Abstract
In lung cancer patients whose tumors harbor activating mutations in the epidermal growth factor receptor (EGFR), increased responses to platinum-based chemotherapies are seen compared to wild-type cancers. However, the mechanisms underlying this association have remained elusive. Here, we describe a cellular phenotype of crosslinker sensitivity in a subset of EGFR-mutant lung cancer cell lines that is reminiscent of the defects seen in cells impaired in the Fanconi Anemia pathway, including a pronounced G2/M cell-cycle arrest and chromosomal radial formation. We identified a defect downstream of FANCD2 at the level of recruitment of FAN1 nuclease and DNA interstrand crosslink (ICL) unhooking. The effect of EGFR mutation was epistatic with FANCD2. Consistent with the known role of FANCD2 in promoting RAD51 foci formation and homologous recombination repair (HRR), EGFR-mutant cells also exhibited an impaired RAD51 foci response to ICLs, but not to DNA double-strand breaks. EGFR kinase inhibition affected RAD51 foci formation neither in EGFR mutant nor wild-type cells. In contrast, EGFR depletion or overexpression of mutant EGFR in wild-type cells suppressed RAD51 foci, suggesting an EGFR kinase-independent regulation of DNA repair. Interestingly, EGFR-mutant cells treated with the PARP inhibitor olaparib also displayed decreased FAN1 foci induction, coupled with a putative block in a late HRR step. As a result, EGFR-mutant lung cancer cells exhibited olaparib sensitivity in-vitro and in-vivo. Our findings provide insight into the mechanisms of cisplatin and PARP inhibitor sensitivity of EGFR-mutant cells, yielding potential therapeutic opportunities for further treatment individualization in this genetically defined subset of lung cancer.
Keywords: epidermal growth factor receptor, Fanconi Anemia, cisplatin, PARP, non-small cell lung cancer
Introduction
Lung cancer is the most common cause of cancer death worldwide. Five-year overall survival of patients remains at a rate of only ∼15%, stressing the need for the development of novel treatment approaches (1). The identification in recent years of molecular lung cancer subsets characterized by targetable oncogenic driver mutations has revolutionized therapy. For example, activating mutations in the epidermal growth factor receptor (EGFR) are associated with tumor response rates to small molecule tyrosine kinase inhibitors (TKI) of around 70% (2). Despite often impressive tumor responses, however, virtually all patients eventually experience progression. Notably, EGFR-mutant lung cancers appear to be more responsive to platinum-based chemotherapy than wild-type tumors in clinical trials (3-5), but the mechanisms underlying this finding remain to be elucidated.
The EGFR has been implicated in the repair of DNA double-strand breaks (DSB) via DNA-PKcs-dependent non-homologous end-joining (NHEJ) (6-8). However, NHEJ is not required for the removal of platinum-induced DNA damage from the genome (9, 10). Homologous recombination repair (HRR) is a pathway critical for several cellular processes including the error-free repair of DSB and the recovery of stalled or collapsed DNA replication forks (11). HRR-defective cells are hypersensitive to DNA lesions that block replication forks, such as DNA inter-strand crosslinks (ICL) produced by cisplatin or mitomycin C (MMC) (12-15). In addition, impaired HRR is synthetically lethal with inhibitors of PARP1/2 (13, 16-20). There is currently great interest in exploring the clinical utility of PARP inhibitors in multiple cancer types including lung cancer (11). It is clear that predictive biomarkers of treatment sensitivity will be needed to select patients most likely to benefit. However, in human cancers, HRR may be altered by various genetic, epigenetic, or other mechanisms, which makes it challenging to assess the functional HRR status in a given tumor (11). We recently identified HRR defects in human lung cancer cell lines and tumors, though whether such defects are more frequent in EGFR-mutant cancers has remained unknown (13).
HRR has evolved to be tightly regulated to promote precise DNA repair and limit genomic alterations. This is achieved through cell cycle phase coordination, post-translational modifications, and many accessory factors that either promote or inhibit protein interactions (11). Thus, for cancers, there exists ample opportunity to deregulate this process. How exactly selection pressure arises during carcinogenesis to disrupt HRR pathways is currently unknown. Given the crucial role of HRR for replication fork restart and repair and the possibility of widespread genomic instability if this process fails, it is conceivable that replication-associated HRR is specifically targeted when premalignant cells accumulate oncogenic stress and associated DNA damage (11).
Stalled replication forks activate the Fanconi Anemia (FA) pathway, which is composed of 15 identified genes, FANCA through FANCP, known to cause FA in patients when mutated in both alleles (except FANCB) (21-24). The FA proteins together with BRCA1 cooperate in a common biochemical “FA/BRCA” pathway, which is believed to function mainly in the detection, stabilization, and repair of stalled DNA replication forks (15). In response to fork-blocking ICLs, mono-ubiquitinated FANCD2 relocates into chromatin and co-localizes with BRCA2, RAD51, and other DNA damage response proteins; and these protein accumulations can be visualized as subnuclear foci (11). The FANCD2/FANCI complex and associated factors promote nucleolytic incision near an ICL, for example via the recently discovered FAN1 nuclease (25-27). The FA proteins are closely linked to HRR via multiple mechanisms, and FA defects can be associated with reduced homology-mediated repair of DSB and impaired RAD51 foci formation (13, 28-30). Crosslinker sensitivity is a hallmark of defects in the FA/BRCA pathway (12, 14, 15).
Here, we describe an unexpected FA-like cellular phenotype in a subset of cisplatin-treated lung cancer cell lines with mutant EGFR. We find that EGFR mutation is closely linked to altered FAN1 function and RAD51 subnuclear localization downstream of FANCD2, leading not only to cisplatin and MMC sensitivity but also sensitivity to the PARP inhibitor olaparib, thus yielding a potential therapeutic opportunity.
Materials and Methods
Cell lines and cell culture
Cell lines were selected from a published panel (31, 32). The identity of each of the cell lines in the panel was described previously (31). A549, NCI-H1650, and HCC4006 were purchased from ATCC. NIH3T3 mouse embryonic fibroblasts (MEF) stably transfected with a pBabe puromycin resistance expression vector encoding human wild-type EGFR, mutant E746_A750, or mutant L858R (33) were maintained in Dulbecco's modified Eagle's medium (DMEM). NCI-H1650, NCI-H1703, NCI-H1792, NCI-H1975, NCI-H3255, HCC4406, HCC827, PC9, PC14, and KHM-3S cells were maintained in RPMI1640. A549 were grown in DMEM and PC3 in DMEM/F12. PD20 human fibroblasts with mutant or wild-type FANCD2 were described previously (28). All media were supplemented with 10% Bovine Growth Serum (HyClone), 20 mM HEPES, 2 mM L-glutamine, and 1% Penicillin-Streptomycin (all Sigma-Aldrich unless indicated otherwise). Cell lines were assigned arbitrary passage numbers upon receipt in the laboratory and passaged < 20 times (i.e., typically 2 months). No cell line was ever treated for mycoplasma and all lines tested mycoplasma free prior to the experiments (MycoAlert, Lonza).
