Abstract
The thioredoxin system is a promising target when aiming to overcome the problem of clinical radiation resistance. Altered cellular redox status and redox sensitive thiols contributing to induction of resistance strongly connect the ubiquitous redox enzyme thioredoxin reductase (TrxR) to the cellular response to ionizing radiation. To further investigate possible strategies in combating clinical radiation resistance, human radio-resistant lung cancer cells were subjected to a combination of single fractions of γ-radiation at clinically relevant doses and non-toxic levels of a well-characterized thioredoxin reductase inhibitor, the phosphine gold(I) compound [Au(SCN)(PEt3)]. The combination of the TrxR-inhibitor and ionizing radiation reduced the surviving fractions and impaired the ability of the U1810 cells to repopulate by approximately 50%. In addition, inhibition of thioredoxin reductase caused changes in the cell cycle distribution, suggesting a disturbance of the mitotic process. Global gene expression analysis also revealed clustered genetic expression changes connected to several major cellular pathways such as cell cycle, cellular response to stress and DNA damage. Specific TrxR-inhibition as a factor behind the achieved results was confirmed by correlation of gene expression patterns between gold and siRNA treatment. These results clearly demonstrate TrxR as an important factor conferring resistance to irradiation and the use of [Au(SCN)(PEt3)] as a promising radiosensitizing agent.
Keywords: ionizing radiation, radiation resistance, lung cancer, thioredoxin reductase, radiosensitizer
Introduction
Radiation treatment is an important therapeutic strategy in the management of lung malignancies. Despite continuing technical achievements in the field of radiotherapy the tendency of cancer cells to develop resistance remains a major obstacle. Interaction of cellular water with high-energy radiation causes chemically reactive molecules either directly or through chain reactions, including the hydroxyl radical (OH−), hydroxyl ion (OH−), superoxide (O2−−) and hydrogen peroxide (H2O2). This will consequently cause damage to the major classes of biomolecules; DNA, proteins and lipid membranes. Mechanisms behind radiation resistance can be categorically divided into DNA-repair, changes in cellular metabolism and changes in cell interactions [1]. The combination of chemo- and radiotherapy by the use of chemical radiosensitizers (for overview, see [2]) is a well-established field. Altered redox homeostasis has possible effects on cellular response to ionizing radiation and radiation induced cell death, thus up-regulation of antioxidant systems might contribute to the induction of resistance mechanisms [3-7]. This makes the ubiquitous redox enzyme thioredoxin reductase a promising drug target. Previous work has shown the involvement of redox sensitive thiols and especially the thioredoxin system in cellular response to ionizing radiation (IR) [8-11]. The thioredoxin system is a multi-functional redox-active protein disulfide reductase system consisting of thioredoxin (Trx), thioredoxin reductase (TrxR) and NADPH (for review, see [12, 13] and references therein). Trx participates in redox reactions through its conserved active site [14] with two thiol residues (–Cys–Gly–Pro–Cys–) that is restored through reduction by TrxR. Previous work has demonstrated that various gold compounds with therapeutic benefit in cancer, rheumatoid arthritis and inflammatory diseases are efficient inhibitors of mammalian TrxR [15-19]. In particular, phosphine gold(I) complexes such as auranofin [Au(I)(PEt3) (2,3,4,6-tetra-O-acetyl-1-thio-β-D-thioglucose-S)] inhibit TrxR even at nanomolar concentrations [20]. Furthermore, the known inhibitor of TrxR, diferuloylmethane (curcumin), has been shown to act as both a radiosensitizer on cancer cells [21] and as a radioprotective agent on primary cells [22]. With TrxR-inhibition as the aim, a series of gold(I) complexes were recently synthesized and tested for inhibition efficiency and specificity [23]. Among them, [Au(SCN)(PEt3)] had selective and efficient TrxR inhibiting characteristics and was further tested as a potential radiosensitizing agent on human lung cancer cells.
Owing to its redox-modulation properties, the thioredoxin system is central in redox-dependent signalling pathways and protection against damaging effects from excess of reactive oxygen species caused either by stress or by an imbalance in endogenous cellular production. The importance of the system is further emphasized by its other multiple important physiological functions including regeneration of antioxidant enzymes by reduction of oxidized thiols, regulation of transcription factors, peroxidase activity and involvement in DNA replication and repair, as Trx acts as a hydrogen donor to ribonucleotide reductase. A specific example of the involvement in stress-response signalling is the nuclear signalling protein redox factor-1 (Ref-1), which is directly involved in cellular recovery upon exogenous stress, e.g. ionizing radiation, through nuclear DNA repair processes. Ref-1 stimulates several downstream transcription factors such as AP-1, NFκβ, HIF-1α, CREB and p53 (for review, see [24]) by enhancing DNA binding activity. Following exposure to ionizing radiation, Trx undergoes intracellular translocation from the cytoplasm to the nucleus [10, 11] and consequently activates Ref-1 [25]. The signal transduction is dependent on reduced Trx, making TrxR an excellent target for modulation of cellular response to radiation.
In the present study, the gold(I) compound [Au(SCN)(PEt3)] was evaluated as an in vitro radiosensitizer on the resistant non–small cell lung cancer (NSCLC) cell line U1810, with the overall aim to test the hypothesis that TrxR is an important factor in radioresistance.
Materials and methods
Chemicals
Synthesis and characterization of the linear two-coordinate gold(I) phosphine complex [Au(SCN)(PEt3)] have previously been described [23, 26]. Dimethyl sulfoxide was used as solvent for [Au(SCN)(PEt3)]. In cell experiments, the final concentration of dimethyl sulfoxide was <5%o.
Cell culture and irradiation
Experiments were conducted on the non–small cell lung cancer cell line U1810. This cell line has previously been characterized with a pronounced radio-resistant profile [27]. U1096e, which is a radiosensitive sub-cell line of the small cell lung carcinoma cell line U1906 [28] was used as a positive control for radiation effects. Cells were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% foetal bovine serum (Invitrogen) at 37°C and 5% CO2. Cell lines were irradiated in triplets with 2 or 5 Gy using a Cobalt-60 machine in room temperature with a dose rate of 0.51–0.50 Gy/min.
TrxR-inhibition
Approximately 500,000 cells were seeded in 25 cm2 flasks and incubated for 24 hrs. Medium was then exchanged for either fresh medium or medium prepared with 2.5 μM [Au(SCN)(PEt3)] and incubated for 24 hrs prior to radiation treatment. Cells were further incubated for 24 hrs immediately after irradiation and then medium was exchanged with either fresh medium or medium prepared with 0.05 μM [Au(SCN)(PEt3)]. For the purpose of gene expression analysis, cells were harvested 96 hrs after subjection to ionizing radiation and stored in RNAlater (Qiagen, Valencia, CA, USA) at −70°C prior to RNA purification. siRNA suppression of TrxR1 was achieved by reverse transfection of approximately 0.5 χ 106 cells 25 cm2 culture flasks using 10 nm TXNRD1 Silencer® Pre-designed siRNA, ID:111302 and Silencer® Negative Control siRNA #1 (Ambion, Austin, TX, USA). The transfection reagent used was siPORT™ NeoFX™ (Ambion). A 6 μl/flask was mixed with siRNA diluted in Opti-MEM I (Invitrogen) and incubated for 10 min. and then added to a suspension of harvested cells to a final volume of 5 ml.
