. Angiogenic stimulators such as vascular endothelial growth factor (VEGF) are increased while angiogenic inhibitors such as pigment epithelium-derived factor (PEDF) are decreased under the tumourous pathologic conditions, resulting in overproliferation of capillary endothelial cells and abnormal formation of new blood vessels in neoplasm [3]. Therefore, angiogenic inhibitor may hold great therapeutic potential in the treatment of solid tumour [4, 5].
Some proteolytic fragments from human plasminogen are reported as angiogenic inhibitors. Kringle 5 (K5) is the fifth kringle domain of human plasminogen and has been widely demonstrated to inhibit proliferation of endothelial cells [6]. Moreover, its inhibitory activity is more potent than angiostatin that includes the first four kringle modules of plasminogen (kringles1–4) [6, 7]. In addition, K5 also inhibits endothelial cell migration, an important process in angiogenesis [8], and exerts its effect on endothelial cells by inducing cell cycle arrest and apoptosis. We previously demonstrated the anti-angiogenic activity of K5 in rat model of oxygen-induced retinopathy (OIR) [9, 10] and rabbit model of alkali-burn-induced corneal neovascularization [11]. Recently, we proved the anti-tumour activity of K5 in hepatocarcinoma [12]. Down-regulation of VEGF and up-regulation of PEDF, thus leading toward restoration of the balance in angiogenic control, may represent a mechanism for the anti-angiogenic activity of K5 [13].
To find more potent and smaller peptide sequence required for the anti-angiogenic activity and define the structure/function relationship of K5, two deletion mutants of K5 were generated in the present study according to the distribution of three disulfide bonds in intact K5. The amino acid residues outside kringle domain of K5 were deleted to form K5mut1 (Cys462-Cys541). The residue Cys462 was deleted again to form K5mut2 (Met463-Cys541). The disulfide bridging conformation and dimensional structure of recombinant K5 and mutants were predicted by 3D-JIGSAW Comparative Modelling Server (UK) and confirmed by mass spectrometry and circular dichroism (CD) analysis. Their anti-angiogenic and anti-hepatoma activities were evaluated both in vitro and in vivo. Our findings demonstrated for the first time that K5mut1 has more potent anti-angiogenic and anti-hepatoma activities than intact K5 and the complete kringle structure with appropriate folding of three disulfide bonds is required for the activity of K5.
Materials and methods
Construction and production of human plasminogen kringle 5 and its deletion mutants
The cDNAs encoding K5 and mutants were generated by polymerase chain reaction using a human plasminogen cDNA as the template with the following primers: K5 P(+): 5′-TGTGAATTCGCCAGATGTAGAGACTCCTTC-3′, P(-):5′-GGAAAGCTTGGCACACTGAGGGACATCACAG-3′; K5mut1 P(+): 5′-ATGAATTCGTGTATGTTTGGGAATGGG-3′, P(-):5′-GCCAAGCTTACACTGAGGGACATCACAGTAG-3′; K5mut2 P(+): 5′-CGGAATTCCATGTTTGGGAATGGGAAAGG-3′, P(-): 5′-CGGAAGCTTACACTGAGGGACATCACAGT-3′; K5mut3 P(+):5′-ACTGAATTCGCCATCTGTATCGACTCCTTCCTCATCATCCTGTATGTTTGG-3′, P(-): 5′-TGCTGCAAGCTTCGCACA CTGAGGGACATCACAGTAGTC-3′. The PCR products were cloned into the pET-22b(+) at EcoRI and HindIII sites. These constructs were introduced into Escherichia coli BL21 (DE3) (Novagen Co., Madison, WI, USA.) strain for protein expression. The expression and purification followed the protocol recommended by Novagen. The purity and identity of recombinant peptides were examined by SDS-PAGE and Western blot analysis using an antibody specific to His-tag (Novagen Co.).
Molecule mass and disulfide bridging conformation analysis of purified soluble K5 and K5 mutants
Molecule mass analysis of K5 and K5 mutants was performed as described previously [14]. Orthogonal digestion combining with MALDI-Q TOF mass spectrometry was used for disulfide bridging conformation analysis of recombinant K5 and K5 mutants. For single enzyme digest, lyophilized K5 or K5 mutants samples were suspended in digest buffer (50 mM Tris-Cl, 1 mM CaCl2, pH7.6) containing 100 ng of Trypsin Gold (Promega, Madison, WI, USA), and digested overnight. For dual enzyme digest, lyophilized K5 or K5 mutants were first digested overnight with immobilized trypsin (Pierce, Rockford, IL, USA) followed by an overnight digestion with 100 ng of Endoproteinase-Asp-N (Roche Diagnostics, Laval, Quebec, Canada). The singly or dually digested solutions were equated into two aliquots and one aliquot was treated with TCEP in 60°C water for 10 min. to reducing the peptides. Peptide fragment analysis was done with an ETTAN MALDI-TOF Pro mass spectrometer (ETTAN MALDI-TOF Pro, Amersham Biosciences, Sweden). Spectra analysis and peptide identity assignment were done using Ettan MALDI-ToF Pro Control module version 2.01 [15].
Cell culture
Human umbilical vein endothelial cells (HUVEC) were freshly isolated from human umbilical cord veins, as previously described [16, 17] and grown in human endothelial-SFM basal growth medium (GIBCO, Grand Island, NY, USA) supplemented with 20% foetal calf serum, 100 μg/ml streptomycin, 100 U/ml penicillin, 5 μg/ml amphotericin B (GIBCO, Grand Island, NY, USA), 2 mmol/l L-glutamine, 15 mg/l ECGS (Upstate, NY, USA). The identity and purity of HUVEC were determined by the incorporation of acetylated low-density lipoprotein labelled with a fluorescent probe DiI (Biomedical Technologies, Inc., Stoughton, MA, USA) [18].
The liver cell line and hepatocarcinoma cell line Bel7402, HepA were provided by Cell Bank of China Science Academy (Shanghai, China). The cells were maintained in DMEM medium (GibcoBRL, Gaithersburg, MD, USA) supplemented with 10% heat-inactivated foetal bovine serum (FBS) and antibiotic-antimycotic.
Cell proliferation assay
The viable cells were quantified by the 3- [4, 5-dimethylthiazol -2-yl]-2,5-dephenyl tetrazolium bromid MTT (Sigma Chemical Co., St. Louis, MO, USA) colorimetric assay following a protocol recommended by the manufacturer [12, 19]. The inhibitory effects of K5 on cell proliferation were expressed as IC50 values, which were determined from three independent tests.