Treatments
Cisplatin and MMC were obtained from Sigma-Aldrich, and olaparib and erlotinib from LC Laboratories. Cisplatin was dissolved at 4 mM in 0.9% NaCl and protected from light at 4°C for 2 weeks. MMC was dissolved in ddH20 and protected from light at 4°C for 2 weeks. All other drugs were dissolved in dimethyl sulfoxide, aliquoted, and protected from light at −20°C for 6 weeks. X-irradiation was carried out as described (31).
Cell survival assays
Clonogenic survival assays were performed as described (31). Cells were treated with varying concentrations of cisplatin or MMC for 1 hour, or olaparib for 72 hours. Following aspiration of drug, cells were washed once, and media was replaced. Depending on doubling time, cells were left to form colonies for 9-29 days. Non-clonogenic cell survival was assessed by fixing and staining cells with a fluorescent nucleic acid stain (syto60) 72 hours after treatment as described (31).
Immunofluorescence microscopy
Foci staining and analysis were performed as described (13, 28, 31). Briefly, cells were incubated with primary antibodies for 2 hours at room temperature (γ-H2AX, 1:200 in 2% BSA/PBS, ab22551 Abcam), for 3 hours at 37°C (FANCD2, 1:500 in 2% BSA/PBS, NB 100-182, Novus Biologicals) or overnight at 4°C (RAD51, 1:500 in 5% FBS/PBS, GTX70230, GeneTex), and subsequently incubated with secondary antibodies for 1 hour at room temperature (Alexa488 goat anti-mouse or Alexa488 chicken anti-rabbit, Invitrogen). Subnuclear foci were scored by fluorescence microscopy (Olympus BX51).
Fresh tumor tissues from patients with EGFR mutant or wild-type tumors were collected on protocols approved by Institutional Review Boards, and subjected to ex-vivo treatment as described (13, 34). Tumor samples were aliquoted and placed in RPMI medium, and either mock-treated or treated with 10 μM olaparib for 24 hours, followed by snap freezing in OCT (Sigma-Aldrich). Cryosections were thawed at room temperature, permeabilized with 0.1% Triton X-100 in PBS for 5 minutes, and subsequently fixed with 2% PFA for 15 minutes. This was followed by another permeabilization step with 0.5% TritonX at room temperature for 5 minutes. Blocking ensued by covering the tissue with 5% goat serum/PBS in a humid chamber at room temperature for 1 hour. Incubation with primary antibody was done at 4°C overnight. Primary antibody solution contained mouse monoclonal anti-γ-H2AX antibody (Abcam ab22551, 1:500 dilution) and rabbit polyclonal anti-PCNA (Abcam ab2426, 1:200) in 3% goat-serum/PBS. Incubation with secondary antibody was done at room temperature in a humid chamber for 1 hour. Secondary antibody solution contained goat anti-mouse IgG (Alexa Fluor488, Invitrogen #A11029, 1:1,000 dilution) and goat anti-rabbit IgG (Alexa Fluor555, Invitrogen #A21429, 1:1,000) in 3% goat-serum/PBS. Nuclei were stained with DAPI (1 μg/ml in ddH2O) for 2 minutes.
Flow cytometry
Cells were treated with MMC (25 ng/ml) for 24 hours. Cells were harvested, fixed with ethanol, and stained with propidium iodide according to standard protocols. Cell-cycle distribution was analyzed at the Ragon Imaging Core as described (31).
Western Blotting
Proteins in whole cell lysates from exponentially growing cell cultures were detected using standard methods. Specific antibodies against human EGFR (2239S, Cell Signaling) and β-actin (Sigma-Aldrich) were used, except for siRNA experiments which utilized a rabbit polyclonal anti-EGFR antibody (sc-03, Santa Cruz Biotechnology). Horseradish peroxidase-conjugated secondary antibodies were used (sc-2031, Santa Cruz Biotechnology). Protein bands were visualized with enhanced chemiluminescence (Invitrogen) followed by autoradiography.
Transfections
Full-length human EGFR expression constructs encoding wild-type or mutant (L858R) proteins were described previously (35). A lentiviral expression vector encoding GFP-flagged FAN1 (PHAGE CMV N-EGFP-FL_FAN1) was a gift from Stephen Elledge (26). Plasmid pEGFP-N1 was obtained from Invitrogen. All transfections were carried out using Metafectene Pro (Biontex) according to the manufacturer's protocol. For siRNA transfections, exponentially growing cells were transfected with validated siEGFR or a scrambled control siRNA (Ambion) using the X-tremeGENE transfection kit (Roche). Western blotting and subsequent experiments were performed 48 hours after transfection.
Modified alkaline Comet Assay
In order to monitor ICL repair, a modified alkaline comet assay (8) was deployed (CometAssay®, #4250-050-K, Trevigen). Tail moment was analyzed using TriTek CometScore™ (see Supplementary Data for details).
Chromatid aberrations
Following treatment of cells with 0.5 μM of MMC for 1 hour, metaphases were scored to determine the fraction of cells with tri- and quadriradials as described previously (36).
Results
EGFR mutation is associated with a FA-like cellular phenotype
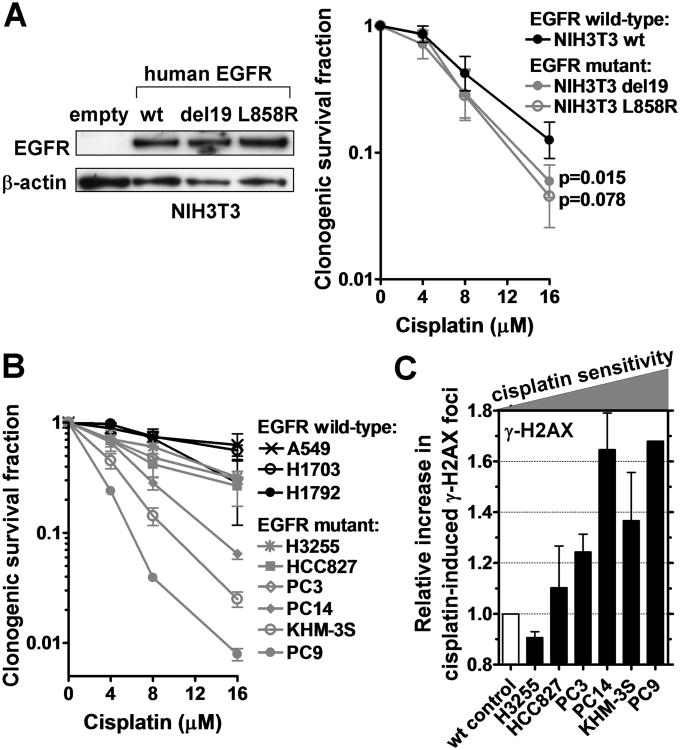
To determine the cellular cisplatin sensitivity associated with EGFR mutation, we first studied isogenic mouse embryonic fibroblasts (MEF) expressing either human wild-type EGFR or EGFR harboring activating mutations (Fig. 1A). For cisplatin concentrations of 8-16 μM, mutant EGFR was associated with a 1.3- to 2.2-fold lower cell survival fraction compared to exogenous wild-type protein. Similarly, overexpressing mutant or wild-type EGFR in lung cancer cells with wild-type or mutant EGFR, respectively, changed cisplatin sensitivity by at least 1.5-fold (Fig. S1A-D). In a panel of lung cancer cell lines, the cell lines with mutant EGFR demonstrated a logarithmic mean survival fraction of 25.8% at 8 μM cisplatin, compared to 73.6% for the wild-type lines (difference of 2.9-fold) (Fig. 1B). In addition, as a surrogate endpoint of cisplatin sensitivity, we observed increased numbers of γ-H2AX foci at 24 hours post-cisplatin treatment in the cisplatin-sensitive EGFR mutant cell lines (Fig. 1C, S1E), consistent with persistent, unrepaired DNA damage (13). Importantly, EGFR-mutant PC9 clones with acquired TKI resistance maintained their cisplatin sensitivity relative to EGFR wild-type cells (Fig. S1F,G).