Assessment of cell repopulation capacity and surviving fractions
The cell lines used in these experiments did not readily form single cell colonies in culture and thus the commonly used method of the clonogenic growth assay was not suitable. As an alternative, cells were monitored over a period of 14 days after seeding and routinely checked with light microscopy and sub-cultured before reaching 100% confluence. At each time of sub-culturing, cells were counted by measuring the optical density (OD) at 600 nm. Absorbance was compared to a standard curve constructed by counting a series of dilutions of cell suspensions from respective cell lines in a Bürker chamber and measuring OD600. The relative cell numbers were then obtained by comparison of subsequent cell counts in relation to sub-cultivation ratios as described previously [29]. The relative numbers of cells after 14 days was evaluated as a measure of repopulation capacity. To corroborate these measurements surviving fractions (SFs) of cells were calculated using graphical extrapolation of growth curves. In short, the obtained numbers of cells at each time of sub-culturing were plotted on a log-linear scale. Linear curves were extrapolated from the plots of each experiment to the time of irradiation. At this point the differences in cell count between irradiated cells and control represents the surviving fraction.
TrxR activity measurement
The amount of active TrxR was determined using insulin as substrate, as described by Holmgren and Bjrnstedt [30] with minor modifications. Samples with 50 μg total protein were incubated with 80 mM HEPES (pH 7.5), 0.9 mg/ml NADPH, 6 mM EDTA, 2 mg/ml insulin and 10 μm Escherichia coli Trx at 37°C for 20 min. A control for every sample with Trx excluded was run in the assay. The reaction was terminated by addition of 500 μl DTNB (0.4 mg/ml) and 6 M guanidine hydrochloride in 0.2 M Tris–Cl (pH 8.0). Absorbance was measured at 412 nm and the control was subtracted. A standard curve was prepared in the range of 0–580 ng/ml TrxR1 in order to perform activity measurements on multiple samples in a 96-well plate.
Quantitative PCR
Purification of mRNA, cDNA synthesis and real-time qPCR was used to determine remaining expression of TrxR1 mRNA following siRNA transfection as described previously [31].
Western blot
A total of 20 μg protein per sample was loaded to a 7.5% Tris–HCl ready-cast gel (Biorad, Hercules, CA, USA) and run at 20 mA for ∼2 hrs followed by semi-dry electroblotting to a nitrocellulose membrane for ∼40 at 100 V. The membrane was probed with anti-TrxR1 (1:500; Upstate, Lake Placid, NY) or anti-actin (1:2000; Sigma-Aldrich, St. Louis, MO, USA) antibodies overnight followed by secondary incubation with HRP-conjugated antibodies (1:1000 or 1:7000 respectively; Dako, Carpinteria, CA, USA). Bound antibodies were detected using chemiluminescence Western lightning kit (PerkinElmer, Boston, MA, USA).
Cell cycle analysis
Approximately 500,000 cells were seeded in 25 cm2 flasks and grown in serum-free medium for 72 hrs. Medium was then exchanged for either fresh medium or medium prepared with 2.5 μM [Au(SCN)(PEt3)] and incubated for 24, 48 and 72 hrs. Cells were harvested and washed with cold PBS and then fixed with 70% ethanol for 15 min. Cells were then incubated for 30 min. in room temperature with 500 μl propidium iodide/RNase solution (50 μg/ml, 0.1 mg/ml, 0.05% Triton in PBS). Samples were run on a BDCalibur flow cytometer (Becton Dickinson, San Jose, CA, USA). Results were analysed with the FlowJo for Windows software (v7.2.5).
Statistics
The Wilcoxon matched pair test was used for statistical analysis of cell experiments. Differences were considered significant at P < 0.05.
RNA isolation and gene array analysis
Total RNA was isolated and purified using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's protocol. Sample purity was analysed by the A260/280 ratio, and RNA quality and integrity were assessed using the Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). Individual samples were hybridized to Affymetrix Human Gene 1.0 ST arrays, containing 19,492 genes. The arrays were scanned using the Affymetrix GeneChip Scanner 3000 7G. RNA isolation and gene array analysis were performed at the Bioinformatics and Expression Analysis (BEA) core facility, Karolinska Institutet.
Data analysis of Affymetrix GeneChip results
Preprocessing of CEL files was performed in Affymetrix Expression Console (EC, version 1.1) using the following methods: (i) summarization: PLIER, (ii) background correction: PM-GCBG and (iii) normalization: Global Median. Expression levels in the treatment groups were compared to the control group using Student's t-test. A two-fold change (FC) in gene expression was used as cut-off for functional analysis. Pearson correlation was used to determine relationships between each treatment category.
Results
Inhibition of TrxR
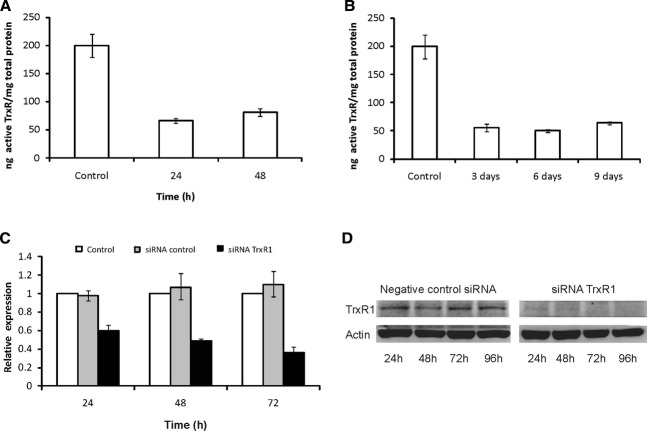
To determine the inhibitory effect of the gold(I) compound [Au(SCN)(PEt3)] on TrxR, cells were cultured with medium containing 2.5 μM [Au(SCN)(PEt3)]. Fractions were harvested after 24 and 48 hrs and assayed for TrxR activity. At both time points activity was decreased by approximately two thirds compared to untreated control cells (Fig. 1A). When cells were continuously cultured with 0.05 μM [Au(SCN)(PEt3)] the activity of TrxR remained suppressed at similar levels (Fig. 1B). The effect of siRNA-inhibition of TrxR1 at mRNA level was measured with qPCR at 24, 48 and 72 hrs after transfection (Fig. 1C). After 24 hrs the remaining expression of TrxR1 was approximately 60% (Fig. 1C). The suppression was subsequently increased at the following time points to levels of ∼50% and 35% remaining expression, respectively. The suppressive effect on TrxR1 at protein level was confirmed by Western blot analysis (Fig. 1D).