Endothelial cell migration assay
Migration assays were performed as described previously [8, 20]. HUVECs in SFM (1×105) were seeded into the top chamber with polycarbonate membrane (8 μm pore sizes, Corning Costar Corp, Cambridge, MA, USA) precoated with 10 μg/ml fibronectin. Top chambers containing HUVEC were placed into a 24-well plate containing 600 μl DMEM with 1% serum and incubated for 30 min. at 37°C. HUVEC migration was stimulated by addition of the VEGF (10 ng/ml) to the lower well of the Boyden chamber. A total of 640 nM of K5, K5mut1 and K5mut2 were added to the lower chamber, respectively. After 6 hrs, the chemotaxis chamber was dismantled and the polycarbonate membranes of top chambers were fixed in formaldehyde for 10 min. The non-migrated cells were scraped off the upper surface of the membrane and the migrated cells on the lower surface of the membrane were stained with haematoxylin and eosin. Each sample was tested in triplicate and the average number of migrating cells per field was assessed by counting three random high-power fields per filter.
Quantitative analysis of apoptosis by flow cytometry
Apoptosis analysis was carried out as described previously [12, 21, 22]. Briefly, HUVEC cells were treated with 320 nM K5 and K5 mutants for 24 hrs. Prior to analysis, cells were washed again with PBS and incubated for 30 min. in propidium iodide staining solution. The suspension was analysed by flow cytometry, using a Coulter (Hialeah, FL, USA) Epic Elite apparatus.
Animals
Care, use and treatment of all animals in this study were in strict agreement with the institutionally approved protocol according to the USPHS Guide for the care and use laboratory animals, as well as the guidelines set forth in the Care and Use of Laboratory Animals by the Sun Yat-sen University. Male 5–6-week-old Kunming mice and male 4–5-week-old athymic nude mice were obtained from the Laboratory Animal Center of Guangdong, China and the animal license number is SCXK(YUE)2004-0011.
Inhibition of liver cancer growth
Hepatocarcinoma mouse model was established as described previously by subcutaneously injection of mouse hepatoma cells (1×106) into the oxter of mice [12]. When the tumour grew to about 200 mm3, mice were then randomized and divided into different groups and received three times peritoneal injection with 72 hrs alternation of PBS, K5 and K5 mutants, respectively. The total amount is 7.5 mg/kg. Two weeks later from the first injection, the mice were executed and tumours were dissected, weighed.
Similarly, male athymic nude mice (4–5-week-old) were used for xenografted hepatocarcinoma model. A single human hepatoma Bel7402 cell suspension of 1.5×105 was implanted subcutaneously in the middle dorsum of each animal. When the tumours became visible (about 9 days after inoculation), mice were randomized into different groups. Animals received five times peritoneal injection with 48 hrs alternation with K5 and K5 mutants at a total amount of 12.5 mg/kg, respectively. Tumour volume was calculated by the following formula: tumour volume (mm3) = (a×b2)/2, where a = length in mm and b = width in mm [23, 24]. One month later from the first injection, the mice were executed and tumours were dissected, weighed.
Microvessel density assay
Immunostaining was performed as described previously with minor modification [16]. Briefly, frozen tissues were cut into 10-μm sections, fixed in acetone at 4°C for 5 min., and blocked for endogenous peroxidase. Tumour vasculature was stained using a monoclonal antibody against mouse CD31 at 1:100 dilutions. Tumour microvessel density was quantified by the Weidenr’s method. In negative-control staining, the primary antibodies were omitted.
Circular dichroism spectroscopy
The purified proteins were dialysed against 25 mmol/l boric acid (pH7.4) for 8 hrs. The CD spectra were measured with a JASCO-J-810 spectropolarimeter under nitrogen flush in 0.2 cm path length cells at 25°C. The spectra were recorded between 260 nm and 200 nm and the average of three recordings was taken. The calibration was carried out with 25 mmol/l boric acid (pH7.4). The proteins were scanned at concentrations of 0.50 mg/ml. The optical activity was normalized to molar ellipticity.
Statistical analysis
Means and SDs were calculated for continuous variables. For two-group comparison, the t-test method was used. For more than two groups’ comparison, one-way ANOVA was used firstly to detect the difference amongst these groups. If the P-value was less than 0.05, then multiple comparison was performed using LSD-t test. All statistical tests were 2-sided, with P-value less than 0.05 considered significant.
Results
Expression and purification of K5 and its deletion mutants
Two deletion mutants were designed according to the structure and disulfide bond distribution of K5 (Pro452-Ala542). K5mut1 (Cys462-Cys541) retained the intact kringle domain and the amino acid residues outside kringle domain of K5 were deleted. The residue Cys462 was deleted again to form K5mut2 (Met463-Cys541). K5mut2 opened the first disulfide bond (Fig. 1B). Recombinant K5 and its mutants were expressed in E. coli. and purified to homogeneity with metal affinity chromatography. The purified recombinant protein K5, K5mut1, K5mut2 showed an apparent molecular weight of 16, 14 and 14 kD, respectively (Fig. 1C). Accurate molecular weight of K5mut1, K5mut2 obtained by MALDI-TOF MS analysis matched the calculated molecular weight from the sequence. The identity of the band was confirmed by Western blot analysis using an anti-His tag antibody (Fig. 1C). An average of 15 mg of purified protein per litre culture and over 90% purity were achieved for recombinant K5 and mutants and being used in this study.
Fig 1.
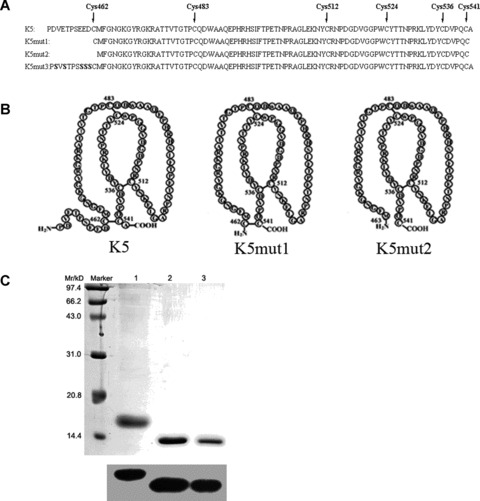
Schematic structure and generation of K5 and its deletion mutants. (A) The primary sequence of K5 and its mutants. (B) The schematic structure of K5 and its mutants. Two deletion mutants were designed according to the structure and disulfide bond distribution of K5 (Pro452-Ala542). K5mut1 (Cys462-Cys541) retained the intact kringle domain and deleted the amino acid residues of both terminals outside kringle domain of K5. K5mut2 (Met463-Cys541) opened the first disulfide bond by deleting the amino acids of Cys462 on the basis of K5mut1 structure. (C) The production and identification of recombinant K5 and its mutant proteins. The top panel is SDS-PAGE with Coomassie blue staining while the bottom panel is Western blot analysis with antibody specific to His-tag; Marker: Protein marker; Lane1: Purified recombinant protein of K5; Lane2: Purified recombinant protein of K5mut1; Lane3: Purified recombinant protein of K5mut2.