Figure 1.
Cisplatin sensitivity of EGFR-mutant cell lines. A, Left panel, Western blot demonstrating expression of human EGFR in NIH3T3 MEFs transfected with wild-type (wt) or mutant E746_A750 (del19) or L858R constructs. Right panel, Clonogenic survival of MEF clones after 1 hour treatment with cisplatin. B, Clonogenic survival of lung cancer cell lines analogous to panel A. C, Relative increase in the fraction of cells with ≥20 γ-H2AX foci 24 hours after cisplatin treatment (8 μM). EGFR-mutant cell lines are ranked according to their relative cisplatin sensitivity with wild-type (wt) A549 as control. All data show mean ± standard error based on 2-3 biological repeats.
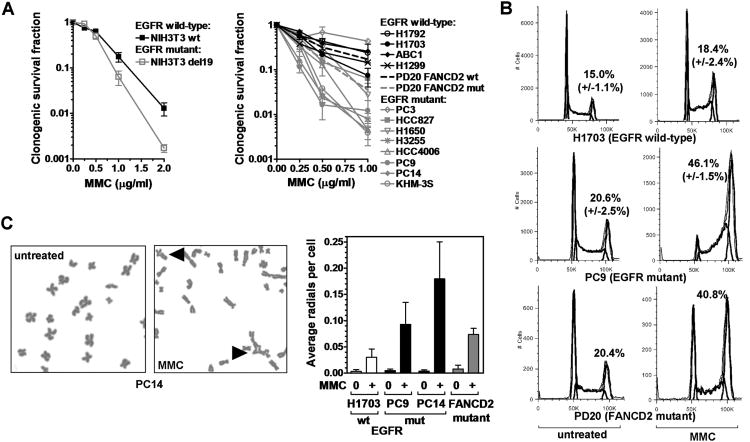
Cisplatin sensitivity can be associated with genetic or epigenetic defects in the FA pathway (11). Strikingly, we observed the hallmarks of the FA phenotype in cells with mutant EGFR. In MEFs expressing mutant EGFR, there was increased MMC sensitivity compared to isogenic cells with wild-type EGFR (Fig. 2A). Furthermore, EGFR-mutant lung cancer cell lines were on average 3.9-fold more MMC sensitive than wild-type lines, based on a logarithmic mean survival fraction (at 0.5 μg/ml) of 8.0% (n=8) versus 30.8% (n=4), respectively. Follow-up analysis in EGFR-mutant PC9/PC14 cells revealed a pronounced damage-induced G2/M cell-cycle arrest (Fig. 2B, S2A) and chromosomal radial formation (Fig. 2C), comparable to the effects of FANCD2 mutation (Fig. 2C, S2A) (36). In addition, as FA cells have been reported to demonstrate increased ATM activity (37), we observed an elevated number of phosphorylated ATM foci (Fig. S2B).
Figure 2.
Hallmarks of the Fanconi Anemia phenotype in EGFR-mutant cell lines. A, Left panel, Clonogenic survival of isogenic MEF pair expressing wild-type (wt) or E746_A750 (del19) EGFR after 1 hour treatment with mitomycin C (MMC). Right panel, Clonogenic survival of lung cancer cell lines. Previously published survival of PD20 fibroblasts with wt or mut FANCD2 is also indicated (dotted lines) (12). B, Cell cycle distribution with and without MMC treatment (25 ng/ml, 24 hours). The fraction of cells in G2/M is indicated. C, Left panel, Representative metaphase spread with MMC-induced chromosomal radials indicated by triangles. Right panel, Average number of radials per cell. FANCD2-mutant PD20 fibroblasts are included as a control. All data depict mean ± standard error based on 2-6 biological repeats.
FANCD2 downstream defect in EGFR-mutant cells
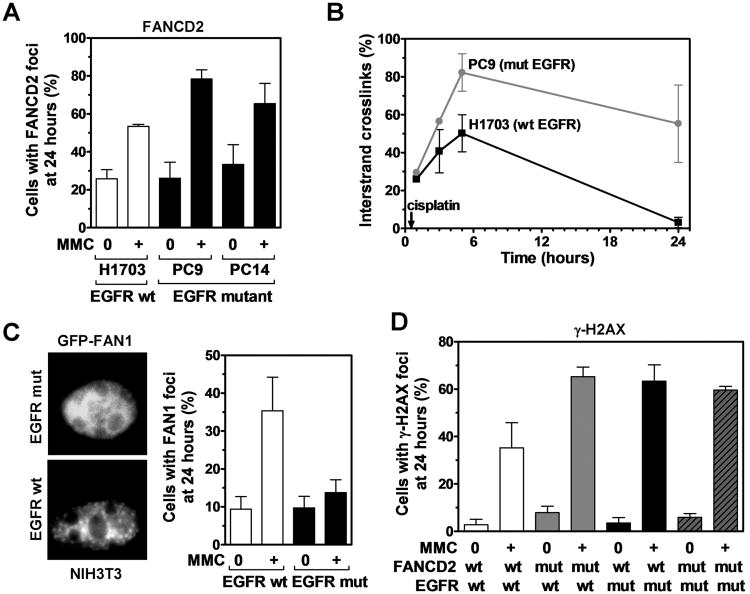
To screen for defects in the FA pathway, we determined the ability of EGFR-mutant cells to form subnuclear foci of FANCD2. Foci formation in EGFR-mutant cells was normal or even elevated compared to wild-type cells (Fig. 3A, S3A). In conjunction with intact FANCD2 mono-ubiquitination (Fig. S3B), this argues against a defect in the nuclear FA core complex or BRCA1, which are upstream of FANCD2 foci formation. Consistent with the latter, BRCA1 foci formed normally (Fig. S3C). To investigate downstream components, we first determined the ability of EGFR-mutant cells to unhook ICLs using a modified alkaline comet assay (8) (Fig. 3B). These cells were dramatically impaired in ICL unhooking at 3-5 hours post-cisplatin exposure (Fig. 3B, S4A). There was also an increase in foci of replication protein A which accumulates on single-stranded DNA exposed at stalled or collapsed replication forks (Fig. S4B).
