Abstract
The causal relationship between persistent infection with high-risk HPV and cervical cancer has resulted in the development of HPV DNA detection systems. The widely used MY09/11 consensus PCR targets a 450bp conserved sequence in the HPV L1 gene, and can therefore amplify a broad spectrum of HPV types. However, limitations of these consensus primers are evident, particularly in regard to the variability in detection sensitivity among different HPV types. This study compared MY09/11 PCR with type-specific PCRs in the detection of oncogenic HPV types. The study population comprised 15, 774 patients. Consensus PCR failed to detect 522 (10.9%) HPV infections indicated by type-specific PCRs. A significant correlation between failure of consensus PCR and HPV type was found. HPV types 51, 68 and 45 were missed most frequently. The clinical relevance of the HPV infections missed by MY09/11 PCR was reflected in the fraction of cases with cytological abnormalities and in follow-up, showing 104 (25.4%) CIN2+ cases. The MY09/11 false negativity could be the result of poor sensitivity, mismatch of MY09/11 primers or disruption of L1 target by HPV integration or DNA degradation. Furthermore, MY09/11 PCR lacked specificity for oncogenic HPVs. Diagnostic accuracy of the PCR systems, in terms of sensitivity (MY09/11 PCR: 87.9%; type-specific PCRs: 98.3%) and specificity (MY09/11 PCR: 38.7%; type-specific PCRs: 76.14%), and predictive values for histologically confirmed CIN2+, suggest that type-specific PCRs could be used in a clinical setting as a reliable screening tool.
Keywords: human papillomavirus, cervical cancer screening, Belgium
Introduction
Cervical carcinoma is the second most common cancer in women worldwide. In Europe, it is only the 10th most common cause of cancer death in women as a result of organized screening programs [1]. Since cervical cancer is the only cancer that is almost completely preventable through regular screening, further implementation of effectively organized screening programs and improvement of existing screening strategies and technologies would inevitably decrease the burden of this disease [2].
There is now overwhelming evidence that infection with certain types of the human papillomavirus (HPV) is the primary risk factor for cervical cancer and its precursor lesions [3, 4]. The strong causal relationship between persistent infection with high-risk HPV (HR-HPV) and cervical cancer has resulted in the development of HPV DNA detection systems. The idea that HPV testing could play a crucial role in cervical cancer screening programs becomes more universal [5]. Several applications for HPV DNA detection have been proposed: triage of women with equivocal cytological results [6], follow-up of women treated for cervical intraepithelial neoplasia (CIN) to evaluate treatment [7] or primary screening for oncogenic HPV types alone or in combination with cytology [2, 8–10]. Considering the rising importance of HPV testing, the performance of HPV tests should be carefully assessed and validated. Consensus polymerase chain reaction (PCR) assays have been devised to amplify the most relevant genital types in one reaction. The widely used MY09/11 consensus primer set targets a 450bp conserved sequence in the HPV L1 gene, and is therefore able to amplify a broad spectrum of genital HPV types [11, 12]. However, limitations of this consensus primer system are evident, particularly in regard to the variability of detection sensitivity among specific HPV types [13, 14]. The aim of the study is to compare the performance of the MY09/11 consensus PCR with that of type-specific PCRs for oncogenic HPV detection in a large number of clinical samples. The study includes an assessment of the sensitivity, specificity and predictive values (PV) of both PCR systems to detect biopsy-proven moderate and severe CIN (CIN2+) cases.
Materials and methods
Sample population
Between January 2000 and December 2005 the Laboratory for Clinical Pathology (Labo Lokeren, campus RIATOL, Antwerp, Belgium) received 524,000 cervical samples for cytological evaluation, taken by general practitioners and gynecologists in Flanders (Belgium) during routine screening or gynecological examination. During this period, all samples with cytological abnormalities were automatically tested by a series of PCR analyses according to an algorithm based on MY09/11 triage for the detection and typing of HPV [15]. Samples with normal cytology were only tested in view of quality control or at specific request of the clinician. In this study, only samples subjected to HPV detection by both MY09/11 PCR and HPV typing with all type-specific PCRs were included, yielding a study population of 15, 774 patients.
Retrospective follow-up of these patients was done regardless of the PCR outcome. As histological examination of biopsy material disclosed the true disease status, the database of the Laboratory for Clinical Pathology was searched for available histological results, submitting women to gold standard verification. A CIN2+ histological outcome was considered as a positive follow-up result. Cytological follow-up was only included when at least two consecutive negative smears were available, which was considered as absence of CIN2+.
The study was performed in accordance with the guidelines of the local ethical committee. Informed consent was not obtained because the study posed no risks. Study-specific patient identification codes were assigned and transmitted in accordance with patient confidentiality standards. All investigations were conducted in the Laboratory for Clinical Pathology, a private laboratory member of the AML-RIATOL group, which has been using liquid-based cytology (LBC) in combination with the Cervex-Brush® (Rovers, Oss, the Netherlands) since 1998.
Sample processing and cytological procedure
Cervical cells were collected using the Cervex-Brush® and transferred to ethanol-based preservative (SurePath, Tripath Imaging Inc., Burlington, NC, USA). Thin-layer LBC preparations were made with the fully robotic Autocyte PREP system (Tripath Imaging Inc., Burlington, NC, USA). The cytological results were classified according to the Bethesda system 2001, using the classes negative for intraepithelial lesions, atypical squamous cells of undetermined significance (ASC-US), atypical squamous cells of undetermined significance cannot exclude high-grade squamous intraepithelial lesions (ASC-H), low-grade squamous intraepithelial lesions (L-SIL), high-grade squamous intraepithelial lesions (H-SIL) and atypical glandular cells (AGC) [16]. Cytology was performed without knowledge of the patient's HPV DNA status.