Recombinant K5 and mutants possess consistent disulfide numbers and folding with the corresponding native peptides as displayed within plasminogen
To identify the disulfide bridging in recombinant K5 and mutants, an orthogonal digestion was carried out either with trypsin alone or with agarose-immobilized trypsin followed by Asp-N endopeptidase under reducing and nonreducing conditions. MALDI-Q TOF mass spectrometry of K5 and mutants protease digests confirmed the presence of three disulfide bridges between the cysteine pairs of Cys462:541, Cys483:524 and Cys512:536 in K5 and K5mut1 (Fig. 2A and B), but the presence of only two disulfide bridges between the cysteine pairs of Cys483:524 and Cys512:536 in K5mut2 (Fig. 2C). These findings confirmed that the disulfide bridges present in recombinant K5 and K5mut1 are in the same conformation as displayed within plasminogen whereas the K5mut2 preserved only two disulfide bridges (Fig. 2).
Fig 2.
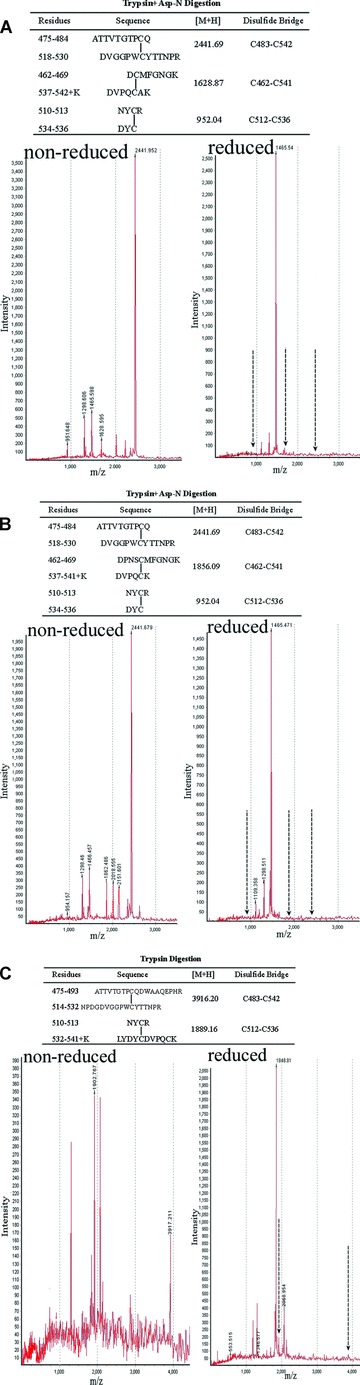
Recombinant K5 and mutants possess consistent disulfide numbers and folding with the native peptides by orthogonal digestion combining with MALDI-Q TOF mass spectrometry analysis. (A) Expected peptide fragments generated for K5 protein digested with trypsin followed by Asp-N endopeptidase treatment under non-reducing condition (top). Spectra confirming Cys483:524, Cys462:541 and Cys512:536 disulfide bond under non-reducing (bottom left) and reducing conditions (bottom right). (B) Expected peptide fragments generated for K5mut1 protein digested with trypsin followed by Asp-N endopeptidase treatment under non-reducing condition (top). Spectra confirming Cys483:524, Cys462:541 and Cys512:536 disulfide bond under non-reducing (bottom left) and reducing conditions (bottom right). (C) Expected peptide fragments generated for K5mut2 protein digested with trypsin alone under non-reducing condition (top). Spectra confirming the disulfide bond of Cys483:524 and Cys512:536 under non-reducing (bottom left) and reducing conditions (bottom right).
More potent inhibiting effect of K5mut1 on endothelial cell proliferation
Both K5 and K5mut1 but not K5mut2 exhibited inhibition of endothelial cell proliferation. The effect appeared to be concentration-dependent. At a concentration as low as 10 nmol/l, K5mut1 treatment resulted in significantly fewer viable cells than the PBS control group and K5-treated group (P < 0.05). K5mut1 showed an IC50 of approximately 35 nmol/l while intact K5 has an IC50 of approximately 70 nmol/l in inhibiting HUVEC proliferation (Fig. 3A). K5mut1, compared with intact K5, has a two-fold enhanced effect (P < 0.05), suggesting that K5mut1 is a more potent inhibitor for endothelial cell proliferation.
Fig 3.
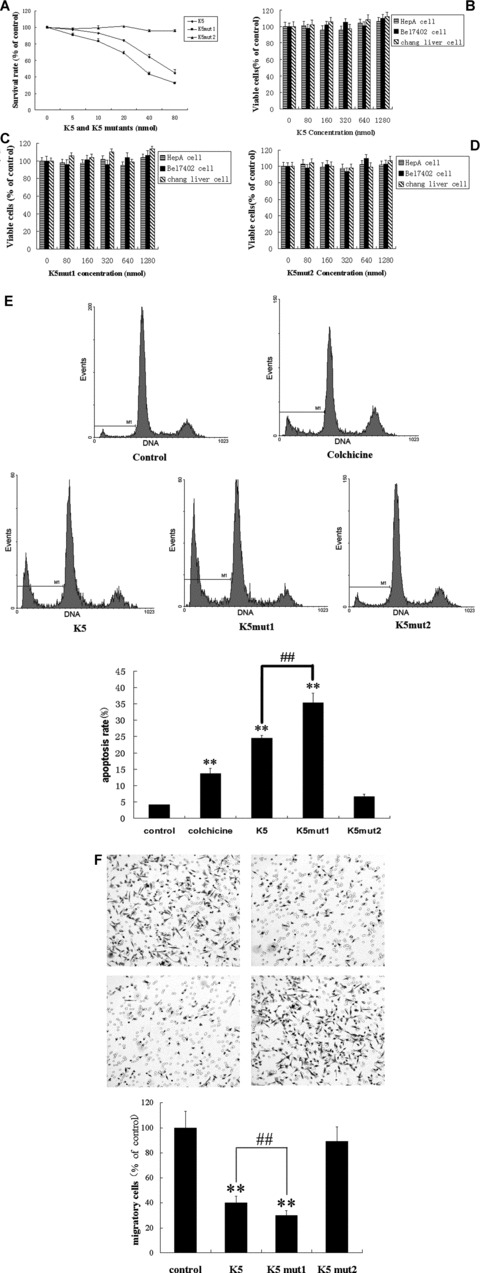
Effects of K5 and its deletion mutants on endothelial cell proliferation, apoptosis, migration and other cells proliferation. Cells were treated with the recombinant K5, K5mut1 and K5mut2 at concentrations as indicated for 72 hrs. The viable cells were quantified using MTT assay (A, B, C). (A) Effect of K5, K5mut1 and K5mut2 on the proliferation of primary HUVEC. (B) Effect of K5 on the proliferation of HepA cells, Bel7402 cells and Chang liver cells. (C) Effect of K5mut1 on the proliferation of HepA cells, Bel7402 cells and Chang liver cells. (D) Effect of K5mut2 on the proliferation of HepA cells, Bel7402 cells and Chang liver cells. (E) Quantitative analysis of HUVEC apoptosis induced by K5, K5mut1 and K5mut2. Apoptotic cell were quantified by flow cytometry with the negative control (PBS group) and the positive control (colchicines group). (F) Effect of K5, K5mut1 and K5mut2 on migration of primary HUVEC (top left: control; top right: K5; bottom left: K5mut1; bottom right: K5mut2). A modified Boyden Chamber-based assay was performed (8 μm pore sizes chamber was used). Values represent the mean of three determinations (±SD) as percentages of control (**: P < 0.05 versus control; ##: P < 0.05 K5 versus K5mut1).