Figure 3.
Relationship between EGFR and the FANCD2 pathway. A, Fraction of nuclei with ≥ 10 FANCD2 foci in wild-type (wt) and mutant (mut) EGFR cell lines 24 hours after MMC treatment (0.5 μg/ml, 1 hour). B, Comparison of ICL repair in wt and mut EGFR cell lines using a modified alkaline comet assay. Cells were treated with 50 μM cisplatin for 1 hour. Immediately before analysis, cells were irradiated with 12.5 Gy to produce a fixed number of DSBs. Results are expressed as a percentage decrease in tail moment (see also Fig. S4A). C, Left panel, Representative immunofluorescence microscopy images for an isogenic MEF pair with wt or E746_A750 (mut) EGFR transfected with GFP-tagged FAN1. Cells were treated with 80 ng/ml MMC for 24 hours. Right panel, Fraction of cells with ≥ 5 foci FAN1. D, Fraction of cells with ≥ 20 γ-H2AX foci in PD20 cells with wt or mut FANCD2 transfected with wt or mut EGFR. Cells were treated with 1 μg/ml MMC for 1 hour and foci scored after 24 hours. All data represent mean ± standard error based on 2-3 biological repeats.
To screen for candidate nucleases involved in ICL incision that might be defective in EGFR-mutant lung cancer cells, we selectively examined the gene expression of SLX4/FANCP, MUS81, EME1, FAN1, and ERCC1 (Fig. S4C). Interestingly, EGFR mutation was correlated with reduced expression of the FANCD2-associated nuclease FAN1 (Fig. S4C,D). We, therefore, wished to determine if FAN1 function was altered in the presence of mutant EGFR. As FAN1 is known to form subnuclear foci in response to ICLs (26), we transfected GFP-tagged FAN1 into isogenic MEFs expressing mutant or wild-type EGFR. Strikingly, the ability of cells to form FAN1 foci was essentially abrogated in EGFR-mutant cells (Fig. 3C). Whether downregulated FAN1 expression was the cause or a result of impaired FAN1 localization and function was not established. Taken together, the findings in Fig. 3A-C suggested a functional defect at the level of ICL unhooking downstream of FANCD2.
We, therefore, predicted that the repair defect associated with mutant EGFR is epistatic with FANCD2. MMC treatment of FANCD2-deficient cells in the presence of wild-type EGFR caused increased γ-H2AX foci, i.e., on average 57.3%, compared to 32.5% for FANCD2 wild-type complemented cells (Fig. 3D, S4E). Transfection of mutant EGFR into FANCD2 wild-type complemented cells also increased MMC-induced γ-H2AX foci, i.e., to 59.7%, but there was no further increase in the context of mutant FANCD2 status (52.7%).
RAD51 foci formation defect in EGFR mutant cells treated with crosslinking drugs
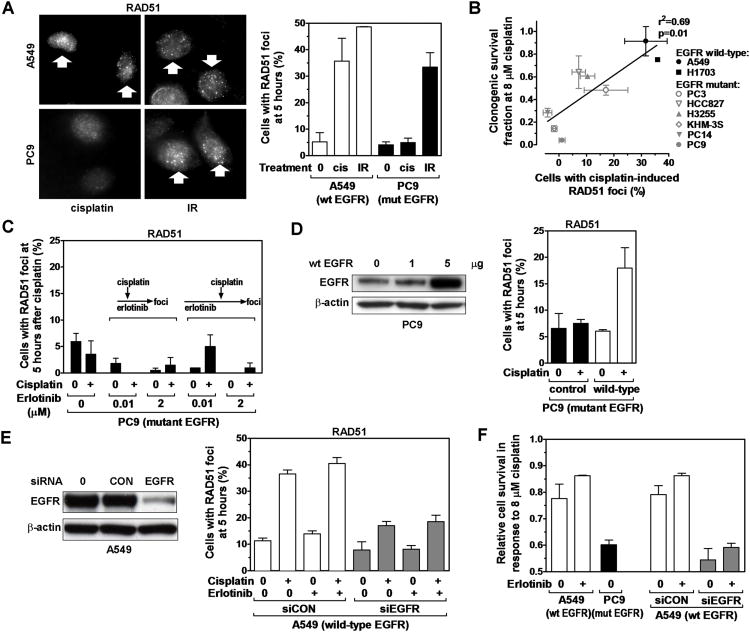
Disruption of the FANCD2-controlled pathway impairs aspects of HRR which can be visualized by reduced RAD51 foci formation at 5-24 hours after MMC or cisplatin treatment (13, 28) (Fig. S5A, and data not shown). Interestingly, EGFR-mutant cancer cells also exhibited a RAD51 foci defect in response to cisplatin but not to DSB caused by ionizing radiation (Fig. 4A, S5B,C) while RAD51 protein levels were normal (Fig. S5D). In addition, homology-dependent repair of DSB generated by the I-SceI endonuclease was intact (Fig. S5E), indicating that there is no global HRR defect associated with mutant EGFR. Remarkably, the ability of EGFR-mutant and wild-type cell lines to form RAD51 foci correlated closely with cisplatin survival (p=0.01) (Fig. 4B).
Figure 4.
RAD51 foci formation as a function of EGFR status. A, Left panel, Representative images of nuclei with RAD51 foci induced 5 hours after treatment with cisplatin (cis) (8 μM) or ionizing radiation (IR) (8 Gy). Right panel, Fraction of cells with ≥ 10 RAD51 foci. B, Correlation of clonogenic cisplatin survival with fraction of cells with induced RAD51 foci. Line represents result of linear regression analysis. C, RAD51 foci formation in erlotinib-treated EGFR-mutant PC9 cells. Cells were exposed to erlotinib for 2 hours (illustrated by arrows in the left figure insert) or 19 hours (right insert) prior to adding cisplatin. D, Left panel, Protein levels of EGFR in EGFR-mutant PC9 following transfection of 1 or 5 μg expression vector encoding wild-type (wt) EGFR. Right panel, Fraction of cells with ≥ 10 RAD51 foci 5 hours after cisplatin treatment and 48 hours after transfection with either 5 μg of wild-type EGFR vector or an empty control. Data not corrected for ∼50% transfection efficiency (Fig. S1B). E, Left panel, Western blot of A549 cells transfected with scrambled control (CON) siRNA or siRNA against EGFR. Right panel, Fraction of cells with RAD51 foci analogous to panel D. F, Cell survival determined by syto60 staining for the cell lines indicated on the x-axis. All data represent mean ± standard error based on 2-4 biological repeats.
