Abstract
Oxidative stress is an imbalance between the level of antioxidants and oxidants in a cell. Oxidative stress has been shown in brain of subjects with mild cognitive impairment (MCI) as well Alzheimer's disease (AD). MCI is considered as a transition phase between control and AD. The focus of the current study was to identify nitrated proteins in the hippocampus and inferior parietal lobule (IPL) brain regions of subjects with amnestic MCI using proteomics. The identified nitrated proteins in MCI brain were compared to those previously reported to be nitrated and oxidatively modified in AD brain, a comparison that might provide an invaluable insight into the progression of the disease.
Keywords: oxidative stress, mild cognitive impairment, protein nitration, hippocampus, inferior parietal lobule, redox proteomics
Introduction
Protein oxidation leads to loss of protein function and often cell death via necrotic or apoptotic processes [1]. Nitric oxide (NO), produced from the conversion of L-arginine to L-citrulline by the enzyme NO synthase, reacts with superoxide radical anion (O2–.) forming the reactive product, peroxynitrite (ONOO –). There are at least three different forms of NO synthase (iNOS, eNOS and nNOS). Peroxynitrite, in the presence of CO2, can decompose to reactive intermediates that then combine with a tyrosine residue at the meta position to form 3-nitrotyrosine (3-NT) [2]. Previous research has shown that peroxynitrite can interact with proteins [3, 4], lipids [5], DNA [6, 7] and RNA [8] to promote damage in these biological molecules. Tyrosine nitration is associated with Alzheimer's disease (AD) [9] as well as Parkinson's Disease [10]. γ-glutamylcysteinylethyl ester (GCEE), a derivative of -glutamyl-cysteine, can cross the blood–brain barrier (BBB) and has been proven to prevent peroxynitrite-induced damage by up-regulating glutathione production [11]. Lipoic acid has also been used to combat protein nitration [12] as has -tocopherol [13]. Increased protein nitration also can lead to an elevated release of reactive nitrogen species (RNS) and detrimental cellular effects. Nitration of proteins results in the inactivation of several important mammalian proteins, such as MnSOD [14–17], glyceraldehyde 3-phosphate dehydrogenase [18, 19], actin, [20, 21], synaptic proteins [22] and tyrosine hydroxylase [23, 24], among others.
Mild cognitive impairment (MCI) is seen as a transitional stage between the cognitive changes of normal aging and the more serious problems caused by AD [25, 26]. MCI can be divided into two broad subtypes: amnestic (memory-affecting) MCI or non-amnestic MCI [27, 28]. Other functions, such as language, attention and visuospatial skills, may be impaired in either type. Pathologically, MCI brain shows mild degradation of the hippocampus, sulci and gyri using magnetic resonance imaging technology [29]. AD patients have considerably greater degradation in these aforementioned areas. Since the hippocampus is the region of the brain primarily responsible for processing memory, it is clearly understandable why those persons with AD and MCI have memory loss. The rate of amnestic (memory-related) MCI conversion to AD is roughly 10–15% per year; however, in some cases MCI individuals can revert to normal [26]. Recent work has shown that protein synthesis in MCI brain may also be affected [30].
Brain from subjects with MCI has elevated levels of 3-NT [31]. By using a redox proteomics-based approach [32], the brain proteins that are nitrated in MCI patients can be identified. A previous study from our laboratory identified carbonylated proteins in the MCI hippocampus [33]. The aim of this study was to identify the nitrated proteins in the hippocampus and inferior parietal lobule of amnestic MCI subjects relative to control brain regions and compare these nitrated proteins to those that are known to be nitrated in AD [2].
Materials and methods
Subjects and materials
Six MCI samples and their age-matched controls were provided by the Rapid Autopsy Program of the University of Kentucky Alzheimer's Disease Clinical Center (UK ADC). Demographic data for all subjects are shown in Table 1. As also indicated in Table 1, the postmortem interval (PMI) prior to the acquisition of brain samples was approximately 3 hrs, an added advantage in studies of human brain. All subjects came from the UK ADC longitudinally followed normal control group and had annual neuropsychological testing and neurological and physical examinations every 2 years. Control subjects had no cognitive complaints, normal cognitive test scores, normal objective memory test scores and normal neurological examinations. MCI patients met the following criteria: a memory complaint confirmed by an associate; objective memory test impairment (age and education adjusted); general normal global intellectual function and Clinical Dementia Rating score of 0.0 to 0.5 (no dementia); and a clinical evaluation that revealed no other cause for memory decline [34].
1.
Demographic data of the control and MCI subjects
| Samples N = 6 | Post-mortem Interval (h) | Age (years) | Sex | Brain Weight (g) |
|---|---|---|---|---|
| Control | 2.9 ± 0.5 | 82 ± 2.6 | 4F, 2M | 1259 ± 44 |
| MCI | 3.1 ± 0.4 | 88 ± 1.5 | 4F, 2M | 1121 ± 25 |
All chemicals were of the highest purity and were obtained from Sigma-Aldrich (St. Louis, MO, USA). The anti-rabbit IgG (secondary antibody), and anti-nitrotyrosine antibody were purchased from Sigma-Aldrich.
Sample preparation
Protein samples (250 μg) were precipitated by addition of ice-cold 100% trichloroacetic acid (TCA) to a final concentration of 15% for 10 min on ice. Precipitates were centrifuged for 2 min at 14,000 x g at 4°C. The pellet was retained and washed three times with 1 ml of 1:1: (v/v) ethyl acetate/ethanol three times. The final pellet was dissolved in rehydration buffer (8M urea, 2M thiourea, 2% CHAPS, 0.2% (v/v) biolytes, 50 mM dithiothreitol (DTT) and bromophenol blue). Samples were sonicated in rehydration buffer three times for 15 sec intervals.
Two-dimensional gel electrophoresis
Two-dimensional polyacrylamide gel electrophoresis was performed with a Bio-Rad system using 110-mm pH 3–10 immobilized pH gradients (IPG) strips and Criterion 8–16% resolving gels. IPG strips were actively rehydrated at 50V 20°C followed by isoelectric focusing: 800 V for 2 hrs linear gradient, 1200 V for 4 hrs slow gradient, 8000 V for 8 hrs linear gradient and 8000 V for 10 hrs rapid gradient. Gel strips were equilibrated for 10 min prior to second- dimension separation in solution A (0.375M Tris-HCl, pH 8.8 containing 6 M urea (Bio-Rad, Hercules, CA, USA), 2% (w/v) sodium dodecyl sulfate (SDS), 20% (v/v glycerol, and 0.5% DTT (Bio-Rad)) followed by re-equilibration for 10 min in solution B (0.375M Tris-HCl pH 8.8 containing 6 M urea (Bio-Rad, Hercules, CA), 2% (w/v) SDS, 20% (v/v glycerol, and 4.5% iodoacetamide (IA) (Bio-Rad)). Control and MCI strips were placed on the Criterion gels, unstained molecular standards were applied, and electrophoresis was performed at 200 V for 65 min.