Inhibition of TrxR. (A) To determine the inhibitory effect of [Au(SCN)(PEt3)] on TrxR, a control experiment was performed where cell line U1810 was cultured with medium containing the gold(I) compound. Cells grown with 2.5 μM [Au(SCN)(PEt3)] were harvested at 24 and 48 hrs and TrxR activity was measured and compared to untreated controls. The y-axis denotes ng-active TrxR/mg total protein. Error bars show standard deviation of triplicates. (B) Cell line U1810 was also continuously cultured with 0.05 μM [Au(SCN)(PEt3)] after the initial 48 hrs treatment. Cells were harvested after 3, 6 and 9 days and TrxR activity measured and compared to untreated control. The y-axis denotes ng-active TrxR/mg total protein. Error bars show standard deviation of triplicates. (C) Cell line U1810 was transfected with siRNA directed at TrxR1 and mRNA levels measured with qPCR after 24, 48 and 72 hrs (black bars). A scrambled siRNA sequence was used as negative control (grey bars). Levels of mRNA were normalized to levels of untreated controls (white bars). Error bars show standard deviation of triplicates. (D) The effect of siRNA transfection on protein level was confirmed with Western blot analysis.
TrxR-inhibition and cell growth
Exposure of cell line U1810 to 2.5 μM [Au(SCN)(PEt3)] for 48 hrs showed no long-term toxic effect as shown by the proliferation curve (Fig. 2A). No direct effects on cell growth with TrxR-inhibition could be seen, however, when the cells were continuously treated with 0.05 μM [Au(SCN)(PEt3)] after the initial 48 hrs, the ability to repopulate was substantially affected. A control experiment with siRNA inhibition confirmed the negative effect on long-term cell proliferation with TrxR1 suppression (Fig. 2B).
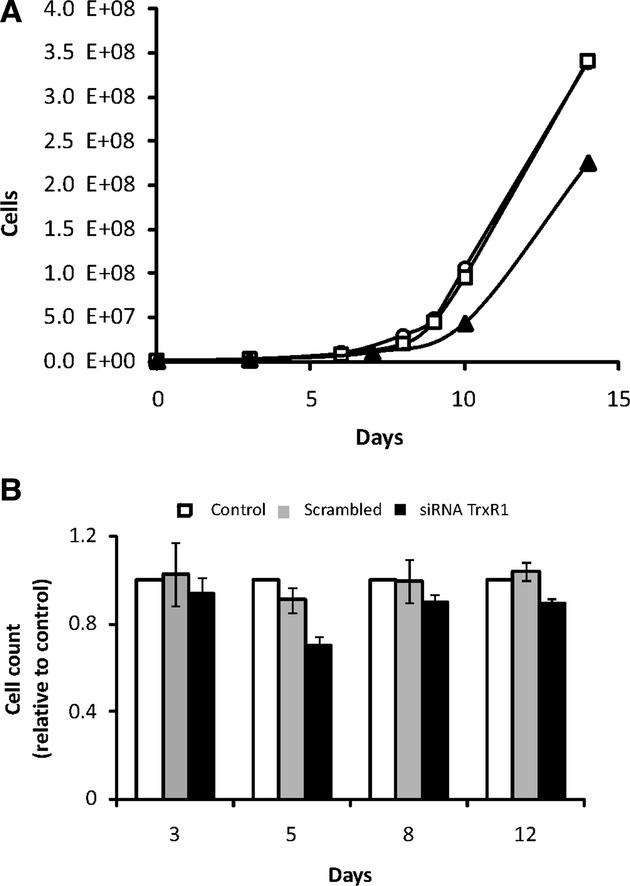
Effects of TrxR-inhibition on cell growth. (A) Growth curves for cell line U1810 without radiation exposure. {○: control, □: 48 hrs treatment with 2.5 μM [Au(SCN)(PEt3)] and ▴: continuous treatment with 0.05 μM [Au(SCN)(PEt3)]}. (B) A control experiment with siRNA transfection against TrxR1 (black bars) and negative control (grey bars) was performed to confirm the suppressive effect on cell proliferation. The figure shows the end-point number of cells normalized to untreated control (white bars). Error bars show standard deviation of triplicates.
Cell cycle modulation by TrxR-inhibition
To further study effects of TrxR-inhibition, U1810 cells were incubated with 2.5 μM [Au(SCN)(PEt3)] and then analysed by flow cytometry. After 24 hrs incubation with the TrxR inhibitor a shift in cell cycle distribution in live cell population was observed, with fewer cells in G1 and a larger ratio of cells exhibiting secondary cycling (n > 2) (Fig. 3A). Incubation periods of 48 and 72 hrs showed similar or greater shifts in cell cycle distribution in live cell population with a larger increase in the ratio of cells exhibiting secondary cycling as compared to untreated control (Fig. 3B and C). This observation was supported by analysis of gene expression data from the treatment group of TrxR-inhibition only, with 20 differentially up-regulated genes (within the selection criteria) connected to the cell cycle (Table 1).
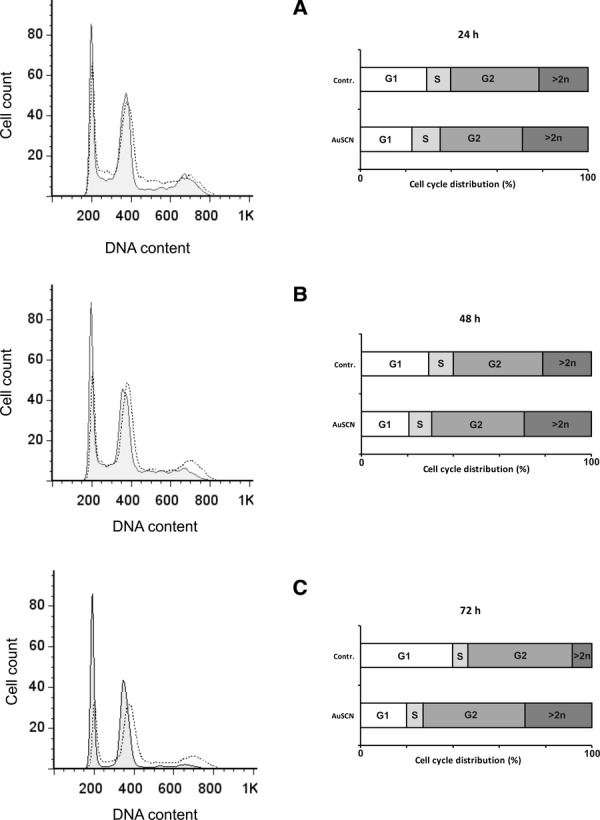
Cell cycle modulation by TrxR-inhibition. Cells were treated with 2.5 μM [Au(SCN)(PEt3)] for 24, 48 and 72 hrs and analysed with flow cytometry. The left graphs display DNA content of live cell populations as measured by PI-staining. Solid lines represent untreated control cells and superimposed dotted lines represent cells treated with TrxR-inhibitor. Left graphs show the cell cycle distribution in percentage of analysed cells. The distribution was modulated at all time points as compared to untreated controls with a larger fraction of cells displaying secondary cycling (n > 2), i.e. polyploidy with inhibition of TrxR.
Table 1.
