Abstract
Glycinamide ribonucleotide transformylase (GAR Tfase) is a folate-dependent enzyme in the de novo purine biosynthesis pathway, which has long been considered a potential target for development of anti-neoplastic therapeutics. Here we report the biological and X-ray crystallographic evaluations of both independent C10 diastereomers 10S and 10R methylthio-DDACTHF bound to human GAR Tfase, including the highest resolution apo GAR Tfase structure to date (1.52 Å). Both diastereomers are potent inhibitors (10R Ki = 210 nM, 10S Ki = 180 nM) of GAR Tfase and exhibit effective inhibition of human leukemia cell growth (IC50 = 80 and 50 nM, respectively). Their inhibitory activity was surprisingly high and these lipophilic C10-substituted analogs show distinct advantages over their hydrophilic counterparts, most strikingly in retaining potency in mutant human leukemia cell lines that lack reduced folate carrier protein activity (IC50 = 70 and 60 nM, respectively). Structural characterization reveals a new binding mode for these diasterisomers, in which the lipophilic thiomethyl groups penetrate deeper into a hydrophobic pocket within the folate-binding site. In silico docking simulations of three other sulfur-containing folate analogs also indicates that this hydrophobic cleft represents a favorable region for binding lipophilic substituents. Overall, these results suggest sulfur and its substitutions play an important role in not only the binding of anti-folates to GAR Tfase, but also in selectivity and cellular activity (growth inhibition), thereby presenting new possibilities for future design of potent and selective anti-folate drugs that target GAR Tfase.
Keywords: De novo purine biosynthesis, glycinamide ribonucleotide transformylase, (6R)-N10-formyltetrahydrofolic, C10 stereochemistry anti-folate therapeutics, x-ray crystallography, in silico modeling
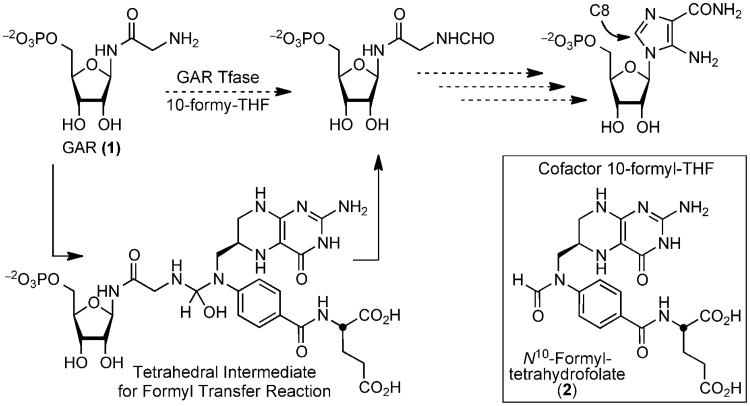
Glycinamide ribonucleotide transformylase (GAR Tfase) is a folate-dependent enzyme in the de novo purine biosynthesis pathway(1-4). GAR Tfase transfers a formyl group to the primary amine of its substrate, β-glycinamide ribonucleotide (β-GAR, 1) through the use of the cofactor (6R)-N10-formyltetrahydrofolic acid (10-formyl-THF, 2) in the third step of the pathway (Figure 1). This one-carbon transfer incorporates the C8 carbon of purines and is the first of two formyl transfer reactions that leads to inosine monophosphate (IMP) and ultimately to purines(1). The GAR Tfase mechanism has been the subject of much study in recent years for the ease with which it catalyzes the formyl transfer reaction(5-7), its biological role in DNA precursor synthesis(8), as an important target for chemotherapeutic drug design(9-20) and, more recently, in Mycobacterium tuberculosis where it has been targeted for drug discovery(21).
Figure 1.
The formyl transfer reaction catalyzed by GAR Tfase, with the proposed tetrahedral intermediate formed between the substrate β-GAR (1) and co-factor 10-formyl-THF (2).
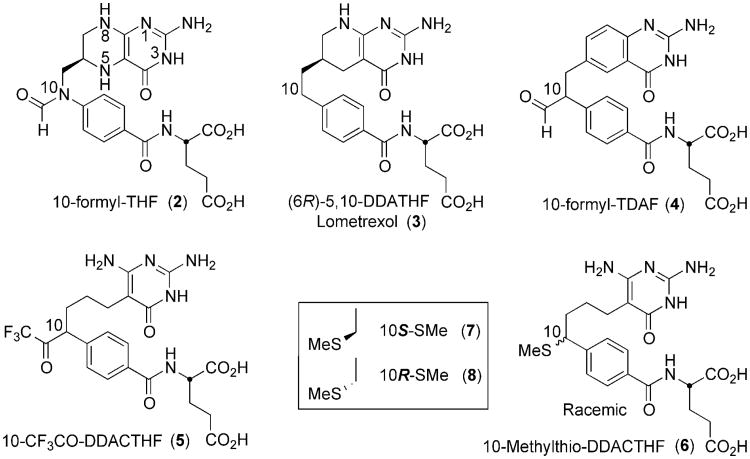
Inhibitors of folate metabolism and the enzymes responsible for the biosynthesis of nucleic acid precursors have long been considered important agents and targets for cancer chemotherapy(22). GAR Tfase was validated over 30 years ago as an anti-cancer target with the discovery of the first potent and selective inhibitor, 5,10-dideaza-5,6,7,8-tetrahydrofolic acid (DDATHF)(9). The compound was effective in vivo against solid murine and human tumors that did not respond to methotrexate. The potent activity of DDATHF was attributed to the reliance of tumor cells on de novo purine biosynthesis, whereas normal cells predominantly use salvage pathways of uridine or cytidine(23). Lometrexol, the 6R diastereomer of DDATHF, (3, Ki = 60 nM, Figure 2) was taken forward for further clinical development. Although some patients suffered cumulative myelosuppression, which was alleviated by co-supplementation folic acid(24, 25), Lometrexol was later withdrawn for economic reasons.
Figure 2.
Chemical structures of the natural cofactor 10-fTHF (2), and representative anti-folate analog inhibitors.
Structural biology has played a pivotal role in the design and evaluation of folate-based inhibitors for dihydrofolate reductase (DHFR)(26, 27), 5,10-methylenetetrahydrofolate reductase (MTHFR)(28), thymidylate synthetase (TS)(29, 30), 5-aminoimidazole-4- carboxamide-ribonucleotide (AICAR Tfase)(31-38) and GAR Tfase(7, 39-41). Previously, we reported on folate-based inhibitors that were designed to incorporate an electrophilic functional group at the C10 position and could potentially interact either with active site nucleophiles or the substrate amine of β-GAR to form adducts. The GAR Tfase inhibitors 10-formyl-TDAF, 4(10, 40) and 10-CF3CO-DDACTHF 5((41, 42)) both contain a nontransferable formyl or trifluoroacetyl group and have proven to be potent inhibitors of GAR Tfase (4, Ki = 260 nM, 5, Ki = 15 nM, Figure 2). X-ray and NMR studies on both 4 with Escherichia coli GAR Tfase (eGAR Tfase) and 5 with human GAR Tfase (hGAR Tfase) have shown they bind as their hydrated gem-diols(40, 41). In both cases, the C10 center rapidly racemizes in solution prohibiting the evaluation of each independent diastereomer. However, the formation of these hydrophilic gem-diol structures mimics the formyl transfer reaction intermediate and provides strong stabilizing interactions between the inhibitor and the catalytic residues of the protein, leading to the highest affinity inhibitors of GAR Tfase tested to date. This led to the discovery of the tetrahedral intermediate mimic(40, 41) and provided a unique design feature that affords selectivity over all other folate-dependent enzymes that do not use a formyl-transfer reaction.
While further investigating the affect of various substitutions at the C10 position, 10R/S-methylthio-DDACTHF 6 was discovered to be a modestly potent, sulfur-containing inhibitor of hGAR Tfase (Ki = 250 nM)(17) (Figure 2). The activity of this compound, both in vitro and in vivo was unexpected, as it does not contain the electrophilic functional group at the C10 position of other potent inhibitors, such as 4 and 5. Instead, the thiomethyl moiety is a weak hydrogen bond acceptor and presents a soft hydrophobic substituent for active site binding. Despite large differences in electrostatic potentials at the C10 position between these inhibitors, both are able to bind to hGAR Tfase with high affinity raising important questions about the activity and selectivity that substitutions at the C10 position can confer.
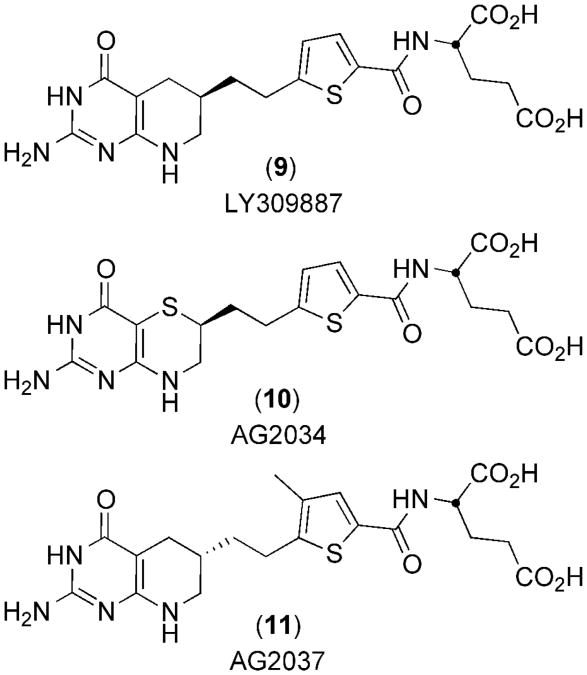
Finally, although not as a C10 substitution, the incorporation of sulfur in hGAR Tfase inhibitors is already well established, with Eli Lilly's LY309887 9(43) and Agouron's AG2034 10(44) and AG2037 11(45), all of which are highly potent (Ki between 5–50 nM) (Figure 3). However, no crystal structures of these are available to elucidate the role of sulfur in binding of these inhibitors to hGAR Tfase.