Enhanced effect of K5mut1 on endothelial cell apoptosis
As shown in Fig. 3E, K5 and K5mut1 both induced apoptosis of HUVECs. Average percentages of apoptotic cells in negative control, positive control, K5 treated and K5mut1 treated cells were 4.1%± 1.11%, 13.7%± 1.72%, 24.5%± 0.85%, 35.3%± 2.95%, respectively. The results showed that K5mut1, compared with intact K5, has an enhanced effect on apoptosis of endothelial cells (P < 0.05).
More potent inhibiting effect of K5mut1 on endothelial cell migration
As shown in Fig. 3F, K5 and K5mut1 exhibited inhibition of HUVEC migration with an IC50 of approximately 300 nmol/l and 240 nmol/l, respectively. These results suggested that K5mut1, in comparison with intact K5, also has an enhanced inhibiting effect on endothelial cell migration (P < 0.05).
Taken together, these data demonstrate that K5mut1, compared with intact K5, has enhanced anti-endothelial cell activities in vitro.
Effects of K5 and K5mut1 on non-endothelial cell proliferation
The effects of K5 and K5mut1 on the proliferation of non-endothelial cell lines, including the normal liver cell line, human hepatocarcinoma cell lines Bel7402 and mouse hepatocarcinoma cell lines HepA was assayed. The results showed that both K5 and K5mut1 did not result in any significant inhibition of the three non-endothelial cell lines even in the higher concentration range (Fig. 3B–D), suggesting their endothelial cell-specific inhibition.
K5mut1 exhibits more potent anti-hepatoma effect
The anti-tumour activity of K5mut1 in vivo was determined in two kinds of animal models of liver cancer: the HepA-grafted hepatocarcinoma mice model and Bel7402-xenografted hepatocarcinoma athymic model. Compared with PBS injection control group, an average of 75% suppression of primary tumour growth was observed in the K5mut1-treated mice while intact K5 had 62% growth suppression rate in HepA-grafted hepatocarcinoma mice model (Fig. 4A and B). The effect of K5 and mutants on tumour angiogenesis was evaluated by CD31 immunostaining for capillaries in tumour tissues. The CD31 stained microvessel density in tumours treated with K5mut1 was lower than that in K5 group (Fig. 4C and D). Similarly, an average of 77% suppression of primary tumour growth was observed in the K5mut1-treated mice and 68% suppression in intact K5-treated group in Bel7402-xenografted hepatocarcinoma athymic model (Fig. 5A and B). Tumour angiogenesis was also evaluated in xenografted hepatocarcinoma athymic model. As same as that in HepA-grafted hepatocarcinoma, the CD31 stained microvessel density in xenografted tumours treated with K5mut1 was lower than that in K5 group (Fig. 5C and D). These results show that K5mut1, in comparison with intact K5, has an enhanced anti-hepatoma activity in two kinds of animal models of liver cancer. The more potent anti-hepatoma effect of K5mut1 might depend on the enhanced anti-angiogenic activity.
Fig 4.
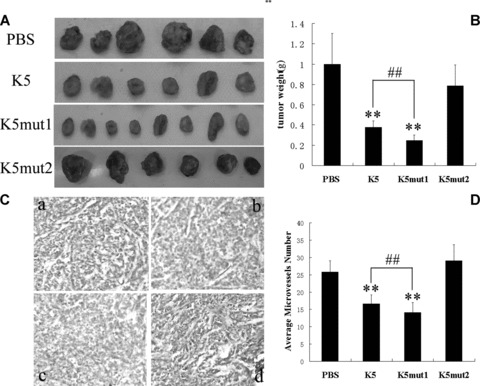
K5mut1 exhibits more potent anti-hepatoma effect in grafted hepatocarcinoma mouse model. (A) Tumour tissues at day 14 treated with PBS, K5, K5mut1 and K5mut2, respectively. (B) An average of 62%, 75% and –7% suppression of primary tumour growth was observed in the K5-treated, K5mut1-treated and K5mut2-treated group compared with control group, respectively. Data are presented as mean ± SD. Values significantly lower than control are indicated (**: P < 0.05 versus control; ##: P < 0.05 K5 versus K5mut1). (C) Representative immunohistochemical data for CD31 immunoreactivity (×200 magnification). At the end of the study, a portion of each tumour from control and drug-treated (K5, K5mut1 and K5mut2) groups was processed for immunohistochemical staining for CD31 as described in ‘Materials and Methods’. a: PBS; b: K5; c: K5mut1; d: K5mut2. (D) Quantitative analysis. Data are the mean ± SD. Microvessels were counted from five randomly selected fields in tumours from three mice of each group (**: P < 0.05 versus control; ##: P < 0.05 K5 versus K5mut1).
Fig 5.