TrxR-inhibition and genes connected to the cell cycle. Analysis of gene expression in cells treated with the TrxR-inhibitor only revealed 20 differentially up-regulated genes connected to the cell cycle (GO:0007049). TrxR-suppression by siRNA was not included in this analysis because it was represented by control experiment with a single sample and consequently not within the selection criteria.
| Probeset ID | Gene assignment | Symbol | Name | Location | Gene ID |
|---|---|---|---|---|---|
| 7900699 | NM_001255 | CDC20 | Cell division cycle 20 homologue (Saccharomyces cerevisiae) | 1p34.1 | 991 |
| 7901010 | NM_006845 | KIF2C | Kinesin family member 2C | 1p34.1 | 11004 |
| 7909708 | NM_016343 | CENPF | Centromere protein F, 350/400 kDa (mitosin) | 1q32–q41 | 1063 |
| 7924096 | NM_002497 | NEK2 | NIMA (never in mitosis gene a)-related kinase 2 | 1q32.2–q41 | 4751 |
| 7929334 | NM_018131 | CEP55 | Centrosomal protein 55 kDa | 10q23.33 | 55165 |
| 7955736 | NM_012291 | ESPL1 | Extra spindle pole bodies homologue 1 (S. cerevisiae) | 12q | 9700 |
| 7960702 | NM_031299 | CDCA3 | Cell division cycle associated 3 | 12p13 | 83461 |
| 7982889 | NM_016359 | NUSAP1 | Nucleolar and spindle associated protein 1 | 15q15.1 | 51203 |
| 7985829 | NM_001113378 | FANCI | Fanconi anaemia, complementation group I | 15q26.1 | 55215 |
| 7991406 | NM_003981 | PRC1 | Protein regulator of cytokinesis 1 | 15q26.1 | 9055 |
| 7994109 | NM_005030 | PLK1 | Polo-like kinase 1 (Drosophila) | 16p12.2 | 5347 |
| 8010260 | NM_001168 | BIRC5 | Baculoviral IAP repeat-containing 5 | 17q25 | 332 |
| 8013671 | NM_006461 | SPAG5 | Sperm associated antigen 5 | 17q11.2 | 10615 |
| 8043602 | NM_015341 | NCAPH | Non-SMC condensin I complex, subunit H | 2q11.2 | 23397 |
| 8059838 | NM_018410 | HJURP | Holliday junction recognition protein | 2q37.1 | 55355 |
| 8062571 | NM_030919 | FAM83D | Family with sequence similarity 83, member D | 20q11.22–q12 | 81610 |
| 8077731 | NM_033084 | FANCD2 | Fanconi anaemia, complementation group D2 | 3p26 | 2177 |
| 8094278 | NM_022346 | NCAPG | Non-SMC condensin I complex, subunit G | 4p15.33 | 64151 |
| 8105828 | NM_031966 | CCNB1 | Cyclin B1 | 5q12 | 891 |
| 8145418 | NM_152562 | CDCA2 | Cell division cycle associated 2 | 8p21.2 | 157313 |
Cell repopulation after ionizing radiation with or without TrxR-inhibition
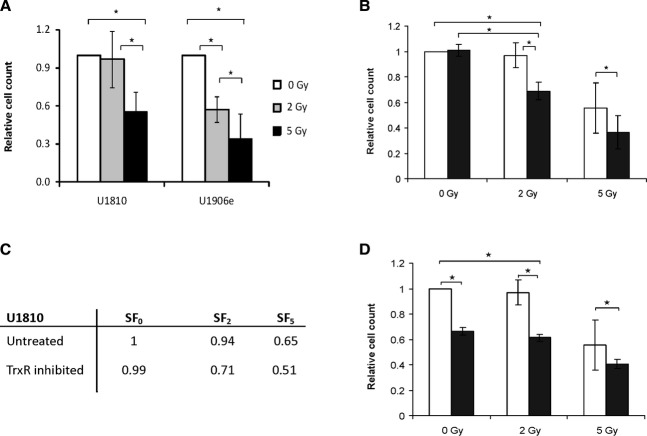
To establish the response of the cell lines to ionizing radiation alone or in combination with TrxR-inhibition, cells were exposed to a single fraction of 2 or 5 Gy and then monitored for a period of 14 days. During this period cells were routinely sub-cultured and counted in order to assess their ability to repopulate. The radiosensitive sub-cell line U1906e was included in the experiments as a positive control for the cell killing effects of radiation treatment. U1906e showed significant sensitivity at both 2 and 5 Gy doses (Fig. 4A). The ability of cell line U1810 to repopulate after irradiation was only affected at the higher dose of 5 Gy (Fig. 4A). A 48-hr treatment with [Au(SCN)(PEt3)] alone showed no significant effect on cell repopulation (Fig. 4B). However, with a combination of the lower radiation dose of 2 Gy and the TrxR-inhibitor [Au(SCN)(PEt3)] the cell growth was significantly impaired (Fig. 4B), both compared to control cells and cells treated only with the TrxR inhibitor. Even though the higher radiation dose of 5 Gy had a strong effect on cell growth by itself, the effect was significantly improved by the addition of the TrxR-inhibitor (Fig. 4B). To support these findings, a graphical extrapolation of growth curves for U1810 was performed to calculate initial relative surviving fractions of cells after irradiation (Fig. 4C). The calculated SF-values were in concordance with the relative end-point numbers of cells cultivated over 14 days. When TrxR was continuously inhibited during the entire 14-day period of culturing the effect was similar to the shorter treatment when combined with radiation (Fig. 4D), even though the effect did not differ significantly from long-term inhibition of TrxR alone.
Effects of IR and TrxR-inhibition on cell growth. (A) Cell lines U1810 and U1906e were exposed to a single fraction of 2 Gy (grey bars) or 5 Gy (black bars) and then counted and sub-cultured over a 2-week period. Cell counts were normalized against untreated control (white bars). Error bars show standard deviation of three different experiments. (B) Cell line U1810 was exposed to a single fraction of 2 or 5 Gy in combination with a 48-hr treatment with 2.5 μM of the TrxR-inhibitor [Au(SCN)(PEt3)] (black bars). Cells were then counted and sub-cultured over a 2-week period. The figure shows the end-point number of cells normalized to untreated control (white bars). Error bars show standard deviation of three different experiments. (C) Surviving fractions of irradiated cells at 2 and 5 Gy, respectively (SF2 and SF5) as compared to unirradiated controls. (D) Cell line U1810 was exposed to a single fraction of 2 or 5 Gy in combination with a 48-hr treatment with 2.5 μM [Au(SCN)(PEt3)] followed by continuous culturing with 0.05 μM [Au(SCN)(PEt3)] (black bars) for a total period of 14 days. The end-point number of cells was normalized to untreated control (white bars). Error bars show standard deviation of three different experiments.
Gene expression analysis
Effects on global gene expression of the TrxR inhibitor alone and in combination with ionizing radiation were evaluated by gene array analysis. Data obtained from the analysis displays a grouping of regulated genes clearly separating the different treatment variations (selection criteria; fold change ≥±2 and P≤ 0.0002) (Fig. 5A). However, the effects on changes in expression levels were markedly stronger with the combination treatment in comparison to exposure of cells to TrxR-inhibition or IR alone. When presented in a heatmap visualizing expression patterns for all treatment variations (Fig. 5B), the combination of inhibition and ionizing radiation takes an exclusive position with a larger part of analysed genes displaying strongly regulated levels of expression. A statistical correlation between the expression patterns (selection criteria; fold change ≥±2 and P≤ 0.0002) of the different treatment groups was performed (Fig. 5C) and showed firm correlation between the group with enzymatic TrxR suppression and TrxR1 suppression with siRNA. Although the effectiveness and specificity of [Au(SCN)(PEt3)] as a TrxR inhibitor has previously been evaluated [23], siRNA directed at TrxR1 was included in the experiments to verify that observed effects are due to the suppression of TrxR. The negative control scrambled siRNA also had little interference on expression patterns as shown by the correlation to the group of untreated controls.