DNA extraction from cervical cells
DNA extraction was performed as previously described [15, 17]. Briefly, for the remainder of each liquid-based preparation, the height of the cell pellet was measured. After vortexing, 400 μl of the remaining cell suspension was transferred to an Eppendorf tube and cells were pelleted by centrifugation. The pellet was resuspended in 50-μl digestion solution (10 mM Tris, 1 mM EDTA, 200 μg/ml Proteinase K) for pellets <3 mm, 100 μl for pellets >3 and >6 mm and 150 μl for pellets >6 mm and digested for 3 hrs at 55°C. Digestion was followed by 10-min incubation at 95°C to inactivate Proteinase K. The DNA extracts were stored at –20°C until PCR was performed.
HPV detection and typing by PCR
Sensitivity of the MY09/11 consensus PCR and type-specific PCRs was determined by using plasmids containing the entire genome of the different HPV types, together with 30 ng of female human DNA (Promega, Madison, USA), to mimic the complex nucleic acid environment present when amplifying genomic DNA. Dilution series were made from 106 to 1 HPV copies and standard curves were constructed for each of the 16 type-specific PCRs. Specificity of the type-specific PCRs was tested by determining the ability to discriminate against plasmids of different HPV types.
Plasmids containing HPV 16 and 18 were commercially available (Clonit, Milan, Italy). Plasmids containing HPV 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 67 and 68 were kindly supplied by T. Matsukara (National Institute of Infectious Diseases, Tokyo, Japan), A. Lörincz (Digene Corp., Gaithersburg, USA), E.-M. de Villiers (DKFZ, Heidelberg, Germany) and G. Orth (Institut Pasteur, Paris, France) or prepared by cloning from PCR products of clinical samples, estimating the copy numbers for individual plasmid preparations by spectrophotometry [17].
In this study HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 67 and 68 were considered high risk. Therefore, all samples were tested with MY09/11 consensus PCR in combination with type-specific real-time PCRs with primers and probes for the detection of HPV 16 E7, 18 E7, 31 E6, 33 E6, 35 E6, 39 E7, 45 E7, 51 E7, 52 E7, 53 E6, 56 E7, 58 E6, 59 E7, 66 E6, 67 L1, 68 E7 and -globin as described previously [11, 15, 17, 18].
Several precautions were taken to prevent false positive results. Different steps, such as DNA extraction, sample preparation, amplification and post-PCR, were performed in strictly separated rooms. The negative PCR control included all PCR components without template DNA. A positive control sample was used for each HPV type.
Statistical analysis
Data were analyzed using SPSS 14.0 for Windows 2000 (SPSS Inc., Chicago, IL, USA) by descriptive statistics and contingency table analysis. Chi-square statistics were performed to assess independence of two variables. To examine the influence of the number of primer mismatches on the efficiency of PCR amplification of the different HPV types one-way ANOVA was performed.
To assess the clinical use of MY09/11 PCR and type-specific PCRs in predicting biopsy- proven CIN2+, their sensitivity, specificity and PVs were determined.
Results
Study population
Between January 2000 and December 2005,524,000 liquid-based cytology samples were processed in the Laboratory for Clinical Pathology. This study comprised 15,774 samples that were subjected to both MY09/11 consensus PCR and HPV typing with type-specific real-time PCRs for HPV 16 E7, 18 E7, 31 E6, 33 E6, 35 E6, 39 E7, 45 E7, 51 E7, 52 E7, 53 E6, 56 E7, 58 E6, 59 E7, 66 E6, 67 L1 and 68 E7 (Fig. 1).
1.
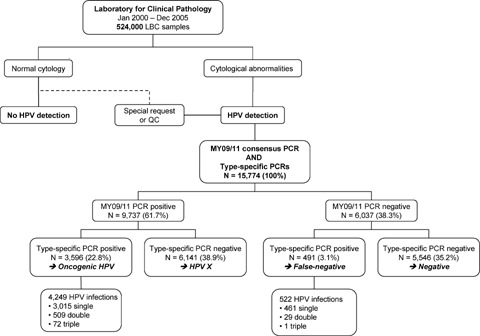
Study overview.
Cytology
Liquid-based cytology was performed on all 15,774 samples: 4,361 (27.6%) showed normal cytology, 7,229 (45.8%) were classified as ASC-US, 319 (2.0%) as ASC-H, 2, 834 (18.0%) as L-SIL and 605 (3.8%) as H-SIL and 426 (2.7%) as AGC. In total 11, 413 samples were classified as abnormal. HPV detection with both PCR methods was predominantly performed on clinically suspicious or abnormal samples, resulting in a study population enriched with clinically abnormal cases.
MY09/11 consensus PCR versus type-specific PCRs
Table 1 shows the characteristics of the different PCR assays. Type-specific PCRs were more sensitive than MY09/11 consensus PCR for all HPV types. In our hands, MY09/11 PCR was able to detect 10 copies of HPV 31, but the detection limit was higher than 102 for all other oncogenic HPV types. For each type-specific PCR a highly significant linear regression was seen between HPV copy number and threshold cycle (CT), defined as the point at which the fluorescence rises appreciably above the background fluorescence. The type-specificity of each PCR was validated. Each PCR exclusively amplified a single HPV genotype.
1.