SYPRO ruby staining
Gels were fixed in a solution containing 10% (v/v) methanol, 7% (v/v) acetic acid for 20 min and stained overnight at room temperature with agitation in 50 ml of SYPRO Ruby gel stain (Bio-Rad). Gels were then destained with 50 ml deionized water overnight.
Immunochemical detection
For immunoblotting analysis, electrophoresis was performed as stated previously, and gels were transferred to a nitrocellulose membrane. The membranes were blocked with 3% bovine serum albumin (BSA) in phosphate-buffered saline containing 0.01% (w/v) sodium azide and 0.2% (v/v) Tween 20 (PBST) overnight at 4°C. The membranes were incubated with anti-nitrotyrosine polyclonal antibody (3-NT) (Sigma-Aldrich), diluted 1:100 in wash blot for 2 hr at room temperature with rocking. Following completion of the primary antibody incubation, the membranes were washed three times in wash blot for 5 min each. An anti-rabbit IgG alkaline phosphatase secondary antibody (Sigma, St. Louis, MO, USA) was diluted 1:3000 in wash blot and incubated with the membranes for 2 hrs at room temperature. The membranes were washed in wash blot three times for 5 min and developed using Sigmafast Tablets (BCIP/NBT substrate) (Sigma) to yield 2D Western blots.
In-gel digestion
Samples were prepared according to the method described by Thongboonkerd et al.[35]. Briefly, the protein spots were cut and removed from the gel with a clean razor blade. The gel pieces were placed into individual, clean 1.5 ml microcentrifuge tubes and kept overnight at –20°C. The gel pieces were thawed and washed with 0.1 M ammonium bicarbonate (NH4HCO3) (Sigma) for 15 min at room temperature. Acetonitrile (Sigma) was added to the gel pieces and incubated for an additional 15 min. The liquid was removed and the gel pieces were allowed to dry. The gel pieces were rehydrated with 20 mM DTT (Bio-Rad) in 0.1 M NH4HCO3 (Sigma) and incubated for 45 min at 56°C. The DTT was removed and replaced with 55 mM IA (Bio-Rad) in 0.1 M NH4HCO3 for 30 min in the dark at room temperature. The liquid was drawn off and the gel pieces were incubated with 50 mM NH4HCO3 at room temperature for 15 min. Acetonitrile was added to the gel pieces for 15 min at room temperature. All solvents were removed and the gel pieces were allowed to dry for 30 min. The gel pieces were rehydrated with addition of a minimal volume of 20 ng/ml modified trypsin (Promega, Madison, WI, USA) in 50mM NH4HCO3. The gel pieces were chopped and incubated with shaking overnight (∼18 hrs) at 37°C.
Analysis of gel images
The analysis of gel maps and membranes compare protein expression and tyrosine nitration content between control and MCI hippocampal samples was performed with PDQuest image analysis software (Bio-Rad). The immunoreactivity of the Western blot was normalized to the actual protein content as measured by the intensity of a SYPRO ruby (Molecular Probes, Eugene, OR, USA) protein stain. Images from SYPRO ruby-stained gels, used to measure protein content, were obtained using a UV transilluminator (ex = 470 nm, em = 618 nm; Molecular Dynamics, Sunnyvale, CA, USA). Comassie blue stained gels and blots were scanned with Adobe Photoshop on a Microtek Scanmaker 4900.
Mass spectrometry
All mass spectra were obtained at the University of Louisville Mass Spectrometry Facility on a Bruker Autoflex MALDI TOF (matrix assisted laser desorption-time of flight) mass spectrometer (Bruker Daltonic, Billerica, MA, USA) operated in the reflectron mode to generate peptide mass fingerprints. Peptides resulting form ingel digestion were analysed on a 384 position, 600 m Anchor-Chip(tm) Target (Bruker Daltonics, Bremen, Germany) and prepared according to AnchorChip recommendations (AnchorChip Technology, Rev. 2; Bruker Daltonics). Briefly, 1 L of tryptic digest was mixed with 1 ml of alpha-cyano-4-hydroxycinnamic acid (0.3 mg/ml in ethanol: acetone, 2:1 ratio) directly on the target and allowed to dry at room temperature. The sample spot was washed with 1 l of 1% TFA solution for approximately 60 sec. The TFA droplet was gently blown off the sample spot with compressed air. The resulting diffuse sample spot was recrystallized (refocused) using 1 l of a solution of ethanol: acetone: 0.1% TFA (6:3:1 ratio). Reported spectra are a summation of 100 laser shots. External calibration of the mass axis was used for acquisition and internal calibration using either trypsin autolysis ions or matrix clusters and was applied after acquisition for accurate mass determination.
Analysis of peptide sequences
Peptide mass fingerprinting was used to identify proteins from tryptic peptide fragments by utilizing the MASCOT search engine (http://www.matrixscience.com) based on the entire National Center for Biotechnology Information (NCBI) protein database. Database searches were conducted allowing for up to one missed trypsin cleavage and using the assumption that the peptides were monoisotopic, oxidized at methionione residues, and carbamiodomethylated at cysteine residues. Mass tolerance of 100 ppm/g was the window of error allowed for matching the peptide mass vales. Probability-based MOWSE scores were estimated by comparison of search results against estimated random match population and were reported as –10 * Log10 (P), where P is the probability that the identification of the protein is not correct. All protein identifications were in the expected size and pI range based on position in the gel.
Immunoprecipitation method
MCI hippocampus samples (250 g) were first pre-cleared using protein A/G–agarose (Pharmacia, USA) for 1 hr at 4°C. The samples were incubated overnight with anti- HSPA8 (HSP70) (Chemicon, Temecula, CA, USA), followed by 1 hr of incubation with protein A/G–agarose then washed three times with buffer B (50 mM Tris–HCl (pH 8.0), 150 mM NaCl, and 1% NP40). Proteins were solubilized in IEF rehydration buffer followed by 2D electrophoresis and 2D Western blot (Bio-Rad). The blots were probed with anti-HSPA8 and were detected by the alkaline-phosphatase-linked secondary antibody (Sigma) as previously described [36].
Statistical analysis
Statistical comparisons of 3-nitrotyrosine protein levels on 2D gels and corresponding matched anti-3NT positive spots, respectively, on 2D-Western blots from MCI hippocampal samples and age-matched control samples, were performed using ANOVA. Pvalues of < 0.05 were considered to be significant. Similarly, after protein identification using the NCBI database, a MOWSE score equal to or greater than 64 was considered to be significant.