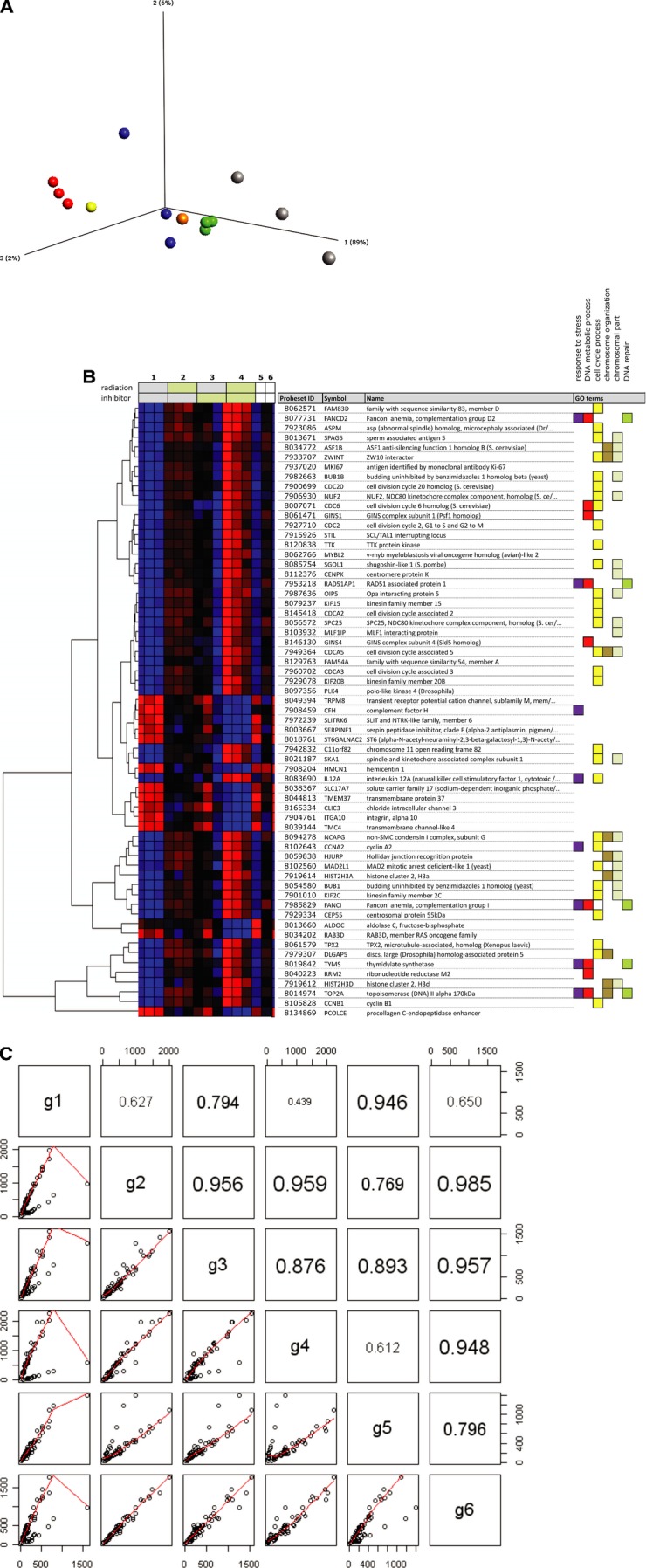
Gene expression analysis. (A) A three-dimensional principal component analysis (PCA) comparing the different treatment groups. Red spheres; control, green spheres; radiation, blue spheres; TrxR-inhibition by [Au(SCN)(PEt3)], grey spheres; combination treatment, orange sphere; TrxR-inhibition by siRNA, yellow sphere; scrambled siRNA sequence. Selection criteria; P≤ 0.0002 and FC ≥±2. Each treatment was performed in triplicates except a parallel control for TrxR-inhibition with siRNA (TrxR1 siRNA and a scrambled sequence, respectively). (B) Heatmap comparing expression patterns for all treatment variations. Selection criteria; P≤ 0.0002 and FC ≥±2. Horizontal rows represent individual genes and vertical columns represent individual samples. Each treatment was performed in triplicates except a parallel control for TrxR-inhibition with siRNA (the last two samples to the right, scrambled and TrxR siRNA, respectively). Coloured boxes to the right denote GO categories associated with the corresponding gene. (C) Pairwise scatterplots with correlation coefficients (Pearson correlation) for the average expression levels of different treatment groups. (g1 – untreated control, g2 – IR 5 Gy, g3 − 2.5 μM [Au(SCN)(PEt3)], g4 – IR+ inhibitor, g5 – Scrambled siRNA sequence and g6 – TrxR1 siRNA). Groups (g1–g6) correspond to numbered sets of vertical columns in (B).
Differential gene expression combining ionizing radiation and TrxR-inhibition
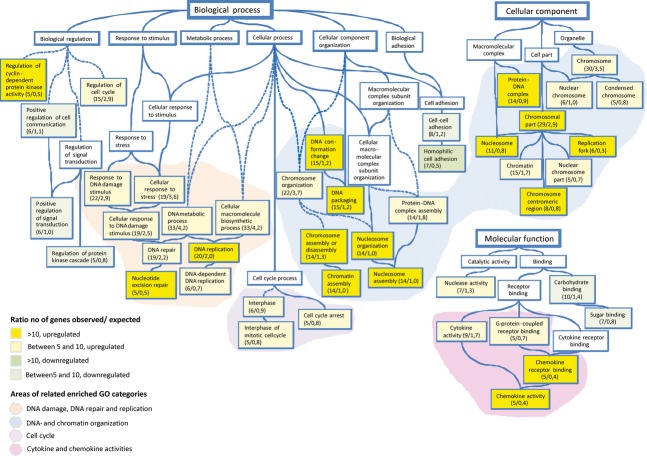
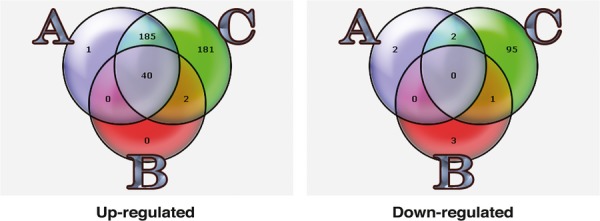
Further analysing gene expression data revealed that the combination treatment had the markedly lowest correlation compared to untreated controls, showing that this group is undoubtedly the most affected. In a functional testing (selection criteria; fold change ≥±2 and P≤ 0.02) comparing the different treatment groups, several overlapping expressions were observed. Strikingly, the combination treatment resulted in 181 uniquely up-regulated and 95 down-regulated genes (listed in Table 1, supplementary material) as compared to infrequent unique (0–3 genes) changes with the single treatments (Fig. 6). In order to facilitate a more comprehensive visualization of observed effects in a context of cell biology within this specific treatment group, the Gene Ontology (GO) vocabulary (www.geneontology.org) was used for functional classification and enrichment testing. When the affected genes were subjected to enrichment testing using Gene Ontology Tree Machine (GOTM, http://bioinfo.vanderbilt.edu/gotm/), several GO terms emerged as significantly enriched. Using this quantitative approach the observed number of differentially regulated genes is compared with the expected number within each GO-category (selection criteria; fold change ≥±2 and P≤ 0.02). The relation between different enriched GO Categories is presented in a Directed Acyclic Graph (Fig. 7). Shaded areas represent clusters of associated GO-categories (boxes) harbouring an over-representation of differentially expressed genes within the treatment group. Among the most significantly enriched categories are DNA replication (GO:0006260), nucleotide excision repair (GO:0006289), DNA conformation change (GO:0071103), chromosomal part (GO:0044427), chemokine receptor binding and activity (GO:0042379, 0008009). Four different clusters of related enriched GO categories could be identified; DNA-damage, DNA-repair and replication, DNA and chromatin organization, cell cycle and finally cytokine and chemokine activities.