Figure 3.
Chemical structures of anti-folate analog inhibitors of hGAR Tfase that contain sulfur atoms.
The inability of the thiomethyl group to epimerize in solution was recently exploited for the successful asymmetric synthesis of 6. This allowed for the investigation of the individual diastereomers (10S 7 and 10R 8) in assessing both the importance of C10 substitution, stereochemistry and the contribution of sulfur to binding in this important class of hGAR Tfase inhibitors(46). Here we report the biological and X-ray crystallographic evaluations of both independent C10 diastereomers 10S 7 and 10R 8 bound to hGAR Tfase, which reveal a new binding mode for these important anti-folates. Furthermore, to investigate the overall contribution of sulfur in binding and GAR Tfase inhibition, three other sulfur-containing folate analogs 9, 10 and 11 were computationally docked into the folate-binding site of hGAR Tfase. These combined results shed light on C10 substitution, stereochemistry and a new role for sulfur in binding within the folate-binding site of hGAR Tfase. In addition, we also provide the highest resolution apo hGAR Tfase structure to date (1.52 Å). This high-resolution structure reveals the conformation of the previously unseen folate-binding loop and provides an excellent template for future ligand design.
Materials and Methods
Materials
Residues 808-1010 for the hGAR Tfase domain (purN) from the native human trifunctional enzyme (purD-purM-purN) were cloned into pET22b encoding a C-terminal hexahistidine tag, as previously described(7). Cofactor 2 was synthesized as previously described(47). All other common reagents and buffers were obtained from Sigma-Aldrich Corp., (St. Louis, MO, USA).
Biological activity against hGAR Tfase and AICAR Tfase
Enzyme activity assays of recombinant hGAR Tfase and recombinant human aminoimidazole carboxamide ribonucleotide transformylase (hAICAR Tfase) were performed as previously described(15, 41). Kinetics of the enzyme reactions were monitored for 2 min after reaction initiation. All inhibition constants (Ki) were calculated using Dixon plots.
Growth inhibition assay
The growth inhibition activity of the compounds was measured using CCRF-CEM human leukemia cell lines as previously described(15, 41, 48).
Protein expression
Stable and soluble expression of the hGAR Tfase domain (PurN) was achieved using transfected plasmid into BL21 (De3) E. coli as the expression host. One liter cultures of LB containing ampicillin (100 μg/mL) were grown at 37 °C to an OD595 of between 0.8–1.0, at which time cells were induced with 0.5 mM IPTG and incubated for a further for 5 hr at 30 °C.
Protein purification
Cells were lysed using an EmulsiFlex C-3 cell disruptor (Avestin, Canada) at 15 kpsi) at 4 °C in binding buffer (100 mM Tris, 500 mM NaCl, 40 mM imidazole, 5 mM beta-mercaptoethanol (β-Me) at pH 8.0)). The lysate was then clarified by centrifugation at 20,000× g for 20 min at 4 °C. The clear supernatant was then passed over a 5 mL Nickel HiTrap IMAC HP column (GE Healthcare, San Diego, CA, USA), followed by a wash of five column volumes of binding buffer. The bound protein was eluted by adding one column volume of elution buffer (100 mM Tris, 500 mM NaCl, 500 mM imidazole, 5 mM β-Me at pH 8.0) five times, and each fraction was analyzed by SDS-PAGE. The hGAR Tfase-containing fractions were pooled and applied to a Superdex™ 75 size exclusion column (Amersham Pharmacia, Pistcataway, NJ, USA) and eluted using 20 mM Tris, 200 mM NaCl, 5 mM DTT at pH 8.0 in 2 mL fractions. Protein purity was assessed by SDS-PAGE and those fractions containing protein of >95% purity were pooled for further use.
Crystallization and data collection
The hGAR Tfase was concentrated to 15 mg/mL in 20 mM Tris, 200 mM NaCl, 5 mM DTT at pH 8.0 and was either crystallized alone or co-crystallized with inhibitors at a 5-fold molar excess (inhibitors solubilized as 500-fold stocks in dimethyl sulfoxide), using the vapor-diffusion sitting drop method. For crystallization, an equal volume (2 uL) of protein and the well condition were mixed and left to equilibrate at 4 °C. Crystals grew from 0.1 M phosphate/citrate buffer, 1.5-2.0 M ammonium sulfate at pH 4.2, 25 % v/v glycerol added as a cryoprotectant. All data were collected at beam line 11-1 at the Stanford Synchrotron Radiation Lightsource (SSRL) at a wavelength of 0.9795 Å. All data sets were integrated and scaled using HKL2000(49). The diffraction data were indexed in space group P6522 with unit cell dimensions a=b=78.2 Å, c=229.3 Å and one monomer per asymmetric unit. The Matthews' coefficient(50) for each structure was approximately 4.44 Å3 Da-1, which translates to a relatively high solvent content of 72 %.
Structure solution and refinement
The structures of hGAR Tfase in both apo and ligand-bound forms were determined by molecular replacement in Phaser(51) using the previously solved low pH structure of unliganded hGAR Tfase (1MEO)(7) as the search model. Further model building and refinement were completed using Coot(52) and Refmac5(53) respectively. After initial rounds of refinement, the location of the inhibitors were determined from Fc-Fo omit maps (supplemental Figure S1). All ligand coordinates and stereochemical library files were generated using PRODRG(54). Riding hydrogens were added and anisotropic B-values assigned during refinement. Final resolutions and Rcryst/Rfree values were 1.52 Å and 17.3 %/19.3 % for apo GAR Tfase; 1.6 Å and 17.5 %/20.5 % for GAR Tfase in complex with 10S7; 1.7 Å and 20.1 %/21.6 % for GAR Tfase in complex with 10R8. Final models were validated using the JCSG quality control server (http://jcsgsrv2/QC) incorporating Molprobity(55), ADIT (http://rcsb-deposit.rutgers.edu/validate), WHATIF(56), Resolve(57) and Procheck(58).
In silico modeling of sulfur-containing hGAR Tfase inhibitors
Docking of the three sulfur-containing folate analogues 9, 10 and 11 was completed using the Dock simulation in the program MOE (Molecular Operating Environment 2011.10, Chemical Computing Group, Montréal, Canada). The high-resolution coordinates for 10S 7 bound to hGAR Tfase were used as the template with the two conserved structural water molecules, which play a vital role in binding the pteridine ring(7), being retained in the simulation. The target pocket was defined as atoms within 5 Å of the bound ligand 10S 7, which was removed prior to the docking simulation. For full details of the docking methodology, see supplemental info.
Results and Discussion
Biological activity
Remarkably, both diastereomers of the thiomethyl derivative exhibited potent activity against hGAR Tfase with 10S 7 being slightly more potent than 10R 8 (Ki = 180 nM vs Ki = 210 nM respectively, Table 1)(46). This similarity in the activity of the two C10-diastereomers is consistent with both possessing near equivalent capabilities for binding at the hGAR Tfase active site. Neither isomer possessed activity against AICAR Tfase (Ki >100 μM), suggesting the compounds act selectively on hGAR Tfase within the purine biosynthesis pathway. This selectivity appears to come directly from the C10 substitution, as the unsubstituted DDACTHF, shows moderate inhibition of AICAR Tfase (Ki = 20 μM) (Table 1)(46). The compounds were also examined for the ability to suppress growth of the human leukemia CCRF-CEM cell line (growth inhibition), both in the presence (+) and absence (™) of added hypoxanthine (purine) or thymidine (pyrimidine). Both 10S 7 and 10R 8 exhibited potent activity in the cell-based assay with an IC50 of 50 nM and 80 nM respectively, which is in line with Lometrexol 3 that have been reported as low as 2.9 nM(59) and in our hands here at 200 nM. The activity of 10S 7 and 10R 8 is only 3–5 times less than 5 (16 nM), the most potent inhibitor of GAR Tfase that we tested (Table 1)(46). In the presence of thymidine, all inhibitors retained their activity, whereas the inhibitors were inactive in the presence of supplemented hypoxanthine, indicating that these compounds inhibit cell growth by selectively inhibiting an enzyme within the purine biosynthetic pathway.
Table 1. In vitro hGAR Tfase and hAICAR Tfase enzyme inhibition and cellular growth inhibition assays.
| Inhibition Ki, μ M | CCRF−CEM (IC50, μM) | ||||
|---|---|---|---|---|---|
|
|
|||||
| Compound | hGAR Tfase | hAICAR Tfase | (−) T, (−) H | (+) T, (−) H | (−) T, (+) H |
| 10S-7 | 0.21 | > 100 | 0.05 | 0.07 | >10 |
| 10R-8 | 0.18 | > 100 | 0.08 | 0.08 | >10 |
| DDACTHF | 1.7 | 20 | 2.7 | 3.6 | >10 |
| 5 | 0.03 | > 100 | 0.016 | 0.017 | >10 |
| 3 Lometrexol | 0.06a | > 100 | 0.2 | 0.2 | >10 |
Folates and anti-folates are transported into the cell through the dominant and ubiquitously expressed reduced folate carrier protein (RFC)(60). Once in the cell, they are converted to long-chain poly-glutamate derivatives by folypolyglutamate synthetase (FPGS), which maximizes their cellular retention(61), as each additional glutamate adds a negative charge that prevents interaction with the efflux pumps(60). Mutant CCRF-CEM cell lines deficient in FPGS or the RFC were used to investigate the involvement of these proteins in regulating intracellular levels of the compounds. All of the most potent hGAR Tfase inhibitors (compounds 3-8) are substrates of FPGS, as previously demonstrated by loss of activity in cell lines that are deficient in FPGS activity (CCRF-CEM/FPGS-)(10, 17, 41, 46). When applied to these cells, both 10R 7 and 10S 8 lost ∼100-fold activity, indicating that they also benefit from intracellular polyglutamation (Table 2). Most strikingly, both 10R 7 and 10S 8 retained all of their potency in a mutant cell line deficient in RFC activity (CCRF-CEM/MTX IC50 = 70 nM and 60 nM respectively) (Table 2)(46). This result demonstrates that these compounds can cross the plasma membrane, even in the absence of RFC proteins as compared to 3 (Lometrexol) and 5 that both lost activity (>100 μM) 5(41, 42). In both mutant cell lines deficient in either FPGS or especially the RFC, the lipophilic thiomethyl-substituted DDACTHF was more potent than the unsubstituted DDACTHF or 5 containing a hydrophilic C10 substitution, highlighting the enhanced growth inhibition properties that lipophilic C10 substitutions can confer.