Results
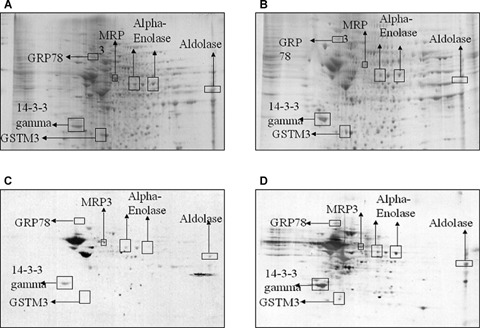
Nitrated proteins in the MCI and control hippocampus and inferior parietal lobule (IPL) were identified immunochemically using 2D-gels and 2D-western blots. These gels and blots were compared using the PDQUEST software package to deduce the individual protein spots that were found to be significantly nitrated in MCI compared to those of control based on 3-NT immunoreactivity (Figs. 1 and 2). As previously reported in other papers by using this approach [2, 37, 38], we confirmed that not all of the protein spots in MCI with increased immunoreactivity are excessively modified proteins compared to control. We identified five proteins that were significantly nitrated in MCI hippocampus Fig. 1D). Malate dehydrogenase (MDH) was identified as one of the protein that showed a trend towards increase in nitration (Table 2). In the MCI IPL, we found five protein spots that showed a significant increase in nitration compared to the age-matched controls (Fig. 2D). These proteins identified in MCI IPL compared to age-matched controls were α enolase (P < 0.05), glucose regulated protein precursor (P < 0.05), aldolase (P < 0.05), glutathione-S-transferases Mu (GSTM3) (P < 0.05), multidrug resistant protein (MRP3) (P < 0.05) and 14-3-3 protein gamma (P < 0.05). In MCI hippocampus, α enolase, MDH, peroxiredoxin 6 (PR VI), dihydropyrminidase like-2 (DRP-2), Fascin 1 and HSPA8 protein were identified to be nitrated in MCI hippocampus compared to age-matched controls. In MCI hippocampus, nitration for the following proteins was increased: α enolase (P < 0.01), PR VI (P < 0.02, DRP-2 (P < 0.01), Fascin 1 (P < 0.04) and HSPA8 (P < 0.05). As in IPL, MDH (P < 0.06) showed a trend toward increase in nitration. Further, the pI and the molecular weight reported in the mass data for all the identified protein spots were consistent with the spot position on the gel. The proteomics-identified nitrated proteins in MCI hippocampus and IPL are shown in the Table 2. Further, to validate the correct identity of the protein, we immunoprecipitated HSPA8 (Fig. 3). The gel map obtained from the immunoprecipitated MCI hippocampus supernatant showed a spot corresponding to HSPA8 missing (Fig. 3B) and the blot probed with anti-3NT antibody (Fig. 3C) showed a spot at the same position of the nitrated HSPA8 protein spot on the nitrated protein blot (Fig. 1D).
1A.
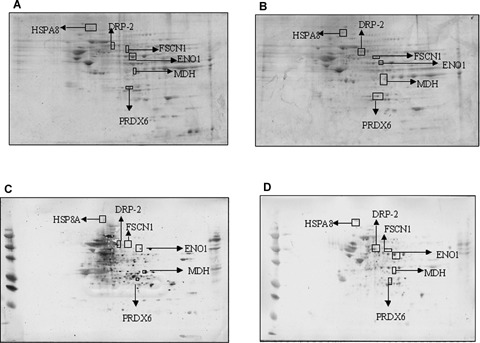
Sypro ruby stained gels from control (A) and MCI hippocampus (B). ‘(C)’ and ‘(D)’ represent Western blots for detection of the level of nitrated proteins from control and MCI hippocampus. In hippocampus, total protein nitration was significantly increased in MCI brain compared to that of control. Protein (250 g) was loaded per gel for detection of protein expression and oxidation. A box is drawn around the area that is enlarged in Figure 1B.
2A.

Sypro ruby stained gels from control (A) and MCI IPL (B). ‘(C)’ and ‘(D)’ represent Western blots for detection of the level of nitrated proteins from control and MCI IPL. In IPL, total protein nitration was significantly increased in MCI brain compared to that of control. Protein (250 g) was loaded per gel for detection of protein expression and oxidation. A box is drawn around the area that is enlarged in Figure 2B. GRP78- Glucose regulated protein, MRP3-Multidrug resistance protein 3.
1B.

Enlarged section of Figure 1A.
2.
Proteomics characteristics
| GI accession number; identity of oxidatively modified proteins in MCI hippocampus and IPL | Brain Region | Protein nitration (% Control) | # Peptides matched of the identified protein | Percent coverage of the protein by the matched peptides | pI, Mr (kDa) | Mowse score |
|---|---|---|---|---|---|---|
| 015438-03-05-00; Multidrug resistance protein 3 (MRP3) | IPL | 189 ± 30 | 7/49 | 34% | 5.22,31.5 | 65 |
| P61981; 14-3-3 protein γ | IPL | 326 ± 38 | 7/30 | 31% | 4.8, 28.3 | 61 |
| P21266-00-00-00; GSTM3 | IPL | 236 ± 23 | 7/37 | 33% | 5.3, 26.8 | 61 |
| P06733; α enolase | IPL | 340 ± 45 | 7/22 | 26% | 6.99,47 | 65 |
| P11021; Glucose regulated protein precursor | IPL | 136 ± 55 | 10/29 | 22% | 5.07,72.4 | 77 |
| P04075-00-01-00; Aldolase | IPL | 297 ± 35 | 8/36 | 36% | 8.55,39.6 | 68 |
| Gi|48257068; HSPA8 | Hippocampus | 576 ± 34.1 | 12/31 | 31% | 5.36, 64.8 | 105 |
| Gi|13623415; Fascin 1 | Hippocampus | 164 ± 49.5 | 7/14 | 18% | 6.84, 55.2 | 80 |
| Gi|77744395; Peroxiredoxin | Hippocampus | 143 ± 11 | 8/27 | 41% | 6.00, 25.1 | 124 |
| Gi|45501286; Dihydropyrminidase like-2 | Hippocampus | 273 ± 9.08 | 10/24 | 27% | 5.95, 62.7 | 146 |
| Gi|15174339; Malate dehydrogenase | Hippocampus | 380 ± 32.6 | 13/38 | 39% | 6.91, 36.6 | 125 |
| Gi|14530765; α enolase | Hippocampus | 830 ± 21.6 | 12/48 | 32% | 7.01, 47.5 | 92 |
2B.
Enlarged section of Figure 2A.
3.
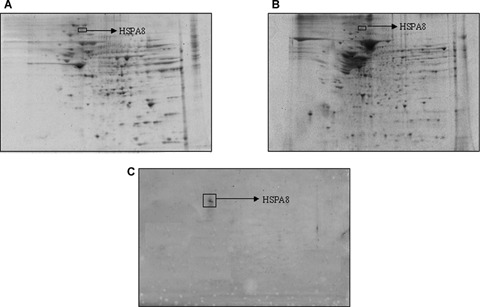
Validation of protein identified by MS using immunoprecipitation and Western blot analysis: ‘A’ represent gel of total hippocampus protein, a box is drawn around the HSPA8, which is one of the nitrated protein. ‘B’ represents gel obtained from the supernatant of immunoprecipitation sample that lacks the HSPA8 protein. Represent blot probed with 3- NT antibody. n = 3.