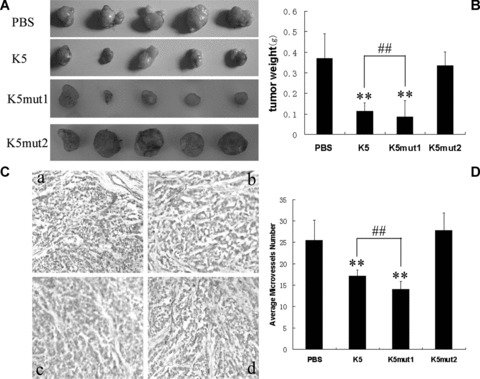
K5mut1 exhibits more potent anti-hepatoma effect in hepatocarcinoma xenograft athymic mouse model. (A) Tumour tissues at day 30 treated with PBS, K5, K5mut1 and K5mut2, respectively. (B) An average of 68%, 77%, and 9% suppression of primary tumour growth was observed in the K5-treated, K5mut1-treated and K5mut2-treated group compared with control group, respectively. Data are presented as mean ± SD. Values significantly lower than control are indicated (**: P < 0.05 versus control; ##: P < 0.05 K5 versus K5mut1). (C) Representative immunohistochemical data for CD31 immunoreactivity (×200 magnification). A portion of each tumour from control and drug-treated (K5, K5mut1 and K5mut2) groups was processed for immunohistochemical staining for CD31 as described in ‘Materials and Methods’. a: PBS; b: K5; c: K5mut1; d: K5mut2. (D) Quantitative analysis. Data are the mean ± SD. Microvessels were counted from five randomly selected fields in tumours from three mice of each group (**: P < 0.05 versus control; ##: P < 0.05 K5 versus K5mut1).
K5mut2 lacks the anti-angiogenesis and anti-tumour activity both in vitro and in vivo
Deletion mutant of K5 (K5mut2) was expressed and purified identically as recombinant K5 and K5mut1 protein (Fig. 1). The activities of K5mut2 were investigated in the cells and the animal models mentioned above with the positive control of K5-treated group. K5mut2 had no significant effect on proliferation, apoptosis and migration of HUVECs even in the higher concentration range compared with intact K5 (Fig. 3A, E and F). Similar deficient effects of K5mut2 were found in hepatocarcinoma animal models mentioned above. In HepA-grafted hepatocarcinoma mice model, –7% suppression of primary tumour growth was observed in the K5mut2-treated group, whereas suppression percentage of K5 was 62% (Fig. 4). Microvessel density in tumours treated with K5mut2 had no difference with that in PBS control group (Fig. 4C and D). Similarly, 9% suppression percentage observed in the K5mut2-treated mice was also markedly lower than intact K5 (68% suppression percentage) in Bel7402-xenografted hepatocarcinoma athymic model (Fig. 5). As same as that in HepA-grafted hepatocarcinoma, K5mut2 had no effect on tumour angiogenesis (Fig. 5C and D). These results show that K5mut2 lost the anti-angiogenic and anti-tumour activities.
Acidic amino acid residues in NH2 terminal outside kringle domain exert shielding effect on the activities of K5
K5mut3 (Pro452-Ala542) retained the intact kringle domain and the five acidic amino acids of the NH2 terminal prior to Cys462 of K5 were substituted by five serine residues (Fig. 6A). K5mut3 was expressed, purified and identified identically as recombinant K5 (Fig. 6B and C). The anti-angiogenic activity of K5mut3 was primarily measured by the analysis of endothelial cell proliferation with the positive control of K5-treated group. K5mut3 exhibited inhibition of endothelial cell proliferation in concentration-dependent manner (Fig. 6D). K5mut3 showed an IC50 of approximately 40 nmol/l while intact K5 has an IC50 of approximately 70 nmol/l in inhibiting HUVEC proliferation (Fig. 6D). The IC50 of K5mut3 is almost same as K5mut1. These results suggested that acidic amino acid residues in NH2 terminal outside kringle domain may shield the activities of K5 and the shielding effect may explain why K5mut1 has higher activity than native K5.
Fig 6.
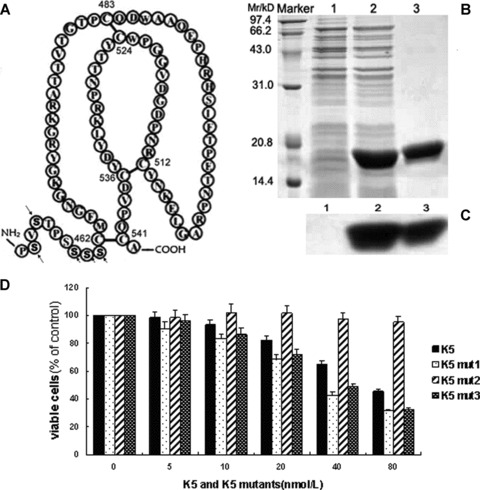
K5mut3 with five serine residues replacing five acidic amino acids in NH2 terminal of K5 has enhanced activity as K5mut1. (A) Schematic diagram of K5mut3. Note that five of the acidic amino acids of K5 were substituted by Serine (arrows). (B) SDS-PAGE analysis of the purified K5mut3 protein with Coomassie blue staining. (C) Western blot analysis of the purified K5mut3 protein with antibody specific to His-tag. (D) Effects of K5mut3 on endothelial cell proliferation. Marker: Protein marker; Lane1: Total cell protein before IPTG induction; Lane2: Total cell protein before IPTG induction; Lane3: Purified recombinant protein of K5mut3.
Conformational analysis of K5 and mutants
To explore the relationship between the structure and function of K5, the secondary structures of K5 and its three mutants were measured by CD. The far UV spectrum show that K5, K5mut1 and K5mut3 have characteristic β-sheet spectrum, while the spectrum of K5mut2 is dominated by the characteristic of random coil structure (Fig. 7). K5 and K5mut3 adopt the shape characteristic of a β-sheet with a single broad peak around 235 nm, suggesting that they share very similar secondary structures. K5mut1 adopt the shape characteristic of a β-sheet with a single broad peak around 230 nm. These results suggested that β-sheet is the structural feature for the activity of K5 and the function will disappear when β-sheet was converted into random coil structure.
Fig 7.
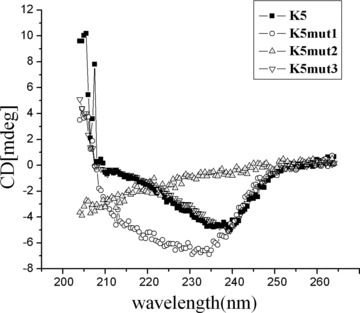
CD spectra show that β-sheet is the structural feature for the active K5 and mutants. The far UV spectra show that K5 (▪)□K5mut1 (○) and K5mut3 (▿) have characteristic β-sheet spectrum. K5 and K5mut3 adopt the shape characteristic of a β-sheet with a single broad peak around 235 nm, while K5mut1 adopts the shape characteristic of a β-sheet with a single broad peak around 230 nm. The spectrum of K5mut2 (▵) is dominated by random coil structure.