Sensitivity of PCR assays and characteristics of standard curves for 16 type-specific PCRs constructed with plasmids containing the entire genome of the different HPV types
| Gene | Sensitivity (HPV copies) | Standard curves | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MY09/11 PCR | Type-specific PCRs | |||||||||||||
| Slope | Efficiency | Value of fit (R2) | ||||||||||||
| HPV 16 E7 | 102 | 10 | 3.4836 | 1.940 | 0.9991 | |||||||||
| HPV 18 E7 | 102 | 10 | 3.3632 | 1.928 | 0.9982 | |||||||||
| HPV 31 E6 | 10 | 1 | 3.4508 | 1.977 | 0.9998 | |||||||||
| HPV 33 E6 | 102 | 10 | 3.4632 | 1.912 | 0.9954 | |||||||||
| HPV 35 E6 | 104 | 10 | 3.3551 | 1.974 | 0.9995 | |||||||||
| HPV 39 E7 | 102 | 10 | 3.5730 | 1.932 | 0.9965 | |||||||||
| HPV 45 E7 | 102 | 1 | 3.1596 | 1.835 | 0.9755 | |||||||||
| HPV 51 E7 | 105 | 1 | 3.3482 | 2.000 | 0.9979 | |||||||||
| HPV 52 E7 | 104 | 10 | 3.3143 | 1.995 | 0.9990 | |||||||||
| HPV 53 E6 | 104 | 10 | 3.4650 | 1.918 | 0.9999 | |||||||||
| HPV 56 E7 | 104 | 10 | 3.1775 | 1.961 | 0.9953 | |||||||||
| HPV 58 E6 | 102 | 10 | 3.5450 | 1.919 | 0.9962 | |||||||||
| HPV 59 E7 | 102 | 10 | 3.6836 | 1.860 | 0.9995 | |||||||||
| HPV 66 E6 | 102 | 10 | 3.5661 | 1.927 | 0.9977 | |||||||||
| HPV 67 L1 | 103 | 102 | 3.4973 | 1.835 | 0.9968 | |||||||||
| HPV 68 E7 | 103 | 10 | 3.2000 | 2.044 | 0.9998 | |||||||||
Using MY09/11 consensus PCR, 9, 737 (61.7%) of a total of 15, 774 samples turned out to be positive for HPV DNA. In 6, 037 (38.3%) cases, consensus PCR failed to detect HPV DNA.
Type-specific PCRs for 16 oncogenic HPV types were performed on both consensus PCR-positive and -negative samples.11, 687 (74.1%) samples did not reveal any oncogenic HPV type; a single HPV type was detected in 3, 476 (22.0%) samples; 538 (3.4%) samples showed double infections and 73 (0.5%) showed triple infections. Multiple infections were separated in constituent types, thus type-specific prevalence represents HPV types present in either single or multiple infections. HPV DNA typing identified 4,771 infections: 771 (16.2%) HPV 16, 431 (2.1%) HPV 18, 338 (7.1%) HPV 31, 359 (7.5%) HPV 33, 456 (9.5%) HPV 35, 361 (7.6%) HPV 39, 129 (2.7%) HPV 45, 563 (11.8%) HPV 51, 258 (5.4%) HPV 52, 79 (1.7%) HPV 53, 366 (7.7%) HPV 56, 66 (1.4%) HPV 58, 88 (1.8%) HPV 59, 368 (7.7%) HPV 66, 37 (0.8%) HPV 67 and 101 (2.1%) HPV 68. A total of 6, 141 samples negative for all type-specific PCRs, but positive for consensus PCR, were considered to contain unidentified HPV genotypes of unknown malignant potential (HPV X).
For each HPV type the fraction of MY09/11 consensus PCR-positive and -negative cases were assessed and are summarized in Table 2.
2.
Fraction of MY09/11 consensus PCR-negative and -positive cases, according to HPV type
| HPV type | MY09/11 consensus PCR | Total | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Negative n | % | Positive n | % | n | % | |||||||||||||||||||||
| 16 | 37 | 4.8% | 734 | 95.2% | 771 | 16.2% | ||||||||||||||||||||
| 18 | 39 | 9.0% | 392 | 91.0% | 431 | 9.0% | ||||||||||||||||||||
| 31 | 18 | 5.3% | 320 | 94.7% | 338 | 7.1% | ||||||||||||||||||||
| 33 | 10 | 2.8% | 349 | 97.2% | 359 | 7.5% | ||||||||||||||||||||
| 35 | 38 | 8.3% | 418 | 91.7% | 456 | 9.6% | ||||||||||||||||||||
| 39 | 46 | 12.7% | 315 | 87.3% | 361 | 7.6% | ||||||||||||||||||||
| 45 | 20 | 15.5% | 109 | 84.5% | 129 | 2.7% | ||||||||||||||||||||
| 51 | 188 | 33.4% | 375 | 66.6% | 563 | 11.8% | ||||||||||||||||||||
| 52 | 24 | 9.3% | 234 | 90.7% | 258 | 5.4% | ||||||||||||||||||||
| 53 | 3 | 3.8% | 76 | 96.2% | 79 | 1.7% | ||||||||||||||||||||
| 56 | 34 | 9.3% | 332 | 90.7% | 366 | 7.7% | ||||||||||||||||||||
| 58 | 5 | 7.6% | 61 | 92.4% | 66 | 1.4% | ||||||||||||||||||||
| 59 | 9 | 10.2% | 79 | 89.8% | 88 | 1.8% | ||||||||||||||||||||
| 66 | 23 | 6.3% | 345 | 93.8% | 368 | 7.7% | ||||||||||||||||||||
| 67 | 1 | 2.7% | 36 | 97.3% | 37 | 0.8% | ||||||||||||||||||||
| 68 | 27 | 26.7% | 74 | 73.3% | 101 | 2.1% | ||||||||||||||||||||
| Total | 522 | 11% | 4,249 | 89% | 4,771 | 100% | ||||||||||||||||||||
Overall, MY09/11 consensus PCR failed to detect 522 (10.9%) of all HPV infections. A significant correlation between failure of consensus PCR and HPV type was found (P < 0.001). HPV 51, HPV 68 and HPV 45 were missed most frequently, in 188 (34.3%), 27 (26.7%) and 20 (15.5%) cases, respectively. False negativity of consensus PCR also occurred for all other HPV types: 4.8% HPV 16, 9.0% HPV 18, 5.3% HPV 31, 2.8% HPV 33, 8.3% HPV 35, 12.7% HPV 39, 9.3% HPV 52, 3.8% HPV 53, 9.3% HPV 56, 7.6% HPV 58, 10.2% HPV 59, 6.3% HPV 66 and 2.7% HPV 67.