Table 2. In vitro growth inhibition.
X-ray structure determination
Crystal structures were determined for apo hGAR Tfase and hGAR Tfase in complex with the independent diastereomers of 10-methylthio-DDACTHF, 10R 7 and 10S 8. All structures were solved to between 1.52-1.8 Å resolution by molecular replacement using the apo hGAR Tfase structure at pH 4.2 (1MEO) as the search model(7). hGAR Tfase crystalized in space group P6522 with one molecule per asymmetric unit, consistent with previous structures(7, 41). The final models included residues 808-1007 from the tri-functional protein, with the last three histidines of the encoded C-terminal hexa-histidine tag not interpretable due to disorder. In previously reported hGAR structures, all contained multiple phosphate and or sulfate ions (or phosphate and glycerol in the case of 1MEJ)(7, 41). Here one phosphate ion is seen bound in the substrate-binding pocket that mimics the phosphate moiety of the substrate 1(7). While the sulfates have no known physiological function, their appearance here is not surprising as the protein was exposed to high concentrations of ammonium sulfate (1.5–2 M) in the crystallization experiments. All structures possess excellent stereo-chemical properties and data collection and refinement statistics are presented in Table 3.
Table 3. Data collection and refinement statistics.
| hGAR Tfase | Apo | 10S-methylthio-DDACTHF | 10R-methylthio-DDACTHF |
|---|---|---|---|
| Data Collection | |||
| Beamline | SSRL 11-1 | SSRL 11-1 | SSRL 11-1 |
| Wavelength (Å) | 0.9795 | 0.9795 | 0.9795 |
| Resolution (Å) | 1.52 (1.52-1.57)a | 1.60 (1.60-1.66)a | 1.70 (1.70-1.76)a |
| Space group | P6522 | P6522 | P6522 |
| a, b, c (Å) | 78.03, 78.03, 230.84 | 78.17, 78.17, 229.34 | 78.12, 78.12, 230.05 |
| No. molecules in a.u. | 1 | 1 | 1 |
| No. observations | 612,892 (47,021)a | 655,997 (65,172)a | 465,880 (35,193)a |
| No. unique reflections | 64,515 (6,187)a | 55,593 (5,431)a | 46,588 (4,512)a |
| Completeness (%) | 99.7 (98.4)a | 99.9 (100)a | 99.9 (99.7)a |
| Rsym (%)b | 4.3 (58.9)a | 4.0 (54.2)a | 4.2 (69.6)a |
| Average I/σ | 42.0 (3.0)a | 57.2 (4.0)a | 38.3 (2.5)a |
| Redundancy | 9.5 (7.6)a | 11.8 (12.0)a | 7.8 (10.0)a |
| Refinement statistics | |||
| Resolution (Å) | 1.52-67.6 | 1.60-67.7 | 1.70-67.7 |
| No. reflections (working set) | 61,124 (4,318)a | 52,652 (3,807)a | 44,103 (3,180)a |
| No. reflections (test set) | 3,263 (229)a | 2,814 (208)a | 2,349 (172)a |
| Rcryst (%)c | 17.3 (29.6)a | 18.0 (22.2)a | 19.9 (30.7)a |
| Rfree (%)d | 19.3 (33.1)a | 21.0 (22.8)a | 21.5 (32.8)a |
| hGAR/ligand/water atoms | 1,532/0/217 | 1,532/33/282 | 1,532/33/173 |
| No. of SO4/PO4 groups | 3/1 | 2/1 | 2/1 |
| Average B-values | |||
| hGAR Tfase | 19.8 | 18.9 | 22.4 |
| Ligand | N/A | 34.6 | 38.7 |
| Wilson B-value | 23.7 | 24.8 | 28.9 |
| Ramachandran plot | |||
| Most favored (%) | 92.8 | 92.3 | 93.4 |
| Additionally allowed (%) | 6.6 | 7.2 | 6.6 |
| Generously allowed (%) | 0.6 | 0.6 | 0 |
| Disallowed (%) | 0 | 0 | 0 |
| R.M.S deviations | |||
| Bond lengths (Å) | 0.018 | 0.017 | 0.018 |
| Angles (°) | 1.64 | 1.67 | 1.76 |
Numbers in parentheses are for highest resolution shell of data.
Rsym = Σhkl | I-<I> | Σhkl I
Rcryst = Σhkl | Fo-Fc| Σhkl Fo
Rfree is the same as Rcryst but for 5% of data excluded from the refinement.
Overall structure
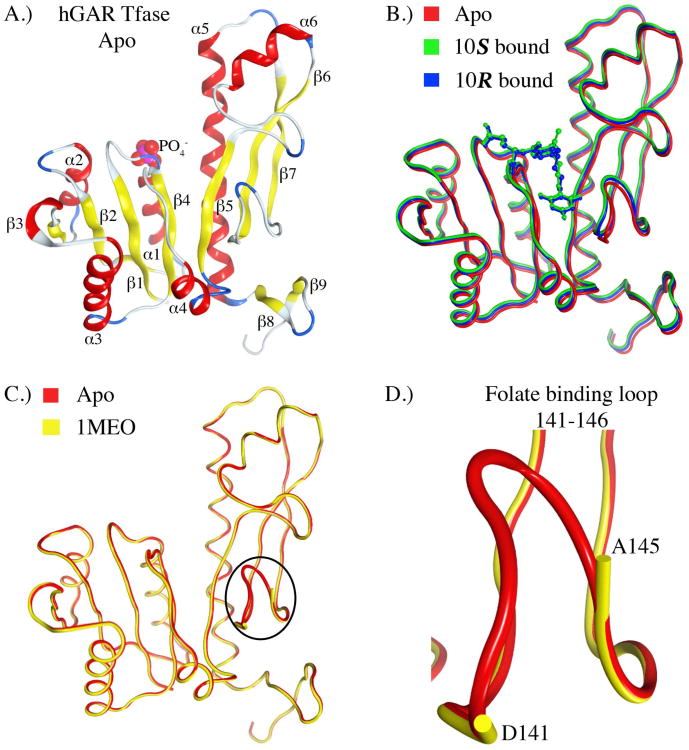
The structure of hGAR Tfase can be divided into two distinct sub-domains, N- and C-terminal, connected through a central parallel, seven-stranded β-sheet. The N-terminal sub-domain contains a Rossmann-type mononucleotide fold; at the C-terminal end of the β-sheet and positioned atop α1 is where the phosphate of the substrate β-GAR 1 would sit, and which is replaced here by an inorganic phosphate ion in the absence of substrate(7) (Figure 4A). Both the ligand-bound and apo forms of hGAR Tfase superpose almost identically across all α-carbons, suggesting ligand binding does not alter the protein conformation (Figure 4B), The overall topologies for both the apo and complexed forms of hGAR Tfase are very similar to that previously reported for the apo form at pH 4.2 (1MEO, Figure 4C). Although hGAR Tfase is inactive at pH 4.2, due to conformational isomerism in the substrate binding loop that precludes binding of the substrate(7)1, the unliganded structure reported here at pH 4.2 and the previously solved structure at pH 8.5 (1MEJ) are very similar, with an RMSD of 1.16Å across all α-carbons (supplemental Figure S1). Residues 141 to 146 of the folate-binding loop were absent in the previously reported low pH, unliganded structure(7) at 1.75 Å, presumably due to disorder or conformational heterogeneity. In the higher resolution structure presented here, we were able to model these residues revealing the conformation of this important loop at low pH (Figure 4D). However, this loop has higher B-values (46 Å(2)) than that of the rest of the protein structure (20 Å(2)), reconfirming that this region is indeed very flexible. This high-resolution structure should serve as an excellent template for further drug discovery of ligands that bind to this pocket.
Figure 4.
The crystal structure of hGAR Tfase in both apo and 10R-, 10S-methylthio-DDACTHF liganded forms. A) Ribbon diagram of the overall topology of unliganded hGAR Tfase. Helices and β-sheets are shown as red coils and yellow strands, respectively. B) Superposition of apo hGAR Tfase (red) with hGAR Tfase in complex with 10S-methylthio-DDACTHF (green) and 10R-methylthio-DDACTHF (blue). C) Superposition of new high-resolution apo hGAR Tfase at 1.52 Å with previous apo GAR Tfase structure at 1.75 Å (1MEO)(7) showing the conformation of the previously disordered folate-binding loop 141-146. D) Close up view of the folate-binding loop 141-146 as determined in the apo hGAR Tfase structure at 1.52 Å. Figures generated using MOE 2011.10. (Chemical Computing Group, Canada).