Discussion
As noted above, our laboratory recently showed that the level of the protein nitration is elevated in hippocampus and IPL brain regions of subjects with amnestic MCI compared to control brain [31]. However, the present paper is the first to report the identity of individual brain proteins specifically nitrated in amnestic MCI brain. In the present study, we identified nitrated proteins in MCI hippocampus and IPL compared to their respective controls, in order to better understand the regional effects of nitration in MCI and to gain insight into the mechanisms of progression of MCI to AD. Previous studies from our laboratory have identified the specifically nitrated proteins in AD brain [2, 39].
The proteins that were found to be specifically nitrated in hippocampus include: HSPA8 protein, Fascin 1, Peroxiredoxin 6, Dihydropyrminidase like-2, MDH, Enolase 1 (α). In IPL, we found α enolase, glucose regulated protein precursor, aldolase, GSTM3 and MRP3 proteins, and 14-3-3 protein to be excessively nitrated compared to control. Some of these proteins were previously reported to be nitrated in AD brain [2, 39], and this difference conceivably could be attributed to different levels of amyloidpeptide and/or oxidative stress. These proteins are involved in regulation of a number of important cellular functions including: energy metabolism, cellular signalling, antioxidant and detoxification, in addition to regulating structural functions of brain cells (Table 3).
3.
Functionalities of identified nitrated proteins in MCI hippocampus and IPL
| Functions | Proteins involved |
|---|---|
| Energy or mitochondrial dysfunction | α enolase |
| Glucose regulated | |
| protein precursor | |
| Aldolase | |
| Malate dehydrogenase | |
| Antioxidant | GSTM3 |
| Defense/Detoxification system dysfunction | MRP3 protein |
| Peroxiredoxin | |
| Heat shock | |
| protein 70 (HSPA8) | |
| Structural dysfunction | Dihydropyrminidase like-2 |
| Fascin 1 | |
| Cell signaling dysfunction | 14-3-3 protein γ |
Energy or mitochondrial dysfunction
α enolase, glucose regulated protein precursor and aldolase are the energy-related proteins that were identified to be nitrated in MCI IPL. We found enolase and MDH as nitrated proteins in MCI hippocampus. The only common target protein of nitration in MCI hipocampus and IPL, the brain regions that are severely affected in AD, is α enolase. Recently, we reported the oxidation of enolase in the MCI hippocampus indexed by protein carbonyls [33], and in addition previous studies of AD and cell culture models of AD showed an increased oxidation of enolase [2, 38–42]. ATP, the energy source of the cell, is extremely important at nerve terminals for normal neural communication. Decreased levels of cellular ATP at nerve terminals may lead to loss of synapses and synaptic function, and may ultimately contribute to memory loss in amnestic MCI patients. The nitration of these proteins that are involved in energy metabolism may disrupt neuronal energy metabolism and ion homeostasis thereby impairing the function of membrane ionmotive ATPases and glucose and glutamate transporters [43, 44], loss of membrane asymmetry and signal transduction. Such oxidative and metabolic compromise may thereby render neurons vulnerable to excitotoxicity and apoptosis. Our finding of the nitration of the proteins that are involved in energy metabolism correlates with the altered energy metabolism reported in brain in an advanced stage of MCI [45–47]. Positron emission tomography (PET) studies also show a pattern consistent with the reduced cerebral glucose utilization in AD brain [48, 49].
Antioxidant or detoxification system dysfunction
GSTM3 and MRP3 proteins were found to be nitrated in MCI IPL. Peroxiredoxin 6 and heat shock protein 70kD isoforms 8 (HSPA8) are found to be nitrated in MCI hippocampus. These four proteins play an important role in regulating cellular process by decreasing the levels of oxidants or by removing toxic compounds that are generated in the cell. In MCI hippocampus and IPL increased levels of protein-bound 4-hydroxy-2-trans-nonenal (HNE), a highly reactive lipid peroxidation product have been found [50]. In AD brain, GST protein levels and activity were reported to be decreased; in addition, GST was the found to be oxidatively modified by HNE [36].
Peroxiredoxin can reduce peroxynitrite at a high catalytic rate, which may modulate protein nitration and cell damage [51]. In addition, peroxiredoxin plays roles in cell differentiation and apoptosis. The decrease in the activity of this enzyme may also lead to decreased phospholipase A2 activity, one of the target proteins regulated by peptidyl prolyl cis/trans isomerase (Pin 1), a protein that has been reported to be down-regulated and have decreased activity in AD brain [52–54].
Ralat et al.[55] have shown that the enzyme GST forms a complex with Prx VI in order to alter both enzyme activities. These proteins work in coordination with one another either directly or indirectly thereby protecting the cell from toxicants. These results provide insight into how the changes of these proteins may contribute to tau hyperphosphorylation and neurofibrillary tangle formation, in addition to development of oxidative stress.
HSPA8 is a member of the heat shock protein family. The main function of heat shock proteins/cognates is to act as chaperone proteins by repairing misfolded proteins. Numerous heat shock proteins have been found to be oxidatively modified in disease or disease models, including AD [41] and Huntington's disease [56], including Hsc71, Hsp90 and Hsp60, respectively. Impairment of HSPA8 may exacerbate protein misfolding and protein aggregation, leading to reduced effective proteosomal activity. Aβ peptide aggregates are the major components of senile plaques, which are a hallmark of AD pathology. A-treated synaptosomes show that heat shock proteins are oxidatively modified [57], further illustrating the importance of functioning heat shock proteins in the cell.
Cell signalling dysfunction
14-3-3-protein gamma is found to be nitrated in MCI IPL. 14-3-3 gamma is a member of the 14-3-3 protein family, and are involved in a number of cellular functions including signal transduction, protein trafficking and metabolism [58, 59]. The levels of 14-3-3 proteins are increased in AD brain [60, 61], AD CSF [62] and in ICV-delivered Aβ and neuronal models of AD [40, 63]. The nitration of 14-3-3 γ could change its conformation, which conceivably could lead to altered binding to two of its normal binding partners, glycogen synthase kinase 3(GSK3β) and tau. One of the isoforms of 14-3-3 can act as a scaffolding protein and simultaneously bind to tau and GSK3βin a multiprotein tau phosphorylation complex [64]. This complex may promote tau phosphorylation and polymerization [65, 66], leading to the formation of tangles and further leading to neurodegeneration in AD.
Structural dysfunction
DRP-2 and Fascin 1 were identified as nitrated proteins in MCI hippocampus. DRP-2 is a member of the dihydropyrimidinase-related protein family that is involved in axonal outgrowth and pathfinding through transmission and modulation of extracellular signals [67, 68]. DRP2 has been reported to be associated with neurofibrillary tangles (NFT), which may lead to decreased levels of cytosolic DRP-2 and eventually lead to shortened neuritic and axonal growth. Such outcomes would accelerate neuritic degeneration in AD [69], which is one the characteristic hallmarks of AD pathology. Increased oxidation [41] and decreased expression of DRP-2 protein was observed in AD. In adult Down's syndrome (DS) [70], fetal DS [71], schizo-phrenia and affective disorders DRP-2 has lower levels in brain. Since memory and learning are associated with synaptic remodelling, nitration and subsequent loss of function of this protein could conceivably be involved in the observed memory decline in MCI. Moreover, the decreased function of DRP-2 could be involved in the shortened dendritic length and synapse loss observed in AD [72].