Gene expression with IR. Effects on global gene expression of radiation with 5 Gy (A), TrxR inhibitor [Au(SCN)(PEt3)] (B) or in combination (C). TrxR-inhibition, IR and combination treatment all display overlapping expression changes. Selection criteria; fold change ≥±2 and P≤ 0.02. An average signal of at least 30 units in at least one of the treatment groups compared were required in order to remove genes with uncertain expression, i.e. close to or below background levels.
Graphical view of enriched GO categories. The GO categories having a statistical overrepresentation (i.e. enrichment) of differentially regulated genes in the treatment group of irradiation and inhibitor in combination have been brought together and visualized as a directed acyclic graph (DAG). A child node represents a more specific instance of a parent node, to which it is connected with a blue line (“is a” relationship). The numbers of observed/expected genes that were differentially regulated are indicated within each box representing an enriched GO-category. The colour of the boxes display the ratio between observed and expected up- or down-regulated genes. White boxes represent non-enriched GO categories that are included in the graph to allow tracing of the interrelationships between the GO categories. Striped lines represent connections where non-enriched GO categories have been omitted. Shaded areas represent related groups of GO categories.
Discussion
Gold compounds such as Auranofin are extensively used both as drugs and within experimental cancer research. However, in order to obtain a more selective cytotoxicity to cancer cells, there is presently a large interest in developing phosphine gold(I) compounds with different ligands. Several complexes with a maintained triethylphosphine synton and different soft ligands have previously been developed. In particular, the [Au(SCN)(PEt3)] complex was shown to exert strong inhibitory action on thioredoxin reductase, where the small NCS− ligand can be easily displaced by the S−/Se− group [23]. Serum concentrations of thiocyanate are relatively high (from 10 to 100 μM) and can be found at even larger concentrations in milk, saliva and tears [32, 33] and it can therefore be considered a “physiological” ligand. In this study the gold(I) compound [Au(SCN)(PEt3)] was tested in non-toxic concentrations as a potential radiosensitizing agent. The compound displayed efficient suppression of TrxR activity with a consequent modulation of the cell cycle.
A radio-resistant lung cancer cell line was treated with the [Au(SCN)(PEt3)] 24 hrs prior to, and 24 hrs following a single fraction of γ-radiation. Alternatively, cells were cultured in a low dose of [Au(SCN)(PEt3)] (0.05 μM) after the initial 48 hrs. The capacity of the cells to repopulate was then evaluated. A 48-hr treatment with the gold compound only showed no signs of cytotoxicity. However, prolonged treatment with the gold compound or TrxR1 siRNA repressed proliferation. With the lower dose of radiation (2 Gy) cells without TrxR-inhibition showed no difference in regrowth compared to unirradiated controls. However, in cells with the 48-hr inhibition treatment repopulation was decreased by approximately 30%, clearly demonstrating a sensitizing effect of TrxR-inhibition. A plausible explanation for these results could be a decrease in cellular antioxidant capacity, as a result of inhibition of the thioredoxin system, in combination with intracellular production of ROS caused by ionizing radiation. Although the higher radiation dose (5 Gy) had a strong effect on cell growth by itself, the effect was significantly pronounced in combination with TrxR-inhibition.
To investigate the combinatory effects of ionizing radiation and TrxR-inhibition further, a global gene expression analysis was performed. The expression analysis revealed a number of genes, either up- or down-regulated, only affected in the treatment group of IR and TrxR-inhibition in combination. The specificity of the effects achieved with TrxR-inhibition was verified by the use of siRNA. A heatmap analysis, supported by a statistical correlation, comparing the different treatment variations visualizes matching expression patterns between TrxR-inhibition by the gold compound and siRNA, which is encouraging considering it is a comparison between inhibition at mRNA and protein level. Within the genetic groupings uniquely affected by the combinatory treatment, four different clusters of related genetic functions displaying marked effects on expression levels were further identified. In general, these groupings (DNA-damage, DNA-repair and replication, DNA and chromatin organization, cell cycle and cytokine and chemokine activities) are of importance in the processes underlying radiation induced cell killing. The origin of the observed effects on expression levels is very likely the efficient inhibition of TrxR in combination with ionizing radiation.
Cellular response to environmental stress is controlled by tightly regulated signalling cascades, not seldom sensitive to changes in cellular redox status [34]. The combination of TrxR-inhibition and ionizing radiation, which also had the strongest effect on cellular repopulation, affected several clusters of related genetic functions. The cluster of functions connected to the cell cycle is of special interest since the relative radiosensitivity of a cell is connected to the cell cycle phase, with the highest sensitivity in the G2/M phase [35]. Radiation induced alterations in cell cycle transition connected to intrinsic radiosensitivity has previously been demonstrated using the U1810 cell line in comparison with more sensitive cells [36]. Previous investigations using knockdown of TrxR by transfection experiments have also demonstrated effects on the cell cycle with accumulation in the G2/M phase [37] and inhibition of cell growth [38]. In the current experiments, TrxR was efficiently inhibited with a subsequent modulation of the cell cycle as demonstrated by FACS-analysis and investigation of gene expression in cells treated with the TrxR-inhibitor only. The inhibitor has also been previously extensively characterized and found not to cause any aggregation of cells that could explain this modulation [23]. This effect was enhanced in combination with IR as shown by supporting microarray data.
In these experiments no striking expression changes related to oxidative stress were observed with the present selection criteria. Although this would be a rational expectation from suppression of an important cellular redox system, the specific conditions of the experiments need to be considered. Microarray analysis was performed 96 hrs after irradiation and 72 hrs after removal of the TrxR-inhibitor. Cellular response to redox-mediated stress is by necessity relatively swift, and changes in related expressions will consequently not be seen under these experimental conditions. However, downstream effects of radiation induced cellular damage that are manifested after the time of irradiation, and as hypothesized enhanced by TrxR-inhibition, are likely to be visible in this context.