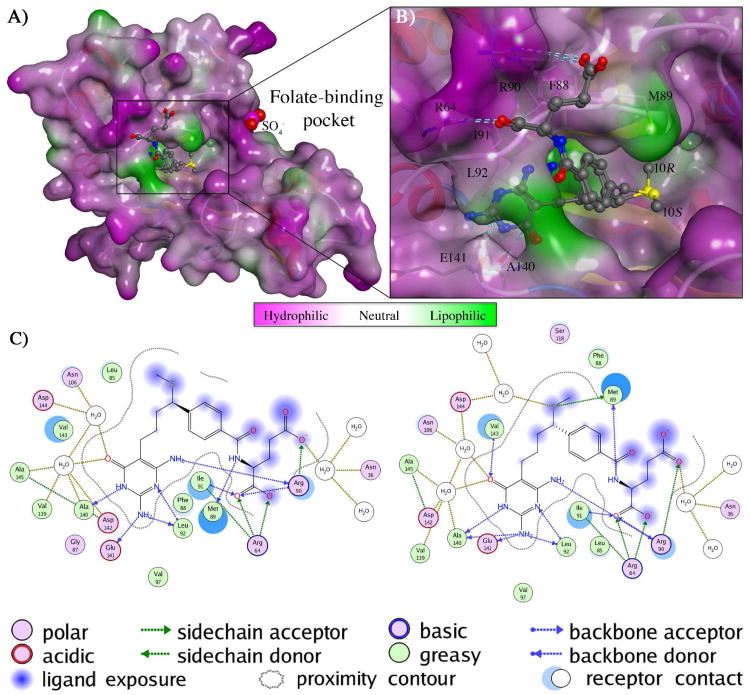
In both the complex structures, the Fo-Fc density maps show the inhibitors bind in the folate-binding pocket with clear, strong and interpretable electron density (supplemental Figure S2). The refined B-values of the rigid pteridine ring were similar to those of neighboring protein atoms suggesting full occupancy of the binding site. This unambiguous electron density made it possible to resolve the structures of each individual diastereomer and investigate the importance of stereochemistry at the C10 position. The folate-binding pocket is positioned at the interface of the N-terminal and C-terminal domains of hGAR Tfase. The binding pocket can be delineated into three parts: the pteridine-binding cleft, the formyl transfer region, and the benzoylglutamate region, described in detail below.
Pteridine-binding pocket
This region is responsible for the binding of the pteridine head of the natural cofactor 2. The pteridine-binding pocket is predominantly lined with hydrophobic residues Leu85, Ile91, Leu92, Phe96 and Val97 at one end, and the folate-binding loop (residues 141–146) making up the other. The diaminopyrimidinone rings of both diastereomers make a hydrogen bond between N2 of the ring and carbonyl of the Glu141 backbone (approx. 3.1Å) that is not seen in the quinazoline ring of 4 in complex with GAR Tfase(40). The diaminopyrimidinone ring makes up to six hydrogen bonds to the main-chain amides and carbonyl oxygens of Arg90, Leu92, Ala140, Glu141 and two hydrogen bonds with structurally ordered waters in a water-mediated network (Figure 5). Previous studies and in vitro growth inhibition experiments showed that the diaminopyrimidinone ring (seen in compounds 5–8) is favored over the quinazoline ring (seen in 4)(15). The diaminopyrimidinone ring of 10R 7 and 10S 8 contains a nitrogen atom at the N8 position, similar to that of the natural cofactor 2 and 3 (also conserved in 9–11). This ring nitrogen plays a key role in recognition and interaction with folate binding enzymes forming one end of a hydrogen bond donor-acceptor array. Although replacement of this N8 nitrogen (for example with carbon in 4) does not preclude binding to hGAR Tfase, its presence has been correlated with recognition by the RFC and/or FPGS(15). The importance of the N8 nitrogen was further demonstrated in the superior biological profile of 5 and in its complex structure (1NJS) where it hydrogen bonds to the carbonyl oxygen of Arg90(41). This important hydrogen bond (∼2.8 Å) is also conserved here in both diastereomers 10R 7 and 10S 8 at (Figure 5).
Figure 5.
Structural analysis of 10S-, 10R-methylthio-DDACTHF bound to hGAR Tfase A) Structure of hGAR Tfase with a molecular surface applied and colored by lipophilic potential as calculated from the Wildman and Crippen SlogP parameters(62) showing the bound conformations of both 10S-methylthio-DDACTHF (7) and 10R-methylthio-DDACTHF (8) in the folate binding pocket B) Close up view of the folate binding pocket showing the thiomethyl of 10S-(7) and 10R-(8) of methylthio-DDACTHF positioned in the lipophilic cleft that is lined by Phe88, Met89, Arg90, ArgI91 and Leu92. C) Schematic 2D projection of the ligand interactions of 10S-methylthio-DDACTHF (7) and 10R-methylthio-DDACTHF (8) with the pteridine-binding cleft, formyl transfer region, and benzoylglutamate region of hGAR Tfase. Legend includes amino acid type, hydrogen bond donor/acceptor origin, ligand exposure and proximity contour of ligand/receptor contacts. Bound water molecules that mediate contacts between ligand and receptor are also shown. Figures generated using MOE 2011.10. (Chemical Computing Group, Canada).
Formyl Transfer Region
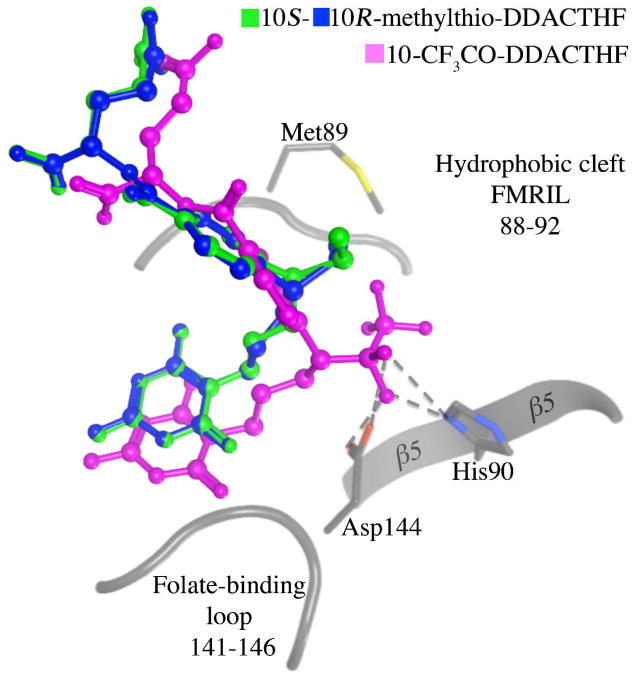
The formyl group of natural cofactor 2 is positioned at the N10 position on the short aliphatic linker between the pteridine head and the benzoylglutamate tail. The natural cofactor 2 and ligands 3 and 4, all possess a fused bicyclic quinazoline ring system, which is replaced by the monocyclic diaminopyrimidinone in both thiomethyl diastereomers 10R 7 and 10S 8. This monocyclic diaminopyrimidinone removes the chiral center at the C6 position and was originally designed to afford flexibility in compound 5 to optimize gem-diol binding with the formyl transfer region(41). In the complex structures with 4 and 5, hydroxyls of the gem-diol structures interact extensively with the formyl transfer region, primarily with Asp144 of the folate binding loop and His108 on the β5 sheet, which are two essential residues involved in the formyl transfer reaction(7, 40, 41). This interaction with the usually highly flexible folate-binding loop (supplemental Figure S3) tethers and stabilizes the region and affords the ligand much of its extra binding capacity over 3, which is devoid of any C10 substituent. However, the thiomethyl moiety of 6 makes no interactions with the folate-binding loop; instead, the sulfur atom is positioned deeper in to a hydrophobic cleft partially occupied by the benzoyl glutamate ring (Figure 5). Application of a molecular surface colored by lipophilic potential (as calculated from the Wildman and Crippen SlogP parameters(62)) shows that this hydrophobic cleft, composed of Phe88, Met89, Ile91 and Val143, is highly lipophilic (Figure 5). Interestingly, the same flexibility designed to optimize the interaction of the gem-diols of the electrophile-containing compounds allows the C10 sulfur atom of both diastereomers to occupy almost identical positions here, superposing to 0.52 Å RMSD (Figure 5). This similarity in binding mode is also reflected in the kinetic data, with each diastereomer being almost equipotent (Table 1). These data suggest that the thiomethyl positioned in this lipophilic cleft is a favorable interaction that accounts for why the ligand is able to bind to hGAR Tfase with high affinity, contrary to our initial assumptions. The position of the thiomethyl in a lipophilic cleft is consistent with previous reports where the molecular modeling program GRID was used to predict possible binding sites for the sulfur atoms(44) and is further backed up by biochemical evidence in assays where the 10R and 10S hydroxy and methoxy C10-substitutions of DDACTHF were not as potent in inhibiting hGAR Tfase(46). The overall binding mode displayed here by both diastereomers 10R 7 and 10S 8 is distinctly different from those observed with any of the electrophile-containing analogs (Figure 6). This highlights the complex array of binding opportunities within the hGAR Tfase folate-binding pocket that could be used to target for both potency and selectivity.
Figure 6.
Superposition of bound ligand conformations of 10S-methylthio-DDACTHF (7), 10R-methylthio-DDACTHF (8) and 10-CF3CO-DDATHF from 1NJS (5) to hGAR Tfase folate-binding site. The thiomethyl of 10S-(7), 10R-methylthio-DDACTHF (8) interacts with the lipophilic cleft made up by Phe88, Met89, Arg90, Ile91 and Leu92, whereas 10-CF3CO-DDATHF (5) interacts with Asp144 of the folate binding loop (141-146) and the catalytic His 90 of strand β5. Figure generated using MOE 2011.10. (Chemical Computing Group, Canada).