Fascin 1 (FSCN1) is a structural protein also known as p55 [73] and is involved in cell adhesion [74] and cell motility [75]. It is a marker for dendritic functionality [76]. Addition of p55 has been shown to protect cells from oxidative stress produced by an insult [77]. The identification of this protein as nitrated in MCI brain is consistent with the notion that loss of function of this protein lessens protection against oxidative damage and could be an important event in the transition of MCI to AD. FSCN1 has also shown to interact with protein kinase Cα(PKCα), which regulates focal adhesions [78]. Impairment of this protein can be related to faulty neurotransmission from the affected dendritic projections. Although there is no known research relating FSCN1 to neurodegenerative diseases, FSCN1 has been viewed as a potential biomarker for certain cancers [79].
In conclusion, the redox proteomics-identified nitrated proteins in MCI brain play important roles in different neuronal functions and are directly or indirectly linked to AD pathology. Comparative analysis of nitrated proteins between MCI IPL, and hippocampus brain regions showed enolase as a common target of nitration. This suggests that energy metabolism may be among the first cellular properties that become severely affected in MCI. A similar sensitivity to energy metabolism was observed in AD brain [2, 38, 39, 41]. Further, we identified DRP-2 as a commonly nitrated protein in AD and MCI brain. In addition, we also found different isoforms of GST and MRP-1 as nitrated in MCI compared to oxidative damage in AD. All these data suggest that nitration of these proteins could be one of the mechanisms that may trigger the conversion of MCI to AD. Future studies using animal models of the different stages of this dementing disorder should help in further delineating the mechanisms of MCI pathogenesis and to develop effective therapies to combat conversion of MCI to AD.
Acknowledgments
The authors thank the University of Kentucky ADC Clinical and Neuropathology Cores for providing the brain specimens used for this study. This research was supported in part by grants from NIH [AG-10836; AG-05119].
References
- 1.Butterfield DA, Stadtman ER. Protein oxidation processes in aging brain. Adv Cell Aging Gerontol. 1997;2:161–91. [Google Scholar]
- 2.Castegna A, Thongboonkerd V, Klein JB, Lynn B, Markesbery WR, Butterfield DA. Proteomic identification of nitrated proteins in Alzheimer's disease brain. J Neurochem. 2003;85:1394–01. doi: 10.1046/j.1471-4159.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- 3.Hazen SL, Gaut JP, Hsu FF, Crowley JR, D'Avignon A, Heinecke JW. p-Hydroxyphenylacetaldehyde, the major product of L-tyrosine oxidation by the myeloper-oxidase-H2O2-chloride system of phagocytes, covalently modifies epsilon-amino groups of protein lysine residues. J Biol Chem. 1997;272:16990–8. doi: 10.1074/jbc.272.27.16990. [DOI] [PubMed] [Google Scholar]
- 4.Smith MA, Richey Harris PL, Sayre LM, Beckman JS, Perry G. Widespread peroxynitrite-mediated damage in Alzheimer's disease. J Neurosci. 1997;17:2653–57. doi: 10.1523/JNEUROSCI.17-08-02653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Botti H, Trostchansky A, Batthyany C, Rubbo H. Reactivity of peroxynitrite and nitric oxide with LDL. IUBMB Life. 2005;57:407–12. doi: 10.1080/15216540500137701. [DOI] [PubMed] [Google Scholar]
- 6.Szabo C. DNA strand breakage and activation of poly-ADP ribosyltransferase: a cytotoxic pathway triggered by peroxynitrite. Free Radic Biol Med. 1996;21:855–69. doi: 10.1016/0891-5849(96)00170-0. [DOI] [PubMed] [Google Scholar]
- 7.Niles JC, Wishnok JS, Tannenbaum SR. Peroxynitrite-induced oxidation and nitration products of guanine and 8-oxoguanine: structures and mechanisms of product formation. Nitric Oxide. 2006;14:109–21. doi: 10.1016/j.niox.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Masuda M, Nishino H, Ohshima H. Formation of 8-nitroguanosine in cellular RNA as a biomarker of exposure to reactive nitrogen species. Chem Biol Interact. 2002;139:187–97. doi: 10.1016/s0009-2797(01)00299-x. [DOI] [PubMed] [Google Scholar]
- 9.Good PF, Werner P, Hsu A, Olanow CW, Perl DP. Evidence of neuronal oxidative damage in Alzheimer's disease. Am J Pathol. 1996;149:21–8. [PMC free article] [PubMed] [Google Scholar]
- 10.Good PF, Hsu A, Werner P, Perl DP, Olanow CW. Protein nitration in Parkinson's disease. J Neuropathol Exp Neurol. 1998;57:338–42. doi: 10.1097/00005072-199804000-00006. [DOI] [PubMed] [Google Scholar]
- 11.Drake J, Kanski J, Varadarajan S, Tsoras M, Butterfield DA. Elevation of brain glutathione by gamma-glutamylcysteine ethyl ester protects against peroxynitrite-induced oxidative stress. J Neurosci Res. 2002;68:776–84. doi: 10.1002/jnr.10266. [DOI] [PubMed] [Google Scholar]
- 12.Whiteman M, Tritschler H, Halliwell B. Protection against peroxynitrite-dependent tyrosine nitration and alpha 1-antiproteinase inactivation by oxidized and reduced lipoic acid. FEBS Lett. 1996;379:74–6. doi: 10.1016/0014-5793(95)01489-6. [DOI] [PubMed] [Google Scholar]
- 13.Christen S, Woodall AA, Shigenaga MK, Southwell-Keely PT, Duncan MW, Ames BN. gamma-tocopherol traps mutagenic electrophiles such as NO(X) and complements alpha-tocopherol: physiological implications. Proc Natl Acad Sci USA. 1997;94:3217–22. doi: 10.1073/pnas.94.7.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacMillan-Crow LA, Thompson JA. Tyrosine modifications and inactivation of active site manganese superoxide dismutase mutant (Y34F) by peroxyni-trite. Arch Biochem Biophys. 1999;366:82–8. doi: 10.1006/abbi.1999.1202. [DOI] [PubMed] [Google Scholar]