Discussion
This present study shows that K5 and K5mut1 exert an anti-hepatoma activity both in HepA-grafted hepatocarcinoma mice model and Bel7402-xenografted hepatocarcinoma athymic model (Figs. 4 and 5). Our previous study also demonstrated the anti-angiogenic activity of K5 in rat model of OIR [9, 10] and rabbit model of alkali-burn-induced corneal neovascularization [11]. These results suggest that K5 and K5mut1 may be used in wide spectrum of angiogenic disorders such as solid tumours derived from different origins [15, 25] and ocular neovascularization. K5 and K5mut1 specific inhibit proliferation, migration and induce apoptosis and autophagy of endothelial cells [13, 14, 26] but has no effect on normal liver cells, HepA and Bel7402 hepatoma cell lines even in high range of concentration, suggesting its endothelial cell-specific inhibition (Fig. 3). These universal effects of K5 and K5mut1 may be correlated with the common anti-endothelial cell activity.
Our study further shows that injection of recombinant K5mut1, in comparison with intact K5, has an enhanced inhibitory effect on hepatocellular carcinoma growth and neovascularization both in grafted and xenografted mice (Figs. 4 and 5). K5mut1 not only inhibited the proliferation and induced apoptosis of primary HUVEC but also blocked migration of HUVEC. Migration is an important step in a multi-step process of angiogenesis. In this report, we demonstrate for the first time that K5mut1 is a novel and more potent inhibitor of endothelial cells. These results suggested that K5mut1 may be a promising angiogenesis inhibitor and tumour suppressor with considerable therapeutic potential because of its high efficacy, cell type-selectivity, short amino acid sequence and stability.
Despite its therapeutic significance, however, the structural basis to maintain the anti-angiogenic activity of K5 is still vague and controversial. Cao et al. reported that the anti-endothelial proliferation activity of K5 was markedly abolished after reduction/alkylation [7]. In contrast to Cao’s results, Ji et al. described that alteration of K5 by reduction/alkylation significantly enhanced its anti-migratory potency [8]. In this study, peptide mapping of K5 protease digests by MALDI-TOF mass spectrometry confirmed the presence of disulfide bridges among Cys462-Cys541, Cys483-Cys524 and Cys512-Cys536. K5mut1 preserved the same disulfide bridges as intact K5 and K5mut2 only kept two disulfide bonds of Cys483-Cys524 and Cys512-Cys536 (Fig. 2). Our cysteine bridging pattern data confirm the finding of other studies that characterize the structure of recombinant K5 domain [15, 27]. These results suggest that the recombinant K5 and mutants adopt a structure consistent with native confirmation as displayed within plasminogen and can be used for structure and function investigation.
Remarkably, the present study shows an interesting finding in that deletion of the sequences outside of K5 domain significantly increases activity. These results indicated that the amino acid sequences outside the triple disulfide bonds is not essential for the anti-angiogenic activity and may shield the functional elements of K5 from effectively interacting with endothelial cells. To explore the possible mechanism whereby K5mut1 has more potent anti-angiogenic activity than intact K5, the third mutant of K5 (K5mut3) was designed considering five acidic amino acids in the NH2 terminal, which may be important in potential interactions with other part of K5 thus shielding the binding site of K5. K5mut3 (Pro452-Ala542) retained the intact kringle domain and the five acidic amino acids of the NH2 terminal prior to Cys462 of K5 were substituted by five serine residues (Figs. 1A and 6A). K5mut3 exhibited more potent inhibitory effect on endothelial cell proliferation than intact K5 (Fig. 6D). The IC50 of K5mut3 is almost the same as K5mut1. Furthermore, CD analysis shows that K5, K5mut1 and K5mut3 share similar secondary structures of a β-sheet and deletion of the sequences outside kringke of K5 does not change the conformation (Fig. 7). Recently, it was found that K5 binds with high affinity to endothelial cells through an interaction on the lysine-binding site of K5 and GRP78. Peptides from the lysine-binding site of K5 potently compete with K5 cellular binding and induce endothelial cell apoptosis. Moreover, the Lys82 of the lysine-binding site of K5 was mutated to alanine, which abolished ∼ 90% of its ability to inhibit in vitro migration and to bind to endothelial cells, although the kringle was still correctly folded as determined by NMR analysis [25, 27]. These results suggested that acidic amino acid residues in NH2 terminal may interact on the key alkaline amino acid lysine in binding site pocket, and therefore shield the binding of K5 to the receptors on endothelial cells. The shielding effect of NH2 terminal of K5 may explain why K5mut1 has higher activity than native K5.
In contrast to K5 and K5mut1, K5mut2 that deleted only the amino terminal cysteine of mut1 lost the activities both in endothelial cells and the animal models. These results suggest that intact kringle structure with three disulfide bridges is essential for K5 to maintain its anti- angiogenic and anti-tumour activity. We can also conclude that K5mut1 is the minimal peptide sequence for the anti-angiogenic activity of K5.
To further define the activity differences among K5 and mutants, the secondary structures of K5, K5mut1 and K5mut2 were measured by CD. The far UV spectrum shows that K5 and K5mut1 have characteristic β-sheet spectrum while the spectrum of K5mut2 is dominated by the characteristic of random coil structure (Fig. 7). The similar antiparallel β-sheet or parallel β-sheet structures were also observed via two-dimensional 1H-NMR analysis in the kringle 4 domain of equine and human plasminogen [27], human apolipoprotein(a) kringle IV type 6 [28] and N-terminal kringle 1 of human hepatocyte growth factor [29]. These results suggested that β-sheet is the general structural feature for the intact kringle domain and the function may change when β-sheet was converted into random coil structure.
In addition to K5, individual and multiple kringle fragments of angiostatin also show anti-proliferative activities and the disulfide bond-mediated appropriate folding of these kringle structures is required to maintain its anti-endothelial potency [5]. Kringle structures analogous to those in plasminogen are also found in a variety of other proteins [30–35]. In general, a kringle is composed of 78–80 amino acids interconnected by a triple disulfide-linked loop. The triple disulfide bonds are strictly conserved between kringles [36]. Kringles derived from other molecules such as kringle 2 of prothrombin, kringles of human hepatocyte growth factor and uPA have been found to be angiogenic inhibitor [37–40]. It is unknown whether the disulfide bond-mediated appropriate folding is required for the anti-angiogenic activity in these non-plasminogen kringle proteins or kringle fragments. Based on our study on K5, however, the intact kringle structure may hold universal significance for the functions of these kringle-containing proteins or kringle fragments.