Benzoylglutamate tail region
The biological and structural roles of polyglutamation of folates and their respective analogs is not fully known. In E. coli polyglutamation, there are two distinct enzymatic activities involving amide linkages through either α- or γ-carboxylates. The addition of the first two L-glutamates is catalyzed by dihydrofolate synthetase-folypolyglutamate synthetase to the γ-carboxylate. Further L-glutamate residues (4-8) are added at the α-carboxylate position by another enzyme, folypoly-α-glutamate synthetase(63, 64). The process in eukaryotes is simpler, with FPGS adding all glutamates at the γ-carboxylate(65). The differences in the biological activities of inhibitors between the E. coli and hGAR Tfase can be partially explained by the observation that eGAR Tfase shows no obvious preference for the binding of the glutamate tail, which is able to switch between two distinct conformations. In eGAR Tfase complexes with both BW1476U89(66) and 10-formyl-TDAF(40), the γ-carboxylates form salt bridges with Arg64 with the α-carboxylate facing out whereas, in an epoxide-derived, multi-substrate analog complex structure (1JKX), the α-carboxylate forms the same salt bridge with the γ-carboxylate facing out(39). It is clear that this moiety is an absolute requirement for tight binding, as previous studies showed that 10-CF3CO-DDATHF derivatives lacking the glutamate tail are inactive against both hGAR Tfase and hAICAR Tfase(67).
In these studies, the mono-glutamate forms of 10R 7 and 10S 8 were used for both the in vitro testing and structural studies. The p-aminobenzoate ring is sandwiched within the lipophilic pocket comprised of Val143, Met89 and Phe88 and forms stacking interactions with Ile91. Although the p-aminobenzoate group sits deeper in the pocket than that of 5, its conformation is consistent with previous reports where the aromatic ring and adjacent carbonyl group are both in plane to each other(41). In both complex structures, the glutamate makes two important conserved contacts: a salt bridge between the α-carboxylate and Arg64 and a hydrogen bond with the backbone amide of Ile91 (Figure 5). As the glutamate tail is solvent-exposed, the electron density was sufficiently clear and unambiguous for modeling of the α-carboxylate only (Figure S1). The binding of the α-carboxylate orients the γ-carboxylate into the solvent where the density becomes weaker and disordered, consistent with the solvent-exposed, flexible nature of the polyglutamate tail.
C10 substitution and stereochemistry
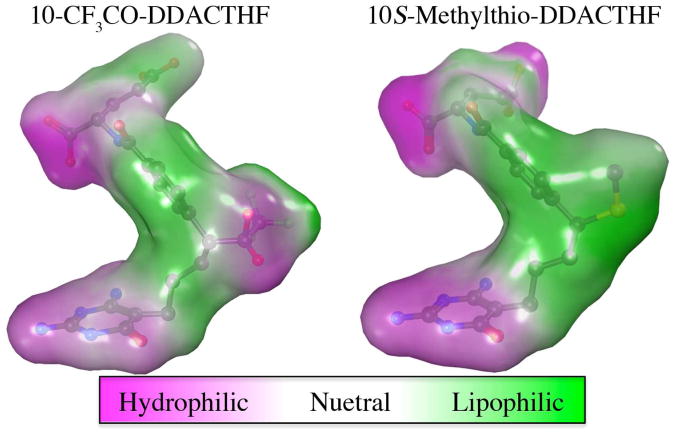
Unlike the N10 position in the natural co-factor 2, the corresponding C10 position can exist as two diasteromers, R or S (Figure 2). The most potent GAR Tfase inhibitors (4 and 5) both contain an electrophilic substitution at the C10 position, which bind as hydrophilic gem-diols mimicking the folate transfer reaction. In both cases, the C10 center rapidly epimerizes, prohibiting the evaluation of each independent diastereomer. In comparison to these transition state analog inhibitors, the biological activity of the lipophilic 10-methylthio-DDACTHF 6 was unexpected, as it does not contain the classical tetrahedral intermediate motif. Despite large differences in electrostatic potentials at the C10 position between these inhibitors (Figure 7), both were able to bind to hGAR Tfase with high affinity, raising important questions about the activity and selectivity that various substitutions at this position can confer.
Figure 7.
Structure and properties of anti-folates CF3CO-DDATHF 5 and 10S-methylthio-DDACTHF 7 as bound to hGAR Tfase. Molecular surface applied and colored by lipophilic potential as calculated from the Wildman and Crippen SlogP parameters(62). Figure generated using MOE 2011.10. (Chemical Computing Group, Canada).
To date, no structural characterization to compare individual C10 stereoisomers bound to hGAR Tfase has been reported. Analysis of Fo-Fc density maps of hGAR Tfase in complex with 4 and 5 revealed that both stereoisomers were present with 4, whereas only one isomer was observed with 5. Subsequent structural analysis and modeling of each isomer of 5 into the structure showed that only the R trifluromethyl ketone isomer can fit, with the alternate S isomer sterically occluded by loop Asp142–Ala145 (see Figure S4). However, the smaller hydrogen of the formyl hydrate can fit either side of this gem-diol structure and, hence, both isomers of 9 are observed in the Fo-Fc electron density maps.
Here, using discrete stereoisomers of 10-methylthio-DDACTHF, 10R 7 and 10S 8, crystallographic analysis showed that both thiomethyl substituents at the C10 position were able to occupy the same position in the lipophilic cleft superposing with an RMSD of 0.52 Å. In the absence of an electrophilic group at the C10 position to interact with the folate-binding loop, the inhibitors appears to adopt a novel binding pose within the folate-binding site that places the C10 position deeper into the lipophilic cleft with no significant stereochemical preference. These structural data are consistent with previous reports supporting the preference for lipophilic C10 substitutions, whereby the corresponding hydroxy and methoxy groups were not as potent while also showing very little difference in activity between the isomers(46).
In silico modeling of sulfur-containing hGAR Tfase inhibitors
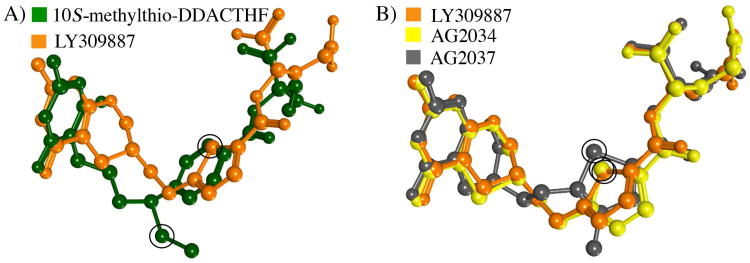
The incorporation of sulfur in hGAR Tfase inhibitors is well established, with compounds 9, 10 and 11 all containing sulfur atoms in thiophene rings located between the pteridine head and glutamate tail (Figure 3). As with 3 (Lometrexol, Ki = 60 nM), inhibitors 9, 10 and 11 also lack a C10 substitution, although they are able to bind to hGAR Tfase with higher affinity than 3 (Ki between 5–50 nM). However, no crystal structures are available demonstrating how these sulfur-containing inhibitors achieve high binding affinity to hGAR Tfase. To address this question, each compound was computationally docked into the folate-binding pocket of hGAR Tfase. With no C10 substitution, the overall binding orientation of each was similar to that of other folate analog inhibitors (Figure 8 and supplemental Figures S5–7). In this conformation, the sulfur-containing thiophene rings of all three analogs were placed into the lipophilic cleft, made up of residues Val143, Phe88, Met89 and Ile91. Inhibitor 10 also contains an extra sulfur atom in the pteridine ring. Docking of 10 placed the sulfur atom at the base of the pteridine-binding pocket, made up of the hydrophobic Leu85, Ile91, Leu92, Phe96 and Val97. Thus, the lipophilic cleft not only appears to be able to accommodate lipophilic C10 substituents, but it is also large enough to accept modified lipophilic ring systems in place of the p-benzoic acid moiety as seen with the 4-methyl-substituted thiophene ring of 11. The placement of the sulfur atoms within these substructures into this lipophilic cleft is most likely the mechanism for which these analogues gain high affinity binding to hGAR Tfase.
Figure 8.
Comparison of bound ligand conformations with sulfur atom positions circled in black. A) Superposition of the docked LY309887 9 and x-ray crystallographically determined 10S-methylthio-DDACTHF conformations as bound to hGAR Tfase 7 B) Superposition of the docked conformations for LY309887 9, AG2034 10 and AG2037 11 as bound to hGAR Tfase. Figures generated using MOE 2011.10. (Chemical Computing Group, Canada).
Conclusions
Herein we presented the highest resolution apo hGAR Tfase structure to date (1.52 Å) with the combined biological and structural evaluation of the two diastereomers, 10R- and 10S-methylthio-DDACTHF. Initially, the biological activity of the lipophilic C10 substitution was surprising(46) given its contrast to previous data for the requirement for hydrophilic gem-diols interacting with the folate-binding loop to achieve high affinity(40, 41). However, structural characterization reveal that the binding mode of both diastereomers, 10R 7 and 10S 8, is different from previous studies with electrophile-containing analogs (Figure 6). The lipophilic C10 thiomethyl penetrates deeper into a lipophilic cleft in the folate-binding site, allowing high affinity to be achieved in the absence of interaction with the folate-binding loop observed in electrophile-containing compounds, such as 4 and 5(40, 41).
Furthermore, in silico docking studies of three sulfur-containing hGAR Tfase inhibitors 9, 10 and 11, which lack a C10 substituent, showed that their sulfur-containing thiophene rings also occupy this lipophilic binding cleft, suggesting a favorable site for binding of sulfur or other lipophilic moieties. A survey of other human folate-binding proteins, such as DHFR (1DHF)(26), TS (1YPV)(68), ATIC (1P4R)(31), methionine synthase (1MSK)(69), mouse serine hydroxymethyltransferase (1EJI)(70) and bacterial MTHFR (1ZP3)(71), show this lipophilic cleft to be unique to GAR Tfase. Future design considerations could use a combination of both lipophilic substitutions to afford it selectivity over other folate-dependent enzymes. The structural characterization of these sulfur-containing anti-folates has therefore shed light on the complex array of new binding opportunities in the folate-binding pocket to improve both potency and selectivity.