Cytological diagnosis was evaluated to assess the clinical importance of the HPV infections missed by MY09/11 consensus PCR. Forty-two (8.6%) showed normal cytology, 186 (37.9%) were diagnosed ASC-US, 25 (5.1%) ASC-H, 170 (34.6%) L-SIL, 54 (11.0%) H-SIL and 14 (2.9%) AGC as summarized in Table 3. Focusing on squamous cell abnormalities, MY09/11 consensus PCR was unable to detect the oncogenic HPV type present in 79 (16.1%) cases with severe cervical dysplasia (ASC-H and H-SIL).
3.
Results of MY09/11 consensus PCR and type-specific PCRs and overview of follow-up for different cytological groups
| MY09/11 PCR | + | − | + | − | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Type-specific PCRs | + | + | − | − | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HPV | HR-HPV | HR-HPV | HPV X | Negative | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Initial diagnosis | N | Follow-up | CIN2+ | n | Follow-up | CIN2+ | n | Follow-up | CIN2+ | N | Follow-up | CIN2+ | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| % | n | n | % | n | n | % | n | n | % | n | n | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Squamous lesions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Negative | 401 | 11.2 | 367 | 80 | 42 | 8.6 | 34 | 6 | 1, 776 | 28.9 | 1, 349 | 0 | 2, 142 | 38.6 | 1, 626 | 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ASC-US | 1, 217 | 33.8 | 1, 069 | 173 | 186 | 37.9 | 142 | 24 | 2, 965 | 48.3 | 2, 253 | 2 | 2, 861 | 51.6 | 2, 166 | 0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ASC-H | 93 | 2.6 | 81 | 17 | 25 | 5.1 | 23 | 8 | 139 | 2.3 | 107 | 0 | 62 | 1.1 | 49 | 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| L-SIL | 1, 418 | 39.4 | 1, 357 | 265 | 170 | 34.6 | 142 | 20 | 995 | 16.2 | 762 | 2 | 251 | 4.5 | 193 | 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| H-SIL | 427 | 11.9 | 356 | 236 | 54 | 11.0 | 54 | 40 | 107 | 1.7 | 86 | 7 | 17 | 0.3 | 16 | 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Glandular lesions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AGC | 40 | 1.1 | 18 | 0 | 14 | 2.9 | 14 | 6 | 159 | 2.6 | 122 | 0 | 213 | 3.8 | 164 | 0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Total | 3,596 | 100 | 3,248 | 771 | 491 | 100 | 409 | 104 | 6,141 | 100 | 4,679 | 11 | 5, 546 | 100 | 4,214 | 4 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Follow-up | % | 90.3 | % | 83.3 | % | 76.2 | % | 76.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CIN2+ | % | 23.7 | % | 25.4 | % | 0.2 | % | 0.1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The cytological results of cases with HPV X were compared with those of cases with oncogenic HPV types. The latter showed significantly more squamous cell abnormalities (n = 4, 087; 2.9% ASC-H + 38.9% L-SIL + 11.8% H-SIL = 53.6%) than cases with HPV X (n = 6, 141; 2.3% ASC-H + 16.2% L-SIL + 1.7% H-SIL = 20.2%) (P < 0.001) (Table 3).
Analysis of the primer-target sequence homology for the MY09/11 primers showed a significant (P < 0.05) association between inefficient amplification of HPV types and the number and position of mismatches, as shown in Table 4 and Figure 2.
4.