With regard to the mechanisms by which mutant EGFR may affect the FANCD2 pathway, it has been suggested that in response to cisplatin wild-type but not mutant EGFR translocates into the nucleus where it promotes DSB repair, possibly through DNA-PKcs (6-8). However, we observed that only ∼6% of cisplatin-treated EGFR wild-type cells contain nuclear EGFR (Fig. S6A), which is not consistent with the magnitude of the FAN1 and RAD51 foci defects seen in Fig. 3C and Fig. 4A. Furthermore, DNA-PKcs had no role in the cisplatin sensitivity of EGFR-mutant cells (Fig. S6B).
Next, we wished to determine whether signal transduction pathways downstream of mutant EGFR were involved in blocking RAD51. For example, Akt has been reported to suppress HRR (38). However, we did not observe rescue of RAD51 foci formation upon pharmacological inhibition of the PI3K-Akt axis (Fig. S7A,B). Similarly, there was no stimulation of cisplatin-induced RAD51 foci in PC9 cells by short-term (2 hours) or long-term (19 hours) EGFR tyrosine kinase inhibition with erlotinib (Fig. S7C, 4C). Vice versa, erlotinib neither sensitized EGFR wild-type A549 cells to cisplatin nor suppressed RAD51 foci (Fig. S7D,E), suggesting that EGFR affects the FANCD2/RAD51 pathway through a kinase-independent function. In support of this notion, overexpression of wild-type EGFR in PC9 cells was able to rescue RAD51 foci formation (Fig. 4D) whereas transfection of mutant EGFR into A549 wild-type cells reduced RAD51 foci (Fig. S7E), which is consistent with competition between wild-type and mutant EGFR proteins (Fig. S7F, and see Discussion). Lastly, as predicted from these data, depletion of wild-type EGFR in A549 cells by siRNA disrupted RAD51 foci formation (Fig. 4E) and caused cisplatin sensitivity (Fig. 4F).
PARP inhibitor sensitivity of EGFR-mutant cells
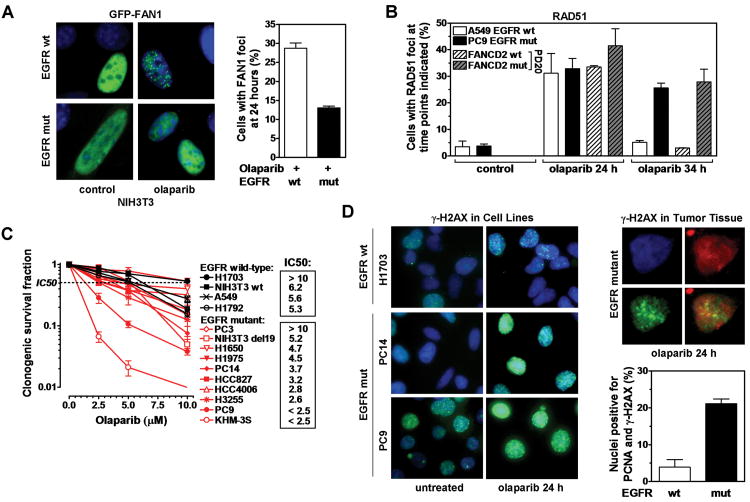
In search for novel treatment approaches in EGFR-mutant lung cancer, we considered that FANCD2-deficient cells are PARP inhibitor sensitive and that FAN1 may have functions in HRR beyond ICL unhooking, such as Holliday junction resolution (17, 25). We thus reasoned that EGFR-mutant cells may be impaired in their ability to form FAN1 foci in response to the PARP inhibitor olaparib, and this is demonstrated in Fig. 5A. Unexpectedly, olaparib-treated EGFR-mutant PC9 cells formed RAD51 foci at levels comparable to wild-type A549 cells up to 24 hours (Fig. 5B). However, at later time points (here shown for 34 hours, Fig. 5B), there was a pronounced persistence of RAD51 foci in EGFR-mutant but not wild-type cells, mirroring the effect of mutant FANCD2 in the PD20 cell pair and suggesting the presence of a block in a late step of HRR.
Figure 5.
PARP inhibitor sensitivity of cells with mutant EGFR. A, Left panel, Representative images of subnuclear GFP signal (green) overlaid with DAPI (blue) in an isogenic MEF pair with wild-type (wt) or mutant (mut) EGFR transfected with GFP-tagged FAN1. Cells were treated with 10 μM olaparib for 24 hours. Right panel, Fraction of cells with ≥ 5 FAN1 foci. B, RAD51 foci formation in cell lines as indicated, treated with 10 μM olaparib for 24 or 34 hours. C, Clonogenic survival of lung cancer and NIH3T3 cell lines after treatment with olaparib for 72 hours. Estimated IC50 values for each cell line are indicated. D, Left panel, Representative images of γ-H2AX signal in cell lines treated with olaparib for 24 hours. Right upper panel, Representative nucleus after counterstaining tumor tissue derived from a lung cancer patient with DAPI (blue), PCNA (red), and γ-H2AX (green). Right lower panel, Fraction of PCNA-positive nuclei containing γ-H2AX foci following ex-vivo treatment of EGFR mutant versus wild-type tumor tissue with 10 μM olaparib for 24 hours. All data represent mean ± standard error based on 2-5 biological repeats, except in D bars represent average number of positive nuclei ± 95% confidence intervals.
The EGFR-mutant cell lines (except PC3) exhibited olaparib sensitivity to a varying degree, with an average of 8.5% (SE +/− 2.8%) survival at 10 μM olaparib, compared to 29.0% (+/− 8.6%) for the wild-type cell lines (difference of 3.4-fold) (Fig. 5C). The survival difference in isogenic MEFs with/without mutant EGFR was 3.8-fold (Fig. 5C) and 1.2-fold in A549 cells with/without EGFR depletion (Fig. S8A) (at 10 μM olaparib). EGFR-mutant and wild-type lines could also be separated based on estimates of IC50 (inhibitory concentration of drug to achieve 50% survival) (Fig. 5C). To determine whether olaparib sensitivity can also be observed in-vivo, we considered that PARP inhibition causes increased γ-H2AX staining. In our olaparib-sensitive EGFR-mutant cell lines, we observed considerably elevated levels of γ-H2AX foci after 24 hours of olaparib treatment, thus further validating this surrogate endpoint (Fig. 5D, Fig. S8B). Note that untreated EGFR-mutant PC9 cells have already higher levels of γ-H2AX signal compared to other cell lines, which is consistent with baseline genomic instability, but there is strong damage induction upon PARP inhibition.
For comparison, fresh lung cancer tissues from patients were exposed to olaparib in the laboratory using a previously established ex-vivo foci protocol (Fig. S8C) (13, 34). Because DSB form in S-phase and the S-phase fraction of lung cancers in patients is much lower than in cell lines (13), we employed co-staining with PCNA to identify cells with an olaparib-specific γ-H2AX signal. As predicted, PCNA-positive cells from an EGFR-mutant tumor exhibited substantially more γ-H2AX staining than wild-type cells, i.e., 21% versus 4% (p=0.03, Fisher Exact) (Fig. 5D, S8D), suggesting that this kind of assay could be adapted to guide the identification of EGFR-mutant NSCLC patients who may benefit from PARP inhibitor treatment.