Folate transport and intracellular retention are also very important aspects of anti-folate design as these are actively transported into the cell through the dominant reduced folate carrier protein (RFC)(60) where they are converted to long-chain poly-glutamate derivatives to increase their cellular retention(61). When designing anti-folates, the resulting compounds must be substrates of these two folate transport proteins in order to achieve maximum anti-tumor activity. It is well established that down regulation of RFC activity is a mechanism of tumor resistance to anti-folate drugs(72). Long-term exposure to anti-folates, such as methotrexate or pemetrexed, can also select for variants with decreased expression or activity of FPGS and thereby reduce retention of folates(73, 74). A study of 14 anti-folate resistant human leukemia cell lines showed most had 90– 99% loss of FPGS activity. However, due to the major contraction in cellular folate pools, these cells exhibited a marked hypersensitivity to lipid soluble anti-folates, such as neutrexin (trimetrexate) and AG377, which can cross freely into the cell(75). The inclusion of lipophilic sulfur substituents at the C10 position has a significant affect on cellular activity with 7 and 8 able to retain their potency in tumor cell lines deficient in the RFC proteins. This suggests that these compounds are able to either freely pass through the membrane environment or advantageously make use of other folate pathways such as the proton coupled mediated folate transporters (PCFT)(76). Utilizing the PCFT folate transport pathway has been seen as important in targeting solid tumors where the acidic micro-environment results in optimal PCFT activity and folate transfer rates over that of the RFC(77). Recently a novel series of 6-substituted classical pyrrolo[2,3,-d]pyrimidine anti-folates were reported to target GAR Tfase and derived their potent anti-tumor activity through the efficient membrane transport by the PCFT(78, 79). Overall, these results suggest sulfur and/or its specific substitutions have an important role in the binding of anti-folates to GAR Tfase, selectivity over other folate-binding enzymes and in cellular transport and growth inhibition. These findings present further expand our knowledge and possibilities for design of potent and selective anti-folate drugs that target GAR Tfase in de novo purine biosynthesis.
Supplementary Material
Acknowledgments
The authors would also like to thank Drs. Robyn Stanfield and Xiaoping Dai in the Wilson laboratory for assistance in data collection and structure solution. This paper is dedicated to Fay Connelly for her love and lifelong support, and who lost her brave battle to pancreatic cancer on the 22nd of February 2010.
Funding Sources: We are grateful for the financial support of the NIH (I.A.W), and the Skaggs Institute of Chemical Biology (I.A.W & D.L.B) and J.K.D who is a Skaggs Research Fellow. We are also grateful to the Susan G Komen for the Cure®, which provided funding for a post-doctoral fellowship (to S.C., SGKBCF PDF0402775). Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.
Abbreviations
- GAR Tfase
Glycinamide ribonucleotide transformylase
- β-GAR
β-glycinamide ribonucleotide
- 10-fTHF
(6R)-N10-formyltetrahydrofolic acid
- IMP
inosine monophosphate
- DHFR
dihydrofolate reductase
- MTFR
5,10-methylenetetrahydrofolate reductase
- TS
thymidylate synthetase
- ATIC
5-aminoimisazole-4-carboxamide-ribonucleotide
- AICAR
aminoimidazole carboxamide ribonucleotide transformylase
- β-Me
beta-mercaptoethanol
- SSRL
Stanford Synchrotron Radiation Lightsource
- RFC
reduced folate carrier protein
- FPGS
folypolyglutamate synthetase
Footnotes
Supporting Information.: In silico docking methodology and figures illustrating omit map 2Fo-Fc density for 7 and 8, superposition of apo hGAR Tfase at pH 4.2 and 8.5, comparison of possible C10 diastereomers of 5 bound to hGAR Tfase and schematic 2D projections of the ligand interactions of compounds 5, 9, 10 and 11 can be found in the supporting information. This material is available free of charge via the Internet at http://pubs.acs.org.
Accession codes: Atomic coordinates for the experimentally determined hGAR structures have been deposited in the RCSB Protein Data Bank (www.pdb.org) and are available under accession codes 4EW1, 4EW2 and. 4EW3.
Author Contributions: The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Notes: The authors declare no competing financial interest.
References
- 1.Dev IK, Harvey RJ. N10-Formyltetrahydrofolate is the formyl donor for glycinamide ribotide transformylase in Escherichia coli. J Biol Chem. 1978;253:4242–4244. [PubMed] [Google Scholar]
- 2.Caperelli CA. Mammalian glycinamide ribonucleotide transformylase. Kinetic mechanism and associated de novo purine biosynthetic activities. J Biol Chem. 1989;264:5053–5057. [PubMed] [Google Scholar]
- 3.Inglese J, Johnson DL, Shiau A, Smith JM, Benkovic SJ. Subcloning, characterization, and affinity labeling of Escherichia coli glycinamide ribonucleotide transformylase. Biochemistry. 1990;29:1436–1443. doi: 10.1021/bi00458a014. [DOI] [PubMed] [Google Scholar]
- 4.Wolfenden R. Transition state analog inhibitors and enzyme catalysis. Annu Rev Biophys Bioeng. 1976;5:271–306. doi: 10.1146/annurev.bb.05.060176.001415. [DOI] [PubMed] [Google Scholar]
- 5.Shim JH, Benkovic SJ. Evaluation of the kinetic mechanism of Escherichia coli glycinamide ribonucleotide transformylase. Biochemistry. 1998;37:8776–8782. doi: 10.1021/bi980244k. [DOI] [PubMed] [Google Scholar]
- 6.Shim JH, Benkovic SJ. Catalytic mechanism of Escherichia coli glycinamide ribonucleotide transformylase probed by site-directed mutagenesis and pH-dependent studies. Biochemistry. 1999;38:10024–10031. doi: 10.1021/bi9904609. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Desharnais J, Greasley SE, Beardsley GP, Boger DL, Wilson IA. Crystal structures of human GAR Tfase at low and high pH and with substrate β-GAR. Biochemistry. 2002;41:14206–14215. doi: 10.1021/bi020522m. [DOI] [PubMed] [Google Scholar]
- 8.Poch MT, Qin W, Caperelli CA. The human trifunctional enzyme of de novo purine biosynthesis: heterologous expression, purification, and preliminary characterization. Protein Expr Purif. 1998;12:17–24. doi: 10.1006/prep.1997.0799. [DOI] [PubMed] [Google Scholar]
- 9.Taylor EC, Harrington PJ, Fletcher SR, Beardsley GP, Moran RG. Synthesis of the antileukemic agents 5,10-dideazaaminopterin and 5,10-dideaza-5,6,7,8-tetrahydroaminopterin. J Med Chem. 1985;28:914–921. doi: 10.1021/jm00145a012. [DOI] [PubMed] [Google Scholar]
- 10.Boger DL, Haynes NE, Kitos PA, Warren MS, Ramcharan J, Marolewski AE, Benkovic SJ. 10-Formyl-5,8,10-trideazafolic acid (10-formyl-TDAF): a potent inhibitor of glycinamide ribonucleotide transformylase. Bioorg Med Chem. 1997;5:1817–1830. doi: 10.1016/s0968-0896(97)00120-x. [DOI] [PubMed] [Google Scholar]
- 11.Boger DL, Haynes NE, Warren MS, Gooljarsingh LT, Ramcharan J, Kitos PA, Benkovic SJ. Functionalized analogues of 5,8,10-trideazafolate as potential inhibitors of GAR Tfase or AICAR Tfase. Bioorg Med Chem. 1997;5:1831–1838. doi: 10.1016/s0968-0896(97)00121-1. [DOI] [PubMed] [Google Scholar]
- 12.Boger DL, Haynes NE, Warren MS, Ramcharan J, Kitos PA, Benkovic SJ. Multisubstrate analogue based on 5,8,10-trideazafolate. Bioorg Med Chem. 1997;5:1853–1857. doi: 10.1016/s0968-0896(97)00124-7. [DOI] [PubMed] [Google Scholar]
- 13.Boger DL, Haynes NE, Warren MS, Ramcharan J, Kitos PA, Benkovic SJ. Functionalized analogues of 5,8,10-trideazafolate: development of an enzyme-assembled tight binding inhibitor of GAR Tfase and a potential irreversible inhibitor of AICAR Tfase. Bioorg Med Chem. 1997;5:1839–1846. doi: 10.1016/s0968-0896(97)00122-3. [DOI] [PubMed] [Google Scholar]
- 14.Boger DL, Haynes NE, Warren MS, Ramcharan J, Marolewski AE, Kitos PA, Benkovic SJ. Abenzyl 10-formyl-trideazafolic acid (abenzyl 10-formyl-TDAF): an effective inhibitor of glycinamide ribonucleotide transformylase. Bioorg Med Chem. 1997;5:1847–1852. doi: 10.1016/s0968-0896(97)00123-5. [DOI] [PubMed] [Google Scholar]
- 15.Marsilje TH, Labroli MA, Hedrick MP, Jin Q, Desharnais J, Baker SJ, Gooljarsingh LT, Ramcharan J, Tavassoli A, Zhang Y, Wilson IA, Beardsley GP, Benkovic SJ, Boger DL. 10-Formyl-5,10-dideaza-acyclic-5,6,7,8-tetrahydrofolic acid (10-formyl-DDACTHF): a potent cytotoxic agent acting by selective inhibition of human GAR Tfase and the de novo purine biosynthetic pathway. Bioorg Med Chem. 2002;10:2739–2749. doi: 10.1016/s0968-0896(02)00102-5. [DOI] [PubMed] [Google Scholar]