In summary, the present study demonstrated for the first time that the functions of K5 depend on the intact kringle structure and the β-sheet is the common structural feature for the active K5 and mutants. Moreover, K5mut1 exhibited more potent anti-neovascularization and anti-hepatoma activities than intact K5. The shielding effect of five acidic amino acids in NH2 terminal outside kringle domain may explain why K5mut1 has higher activity. K5mut1 is the minimal active peptide sequence of K5 and may be a promising angiogenesis inhibitor and tumour suppressor.
Acknowledgments
We thank Dr. Yi Sheng in York University of Toronto and Dr. Simon Huang in British Columbia University of Vancouver for the manuscript proofreading. This study was supported by National Nature Science Foundation of China, Grant Number: 30370313, 30570372, 30600724, 30700120, 30872980, 30971208, 30973449; Program for Doctoral Station in University, Grant Number: 20070558209, 20070558215; Key Sci-tech Research Project in University, Grant Number: 108104; Team Project of Nature Science Foundation of Guangdong Province, China, Grant Number: 06201946; Key Sci-tech Research Project of Guangdong Province, China, Grant Number: 2008B080703027; Key Sci-tech Research Project of Guangzhou Municipality, China, Grant Number: 2007Z3-E5041, 2008Z1-E231; National Key Sci-Tech Special Project of China, Grant Number: 2008ZX10002-019, 2009ZX09103-642.
References
- 1.Carmeliet P. Angiogenesis in health and disease. Nat Med. 2003;9:653–60. doi: 10.1038/nm0603-653. [DOI] [PubMed] [Google Scholar]
- 2.Folkman J. Role of angiogenesis in tumor growth and metastasis. Semin Oncol. 2002;29:15–8. doi: 10.1053/sonc.2002.37263. [DOI] [PubMed] [Google Scholar]
- 3.Gao G, Li Y, Fant J, et al. Difference in ischemic regulation of vascular endothelial growth factor and pigment epithelium-derived factor in brown Norway and Sprague-Dawley rats contributing to different susceptibilities to retinal neovascularization. Diabetes. 2002;51:1218–25. doi: 10.2337/diabetes.51.4.1218. [DOI] [PubMed] [Google Scholar]
- 4.Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med. 1971;285:1182–6. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 5.Cao Y. Antiangiogenic cancer therapy. Semin Cancer Biol. 2004;14:139–45. doi: 10.1016/j.semcancer.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 6.Cao Y, Chen A, An SS, et al. Kringle 5 of plasminogen is a novel inhibitor of endothelial cell growth. J Biol Chem. 1997;272:22924–8. doi: 10.1074/jbc.272.36.22924. [DOI] [PubMed] [Google Scholar]
- 7.Cao Y, Ji RW, Davidson D, et al. Kringle domains of human angiostatin. Characterization of the anti-proliferative activity on endothelial cells. J Biol Chem. 1996;271:29461–7. doi: 10.1074/jbc.271.46.29461. [DOI] [PubMed] [Google Scholar]
- 8.Ji WR, Barrientos LG, Llinás M, et al. Selective inhibition by kringle 5 of human plasminogen on endothelial cell migration, an important process in angiogenesis. Biochem Biophys Res Commun. 1998;247:414–9. doi: 10.1006/bbrc.1998.8825. [DOI] [PubMed] [Google Scholar]
- 9.Zhang D, Kaufman PL, Gao G, et al. Intravitreal injection of plasminogen kringle 5, an endogenous angiogenic inhibitor, arrests retinal neovascularization in rats. Diabetologia. 2001;44:757–65. doi: 10.1007/s001250051685. [DOI] [PubMed] [Google Scholar]
- 10.Zhang SX, Sima J, Shao C, et al. Plasminogen kringle 5 reduces vascular leakage in the retina in rat models of oxygen-induced retinopathy and diabetes. Diabetologia. 2004;47:124–131. doi: 10.1007/s00125-003-1276-4. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z, Ma JX, Gao G, et al. Plasminogen kringle 5 inhibits alkali-burn-induced corneal neovascularization. Invest Ophthalmol Vis Sci. 2005;46:4062–71. doi: 10.1167/iovs.04-1330. [DOI] [PubMed] [Google Scholar]
- 12.Yang X, Cheng R, Li C, et al. Kringle 5 of human plasminogen suppresses hepatocellular carcinoma growth both in grafted and xenografted mice by anti-angiogenic activity. Cancer Biol Ther. 2006;5:399–405. doi: 10.4161/cbt.5.4.2511. [DOI] [PubMed] [Google Scholar]
- 13.Gao G, Li Y, Gee S, et al. Down-regulation of vascular endothelial growth factor and up-regulation of pigment epithelium-derived factor: a possible mechanism for the anti-angiogenic activity of plasminogen kringle 5. J Biol Chem. 2002;277:9492–7. doi: 10.1074/jbc.M108004200. [DOI] [PubMed] [Google Scholar]
- 14.Cai W, Ma J, Li C, et al. Enhanced anti-angiogenic effect of a deletion mutant of plasminogen kringle 5 on neovascularization. J Cell Biochem. 2005;96:1254–61. doi: 10.1002/jcb.20601. [DOI] [PubMed] [Google Scholar]
- 15.Perri SR, Nalbantoglu J, Annabi B, et al. Plasminogen kringle 5-engineered glioma cells block migration of tumor-associated macrophages and suppress tumor vascularization and progression. Cancer Res. 2005;65:8359–65. doi: 10.1158/0008-5472.CAN-05-0508. [DOI] [PubMed] [Google Scholar]
- 16.Cooke BM, Usami S, Perry I, et al. A simplified method for culture of endothelial cells and analysis of adhesion of blood cells under conditions of flow. Microvasc Res. 1993;45:33–45. doi: 10.1006/mvre.1993.1004. [DOI] [PubMed] [Google Scholar]
- 17.Houliston RA, Pearson JD, Wheeler-Jones CP. Agonist-specific cross talk between ERKs and p38(mapk) regulates PGI(2) synthesis in endothelium. Am J Physiol Cell Physiol. 2001;281:C1266–76. doi: 10.1152/ajpcell.2001.281.4.C1266. [DOI] [PubMed] [Google Scholar]
- 18.Grant MB, Guay C. Plasminogen activator production by human retinal endothelial cells of nondiabetic and diabetic origin. Invest Ophthalmol Vis Sci. 1991;32:53–64. [PubMed] [Google Scholar]