Positions and number of mismatches between MY09/11 primers and different oncogenic HPV types
| HPV type | Primer mismatch | |||||||
|---|---|---|---|---|---|---|---|---|
| MY09 Forward primer | MY11 Reversed primer | n | ||||||
| 5′-CGT CCM ARR GGA WAC TGA TC-3′ | 5′-GCM CAG GGW CAT AAY AAT GG-3′ | mismatches | ||||||
| 16 | … ..T ………… .. | …… ..C ..C …… .. | 3 | |||||
| 18 | ………… ..T … .. | ……………… .. | 1 | |||||
| 31 | ..A … ..T ……… .. | ..T …… ..C …… .. | 4 | |||||
| 33 | ……………… .. | … ..A ………… .. | 1 | |||||
| 35 | ..G … ..C ……… .. | … ..A ..C ……… .. | 4 | |||||
| 39 | ……… ..G ..T … .. | …… ..C ..C …… .. | 4 | |||||
| 45 | ..A ……… ..T … .. | ..C ……… .. | 3 | |||||
| 51 | ..A … ..T .C…T ..G .. | ..G …… ..C …… .. | 7 | |||||
| 52 | .TA ..T ………… .. | ..G … ..C ..C …… .. | 6 | |||||
| 53 | .TG …………… .. | ……………… .. | 2 | |||||
| 56 | .TA … ..T … ..T … .. | … ..A ..C ……… .. | 6 | |||||
| 58 | ……………… .. | … ..A ..C ……… .. | 2 | |||||
| 59 | ……………… .. | ..T …… TTA …… .. | 4 | |||||
| 66 | .TA …………… .. | …… ..C ……… .. | 3 | |||||
| 67 | .TA ..T …… ..T … .. | … ..A ………… .. | 5 | |||||
| 68 | … ..T ..T ..G ..T ..G .. | ……… ..C …… .. | 6 | |||||
Nucleotide homology is indicated with a period and mismatches are indicated with the nucleotide change in the corresponding sequence. The degenerate base code is as follows: M = A or C, W = A or T, Y = C or T, and R = A or G.
2.
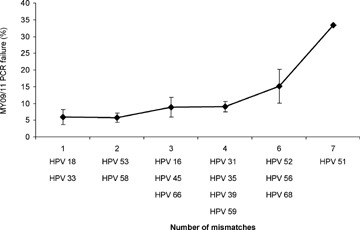
Association between inefficient amplification of different oncogenic HPV types with MY09/11 consensus PCR and the number of mismatches between MY09/11 primers and target sequences. MY09/11 primers showed five mismatches with HPV 67, which was only missed once (2.7%). This result, however, was not included in the analysis because of the low prevalence of HPV 67 and because this was the only type for which the type-specific PCR was directed against L1, showing a lower sensitivity than the other type-specific PCRs.
Follow-up
Histological (n = 3, 124; mean follow-up period = 256 ± 245 days; median follow-up period = 127 days; range = 0–1978 days) or cytological (n = 9, 426; mean follow-up period = 709 ± 399 days; median follow-up period = 602 days; range = 25–2133 days) follow-up was available for 12, 550 (79.6%) of the 15,774 cases. Cytological follow-up was partially based on a reliable estimation of the prevalence of at least two consecutive smears within the different HPV groups. Table 3 shows the follow-up results according to the initial cytological diagnosis. Biopsy showed 771 (23.7%) CIN2+ cases in the MY09/11 and type-specific PCR-positive HR-HPV group, 104 (25.4%) in the MY09/11-negative, type-specific PCR-positive HR-HPV group, 11 (0.2%) in the HPV X group and 4 (0.1%) in the HPV-negative group.
Diagnostic accuracy and predictive values
Type-specific PCRs showed a sensitivity of 98.3% and a specificity of 76.1% to detect biopsy-proven CIN2+. Positive and negative PVs were 23.9% and 99.8%, respectively.
MY09/11 consensus PCR showed a sensitivity of 87.9% and a specificity of 38.7% to detect biopsy-proven CIN2+. Positive and negative PVs were 9.9% and 97.7%, respectively.
Discussion
Establishment of the critical role of HPV in the car-cinogenesis of cervical carcinoma has led to the development of new applications to identify cancer precursors. Currently, PCR amplification is considered as the most sensitive method for detection of HR-HPV DNA and is highly reproducible, easily monitored, provides an objective test outcome and can easily be automated. Consensus PCR assays have been devised to amplify the most relevant genital HPV types in one reaction. When the MY09/11 PCR was designed, only HPV 6, 11, 16, 18 and 33 were known [12]. The MY09/11 primers were designed in a conserved region of the L1 open reading frame (ORF) with the intent to amplify these five genotypes and any other genital HPVs with shared sequence homology in a single reaction [11]. Because the targets were not entirely homologues, positions with nucleotide base heterogeneity were accommodated by inclusion of degenerate base sites. A mixture of 24 oligonucleotides forms a degenerated pool of primers amplifying a broad spectrum of HPV genotypes with various levels of sensitivity. Insertion of nucleotide bases at positions of degeneracy is a random process, and hence does not ensure an equivalent representation of all primers. Lot-to-lot variations among separate syntheses of primers could result in differences in type-specific amplification efficiencies [13]. In this study, only two different lots of MY09/11 primers were used and lot-to-lot variations in type-specific amplification efficiencies were monitored and considered absent.
This study compared the performance of MY09/11 consensus PCR with that of type-specific PCRs, mainly directed at HPV E6 and E7 genes, which encode oncogenic products.
The set of HR-HPV types varies between studies and here HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 67 and 68 were detected. These include the 13 HPV types detected by the high-risk probe cocktail of the Hybrid Capture II system (HC2, Digene Corp., Gaithersburg, USA), the only HPV test that has been FDA-approved for cervical cancer screening in combination with cytology after the age of 30 [5, 19]. However, this test shows a number of disadvantages such as the inability to identify HPV types and the risk of cross-hybridization of additional HPV types with the probe mix [20, 21].
MY09/11 consensus PCR failed to detect 522 HPV infections indicated by type-specific PCRs (Fig. 1). False negativity of MY09/11 PCR occurred in 10.9% of all HPV infections detected with type-specific PCRs (n = 4,771). Statistical analysis showed a significant correlation between failure of consensus PCR and HPV type. HPV types 51 (33.4%), 68 (26.7%) and 45 (15.5%) were missed most frequently (Table 2). Type-specific prevalence and risk of cervical cancer associated with different HPV types were assessed to evaluate the clinical importance of PCR false negativity. HPV 45 is the third most common type worldwide with an odds ratio for cervical cancer of 197.6. The prevalence of HPV 51 and 68 is lower, but odds ratios for cervical cancer confirm the high-risk nature of these HPV types [22, 23].