- 16.Varney MD, Palmer CL, Romines WH, 3rd, Boritzki T, Margosiak SA, Almassy R, Janson CA, Bartlett C, Howland EJ, Ferre R. Protein structure-based design, synthesis, and biological evaluation of 5-thia-2,6-diamino-4(3H)-oxopyrimidines: potent inhibitors of glycinamide ribonucleotide transformylase with potent cell growth inhibition. J Med Chem. 1997;40:2502–2524. doi: 10.1021/jm9607459. [DOI] [PubMed] [Google Scholar]
- 17.Cheng H, Chong Y, Hwang I, Tavassoli A, Zhang Y, Wilson IA, Benkovic SJ, Boger DL. Design, synthesis, and biological evaluation of 10-methanesulfonyl-DDACTHF, 10-methanesulfonyl-5-DACTHF, and 10-methylthio-DDACTHF as potent inhibitors of GAR Tfase and the de novo purine biosynthetic pathway. Bioorg Med Chem. 2005;13:3577–3585. doi: 10.1016/j.bmc.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 18.Cheng H, Hwang I, Chong Y, Tavassoli A, Webb ME, Zhang Y, Wilson IA, Benkovic SJ, Boger DL. Synthesis and biological evaluation of N-[4-[5-(2,4-diamino-6-oxo-1,6-dihydropyrimidin-5-yl)-2-(2,2,2-trifluoroac etyl)pentyl]benzoyl]-L-glutamic acid as a potential inhibitor of GAR Tfase and the de novo purine biosynthetic pathway. Bioorg Med Chem. 2005;13:3593–3599. doi: 10.1016/j.bmc.2004.11.049. [DOI] [PubMed] [Google Scholar]
- 19.Chong Y, Hwang I, Tavassoli A, Zhang Y, Wilson IA, Benkovic SJ, Boger DL. Synthesis and biological evaluation of α-and γ-carboxamide derivatives of 10-CF3CO-DDACTHF. Bioorg Med Chem. 2005;13:3587–3592. doi: 10.1016/j.bmc.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Desmoulin SK, Cherian C, Polin L, White K, Kushner J, Fulterer A, Chang MH, Mitchell-Ryan S, Stout M, Romero MF, Hou Z, Matherly LH, Gangjee A. Synthesis, biological, and antitumor activity of a highly potent 6-substituted pyrrolo[2,3-d]pyrimidine thienoyl antifolate inhibitor with proton-coupled folate transporter and folate receptor selectivity over the reduced folate carrier that inhibits β-glycinamide ribonucleotide formyltransferase. J Med Chem. 2011;54:7150–7164. doi: 10.1021/jm200739e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z, Caradoc-Davies TT, Dickson JM, Baker EN, Squire CJ. Structures of glycinamide ribonucleotide transformylase (PurN) from Mycobacterium tuberculosis reveal a novel dimer with relevance to drug discovery. J Mol Biol. 2009;389:722–733. doi: 10.1016/j.jmb.2009.04.044. [DOI] [PubMed] [Google Scholar]
- 22.Hagner N, Joerger M. Cancer chemotherapy: targeting folic acid synthesis. Cancer Manag Res. 2010;2:293–301. doi: 10.2147/CMR.S10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson RC, Harkrader RJ. In Nucleosides and Cancer Treatment. Sydney: Academic Press; 1981. pp. 18–31. [Google Scholar]
- 24.Laohavinij S, Wedge SR, Lind MJ, Bailey N, Humphreys A, Proctor M, Chapman F, Simmons D, Oakley A, Robson L, Gumbrell L, Taylor GA, Thomas HD, Boddy AV, Newell DR, Calvert AH. A phase I clinical study of the antipurine antifolate lometrexol (DDATHF) given with oral folic acid. Invest New Drugs. 1996;14:325–335. doi: 10.1007/BF00194536. [DOI] [PubMed] [Google Scholar]
- 25.Roberts JD, Poplin EA, Tombes MB, Kyle B, Spicer DV, Grant S, Synold T, Moran R. Weekly lometrexol with daily oral folic acid is appropriate for phase II evaluation. Cancer Chemother Pharmacol. 2000;45:103–110. doi: 10.1007/s002800050017. [DOI] [PubMed] [Google Scholar]
- 26.Davies JF, 2nd, Delcamp TJ, Prendergast NJ, Ashford VA, Freisheim JH, Kraut J. Crystal structures of recombinant human dihydrofolate reductase complexed with folate and 5-deazafolate. Biochemistry. 1990;29:9467–9479. doi: 10.1021/bi00492a021. [DOI] [PubMed] [Google Scholar]
- 27.Oefner C, D'Arcy A, Winkler FK. Crystal structure of human dihydrofolate reductase complexed with folate. Eur J Biochem. 1988;174:377–385. doi: 10.1111/j.1432-1033.1988.tb14108.x. [DOI] [PubMed] [Google Scholar]
- 28.Pejchal R, Campbell E, Guenther BD, Lennon BW, Matthews RG, Ludwig ML. Structural perturbations in the Ala --> Val polymorphism of methylenetetrahydrofolate reductase: how binding of folates may protect against inactivation. Biochemistry. 2006;45:4808–4818. doi: 10.1021/bi052294c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hardy LW, Finer-Moore JS, Montfort WR, Jones MO, Santi DV, Stroud RM. Atomic structure of thymidylate synthase: target for rational drug design. Science. 1987;235:448–455. doi: 10.1126/science.3099389. [DOI] [PubMed] [Google Scholar]
- 30.Montfort WR, Perry KM, Fauman EB, Finer-Moore JS, Maley GF, Hardy L, Maley F, Stroud RM. Structure, multiple site binding, and segmental accommodation in thymidylate synthase on binding dUMP and an anti-folate. Biochemistry. 1990;29:6964–6977. doi: 10.1021/bi00482a004. [DOI] [PubMed] [Google Scholar]
- 31.Cheong CG, Wolan DW, Greasley SE, Horton PA, Beardsley GP, Wilson IA. Crystal structures of human bifunctional enzyme aminoimidazole-4-carboxamide ribonucleotide transformylase/IMP cyclohydrolase in complex with potent sulfonyl-containing antifolates. J Biol Chem. 2004;279:18034–18045. doi: 10.1074/jbc.M313691200. [DOI] [PubMed] [Google Scholar]
- 32.Greasley SE, Horton P, Ramcharan J, Beardsley GP, Benkovic SJ, Wilson IA. Crystal structure of a bifunctional transformylase and cyclohydrolase enzyme in purine biosynthesis. Nat Struct Biol. 2001;8:402–406. doi: 10.1038/87555. [DOI] [PubMed] [Google Scholar]
- 33.Li C, Xu L, Wolan DW, Wilson IA, Olson AJ. Virtual screening of human 5-aminoimidazole-4-carboxamide ribonucleotide transformylase against the NCI diversity set by use of AutoDock to identify novel nonfolate inhibitors. J Med Chem. 2004;47:6681–6690. doi: 10.1021/jm049504o. [DOI] [PubMed] [Google Scholar]
- 34.Wolan DW, Cheong CG, Greasley SE, Wilson IA. Structural insights into the human and avian IMP cyclohydrolase mechanism via crystal structures with the bound XMP inhibitor. Biochemistry. 2004;43:1171–1183. doi: 10.1021/bi030162i. [DOI] [PubMed] [Google Scholar]
- 35.Wolan DW, Greasley SE, Beardsley GP, Wilson IA. Structural insights into the avian AICAR transformylase mechanism. Biochemistry. 2002;41:15505–15513. doi: 10.1021/bi020505x. [DOI] [PubMed] [Google Scholar]
- 36.Wolan DW, Greasley SE, Wall MJ, Benkovic SJ, Wilson IA. Structure of avian AICAR transformylase with a multisubstrate adduct inhibitor β-DADF identifies the folate binding site. Biochemistry. 2003;42:10904–10914. doi: 10.1021/bi030106h. [DOI] [PubMed] [Google Scholar]
- 37.Xu L, Chong Y, Hwang I, D'Onofrio A, Amore K, Beardsley GP, Li C, Olson AJ, Boger DL, Wilson IA. Structure-based design, synthesis, evaluation, and crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase. J Biol Chem. 2007;282:13033–13046. doi: 10.1074/jbc.M607293200. [DOI] [PubMed] [Google Scholar]
- 38.Xu L, Li C, Olson AJ, Wilson IA. Crystal structure of avian aminoimidazole-4-carboxamide ribonucleotide transformylase in complex with a novel non-folate inhibitor identified by virtual ligand screening. J Biol Chem. 2004;279:50555–50565. doi: 10.1074/jbc.M406801200. [DOI] [PubMed] [Google Scholar]
- 39.Greasley SE, Marsilje TH, Cai H, Baker S, Benkovic SJ, Boger DL, Wilson IA. Unexpected formation of an epoxide-derived multisubstrate adduct inhibitor on the active site of GAR transformylase. Biochemistry. 2001;40:13538–13547. doi: 10.1021/bi011482+. [DOI] [PubMed] [Google Scholar]
- 40.Greasley SE, Yamashita MM, Cai H, Benkovic SJ, Boger DL, Wilson IA. New insights into inhibitor design from the crystal structure and NMR studies of Escherichia coli GAR transformylase in complex with β-GAR and 10-formyl-5,8,10-trideazafolic acid. Biochemistry. 1999;38:16783–16793. doi: 10.1021/bi991888a. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Desharnais J, Marsilje TH, Li C, Hedrick MP, Gooljarsingh LT, Tavassoli A, Benkovic SJ, Olson AJ, Boger DL, Wilson IA. Rational design, synthesis, evaluation, and crystal structure of a potent inhibitor of human GAR Tfase: 10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid. Biochemistry. 2003;42:6043–6056. doi: 10.1021/bi034219c. [DOI] [PubMed] [Google Scholar]