- 15.Aoyama K, Matsubara K, Fujikawa Y, Nagahiro Y, Shimizu K, Umegae N, Hayase N, Shiono H, Kobayashi S. Nitration of manganese superoxide dismutase in cerebrospinal fluids is a marker for per-oxynitrite-mediated oxidative stress in neurodegenerative diseases. Ann Neurol. 2000;47:524–7. [PubMed] [Google Scholar]
- 16.Ischiropoulos H, Zhu L, Chen J, Tsai M, Martin JC, Smith CD, Beckman JS. Peroxynitrite-mediated tyrosine nitration catalyzed by superoxide dismutase. Arch Biochem Biophys. 1992;298:431–37. doi: 10.1016/0003-9861(92)90431-u. [DOI] [PubMed] [Google Scholar]
- 17.Anantharaman M, Tangpong J, Keller JN, Murphy MP, Markesbery WR, Kiningham KK, St Clair DK. Beta-amyloid mediated nitration of manganese superoxide dismutase: implication for oxidative stress in a APPNLH/NLH X PS-1P264L/P264L double knockin mouse model of Alzheimer's disease. Am J Pathol. 2006;168:1608–18. doi: 10.2353/ajpath.2006.051223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Souza JM, Radi R. Glyceraldehyde-3-phosphate dehydrogenase inactivation by peroxynitrite. Arch Biochem Biophys. 1998;360:187–94. doi: 10.1006/abbi.1998.0932. [DOI] [PubMed] [Google Scholar]
- 19.Hara MR, Cascio MB, Sawa A. GAPDH as a sensor of NO stress. Biochim Biophys Acta. 2006;1762:502–9. doi: 10.1016/j.bbadis.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 20.Neumann P, Gertzberg N, Vaughan E, Weisbrot J, Woodburn R, Lambert W, Johnson A. Peroxynitrite mediates TNF-induced endothelial barrier dysfunction and nitration of actin. Am J Physiol Lung Cell Mol Physiol. 2006;290:L674–84. doi: 10.1152/ajplung.00391.2005. [DOI] [PubMed] [Google Scholar]
- 21.Clements MK, Siemsen DW, Swain SD, Hanson AJ, Nelson-Overton LK, Rohn TT, Quinn MT. Inhibition of actin polymerization by peroxynitrite modulates neutrophil functional responses. J Leukoc Biol. 2003;73:344–55. doi: 10.1189/jlb.0802401. [DOI] [PubMed] [Google Scholar]
- 22.Di Stasi AM, Mallozzi C, Macchia G, Maura G, Petrucci TC, Minetti M. Peroxynitrite affects exocytosis and SNARE complex formation and induces tyrosine nitration of synaptic proteins. J Neurochem. 2002;82:420–9. doi: 10.1046/j.1471-4159.2002.00980.x. [DOI] [PubMed] [Google Scholar]
- 23.Blanchard-Fillion B, Souza JM, Friel T, Jiang GC, Vrana K, Sharov V, Barron L, Schoneich C, Quijano C, Alvarez B, Radi R, Przedborski S, Fernando GS, Horwitz J, Ischiropoulos H. Nitration and inactivation of tyrosine hydroxylase by peroxynitrite. J Biol Chem. 2001;276:46017–23. doi: 10.1074/jbc.M105564200. [DOI] [PubMed] [Google Scholar]
- 24.Gow A, Duran D, Thom SR, Ischiropoulos H. Carbon dioxide enhancement of peroxynitrite-mediated protein tyrosine nitration. Arch Biochem Biophys. 1996;333:42–8. doi: 10.1006/abbi.1996.0362. [DOI] [PubMed] [Google Scholar]
- 25.Chetelat G, Desgranges B, De La Sayette V, Viader F, Eustache F. At the boundary between normal aging and Alzheimer disease. Rev Neurol. 2004;160:S55–63. doi: 10.1016/s0035-3787(04)70944-3. [DOI] [PubMed] [Google Scholar]
- 26.Petersen RC. Mild cognitive impairment: transition between aging and Alzheimer's disease. Neurologia. 2000;15:93–1. [PubMed] [Google Scholar]
- 27.Petersen RC. Mild cognitive impairment as a diagnostic entity. J Intern Med. 2004;256:183–94. doi: 10.1111/j.1365-2796.2004.01388.x. [DOI] [PubMed] [Google Scholar]
- 28.Portet F, Ousset PJ, Touchon J. Wha? is a mild cognitive impairment? Rev Prat. 2005;55:1891–4. [PubMed] [Google Scholar]
- 29.Jack CR, Jr, Petersen RC, Xu YC, O'Brien PC, Smith GE, Ivnik RJ, Boeve BF, Waring SC, Tangalos EG, Kokmen E. Prediction of AD with MRI-based hippocampal volume in mild cognitive impairment. Neurology. 1999;52:1397–03. doi: 10.1212/wnl.52.7.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding Q, Markesbery WR, Chen Q, Li F, Keller JN. Ribosome dysfunction is an early event in Alzheimer's disease. J Neurosci. 2005;25:9171–5. doi: 10.1523/JNEUROSCI.3040-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Butterfield DA, Reed TT, Perluigi M, De Marco C, Coccia R, Keller JN, Markesbery WR, Sultana R. Elevated levels of 3-Nitrotyrosine in brain from subjects with amnestic Mild Cognitive Impairment: Implications for the role of nitration in the progression of Alzheimer's Disease. Brain Res. 2007;1148:242–8. doi: 10.1016/j.brainres.2007.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dalle-Donne I, Scaloni A, Butterfield DA. Redox Proteomics: From protein modifications to cellular dysfunction and diseases. John Wiley and Son; 2006. Hoboken; NJ; [DOI] [PubMed] [Google Scholar]
- 33.Butterfield DA, Poon HF, St Clair D, Keller JN, Pierce WM, Klein JB, Markesbery WR. Redox proteomics identification of oxidatively modified hippocampal proteins in mild cognitive impairment: Insights into the development of Alzheimer's disease. Neurobiol Dis. 2006;22:223–32. doi: 10.1016/j.nbd.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 34.Morris JC, Cummings J. Mild cognitive impairment (MCI) represents earlystage Alzheimer's disease. J Alzheimers Dis. 2005;7:235–239. doi: 10.3233/jad-2005-7306. [DOI] [PubMed] [Google Scholar]
- 35.Thongboonkerd V, Luengpailin J, Cao J, Pierce WM, Cai J, Klein JB, Doyle RJ. Fluoride exposure attenuates expression of Streptococcus pyogenes virulence factors. J Biol Chem. 2002;277:16599–05. doi: 10.1074/jbc.M200746200. [DOI] [PubMed] [Google Scholar]
- 36.Sultana R, Butterfield DA. Oxidatively modified GST and MRP1 in Alzheimer's disease brain: implications for accumulation of reactive lipid peroxidation products. Neurochem Res. 2004;29:2215–20. doi: 10.1007/s11064-004-7028-0. [DOI] [PubMed] [Google Scholar]