Discussion
We report that mutant EGFR is correlated with a cellular FA phenotype characterized not only by the hallmark of crosslinker sensitivity (cisplatin, MMC) (Fig. 1A,B, 2A), but also PARP inhibitor sensitivity (Fig. 5C) and impaired RAD51 foci formation (Fig. 4A, S5C). Mutant EGFR is epistatic with FANCD2 (Fig. 3D), which is a central chromatin-associated regulator of ICL repair, recruiting and coordinating several repair proteins at stalled replication forks (24). While our current knowledge of FANCD2 function and regulation remains limited and the role of the FA proteins in ICL unhooking is not without controversy (23, 24), it is clear that FANCD2 promotes RAD51 recruitment specifically in response to ICLs (Fig. S5A) (13, 28) and acts upstream of several structure-specific endonucleases such as FAN1 and SLX1-SLX4/FANCP (27, 39). Accordingly, EGFR-mutant cells display a profound ICL unhooking defect as well as impaired RAD51 foci formation after cisplatin treatment (Fig. 3B, 4A). Although we demonstrate impaired FAN1 foci formation (Fig. 3C), we acknowledge that the magnitude of the ICL unhooking defect (Fig. 3B) suggests that other endonucleases may be affected as well.
The extent of RAD51 foci formation at 24 hours in olaparib-treated EGFR-mutant PC9 cells was comparable to cells with wild-type EGFR (Fig. 5B), in contrast to the lack of RAD51 foci induction following cisplatin treatment (Fig. 4A). A possible explanation is that mutant EGFR only disrupts FANCD2-dependent endonuclease recruitment but not RAD51 foci formation, so that after crosslinker treatment RAD51 foci do not form (Fig. 4A) because a proper one-ended DSB substrate at the stalled fork is not being produced. Because RAD51 foci formation appears intact after depletion of either FAN1 or SLX4/FANCP (25), this model predicts the impairment of more than one endonuclease involved in ICL incision. On the other hand, RAD51 can load onto stalled replication forks even before DSB formation, at least in Xenopus extracts (40).
Alternatively, the olaparib data in Fig. 5B can be interpreted as showing a relative RAD51 foci formation defect in EGFR-mutant cells at 24 hours, considering the substantially increased amount of DNA damage at that olaparib concentration compared to wild-type EGFR cells (Fig. 5D, S8B), which should have led to much higher numbers of RAD51 foci. Thus, RAD51 foci recruitment may in fact be impaired after both crosslinker and PARP inhibitor treatment. However, in contrast to other data (8, 41, 42), our findings do not imply that wild-type EGFR promotes HRR of two-ended DSB such as caused by ionizing radiation or I-SceI endonuclease breaks (Fig. 4A, S5E).
Interestingly, RAD51 foci persisted in EGFR-mutant, as well as FANCD2-mutant, cells after >24 hours of olaparib treatment (Fig. 5B). We hypothesize that EGFR and FANCD2 may promote endonucleases, such as FAN1, that not only function in ICL unhooking but also in a Holliday junction resolvase complex, disruption of which will impair the disassembly of RAD51 foci. Whether impaired RAD51 foci formation or disassembly is more important for olaparib toxicity remains to be determined. Altogether, the data suggest a complex model in which EGFR may affect a FANCD2-dependent pathway of replication fork maintenance and HRR through multiple mechanisms. As upstream FANCD2 foci formation and mono-ubiquitination is intact in EGFR-mutant cells (Fig. 3A, S3B), we speculate that EGFR targets yet unknown binding partners of chromatin-associated mono-ubiquitinated FANCD2.
Our data indicate that the impaired ability of EGFR-mutant cells to remove ICLs is not genetically fixed. Either expression of mutant EGFR in a wild-type background or the reverse can modify crosslinker sensitivity (Fig. 1A, 2A, S1A-D). Similarly, expression of wild-type EGFR in PC9 cells but not erlotinib treatment rescues RAD51 foci formation (Fig. 4C, D), while mutant EGFR expression in A549, or depletion of endogenous wild-type protein, suppresses the RAD51 foci response (Fig. 4E, S7E), implying that there is competition between wild-type and mutant proteins. Transfection of human mutant EGFR into MEFs with endogenous wild-type EGFR is thought to reflect the same mechanism (Fig. 1A). We, therefore, hypothesize that wild-type EGFR promotes ICL repair through an EGFR kinase-independent function and that mutant protein disrupts this function in a dominant-negative fashion (model in Fig. S7F). However, the precise biochemical mechanism linking EGFR function to the FANCD2 pathway remains to be established.
To this end, we note that it has been reported that in response to DNA damaging agents, including cisplatin, wild-type but not mutant EGFR translocates into the nucleus where it may promote DNA repair (6-8). However, we found that only ∼6% of nuclei contain wild-type EGFR in response to cisplatin (Fig. S6A), which clearly is not consistent with the magnitude of the repair defect we observe (Fig. 3B, 4A). It has also been suggested that EGFR-mutant NSCLCs have low ERCC1 expression (43) However, we do not see this on our cell lines (Fig. S4C), and it is unclear how ERCC1 deficiency would abrogate RAD51 foci formation or lead to PARP inhibitor sensitivity (44).
A link between altered FA/BRCA function and EGFR signaling is not without precedent. Anecdotally, EGFR-mutant lung cancer has been linked to BRCA germline mutations (45). In addition, BRCA1-mutant breast cancers exhibit increased EGFR expression (46). It is tempting to speculate that altered repair of replication fork-blocking DNA damage may produce an environment favorable to oncogenic EGFR signaling. At least in a glioblastoma model, EGFRvIII was associated with an increased level of oxidative stress, and there is evidence that the FA/BRCA pathway is required for the response to oxidative DNA lesions that pose a barrier to replication fork progression (28, 47, 48). We hypothesize that oxidative DNA damage accumulating in pre-malignant bronchial epithelial cells harboring mutant EGFR may generate selection pressure for impaired FA/BRCA function and perturbed HRR by producing continuous fork stalling and collapse, a concept that mirrors several features of the previously described oncogene-induced DNA damage model in carcinogenesis (49).
We acknowledge that the potential clinical significance of the association of EGFR mutation with cisplatin and PARP inhibitor sensitivity remains to be fully determined as the survival differences in the isogenic models are relatively small and several of our EGFR-mutant lung cancer cell lines appear to have restored HRR and display little or no drug sensitivity. Still, EGFR mutation may be a useful biomarker in a clinical trial to enrich a study population treated with a PARP inhibitor +/− chemotherapy or radiation. In addition, the ex-vivo foci assay that we have described in Fig. 5D and elsewhere (13, 34) should be a useful tool for determining whether a given EGFR-mutant lung cancer is sensitive to PARP inhibition.