As the principal cell killing mechanism of ionizing radiation is the conversion of intracellular H2O to reactive oxygen intermediates (e.g. O2−−, OH− and H2O2) causing damage to DNA [39-41], the emergence of distinctively induced genetic clusters associated with properties relating to DNA (DNA-damage, DNA-repair, DNA-replication and chromatin organization) is decidedly an appealing finding. The up-regulation of the gene encoding the catalytical subunit of ribonucleotide reductase (RRM2) found within the GO-category “DNA metabolic process,” is a specific example of DNA-related functions further strengthening the notion of involvement of the thioredoxin system in radiation resistance.
Another interesting molecular function affected is cytokine activity, which includes important functions for control of survival, growth, differentiation and effector functions of cells and tissues. Included within cytokine activity, a more specific association with immunoregulatory processes is found, namely, the chemokines. This is a group of functional molecules strongly connected to redox functions since they are involved in intramolecular disulfide bond formations through a number of conserved cysteine residues. The thioredoxin system also has chemotactic properties through secretion of Trx [42] and redox-independent mitogenic cytokine effects through the truncated form of Trx (Trx80) [43]. Among the enriched GO categories there also appear down-regulated groups that are interesting from a perspective of tumour biology, namely, “cell-cell adhesion” and “positive regulation of cell communication” that are of great interest for future studies.
Possible clinical applications
The difficulty of delivering clinically relevant doses to kill all tumour cells in patients with lung cancer remains a true clinical problem. This is partly due to the organs at risk, i.e. normal lung and spinal cord that limit the total dose of irradiation that can be delivered to the tumour. Recent technical advancements in radiotherapy such as four-dimensional computed tomography (4DCT) and intensity-modulated radiation therapy (IMRT) have allowed us to augment irradiation doses without compromising patient safety. However, a method to sensitize tumour cells to radiotherapy would enable us to target tumours that otherwise could not be cured.
Conclusions
This study reveals increased radiation sensitivity of cultured lung cancer cells (cell line U1810) with inhibition of the multi-functional redox enzyme TrxR by the gold(I) complex [Au(SCN)(PEt3)] demonstrated by a decreased ability to recover after radiation treatment. Our data clearly demonstrate an important role of TrxR in radiation resistance, and warrant further studies to elucidate the specific mechanisms of TrxR involvement in resistance development. In conclusion, pharmacological inhibition of TrxR is an attractive treatment strategy to optimize the efficacy of radiation therapy.
Acknowledgments
This investigation was supported by Cancerfonden (The Swedish Cancer Society) (M.B.), Cancer och allergifonden (The Swedish Cancer and Allergy Foundation) (M.B. and A.F.), ALF (Stockholm County Council) (MB), Radiumhemmets forskningsfonder (M.B. and A.F.), Hjärt- lung fonden (M.S.), Karolinska Institute research grants (M.B. and A.F.), Åke Wibergs stiftelse (A.F.), Magnus Bergwalls stiftelse (A.F.), Ministero dell'Istruzionedell'Universit e della Ricerca (PRIN20078EWK9B) and CIRCMSB (ConsorzioInteruniversitario di Ricerca in Chimica dei Metalli nei Sistemi Biologici) (V.G., C.M., M.R. and A.B.).
Conflict of interest
The authors declare no conflict of interest.
Supporting Information
References
- 1.Powell SN, Abraham EH. The biology of radioresistance: similarities, differences and interactions with drug resistance. Cytotechnology. 1993;12:325–45. doi: 10.1007/BF00744671. [DOI] [PubMed] [Google Scholar]
- 2.Wardman P. Chemical radiosensitizers for use in radiotherapy. Clin Oncol (R Coll Radiol) 2007;19:397–417. doi: 10.1016/j.clon.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 3.Demizu Y, Sasaki R, Trachootham D, et al. Alterations of cellular redox state during NNK-induced malignant transformation and resistance to radiation. Antioxid Redox Signal. 2008;10:951–61. doi: 10.1089/ars.2007.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diehn M, Cho RW, Lobo NA, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–3. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee HC, Kim DW, Jung KY, et al. Increased expression of antioxidant enzymes in radioresistant variant from U251 human glioblastoma cell line. Int J Mol Med. 2004;13:883–7. [PubMed] [Google Scholar]
- 6.Mirkovic N, Voehringer DW, Story MD, et al. Resistance to radiation-induced apoptosis in Bcl-2-expressing cells is reversed by depleting cellular thiols. Oncogene. 1997;15:1461–70. doi: 10.1038/sj.onc.1201310. [DOI] [PubMed] [Google Scholar]
- 7.Sun J, Chen Y, Li M, et al. Role of antioxidant enzymes on ionizing radiation resistance. Free Radic Biol Med. 1998;24:586–93. doi: 10.1016/s0891-5849(97)00291-8. [DOI] [PubMed] [Google Scholar]
- 8.Biaglow JE, Ayene IS, Koch CJ, et al. Radiation response of cells during altered protein thiol redox. Radiat Res. 2003;159:484–94. doi: 10.1667/0033-7587(2003)159[0484:rrocda]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 9.Husbeck B, Peehl DM, Knox SJ. Redox modulation of human prostate carcinoma cells by selenite increases radiation-induced cell killing. Free Radic Biol Med. 2005;38:50–7. doi: 10.1016/j.freeradbiomed.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 10.Karimpour S, Lou J, Lin LL, et al. Thioredoxin reductase regulates AP-1 activity as well as thioredoxin nuclear localization via active cysteines in response to ionizing radiation. Oncogene. 2002;21:6317–27. doi: 10.1038/sj.onc.1205749. [DOI] [PubMed] [Google Scholar]
- 11.Wei SJ, Botero A, Hirota K, et al. Thioredoxin nuclear translocation and interaction with redox factor-1 activates the activator protein-1 transcription factor in response to ionizing radiation. Cancer Res. 2000;60:6688–95. [PubMed] [Google Scholar]
- 12.Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267:6102–9. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 13.Gromer S, Urig S, Becker K. The thioredoxin system-from science to clinic. Med Res Rev. 2004;24:40–89. doi: 10.1002/med.10051. [DOI] [PubMed] [Google Scholar]
- 14.Holmgren A. Thioredoxin. Annu Rev Biochem. 1985;54:237–71. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 15.Bragadin M, Scutari G, Folda A, et al. Effect of metal complexes on thioredoxin reductase and the regulation of mitochondrial permeability conditions. Ann N Y Acad Sci. 2004;1030:348–54. doi: 10.1196/annals.1329.043. [DOI] [PubMed] [Google Scholar]