- 37.Poon HF, Frasier M, Shreve N, Calabrese V, Wolozin B, Butterfield DA. Mitochondrial associated metabolic proteins are selectively oxidized in A30P alpha-synuclein transgenic mice–a model of familial Parkinson's disease. Neurobiol Dis. 2005;18:492–8. doi: 10.1016/j.nbd.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 38.Sultana R, Boyd-Kimball D, Poon HF, Cai J, Pierce WM, Klein JB, Merchant M, Markesbery WR, Butterfield DA. Redox proteomics identification of oxidized proteins in Alzheimer's disease hippocampus and cerebellum: An approach to understand pathological and biochemical alterations in AD. Neurobiol Aging. 2006;27:1564–76. doi: 10.1016/j.neurobiolaging.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 39.Sultana R, Poon HF, Cai J, Pierce WM, Merchant M, Klein JB, Markesbery WR, Butterfield DA. Identification of nitrated proteins in Alzheimer's disease brain using a redox proteomics approach. Neurobiol Dis. 2006;22:76–87. doi: 10.1016/j.nbd.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 40.Boyd-Kimball D, Sultana R, Poon HF, Lynn BC, Casamenti F, Pepeu G, Klein JB, Butterfield DA. Proteomic identification of proteins specifically oxidized by intracerebral injection of amyloid beta-peptide (1–42) into rat brain: implications for Alzheimer's disease. Neurosci. 2005;132:313–24. doi: 10.1016/j.neuroscience.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 41.Castegna A, Aksenov M, Thongboonkerd V, Klein JB, Pierce WM, Booze R, Markesbery WR, Butterfield DA. Proteomic identification of oxidatively modified proteins in Alzheimer's disease brain. Part II: dihydropyrimidinase-related protein 2, alpha-enolase and heat shock cognate 71. J Neurochem. 2002;82:1524–32. doi: 10.1046/j.1471-4159.2002.01103.x. [DOI] [PubMed] [Google Scholar]
- 42.Poon HF, Castegna A, Farr SA, Thongboonkerd V, Lynn BC, Banks WA, Morley JE, Klein JB, Butterfield DA. Quantitative proteomics analysis of specific protein expression and oxidative modification in aged senescence-accelerated-prone 8 mice brain. Neurosci. 2004;126:915–26. doi: 10.1016/j.neuroscience.2004.04.046. [DOI] [PubMed] [Google Scholar]
- 43.Keller JN, Mark RJ, Bruce AJ, Blanc E, Rothstein JD, Uchida K, Waeg G, Mattson MP. 4-Hydroxynonenal, an aldehydic product of membrane lipid peroxidation, impairs glutamate transport and mitochondrial function in synaptosomes. Neurosci. 1997;80:685–96. doi: 10.1016/s0306-4522(97)00065-1. [DOI] [PubMed] [Google Scholar]
- 44.Mattson MP. Metal-catalyzed disruption of membrane protein and lipid signaling in the pathogenesis of neurodegenerative disorders. Ann N Y Acad Sci. 2004;1012:37–50. doi: 10.1196/annals.1306.004. [DOI] [PubMed] [Google Scholar]
- 45.Geddes JW, Pang Z, Wiley DH. Hippocampal damage and cytoskeletal disruption resulting from impaired energy metabolism. Implications for Alzheimer disease. Mol Chem Neuropathol. 1996;28:65–74. doi: 10.1007/BF02815206. [DOI] [PubMed] [Google Scholar]
- 46.Messier C, Gagnon M. Glucose regulation and cognitive functions: relation to Alzheimer's disease and diabetes. Behav Brain Res. 1996;75:1–11. doi: 10.1016/0166-4328(95)00153-0. [DOI] [PubMed] [Google Scholar]
- 47.Vanhanen M, Soininen H. Glucose intolerance, cognitive impairment and Alzheimer's disease. Curr Opin Neurol. 1998;11:673–7. doi: 10.1097/00019052-199812000-00011. [DOI] [PubMed] [Google Scholar]
- 48.Hoyer S. Causes and consequences of disturbances of cerebral glucose metabolism in sporadic Alzheimer disease: therapeutic implications. Adv Exp Med Biol. 2004;541:135–52. doi: 10.1007/978-1-4419-8969-7_8. [DOI] [PubMed] [Google Scholar]
- 49.Rapoport SI. In vivo PET imaging and postmortem studies suggest potentially reversible and irreversible stages of brain metabolic failure in Alzheimer's disease. Eur Arch Psychiatry Clin Neurosci. 1999;249:46–55. doi: 10.1007/pl00014174. [DOI] [PubMed] [Google Scholar]
- 50.Butterfield DA, Reed T, Perluigi M, De Marco C, Coccia R, Cini C, Sultana R. Elevated protein-boundlevels of the lipid peroxidation product, 4-hydroxy-2-nonenal, in brain from persons with mild cognitive impairment. Neurosci Lett. 2006;397:170–3. doi: 10.1016/j.neulet.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 51.Peshenko IV, Shichi H. Oxidation of active center cysteine of bovine 1-Cys peroxiredoxin to the cysteine sulfenic acid form by peroxide and peroxynitrite. Free Radic Biol Med. 2001;31:292–03. doi: 10.1016/s0891-5849(01)00579-2. [DOI] [PubMed] [Google Scholar]
- 52.Lu KP. Phosphorylation-dependent prolyl isomerization: a novel cell cycle regulatory mechanism. Prog Cell Cycle Res. 2000;4:83–96. doi: 10.1007/978-1-4615-4253-7_8. [DOI] [PubMed] [Google Scholar]
- 53.Lu PJ, Wulf G, Zhou XZ, Davies P, Lu KP. The prolyl isomerase Pin1 restores the function of Alzheimer-associated phosphorylated tau protein. Nature. 1999;399:784–8. doi: 10.1038/21650. [DOI] [PubMed] [Google Scholar]
- 54.Sultana R, Boyd-Kimball D, Poon HF, Cai J, Pierce WM, Klein JB, Markesbery WR, Zhou XZ, Lu KP, Butterfield DA. Oxidative modification and down-regulation of Pin1 in Alzheimer's disease hippocampus: A redox proteomics analysis. Neurobiol Aging. 2006;27:918–25. doi: 10.1016/j.neurobiolaging.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 55.Ralat LA, Manevich Y, Fisher AB, Colman RF. Direct evidence for the formation of a complex between 1-cysteine peroxiredoxin and glutathione S-transferase pi with activity changes in both enzymes. Biochem. 2006;45:360–72. doi: 10.1021/bi0520737. [DOI] [PubMed] [Google Scholar]
- 56.Perluigi M, Poon HF, Maragos W, Pierce WM, Klein JB, Calabrese V, Cini C, De Marco C, Butterfield DA. Proteomic analysis of protein expression and oxidative modification in r6/2 transgenic mice: a model of Huntington disease. Mol Cell Proteomics. 2005;4:1849–61. doi: 10.1074/mcp.M500090-MCP200. [DOI] [PubMed] [Google Scholar]