Supplementary Material
Acknowledgments
The authors wish to thank Stephen Elledge and Matthew Meyerson for kindly providing materials and Simon Powell for stimulating discussion. The excellent technical assistance of Chake Tokadjian and Alexandra Zielinski is acknowledged.
Financial Support: This work was supported by the Dana-Farber/Harvard Cancer Center SPORE in Lung Cancer, NCI P50 CA090578 (J.S., L.Z., H.W.), Department of Defense W81XWH-06-1-0309 (H.W.), the UK Wellcome Trust 086357 (C.H.B., J.S.), and the Federal Share of program income earned by Massachusetts General Hospital on C06 CA059267, Proton Therapy Research and Treatment Center (L.Z., L.S., H.W.).
Footnotes
Conflicts of Interest: Jeffrey Settleman, Ph.D., is employed by Genentech Inc., San Francisco, CA
References
- 1.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12:175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 2.Neal JW, Sequist LV. First-line use of EGFR tyrosine kinase inhibitors in patients with NSCLC containing EGFR mutations. Clin Adv Hematol Oncol. 2010;8:119–126. [PubMed] [Google Scholar]
- 3.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, Nishiwaki Y, Ohe Y, Yang JJ, Chewaskulyong B, Jiang H, Duffield EL, Watkins CL, Armour AA, Fukuoka M. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 4.Janne PA, Wang X, Socinski MA, Crawford J, Stinchcombe TE, Gu L, Capelletti M, Edelman MJ, Villalona-Calero MA, Kratzke R, Vokes EE, Miller VA. Randomized phase II trial of erlotinib alone or with carboplatin and paclitaxel in patients who were never or light former smokers with advanced lung adenocarcinoma: CALGB 30406 trial. J Clin Oncol. 2012;30:2063–2069. doi: 10.1200/JCO.2011.40.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eberhard DA, Johnson BE, Amler LC, Goddard AD, Heldens SL, Herbst RS, Ince WL, Janne PA, Januario T, Johnson DH, Klein P, Miller VA, Ostland MA, Ramies DA, Sebisanovic D, Stinson JA, Zhang YR, Seshagiri S, Hillan KJ. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 6.Das AK, Chen BP, Story MD, Sato M, Minna JD, Chen DJ, Nirodi CS. Somatic mutations in the tyrosine kinase domain of epidermal growth factor receptor (EGFR) abrogate EGFR-mediated radioprotection in non-small cell lung carcinoma. Cancer Res. 2007;67:5267–5274. doi: 10.1158/0008-5472.CAN-07-0242. [DOI] [PubMed] [Google Scholar]
- 7.Dittmann K, Mayer C, Fehrenbacher B, Schaller M, Raju U, Milas L, Chen DJ, Kehlbach R, Rodemann HP. Radiation-induced epidermal growth factor receptor nuclear import is linked to activation of DNA-dependent protein kinase. J Biol Chem. 2005;280:31182–31189. doi: 10.1074/jbc.M506591200. [DOI] [PubMed] [Google Scholar]
- 8.Liccardi G, Hartley JA, Hochhauser D. EGFR nuclear translocation modulates DNA repair following cisplatin and ionizing radiation treatment. Cancer Res. 2011;71:1103–1114. doi: 10.1158/0008-5472.CAN-10-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frankenberg-Schwager M, Kirchermeier D, Greif G, Baer K, Becker M, Frankenberg D. Cisplatin-mediated DNA double-strand breaks in replicating but not in quiescent cells of the yeast Saccharomyces cerevisiae. Toxicology. 2005;212:175–184. doi: 10.1016/j.tox.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 10.Collins AR. Mutant rodent cell lines sensitive to ultraviolet light, ionizing radiation and cross-linking agents: a comprehensive survey of genetic and biochemical characteristics. Mutat Res. 1993;293:99–118. doi: 10.1016/0921-8777(93)90062-l. [DOI] [PubMed] [Google Scholar]
- 11.Willers H, Pfäffle HN, Zou L. Targeting Homologous Recombination Repair in Cancer. Academic Press; Elsevier; 2012. pp. 119–160. [Google Scholar]
- 12.Kachnic LA, Li L, Fournier L, Willers H. Fanconi anemia pathway heterogeneity revealed by cisplatin and oxaliplatin treatments. Cancer Lett. 2010;292:73–79. doi: 10.1016/j.canlet.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 13.Birkelbach M, Ferraiolo N, Gheorghiu L, Pfäffle HN, Daly B, Ebright M, Spencer c, O'Hara C, Whetstine JR, Benes CH, Sequist LV, Zou L, Dahm-Daphi J, Kachnic LA, Willers H. Detection of Impaired Homologous Recombination Repair in NSCLC Cells and Tissues. J Thorac Oncol. 2013;8:279–286. doi: 10.1097/JTO.0b013e31827ecf83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem. 2000;275:23899–23903. doi: 10.1074/jbc.C000276200. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Molecular Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 16.Bryant HE, Helleday T. Inhibition of poly (ADP-ribose) polymerase activates ATM which is required for subsequent homologous recombination repair. Nucleic Acids Res. 2006;34:1685–1691. doi: 10.1093/nar/gkl108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, Ji J, Takeda S, Pommier Y. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012;72:5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 19.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 20.McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, Giavara S, O'Connor MJ, Tutt AN, Zdzienicka MZ, Smith GC, Ashworth A. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–8115. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 21.Cybulski KE, Howlett NG. FANCP/SLX4: a Swiss army knife of DNA interstrand crosslink repair. Cell Cycle. 2011;10:1757–1763. doi: 10.4161/cc.10.11.15818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat Rev Cancer. 2011;11:467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muniandy PA, Liu J, Majumdar A, Liu ST, Seidman MM. DNA interstrand crosslink repair in mammalian cells: step by step. Crit Rev Biochem Mol Biol. 2010;45:23–49. doi: 10.3109/10409230903501819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim H, D'Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012;26:1393–1408. doi: 10.1101/gad.195248.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacKay C, Declais AC, Lundin C, Agostinho A, Deans AJ, MacArtney TJ, Hofmann K, Gartner A, West SC, Helleday T, Lilley DM, Rouse J. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010;142:65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smogorzewska A, Desetty R, Saito TT, Schlabach M, Lach FP, Sowa ME, Clark AB, Kunkel TA, Harper JW, Colaiacovo MP, Elledge SJ. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu T, Ghosal G, Yuan J, Chen J, Huang J. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 2010;329:693–696. doi: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