The clinical relevance of the HPV infections missed by MY09/11 PCR is also reflected in the fraction of cases with cytological abnormalities ASC-H (5.1%) and H-SIL (11.0%) (Table 3). Moreover, follow-up of 409 (83.3%) of the 491 cases with MY09/11 false negativity resulted in 104 (25.4%) CIN2+ cases (Table 3).
The sensitivity of MY09/11 PCR was shown to be significantly lower than that of the type-specific PCRs for all HPV types, leading to false negative results due to a viral load beneath the detection limit of the MY09/11 PCR (Table 1).
PCR failure may be caused by mismatches between MY09/11 primers and the L1 target (Table 4, Fig. 2). A significant association between inefficient amplification of HPV types and the number and position of mismatches was found. For HPV 51 and 68, which were missed most frequently, MY09/11 PCR clearly malfunctions because of the lack of primer specificity (Table 4, Fig. 2). Addition of an extra sequence-specific oligonucleotide (HMB01) directed to the minus strand of HPV may facilitate HPV 51 amplification [24]. Moreover, PGMY09/11 primers were designed to eliminate degeneracies and improve sensitivity across the type spectrum with increased detection of multiple infections and improved reproducibility and specificity [13, 25]. Surprisingly, one report shows similar analytical sensitivities for MY09/11 and PGMY09/11, with better detection of several important HPV types, including HPV 16, by MY09/11. As such, caution should be exerted and reproducibility should be monitored when comparing performances of different primer systems [26].
MY09/11 false negativity could also result from loss of the L1 ORF during integration of the viral DNA into the host genome. Integration is one of the critical contributing factors to malignant transformation and often occurs in the L1 region [4]. This could particularly be the case for HPV 18, which is more often disrupted in the L1 region than other HPV types (Table 2) [27]. PCR primers targeting L1 can therefore be considered less reliable than PCRs directed against the E6 or E7 genes, which encode oncogenic products and always remain intact [28]. Besides the risk of integration, the extensive length of the MY09/11 L1 amplicon promotes false negative results because the efficiency of a PCR reaction generally decreases with increasing amplicon size. Subjecting clinical samples to treatments, such as fixation, is known to degrade DNA. Consequently, the efficiency of PCR primers generating a small product is considerably higher than primers yielding larger amplicons [29]. Overall, the choice of the region used for PCR has important implications for successful HPV detection in clinical diagnosis and management of cervical cancer.
Furthermore, MY09/11 PCR lacks specificity for oncogenic HPVs. A total of 6, 141 samples tested positive for MY09/11 consensus PCR, but negative for all PCRs targeting oncogenic HPV types (Fig. 1). Since MY09/11 PCR can produce nonspecific DNA fragments in the same size range as the HPV L1 product, and since the MY09/11 results are based on gel analysis and not on hybridization with HPV L1 probes, a number of samples could have been falsely diagnosed as HPV-positive. Considering the overall higher sensitivity of the type-specific PCRs, it seems very unlikely that consensus PCR would detect true oncogenic HPVs missed by type-specific PCRs. As such, the samples likely contain unidentified HPV genotypes of unknown malignant potential (HPV X), such as low-risk HPV 6 and 11, which are not likely to induce the carcinogenic process, or such as HPV 26 and 73, which are probably oncogenic but not targeted by this study's type-specific PCRs [19]. This is reflected in the cytology results of cases with HPV X-infection and in their follow-up, yielding a minor fraction of CIN2+ cases (0.2%).
The performance characteristics of both PCR systems in the prediction of CIN2+ lesions further strengthen the clinical importance of our findings. The difference in sensitivity for the detection of CIN2+, 98.3% for the type-specific PCRs versus 87.9% for MY09/11 PCR, is a direct consequence of the difference in detection sensitivity for oncogenic HPVs. The poor specificity of MY09/11 PCR for the detection of CIN2+, 38.7%versus 76.1% for the type-specific PCRs, is a result of its lack of specificity for oncogenic HPV types.
In conclusion, type-specific PCRs for a defined range of HR-HPV types may constitute a more suitable HPV screening test, when compared with a consensus approach, provided that practical issues related to the performance and analysis of multiple PCRs are dealt with. Our current laboratory set-up allows high-throughput, type-specific HPV detection with a turnaround time of less than 72 hrs and better cost-effectiveness than commercial alternatives. Nevertheless, type-specific HPV testing is valuable to address the burden of HPV infections epidemio-logically and to gain more insights into the natural history and dynamics of HPV infections. Therefore, it can be considered as an indispensable tool to monitor the impact of vaccination on the risk of acquisition of individual HPV types. Since type-specific PCRs enable the determination of viral load and integration, which have been suggested as type-dependent risk markers for high-grade cervical lesions and carcinoma, their niche in clinical practice seems undisputable [30–34].
Acknowledgments
We thank the cytotechnologists Karin Francken, Sabrina Van Belle, Kristine Van Belle, Tamara Vandenbroeck and laboratory technicians Katrien Beerden, Sarah Bergmans, Ludo Boels, Isabel De Bradander, Carmen De Maesschalk, Brenda Gabriels, Inge Goeghebeur, Karen Ileghems and Nancy Segers. We also thank Danny Vindevogel for the language review.