- 16.Omata Y, Folan M, Shaw M, et al. Sublethal concentrations of diverse gold compounds inhibit mammalian cytosolic thioredoxin reductase (TrxR1) Toxicol In Vitro. 2006;20:882–90. doi: 10.1016/j.tiv.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 17.Rigobello M, Messori L, Marcon G, et al. Gold complexes inhibit mitochondrial thioredoxin reductase: consequences on mitochondrial functions. J Inorg Biochem. 2004;98:1634–41. doi: 10.1016/j.jinorgbio.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 18.Rigobello MP, Folda A, Baldoin MC, et al. Effect of auranofin on the mitochondrial generation of hydrogen peroxide. Role of thioredoxin reductase. Free Radic Res. 2005;39:687–95. doi: 10.1080/10715760500135391. [DOI] [PubMed] [Google Scholar]
- 19.Rigobello MP, Scutari G, Folda A, et al. Mitochondrial thioredoxin reductase inhibition by gold(I) compounds and concurrent stimulation of permeability transition and release of cytochrome c. Biochem Pharmacol. 2004;67:689–96. doi: 10.1016/j.bcp.2003.09.038. [DOI] [PubMed] [Google Scholar]
- 20.Marzano C, Gandin V, Folda A, et al. Inhibition of thioredoxin reductase by auranofin induces apoptosis in cisplatin-resistant human ovarian cancer cells. Free Radic Biol Med. 2007;42:872–81. doi: 10.1016/j.freeradbiomed.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 21.Chendil D, Ranga RS, Meigooni D, et al. Curcumin confers radiosensitizing effect in prostate cancer cell line PC-3. Oncogene. 2004;23:1599–607. doi: 10.1038/sj.onc.1207284. [DOI] [PubMed] [Google Scholar]
- 22.Srinivasan M, Rajendra Prasad N, Menon VP. Protective effect of curcumin on gamma-radiation induced DNA damage and lipid peroxidation in cultured human lymphocytes. Mutat Res. 2006;611:96–103. doi: 10.1016/j.mrgentox.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Gandin V, Fernandes AP, Rigobello MP, et al. Cancer cell death induced by phosphine gold(I) compounds targeting thioredoxin reductase. Biochem Pharmacol. 79:90–101. doi: 10.1016/j.bcp.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 24.Fishel ML, Kelley MR. The DNA base excision repair protein Ape1/Ref-1 as a therapeutic and chemopreventive target. Mol Aspects Med. 2007;28:375–95. doi: 10.1016/j.mam.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 25.Hirota K, Matsui M, Iwata S, et al. AP-1 transcriptional activity is regulated by a direct association between thioredoxin and Ref-1. Proc Natl Acad Sci USA. 1997;94:3633–8. doi: 10.1073/pnas.94.8.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.El-Etri MM, Scovell WM. Synthesis and spectroscopic characterization of (triethylphosphine)gold(I) complexes AuX(PEt3) (X = Cl, Br, CN, SCN), [AuL(PEt3)+] (L = SMe2, SC(NH22, H2O), and (.mu.-S)[Au(PEt3)]2. Inorg Chem. 1990;29:480–4. [Google Scholar]
- 27.Brodin O, Lennartsson L, Nilsson S. Single-dose and fractionated irradiation of four human lung cancer cell lines in vitro. Acta Oncol. 1991;30:967–74. doi: 10.3109/02841869109088251. [DOI] [PubMed] [Google Scholar]
- 28.Brodin O, Arnberg H, Bergh J, et al. Increased radioresistance of an in vitro transformed human small cell lung cancer cell line. Lung Cancer. 1995;12:183–98. doi: 10.1016/0169-5002(95)00444-6. [DOI] [PubMed] [Google Scholar]
- 29.Johansson L, Nilsson K, Carlsson J, et al. Radiation effects on cultured human lymphoid cells. Analysis using the growth extrapolation method. Acta Radiol Oncol. 1981;20:51–9. doi: 10.3109/02841868109130189. [DOI] [PubMed] [Google Scholar]
- 30.Holmgren A, Bjornstedt M. Thioredoxin and thioredoxin reductase. Methods Enzymol. 1995;252:199–208. doi: 10.1016/0076-6879(95)52023-6. [DOI] [PubMed] [Google Scholar]
- 31.Rundlof AK, Fernandes AP, Selenius M, et al. Quantification of alternative mRNA species and identification of thioredoxin reductase 1 isoforms in human tumour cells. Differentiation. 2007;75:123–32. doi: 10.1111/j.1432-0436.2006.00121.x. [DOI] [PubMed] [Google Scholar]
- 32.Arlandson M, Decker T, Roongta VA, et al. Eosinophil peroxidase oxidation of thiocyanate. Characterization of major reaction products and a potential sulfhydryl-targeted cytotoxicity system. J Biol Chem. 2001;276:215–24. doi: 10.1074/jbc.M004881200. [DOI] [PubMed] [Google Scholar]
- 33.Thomas E. The Lactoperoxidase System, Chemsistry and Biological Significance. New York: Marcel Dekker; 1985. [Google Scholar]
- 34.Mikkelsen RB, Wardman P. Biological chemistry of reactive oxygen and nitrogen and radiation-induced signal transduction mechanisms. Oncogene. 2003;22:5734–54. doi: 10.1038/sj.onc.1206663. [DOI] [PubMed] [Google Scholar]
- 35.Pawlik TM, Keyomarsi K. Role of cell cycle in mediating sensitivity to radiotherapy. Int J Radiat Oncol Biol Phys. 2004;59:928–42. doi: 10.1016/j.ijrobp.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 36.Sirzen F, Heiden T, Nilsson A, et al. Characterisation of the G1/S cell cycle checkpoint defect in lung carcinoma cells with different intrinsic radiosensitivities. Anticancer Res. 1997;17:3381–6. [PubMed] [Google Scholar]
- 37.Yoo MH, Xu XM, Carlson BA, et al. Targeting thioredoxin reductase 1 reduction in cancer cells inhibits self-sufficient growth and DNA replication. PLoS One. 2007;2:e1112. doi: 10.1371/journal.pone.0001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gan L, Yang XL, Liu Q, et al. Inhibitory effects of thioredoxin reductase antisense RNA on the growth of human hepatocellular carcinoma cells. J Cell Biochem. 2005;96:653–64. doi: 10.1002/jcb.20585. [DOI] [PubMed] [Google Scholar]
- 39.Powell S, McMillan TJ. DNA damage and repair following treatment with ionizing radiation. Radiother Oncol. 1990;19:95–108. doi: 10.1016/0167-8140(90)90123-e. [DOI] [PubMed] [Google Scholar]
- 40.Ross GM. Induction of cell death by radiotherapy. Endocr Relat Cancer. 1999;6:41–4. doi: 10.1677/erc.0.0060041. [DOI] [PubMed] [Google Scholar]
- 41.Willers H, Dahm-Daphi J, Powell SN. Repair of radiation damage to DNA. Br J Cancer. 2004;90:1297–301. doi: 10.1038/sj.bjc.6601729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bertini R, Howard OM, Dong HF, et al. Thioredoxin, a redox enzyme released in infection and inflammation, is a unique chemoattractant for neutrophils, monocytes, and T cells. J Exp Med. 1999;189:1783–9. doi: 10.1084/jem.189.11.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pekkari K, Avila-Carino J, Gurunath R, et al. Truncated thioredoxin (Trx80) exerts unique mitogenic cytokine effects via a mechanism independent of thiol oxido-reductase activity. FEBS Lett. 2003;539:143–8. doi: 10.1016/s0014-5793(03)00214-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.