- 28.Willers H, Kachnic LA, Luo CM, Li L, Purschke M, Borgmann K, Held KD, Powell SN. Biomarkers and Mechanisms of FANCD2 Function. Journal of Biomedicine and Biotechnology. 2008:821529. doi: 10.1155/2008/821529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, D'Andrea AD, Wang ZQ, Jasin M. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci U S A. 2005;102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Litman R, Peng M, Jin Z, Zhang F, Zhang J, Powell S, Andreassen PR, Cantor SB. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005;8:255–265. doi: 10.1016/j.ccr.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 31.Wang M, Morsbach F, Sander D, Gheorghiu L, Nanda A, Benes C, Kriegs M, Krause M, Dikomey E, Baumann M, Dahm-Daphi J, Settleman J, Willers H. EGF receptor inhibition radiosensitizes NSCLC cells by inducing senescence in cells sustaining DNA double-strand breaks. Cancer Res. 2011;71:6261–6269. doi: 10.1158/0008-5472.CAN-11-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, Liu Q, Iorio F, Surdez D, Chen L, Milano RJ, Bignell GR, Tam AT, Davies H, Stevenson JA, Barthorpe S, Lutz SR, Kogera F, Lawrence K, McLaren-Douglas A, Mitropoulos X, Mironenko T, Thi H, Richardson L, Zhou W, Jewitt F, Zhang T, O'Brien P, Boisvert JL, Price S, Hur W, Yang W, Deng X, Butler A, Choi HG, Chang JW, Baselga J, Stamenkovic I, Engelman JA, Sharma SV, Delattre O, Saez-Rodriguez J, Gray NS, Settleman J, Futreal PA, Haber DA, Stratton MR, Ramaswamy S, McDermott U, Benes CH. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuza Y, Glatt KA, Jiang J, Greulich H, Minami Y, Woo MS, Shimamura T, Shapiro G, Lee JC, Ji H, Feng W, Chen TH, Yanagisawa H, Wong KK, Meyerson M. Allele-dependent variation in the relative cellular potency of distinct EGFR inhibitors. Cancer Biol Ther. 2007;6:661–667. doi: 10.4161/cbt.6.5.4003. [DOI] [PubMed] [Google Scholar]
- 34.Willers H, Taghian AG, Luo CM, Treszezamsky AD, Sgroi D, Powell SN. Utility of DNA repair protein foci for the detection of putative BRCA1-pathway defects in breast cancer biopsies. Mol Cancer Res. 2009;7:1304–1309. doi: 10.1158/1541-7786.MCR-09-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 36.Kachnic LA, Li L, Fournier L, Ferraiolo N, Dahm-Daphi J, Borgmann K, Willers H. FANCD2 but not FANCA promotes cellular resistance to type II topoisomerase poisons. Cancer Lett. 2011;305:86–93. doi: 10.1016/j.canlet.2011.02.030. [DOI] [PubMed] [Google Scholar]
- 37.Kennedy RD, Chen CC, Stuckert P, Archila EM, De la Vega MA, Moreau LA, Shimamura A, D'Andrea AD. Fanconi anemia pathway-deficient tumor cells are hypersensitive to inhibition of ataxia telangiectasia mutated. J Clin Invest. 2007;117:1440–1449. doi: 10.1172/JCI31245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plo I, Laulier C, Gauthier L, Lebrun F, Calvo F, Lopez BS. AKT1 inhibits homologous recombination by inducing cytoplasmic retention of BRCA1 and RAD51. Cancer Res. 2008;68:9404–9412. doi: 10.1158/0008-5472.CAN-08-0861. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto KN, Kobayashi S, Tsuda M, Kurumizaka H, Takata M, Kono K, Jiricny J, Takeda S, Hirota K. Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proc Natl Acad Sci U S A. 2011;108:6492–6496. doi: 10.1073/pnas.1018487108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Long DT, Raschle M, Joukov V, Walter JC. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science. 2011;333:84–87. doi: 10.1126/science.1204258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li L, Wang H, Yang ES, Arteaga CL, Xia F. Erlotinib attenuates homologous recombinational repair of chromosomal breaks in human breast cancer cells. Cancer Res. 2008;68:9141–9146. doi: 10.1158/0008-5472.CAN-08-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Myllynen L, Rieckmann T, Dahm-Daphi J, Kasten-Pisula U, Petersen C, Dikomey E, Kriegs M. In tumor cells regulation of DNA double strand break repair through EGF receptor involves both NHEJ and HR and is independent of p53 and K-Ras status. Radiother Oncol. 2011;101:147–151. doi: 10.1016/j.radonc.2011.05.046. [DOI] [PubMed] [Google Scholar]
- 43.Gandara DR, Grimminger P, Mack PC, Lara PN, Jr, Li T, Danenberg PV, Danenberg KD. Association of epidermal growth factor receptor activating mutations with low ERCC1 gene expression in non-small cell lung cancer. J Thorac Oncol. 2010;5:1933–1938. doi: 10.1097/JTO.0b013e3181fd418d. [DOI] [PubMed] [Google Scholar]
- 44.Al-Minawi AZ, Lee YF, Hakansson D, Johansson F, Lundin C, Saleh-Gohari N, Schultz N, Jenssen D, Bryant HE, Meuth M, Hinz JM, Helleday T. The ERCC1/XPF endonuclease is required for completion of homologous recombination at DNA replication forks stalled by inter-strand cross-links. Nucleic Acids Res. 2009;37:6400–6413. doi: 10.1093/nar/gkp705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marks JL, Golas B, Kirchoff T, Miller VA, Riely GJ, Offit K, Pao W. EGFR mutant lung adenocarcinomas in patients with germline BRCA mutations. J Thorac Oncol. 2008;3:805. doi: 10.1097/JTO.0b013e31817e4664. [DOI] [PubMed] [Google Scholar]
- 46.Lakhani SR, Reis-Filho JS, Fulford L, Penault-Llorca F, van der Vijver M, Parry S, Bishop T, Benitez J, Rivas C, Bignon YJ, Chang-Claude J, Hamann U, Cornelisse CJ, Devilee P, Beckmann MW, Nestle-Kramling C, Daly PA, Haites N, Varley J, Lalloo F, Evans G, Maugard C, Meijers-Heijboer H, Klijn JG, Olah E, Gusterson BA, Pilotti S, Radice P, Scherneck S, Sobol H, Jacquemier J, Wagner T, Peto J, Stratton MR, McGuffog L, Easton DF. Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin Cancer Res. 2005;11:5175–5180. doi: 10.1158/1078-0432.CCR-04-2424. [DOI] [PubMed] [Google Scholar]
- 47.Nitta M, Kozono D, Kennedy R, Stommel J, Ng K, Zinn PO, Kushwaha D, Kesari S, Inda MD, Wykosky J, Furnari F, Hoadley KA, Chin L, DePinho RA, Cavenee WK, D'Andrea A, Chen CC. Targeting EGFR induced oxidative stress by PARP1 inhibition in glioblastoma therapy. PLoS One. 2010;5:e10767. doi: 10.1371/journal.pone.0010767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson LH, Hinz JM. Cellular and molecular consequences of defective Fanconi anemia proteins in replication-coupled DNA repair: mechanistic insights. Mutat Res. 2009;668:54–72. doi: 10.1016/j.mrfmmm.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.