- 42.Desharnais J, Hwang I, Zhang Y, Tavassoli A, Baboval J, Benkovic SJ, Wilson IA, Boger DL. Design, synthesis and biological evaluation of 10-CF3CO-DDACTHF analogues and derivatives as inhibitors of GAR Tfase and the de novo purine biosynthetic pathway. Bioorg Med Chem. 2003;11:4511–4521. doi: 10.1016/s0968-0896(03)00458-9. [DOI] [PubMed] [Google Scholar]
- 43.Takimoto CH. Antifolates in clinical development. Semin Oncol. 1997;24:S18–51. [PubMed] [Google Scholar]
- 44.Boritzki TJ, Barlett CA, Zhang C, Howland EF. AG2034: a novel inhibitor of glycinamide ribonucleotide formyltransferase. Invest New Drugs. 1996;14:295–303. doi: 10.1007/BF00194533. [DOI] [PubMed] [Google Scholar]
- 45.Chen ZH, Olopade OI, Savarese TM. Expression of methylthioadenosine phosphorylase cDNA in p16-, MTAP-malignant cells: restoration of methylthioadenosine phosphorylase-dependent salvage pathways and alterations of sensitivity to inhibitors of purine de novo synthesis. Mol Pharmacol. 1997;52:903–911. doi: 10.1124/mol.52.5.903. [DOI] [PubMed] [Google Scholar]
- 46.DeMartino JK, Hwang I, Connelly S, Wilson IA, Boger DL. Asymmetric synthesis of inhibitors of glycinamide ribonucleotide transformylase. J Med Chem. 2008;51:5441–5448. doi: 10.1021/jm800555h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rayl EA, Moroson BA, Beardsley GP. The human purH gene product, 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase. Cloning, sequencing, expression, purification, kinetic analysis, and domain mapping. J Biol Chem. 1996;271:2225–2233. doi: 10.1074/jbc.271.4.2225. [DOI] [PubMed] [Google Scholar]
- 48.Bigham EC, Hodson SJ, Mallory WR, Wilson D, Duch DS, Smith GK, Ferone R. Synthesis and biological activity of open-chain analogues of 5,6,7,8-tetrahydrofolic acid--potential antitumor agents. J Med Chem. 1992;35:1399–1410. doi: 10.1021/jm00086a008. [DOI] [PubMed] [Google Scholar]
- 49.Otwinowski Z, M W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276A:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 50.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 51.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 52.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 53.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 54.Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 55.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Cα geometry: Π, ψ and Cβ deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 56.Vriend G. WHAT IF: a molecular modeling and drug design program. J Mol Graph. 1990;8:52–56. 29. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 57.Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr D Biol Crystallogr. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 59.Moran RG, Baldwin SW, Taylor EC, Shih C. The 6S-and 6R-diastereomers of 5, 10-dideaza-5, 6, 7, 8-tetrahydrofolate are equiactive inhibitors of de novo purine synthesis. J Biol Chem. 1989;264:21047–21051. [PubMed] [Google Scholar]
- 60.Matherly LH, Goldman DI. Membrane transport of folates. Vitam Horm. 2003;66:403–456. doi: 10.1016/s0083-6729(03)01012-4. [DOI] [PubMed] [Google Scholar]
- 61.Moran RG. Roles of folylpoly-gamma-glutamate synthetase in therapeutics with tetrahydrofolate antimetabolites: an overview. Semin Oncol. 1999;26:24–32. [PubMed] [Google Scholar]
- 62.Wildman SA, Crippen GA. Prediction of Physicochemical Parameters by Atomic Contributions. J Chem Inf Comput Sci. 1999;39:868–873. [Google Scholar]
- 63.Ferone R, Hanlon MH, Singer SC, Hunt DF. α-Carboxyl-linked glutamates in the folylpolyglutamates of Escherichia coli. J Biol Chem. 1986;261:16356–16362. [PubMed] [Google Scholar]
- 64.Ferone R, Singer SC, Hunt DF. In vitro synthesis of α-carboxyl-linked folylpolyglutamates by an enzyme preparation from Escherichia coli. J Biol Chem. 1986;261:16363–16371. [PubMed] [Google Scholar]
- 65.Moran RG. Characterization of the function of mammalian folylpolyglutamate synthetase (FPGS) Adv Exp Med Biol. 1983;163:327–339. doi: 10.1007/978-1-4757-5241-0_24. [DOI] [PubMed] [Google Scholar]
- 66.Klein C, Chen P, Arevalo JH, Stura EA, Marolewski A, Warren MS, Benkovic SJ, Wilson IA. Towards structure-based drug design: crystal structure of a multisubstrate adduct complex of glycinamide ribonucleotide transformylase at 1.96 Å resolution. J Mol Biol. 1995;249:153–175. doi: 10.1006/jmbi.1995.0286. [DOI] [PubMed] [Google Scholar]
- 67.Marsilje TH, Hedrick MP, Desharnais J, Tavassoli A, Zhang Y, Wilson IA, Benkovic SJ, Boger DL. Design, synthesis, and biological evaluation of simplified α-Keto heterocycle, trifluoromethyl ketone, and formyl substituted folate analogues as potential inhibitors of GAR transformylase and AICAR transformylase. Bioorg Med Chem. 2003;11:4487–4501. doi: 10.1016/s0968-0896(03)00456-5. [DOI] [PubMed] [Google Scholar]
- 68.Lovelace LL, Minor W, Lebioda L. Structure of human thymidylate synthase under low-salt conditions. Acta Crystallogr D Biol Crystallogr. 2005;61:622–627. doi: 10.1107/S0907444905005895. [DOI] [PubMed] [Google Scholar]
- 69.Dixon MM, Huang S, Matthews RG, Ludwig M. The structure of the C-terminal domain of methionine synthase: presenting S-adenosylmethionine for reductive methylation of B12. Structure. 1996;4:1263–1275. doi: 10.1016/s0969-2126(96)00135-9. [DOI] [PubMed] [Google Scholar]
- 70.Szebenyi DM, Liu X, Kriksunov IA, Stover PJ, Thiel DJ. Structure of a murine cytoplasmic serine hydroxymethyltransferase quinonoid ternary complex: evidence for asymmetric obligate dimers. Biochemistry. 2000;39:13313–13323. doi: 10.1021/bi000635a. [DOI] [PubMed] [Google Scholar]
- 71.Pejchal R, Sargeant R, Ludwig ML. Structures of NADH and CH3-H4folate complexes of Escherichia coli methylenetetrahydrofolate reductase reveal a spartan strategy for a ping-pong reaction. Biochemistry. 2005;44:11447–11457. doi: 10.1021/bi050533q. [DOI] [PubMed] [Google Scholar]
- 72.Assaraf YG. Molecular basis of antifolate resistance. Cancer Metastasis Rev. 2007;26:153–181. doi: 10.1007/s10555-007-9049-z. [DOI] [PubMed] [Google Scholar]
- 73.Wang Y, Zhao R, Goldman ID. Decreased expression of the reduced folate carrier and folypolyglutamate synthetase is the basis for acquired resistance to the pemetrexed antifolate (LY231514) in an L1210 murine leukemia cell line. Biochem Pharmacol. 2003;65:1163–1170. doi: 10.1016/s0006-2952(03)00007-8. [DOI] [PubMed] [Google Scholar]
- 74.McCloskey DE, McGuire JJ, Russell CA, Rowan BG, Bertino JR, Pizzorno G, Mini E. Decreased folylpolyglutamate synthetase activity as a mechanism of methotrexate resistance in CCRF-CEM human leukemia sublines. J Biol Chem. 1991;266:6181–6187. [PubMed] [Google Scholar]
- 75.Liani E, Rothem L, Bunni MA, Smith CA, Jansen G, Assaraf YG. Loss of folylpoly-γ-glutamate synthetase activity is a dominant mechanism of resistance to polyglutamylation-dependent novel antifolates in multiple human leukemia sublines. Int J Cancer. 2003;103:587–599. doi: 10.1002/ijc.10829. [DOI] [PubMed] [Google Scholar]
- 76.Desmoulin SK, Hou Z, Gangjee A, Matherly LH. The human proton-coupled folate transporter: Biology and therapeutic applications to cancer. Cancer Biol Ther. 2012;13:1355–1373. doi: 10.4161/cbt.22020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gonen N, Assaraf YG. Antifolates in cancer therapy: structure, activity and mechanisms of drug resistance. Drug Resist Updat. 2012;15:183–210. doi: 10.1016/j.drup.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 78.Deng Y, Wang Y, Cherian C, Hou Z, Buck SA, Matherly LH, Gangjee A. Synthesis and discovery of high affinity folate receptor-specific glycinamide ribonucleotide formyltransferase inhibitors with antitumor activity. J Med Chem. 2008;51:5052–5063. doi: 10.1021/jm8003366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cherian C, Kugel Desmoulin S, Wang L, Polin L, White K, Kushner J, Stout M, Hou Z, Gangjee A, Matherly LH. Therapeutic targeting malignant mesothelioma with a novel 6-substituted pyrrolo[2,3-d]pyrimidine thienoyl antifolate via its selective uptake by the proton-coupled folate transporter. Cancer Chemother Pharmacol. 2013;71:999–1011. doi: 10.1007/s00280-013-2094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Habeck LL, Leitner TA, Shackelford KA, Gossett LS, Schultz RM, Andis SL, Shih C, Grindey GB, Mendelsohn LG. A novel class of monoglutamated antifolates exhibits tight-binding inhibition of human glycinamide ribonucleotide formyltransferase and potent activity against solid tumors. Cancer Res. 1994;54:1021–1026. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.