GB is supported by the Fund for Scientific Research Flanders. CH is supported by the Institute for Promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen). JB is supported by the Fund for Scientific Research Flanders (FWO-Vlaanderen, G.0205.04) and the Belgian Cancer Foundation (Belgische Stichting tegen Kanker).
References
- 1.Ferlay J, Bray F, Pisani P, Parkin DM. GLOBOCAN 2000: cancer incidence, mortality and prevalence worldwide. Version 1.0. Lyon: IARC Press; 2001. [Google Scholar]
- 2.Davies P, Arbyn M, Dillner J, Kitchener HC, Meijer CJ, Ronco G, Hakama M. A report on the current status of European research on the use of human papil-lomavirus testing for primary cervical cancer screening. Int J Cancer. 2006;118:791–6. doi: 10.1002/ijc.21611. [DOI] [PubMed] [Google Scholar]
- 3.Franco EL, Rohan TE, Villa LL. Epidemiologic evidence and human papillomavirus infection as a necessary cause of cervical cancer. J Natl Cancer Inst. 1999;91:506–11. doi: 10.1093/jnci/91.6.506. [DOI] [PubMed] [Google Scholar]
- 4.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Munoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–9. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 5.IARC Working Group on the Evaluation of Cancer Prevention Strategies. Cervix cancer screening. Lyon: IARC Press; 2005. [Google Scholar]
- 6.Arbyn M, Buntinx F, Van Ranst M, Paraskevaidis E, Martin-Hirsch P, Dillner J. Virologic versus cytologic triage of women with equivocal Pap smears: a meta-analysis of the accuracy to detect high-grade intraepithelial neoplasia. J Natl Cancer Inst. 2004;96:280–93. doi: 10.1093/jnci/djh037. [DOI] [PubMed] [Google Scholar]
- 7.Arbyn M, Paraskevaidis E, Martin-Hirsch P, Prendiville W, Dillner J. Clinical utility of HPV-DNA detection: triage of minor cervical lesions, follow-up of women treated for high-grade CIN: an update of pooled evidence. Gynecol Oncol. 2005;99:7–11. doi: 10.1016/j.ygyno.2005.07.033. [DOI] [PubMed] [Google Scholar]
- 8.Clavel C, Masure M, Bory JP, Putaud I, Mangeonjean C, Lorenzato M, Nazeyrollas P, Gabriel R, Quereux C, Birembaut P. Human papillo-mavirus testing in primary screening for the detection of high-grade cervical lesions: a study of 7932 women. Br J Cancer. 2001;84:1616–23. doi: 10.1054/bjoc.2001.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuzick J, Beverley E, Ho L, Terry G, Sapper H, Mielzynska I, Lorincz A, Chan WK, Krausz T, Soutter P. HPV testing in primary screening of older women. Br J Cancer. 1999;81:554–8. doi: 10.1038/sj.bjc.6690730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schiffman M, Herrero R, Hildesheim A, Sherman ME, Bratti M, Wacholder S, Alfaro M, Hutchinson M, Morales J, Greenberg MD, Lorincz AT. HPV DNA testing in cervical cancer screening: results from women in a high-risk province of Costa Rica. JAMA. 2000;283:87–93. doi: 10.1001/jama.283.1.87. [DOI] [PubMed] [Google Scholar]
- 11.Bauer HM, Ting Y, Greer CE, Chambers JC, Tashiro CJ, Chimera J, Reingold A, Manos MM. Genital human papillomavirus infection in female university students as determined by a PCR based method. JAMA. 1991;265:472–7. [PubMed] [Google Scholar]
- 12.Manos MM, Ting MY, Wright DK, Lewis AJ, Broker TR, Wolinsky SM. The use of polymerase chain reaction amplification for the detection of genital human papillomaviruses. Cancer Cells. 1989;7:209–14. [Google Scholar]
- 13.Gravitt PE, Peyton CL, Alessi TQ, Wheeler CM, Coutlee F, Hildesheim A, Schiffman MH, Scott DR, Apple RJ. Improved amplification of genital human papillomaviruses. J Clin Microbiol. 2000;38:357–61. doi: 10.1128/jcm.38.1.357-361.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qu W, Jiang G, Cruz Y, Chang CJ, Ho GY, Klein RS, Burk RD. PCR detection of human papillomavirus: comparison between MY09/MY11 and GP5+/GP6+ primer systems. J Clin Microbiol. 1997;35:1304–10. doi: 10.1128/jcm.35.6.1304-1310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Depuydt CE, Vereecken AJ, Salembier GM, Vanbrabant AS, Boels LA, Van Herck E, Arbyn M, Segers K, Bogers JJ. Thin-layer liquid-based cervical cytology and PCR for detecting and typing human papillomavirus DNA in Flemish women. Br J Cancer. 2003;88:560–6. doi: 10.1038/sj.bjc.6600756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Solomon D, Davey D, Kurman R, Moriarty A, O'Connor D, Prey M, Raab S, Sherman M, Wilbur D, Wright T, Jr, Young N Forum Group Members. Bethesda 2001 Workshop. The 2001 Bethesda System: terminology for reporting results of cervical cytology. JAMA. 2002;287:2114–9. doi: 10.1001/jama.287.16.2114. [DOI] [PubMed] [Google Scholar]
- 17.Depuydt CE, Benoy IH, Bailleul EJ, Vandepitte J, Vereecken AJ, Bogers JJ. Improved endocervical sampling and HPV viral load detection by Cervex-Brush Combi. Cytopathology. 2006;17:374–81. doi: 10.1111/j.1365-2303.2006.00386.x. [DOI] [PubMed] [Google Scholar]