- 19.Carmichael J, DeGraff WG, Gazdar AF, et al. Evaluation of a tetrazolium-based semiautomated colorimetric assay: assessment of chemosensitivity testing. Cancer Res. 1987;47:936–42. [PubMed] [Google Scholar]
- 20.Annabi B, Lachambre MP, Bousquet-Gagnon N, et al. Green tea polyphenol(-)-epigallocatechin 3-gallate inhibits MMP-2 secretion and MT1-MMP-driven migration in glioblastoma cells. Biochim Biophys Acta. 2002;1542:209–20. doi: 10.1016/s0167-4889(01)00187-2. [DOI] [PubMed] [Google Scholar]
- 21.Ormerod MG, Collins MK, Rodriguez-Tarduchy G, et al. Apoptosis in interleukin-3-dependent haemopoietic cells: quantification by two flow cytometric methods. J Immunol Methods. 1992;153:57–65. doi: 10.1016/0022-1759(92)90305-d. [DOI] [PubMed] [Google Scholar]
- 22.Dive C, Gregory CD, Phipps DJ, et al. Analysis and discrimination of necrosis and apoptosis (programmed cell death) by multiparameter flow cytometry. Biochim Biophys Acta. 1992;1133:275–85. doi: 10.1016/0167-4889(92)90048-g. [DOI] [PubMed] [Google Scholar]
- 23.Gr Griscelli F, Li H, Bennaceur-Griscelli A, et al. Angiostatin gene transfer: inhibition of tumor growth in vivo by blockage of endothelial cell proliferation associated with a mitosis arrest. Proc Natl Acad Sci USA. 1998;95:6367–72. doi: 10.1073/pnas.95.11.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Lu H, Griscelli F, et al. Adenovirus-mediated delivery of a uPA/uPAR antagonist suppresses angiogenesis-dependent tumor growth and dissemination in mice. Gene Ther. 1998;5:1105–13. doi: 10.1038/sj.gt.3300742. [DOI] [PubMed] [Google Scholar]
- 25.Davidson DJ, Haskell C, Majest S, et al. Kringle 5 of human plasminogen induces apoptosis of endothelial and tumor cells through surface-expressed glucose-regulated protein 78. Cancer Res. 2005;65:4663–72. doi: 10.1158/0008-5472.CAN-04-3426. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen TMB, Subramanian IV, Kelekar A, et al. Kringle 5 of human plasminogen, an angiogenesis inhibitor, induces both autophagy and apoptotic death in endothelial cells. Blood. 2007;109:4793–802. doi: 10.1182/blood-2006-11-059352. [DOI] [PubMed] [Google Scholar]
- 27.Cox M, Schaller J, Boelens R, et al. Kringle solution structures via NMR: two-dimensional 1H-NMR analysis of horse plasminogen kringle 4. Chem Phys Lipids. 1994:67–68. doi: 10.1016/0009-3084(94)90123-6. [DOI] [PubMed] [Google Scholar]
- 28.Maderegger B, Bermel W, Hrzenjak A, et al. Solution structure of human apolipoprotein(a) kringle IV type 6. Biochemistry. 2002;41:660–8. doi: 10.1021/bi011430k. [DOI] [PubMed] [Google Scholar]
- 29.Ultsch M, Lokker NA, Godowski PJ, et al. Crystal structure of the NK1 fragment of human hepatocyte growth factor at 2.0 A resolution. Structure. 1998;6:1383–93. doi: 10.1016/s0969-2126(98)00138-5. [DOI] [PubMed] [Google Scholar]
- 30.McLean JW, Tomlinson JE, Kuang WJ, et al. cDNA sequence of human apolipoprotein(a) is homologous to plasminogen. Nature. 1987;330:132–7. doi: 10.1038/330132a0. [DOI] [PubMed] [Google Scholar]
- 31.Walz DA, Hewett-Emmett D, Seegers WH. Amino acid sequence of human prothrombin fragments 1 and 2. Proc Natl Acad Sci USA. 1977;74:1969–72. doi: 10.1073/pnas.74.5.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gunzler WA, Steffens GJ, Otting F, et al. The primary structure of high molecular mass urokinase from human urine. The complete amino acid sequence of the A chain. Hoppe Seylers Z Physiol Chem. 1982;363:1155–65. doi: 10.1515/bchm2.1982.363.2.1155. [DOI] [PubMed] [Google Scholar]
- 33.Pennica D, Holmes WE, Kohr WJ, et al. Cloning and expression of human tissue-type plasminogen activator cDNA in E. coli. Nature. 1983;301:214–21. doi: 10.1038/301214a0. [DOI] [PubMed] [Google Scholar]
- 34.Wilson C, Goberdhan DC, Steller H. Dror, a potential neurotrophic receptor gene, encodes a Drosophila homolog of the vertebrate Ror family of Trk-related receptor tyrosine kinases. Proc Natl Acad Sci USA. 1993;90:7109–13. doi: 10.1073/pnas.90.15.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lokker NA, Presta LG, Godowski PJ. Mutational analysis and molecular modeling of the N-terminal kringle-containing domain of hepatocyte growth factor identifies amino acid side chains important for interaction with the c-Met receptor. Protein Eng. 1994;7:895–903. doi: 10.1093/protein/7.7.895. [DOI] [PubMed] [Google Scholar]
- 36.Castellino FJ, Beals JM. The genetic relationships between the kringle domains of human plasminogen, prothrombin, tissue plasminogen activator, urokinase, and coagulation factor XII. J Mol Evol. 1987;26:358–69. doi: 10.1007/BF02101155. [DOI] [PubMed] [Google Scholar]
- 37.Lee TH, Rhim T, Kim SS. Prothrombin kringle-2 domain has a growth inhibitory activity against basic fibroblast growth factor-stimulated capillary endothelial cells. J Biol Chem. 1998;273:28805–12. doi: 10.1074/jbc.273.44.28805. [DOI] [PubMed] [Google Scholar]
- 38.Xin L, Xu R, Zhang Q, et al. Kringle 1 of human hepatocyte growth factor inhibits bovine aortic endothelial cell proliferation stimulated by basic fibroblast growth factor and causes cell apoptosis. Biochem Biophys Res Commun. 2000;277:186–90. doi: 10.1006/bbrc.2000.3658. [DOI] [PubMed] [Google Scholar]
- 39.Kuba K, Matsumoto K, Date K, et al. HGF/NK4, a four-kringle antagonist of hepatocyte growth factor, is an angiogenesis inhibitor that suppresses tumor growth and metastasis in mice. Cancer Res. 2000;60:6737–43. [PubMed] [Google Scholar]
- 40.Kim KS, Hong YK, Joe YA, et al. Anti-angiogenic activity of the recombinant kringle domain of urokinase and its specific entry into endothelial cells. J Biol Chem. 2003;278:11449–56. doi: 10.1074/jbc.M212358200. [DOI] [PubMed] [Google Scholar]