- 57.Boyd-Kimball D, Castegna A, Sultana R, Poon HF, Petroze R, Lynn BC, Klein JB, Butterfield DA. Proteomic identification of proteins oxidized by Abeta(1–42) in synaptosomes: implications for Alzheimer's disease. Brain Res. 2005;1044:206–15. doi: 10.1016/j.brainres.2005.02.086. [DOI] [PubMed] [Google Scholar]
- 58.Dougherty MK, Morrison DK. Unlocking the code of 14-3-3. J Cell Sci. 2004;117:1875–84. doi: 10.1242/jcs.01171. [DOI] [PubMed] [Google Scholar]
- 59.Takahashi Y. The 14-3-3 proteins: gene, gene expression, and function. Neurochem Res. 2003;28:1265–73. doi: 10.1023/a:1024296932670. [DOI] [PubMed] [Google Scholar]
- 60.Frautschy SA, Baird A, Cole GM. Effects of injected Alzheimer beta-amyloid cores in rat brain. Proc Natl Acad Sci USA. 1991;88:8362–66. doi: 10.1073/pnas.88.19.8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Layfield R, Fergusson J, Aitken A, Lowe J, Landon M, Mayer RJ. Neurofibrillary tangles of Alzheimer's disease brains contain 14-3-3 proteins. Neurosci Lett. 1996;209:57–60. doi: 10.1016/0304-3940(96)12598-2. [DOI] [PubMed] [Google Scholar]
- 62.Burkhard PR, Sanchez JC, Landis T, Hochstrasser DF. CSF detection of the 14-3-3 protein in unselected patients with dementia. Neurology. 2001;56:1528–33. doi: 10.1212/wnl.56.11.1528. [DOI] [PubMed] [Google Scholar]
- 63.Sultana R, Newman SF, Abdul HM, Cai J, Pierce WM, Klein JB, Merchant M, Butterfield DA. Protective effect of D609 against amyloid-beta-42-induced oxidative modification of neuronal proteins: redox proteomics study. J Neurosci Res. 2006;84:409–17. doi: 10.1002/jnr.20876. [DOI] [PubMed] [Google Scholar]
- 64.Agarwal-Mawal A, Qureshi HY, Cafferty PW, Yuan Z, Han D, Lin R, Paudel HK. 14-3-3 connects glycogen synthase kinase-3 beta to tau within a brain microtubule-associated tau phosphorylation complex. J Biol Chem. 2003;278:12722–8. doi: 10.1074/jbc.M211491200. [DOI] [PubMed] [Google Scholar]
- 65.Hashiguchi M, Sobue K, Paudel HK. 14-3-3zeta is an effector of tau protein phosphorylation. J Biol Chem. 2000;275:25247–54. doi: 10.1074/jbc.M003738200. [DOI] [PubMed] [Google Scholar]
- 66.Hernandez F, Cuadros R, Avila J. Zeta 14-3-3 protein favours the formation of human tau fibrillar polymers. Neurosci Lett. 2004;357:143–6. doi: 10.1016/j.neulet.2003.12.049. [DOI] [PubMed] [Google Scholar]
- 67.Hamajima N, Matsuda K, Sakata S, Tamaki N, Sasaki M, Nonaka M. A novel gene family defined by human dihydropyrimidinase and three related proteins with differential tissue distribution. Gene. 1996;180:157–63. doi: 10.1016/s0378-1119(96)00445-3. [DOI] [PubMed] [Google Scholar]
- 68.Kato Y, Hamajima N, Inagaki H, Okamura N, Koji T, Sasaki M, Nonaka M. Post-meiotic expression of the mouse dihydropyrimidinase-related protein 3 (DRP-3) gene during spermiogenesis. Mol Reprod Dev. 1998;51:105–11. doi: 10.1002/(SICI)1098-2795(199809)51:1<105::AID-MRD13>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 69.Yoshida H, Watanabe A, Ihara Y. Collapsin response mediator protein-2 is associated with neurofibrillary tangles in Alzheimer's disease. J Biol Chem. 1998;273:9761–8. doi: 10.1074/jbc.273.16.9761. [DOI] [PubMed] [Google Scholar]
- 70.Lubec G, Nonaka M, Krapfenbauer K, Gratzer M, Cairns N, Fountoulakis M. Expression of the dihydropyrimidinase related protein 2 (DRP-2) in Down syndrome and Alzheimer's disease brain is down-regulated at the mRNA and dysregulated at the protein level. J Neural Transm Suppl. 1999;57:161–77. doi: 10.1007/978-3-7091-6380-1_10. [DOI] [PubMed] [Google Scholar]
- 71.Weitzdoerfer R, Dierssen M, Fountoulakis M, Lubec G. Fetal life in down syndrome starts with normal neuronal density but impaired dendritic spines and synaptosomal structure. J Neural Transm Suppl. 2001:59–70. doi: 10.1007/978-3-7091-6262-0_5. [DOI] [PubMed] [Google Scholar]
- 72.Coleman PD, Flood DG. Neuron numbers and dendritic extent in normal aging and Alzheimer's disease. Neurobiol Aging. 1987;8:521–45. doi: 10.1016/0197-4580(87)90127-8. [DOI] [PubMed] [Google Scholar]
- 73.Yamashiro S, Yamakita Y, Ono S, Matsumura F. Fascin, an actin-bundling protein, induces membrane protrusions and increases cell motility of epithelial cells. Mol Biol Cell. 1998;9:993–06. doi: 10.1091/mbc.9.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Adams JC. Formation of stable microspikes containing actin and the 55 kDa actin bundling protein, fascin, is a consequence of cell adhesion to throm-bospondin-1: implications for the anti-adhesive activities of thrombospondin-1. J Cell Sci. 1995;108:1977–90. doi: 10.1242/jcs.108.5.1977. [DOI] [PubMed] [Google Scholar]
- 75.Adams JC. Roles of fascin in cell adhesion and motility. Curr Opin Cell Biol. 2004;16:590–6. doi: 10.1016/j.ceb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 76.Pinkus GS, Lones MA, Matsumura F, Yamashiro S, Said JW, Pinkus JL. Langerhans cell histiocytosis immunohistochemical expression of fascin, a dendritic cell marker. Am J Clin Pathol. 2002;118:335–43. doi: 10.1309/N2TW-ENRB-1N1C-DWL0. [DOI] [PubMed] [Google Scholar]
- 77.Graziewicz MA, Day BJ, Copeland WC. The mitochondrial DNA polymerase as a target of oxidative damage. Nucleic Acids Res. 2002;30:2817–24. doi: 10.1093/nar/gkf392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anilkumar N, Parsons M, Monk R, Ng T, Adams JC. Interaction of fascin and protein kinase Calpha: a novel intersection in cell adhesion and motility. Embo J. 2003;22:5390–02. doi: 10.1093/emboj/cdg521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hashimoto Y, Skacel M, Adams JC. Role? of fascin in human carcinoma motility and signaling: prospects for a novel biomarker? Int J Biochem Cell Biol. 2005;37:1787–04. doi: 10.1016/j.biocel.2005.05.004. [DOI] [PubMed] [Google Scholar]