- 18.Moberg M, Gustavsson I, Gyllensten U. Real-time PCR-based system for simultaneous quantification of human papillomavirus types associated with high risk of cervical cancer. J Clin Microbiol. 2003;41:3221–8. doi: 10.1128/JCM.41.7.3221-3228.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoler MH, Castle PE, Solomon D, Schiffman M American Society for Colposcopy and Cervical Pathology. The expanded use of HPV testing in gyne-cologic practice per ASCCP-guided management requires the use of well-validated assays. Am J Clin Pathol. 2007;127:335–7. doi: 10.1309/RNF3C01JKADQCLKP. [DOI] [PubMed] [Google Scholar]
- 20.Peyton CL, Schiffman M, Lorincz AT, Hunt WC, Mielzynska I, Bratti C, Eaton S, Hildesheim A, Morera LA, Rodriguez AC, Herrero R, Sherman ME, Wheeler CM. Comparison of PCR- and hybrid capture-based human papillomavirus detection systems using multiple cervical specimen collection strategies. J Clin Microbiol. 1998;36:3248–54. doi: 10.1128/jcm.36.11.3248-3254.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vernon SD, Unger ER, Williams D. Comparison of human papillomavirus detection and typing by cycle sequencing, line blotting, and hybrid capture. J Clin Microbiol. 2000;38:651–5. doi: 10.1128/jcm.38.2.651-655.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munoz N, Bosch FX, De Sanjosé S, Herrero R, Castellsagué X, Shah KV, Snijders PJ, Meijer CJ International Agency for Research on Cancer Multicenter Cervical Cancer Study Group. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Eng J Med. 2003;348:518–27. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 23.Clifford GM, Smith JS, Plummer M, Munoz N, Franceschi S. Human papillomavirus types in invasive cervical cancer worldwide: a meta-analysis. Br J Cancer. 2003;88:63–73. doi: 10.1038/sj.bjc.6600688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hildesheim A, Schiffman MH, Gravitt PE, Glass AG, Greer CE, Zhang T, Scott DR, Rush BB, Lawler P, Sherman ME, Kurman RJ, Manos MM. Persistence of type-specific human papillomavirus infection among cytologically normal women. J Infect Dis. 1994;169:235–40. doi: 10.1093/infdis/169.2.235. [DOI] [PubMed] [Google Scholar]
- 25.Coutlee F, Gravitt P, Kornegay J, Hankins C, Richardson H, Lapointe N, Voyer H, Franco E. Use of PGMY primers in L1 consensus PCR improves detection of human papillomavirus DNA in genital samples. J Clin Microbiol. 2002;40:902–7. doi: 10.1128/JCM.40.3.902-907.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castle PE, Schiffman M, Gravitt PE, Kendall H, Fishman S, Dong H, Hildesheim A, Herrero R, Bratti MC, Sherman ME, Lorincz A, Schussler JE, Burk RD. Comparisons of HPV DNA detection by MY09/11 PCR methods. J Med Virol. 2002;68:417–23. doi: 10.1002/jmv.10220. [DOI] [PubMed] [Google Scholar]
- 27.Berumen J, Casas L, Segura E, Amezcua JL, Garcia-Carranca A. Genome amplification of human papillomavirus types 16 and 18 in cervical carcinomas is related to the retention of E1/E2 genes. Int J Cancer. 1994;56:640–5. doi: 10.1002/ijc.2910560506. [DOI] [PubMed] [Google Scholar]
- 28.Morris BJ. Cervical human papillomavirus screening by PCR: advantages of targeting the E6/E7 region. Clin Chem Lab Med. 2005;43:1171–7. doi: 10.1515/CCLM.2005.203. [DOI] [PubMed] [Google Scholar]
- 29.Molijn A, Kleter B, Quint W, Van Doorn LJ. Molecular diagnosis of human papillomavirus infections. J Clin Virol. 2005;32:43–51. doi: 10.1016/j.jcv.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 30.Woodman CB, Collins SI, Young LS. The natural history of cervical HPV infection: unresolved issues. Nat Rev Cancer. 2007;7:11–22. doi: 10.1038/nrc2050. [DOI] [PubMed] [Google Scholar]
- 31.Moberg M, Gustavsson I, Wilander E, Gyllensten U. High viral loads of human papillomavirus predict risk of invasive cervical carcinoma. Br J Cancer. 2005;92:891–4. doi: 10.1038/sj.bjc.6602436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andersson S, Safari H, Mints M, Lewensohn-Fuchs I, Gyllensten U, Johansson B. Type distribution, viral load and integration status of high-risk human papillo-maviruses in pre-stages of cervical cancer (CIN) Br J Cancer. 2005;92:2195–200. doi: 10.1038/sj.bjc.6602648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheung JL, Lo KW, Cheung TH, Tang JW, Chan PK. Viral load, E2 gene disruption status, and lineage of human papillomavirus type 16 infection in cervical neoplasia. J Infect Dis. 2006;194:1706–12. doi: 10.1086/509622. [DOI] [PubMed] [Google Scholar]
- 34.Guo M, Sneige N, Silva EG, Jan YJ, Cogdell DE, Lin E, Luthra R, Zhang W. Distribution and viral load of eight oncogenic types of human papillomavirus (HPV) and HPV 16 integration status in cervical intraepithelial neoplasia and carcinoma. Mod Pathol. 2007;20:256–66. doi: 10.1038/modpathol.3800737. [DOI] [PubMed] [Google Scholar]


