Abstract
Terminal progression of colorectal cancer (CRC) culminates in liver metastasis. To identify genes that are involved in the metastatic phenotype, cDNA microarrays were used to analyse mRNA expression profiles of colorectal carcinoma (CC)531 rat colon adenocarcinoma cells for changes related to their homing into the liver. Briefly, CC531 cells were intraportally implanted into the liver of Wag-Rij rats and re-isolated after 3, 6, 9, 14 and 21 days. Compared to control CC531 cells, claudin1 and claudin4 were among the ≥8-fold initially down-regulated genes. The co-culture of tumour cells with isolated rat hepatocytes and Kupffer cells did not induce down-regulation of either claudin1 or 4. When the environment effective on circulating tumour cells was simulated by cell culture conditions favouring their adhesion, only claudin4 showed augmented expression. Knockdown of claudin1 and claudin4 mediated by small interfering RNA caused significantly increased migration and decreased clonogenic growth of tumour cells (P < 0.05), but had no effect on their proliferation. These experimental results were paralleled by increased claudin1 and claudin4 expression in human CRC samples in Union for International Cancer Control (UICC) stages I–III, as evaluated by real-time PCR. Increased claudin4 levels were correlated with significantly reduced overall survival (log-rank test, P= 0.018). Further, significantly (P < 0.05) reduced expression of claudin1 and claudin4 was observed in stage IV and liver metastasis by immunohistochemistry. In conclusion, sequential biphasic changes in claudin1 and claudin4 expression occur during the homing of rat CC531 CRC cells to the liver. This modulation is reflected by significant changes in claudin expression in human primary and metastatic CRC.
Keywords: CLDN1, CLDN4, CC531 cells, hepatocytes, adhesion, co-culture, microarray
Introduction
Colorectal cancer (CRC) accounts for 11.5% of cancer-related deaths worldwide [1]. CRC progression is characterized by increased growth of the primary carcinoma as well as lymphatic and haematogenic spread. For 80% of patients with recurrent disease, the liver is the predominant site of metastatic spread along the mesenteric circulation [2]. The reported 5 year survival rate ranges from 25% to 44% after resection of colorectal liver metastases [3]. As shown by Fearon and Vogelstein [4], the loss of certain tumour suppressor genes and activation of oncogenes is correlated with progression of CRC. However, this model is an incomplete portrait of the genetic and epigenetic changes that are instrumental in cancer progression. The role of genes contributing to metastasis in the liver, skeleton, lungs and brain has only recently been investigated [5]. Within the multitude of factors that contribute to metastasis, such as growth factors and matrix metalloproteinases [6–8], proteins involved in cell–cell contact have emerged as loss of cell polarity and increased cell permeability are hallmarks of cancerous tissues [9]. In line with this, an increase in tight junction (TJ) permeability in CRC constitutes a decisive event in the progression of this cancer [9].
In multicellular organisms, TJs constitute one type of cell–cell adhesion found in endothelial and epithelial cellular sheets. TJs have many pivotal roles; they form a fence between the apical and basolateral plasma membrane domains [10, 11], act as a diffusion barrier for solutes through the intercellular space [11–15] and draft several signalling and cytoskeletal molecules at their cytoplasmic surface [10, 16]. Claudins (CLDNs) form the structural backbone of TJs, and comprise at least 24 members of integral transmembrane proteins [17] ranging in size between 20 and 27 kD [10]. Recently, the altered expression of various claudins has been implicated in the progression of several human cancers [18–26]. In contrast to the general notion that claudin expression would decrease in tumorigenesis as TJs are lost during cellular transformation, the claudin status seems to change in a tissue-specific manner. For example, overexpression of CLDN2 has been correlated to CRC [27], whereas decreased CLDN7 expression has been reported in head and neck cancer [28], invasive ductal breast carcinoma [29] and metastatic breast cancer [30]. In addition, CLDN3 and CLDN4 have been found repeatedly elevated in a variety of cancers including pancreatic ductal adenocarcinoma [31], ovarian, uterine, prostate and breast cancers [32]. In partial contrast, reduced expression of CLDN4 and CLDN5 was detected in hepatocellular and renal carcinomas [33]. In CRC, both, up- and down-regulation of claudin4 expression have been described [34, 35], as well as aberrant expression of claudin1 [36–39]. A recent study by Dhawan et al. has shown increased CLDN1 expression compared to normal mucosa in human primary CRC and metastasis samples, as well as in cell lines derived from primary and metastatic lesions [37]. For this claudin, the β-catenin/Tcf signalling has been suggested as a potential mechanism underlying Cldn1-dependent changes in CRC [40]. At the transcriptional level, transcription factors such as Snail [41], Cdx-2, hypoxia necrosis factor-alpha (HNF)-α and GATA-sequence-binding transcription factor-4 (GATA-4) [42, 43] can bind to the promoter regions of various claudins including Cldn3 and Cldn4, thereby affecting their expression.
In this study, we describe the expression profile and regulation of Cldn1 and Cldn4 in correlation with CRC progression and liver metastasis formation. We used rat CC531 CRC cells as model, because these cells typically metastasize to and grow in the liver of syngeneic rats. These tumour cells were originally isolated from a 1, 2-dimethylhydrazine-induced colon adenocarcinoma in Wag/Rij rats [44]. For mimicking human liver metastasis, these cells are injected intraportally and grow in the liver by diffuse infiltration [45]. CC531 cells homing to liver tissue were re-isolated after defined time-points and evaluated for alterations in claudin genes’ expression. Claudin1 and claudin4 were significantly down-regulated during early metastasis in our model system, and in consequence, were investigated in greater detail in vitro as well as in human CRC and corresponding liver metastases. Our data suggest that sequential changes in Cldn1 and Cldn4 expression are a typical feature for homing of CC531 CRC cells to the liver. This modulation is reflected by significant changes in claudin expression in human primary and metastatic CRC.
Materials and methods
Cell culture
The rat CC531 colon adenocarcinoma cell line [44] was obtained 1996 by Dr. J Gahlen and maintained under standard conditions (37°C, humidified atmosphere with 5% CO2) in Roswell Park Memorial Institute (RPMI)-1640 medium (Invitrogen, Karlsruhe, Germany), supplemented with 10% foetal calf serum (FCS), L-glutamine (2 mM), penicillin (100 IU/ml) and streptomycin (100 μg/ml, Invitrogen). For isolation and propagation, cells were washed with phosphate-buffered saline (PBS), trypsinized (0.25% trypsin/ethylenediaminetetraacetic acid), pelleted at 1500 rpm for 5 min. and suspended at the desired concentration in RPMI-1640 medium (Invitrogen). The cell line was authenticated by the DSMZ (Braunschweig, Germany) with the STR DNA cell identity test.
Preparation of CC531 tumour cells for injection
CC531 cells were washed with PBS, trypsinized, pelleted and suspended at a concentration of 4 × 106 cells/500 μl (350 μl PBS + 150 μl Biomatrix EHC; Serva, Heidelberg, Germany) and kept on ice.
Tumour cell injection and re-isolation
Six- to eight-week-old male WAG/Rij rats (Charles River, Sulzfeld, Germany) were used for the experiments. They were fed a standard diet ad lib. and were given an adaptation period of 1 week prior to any experimental procedures. All animal experiments were approved by the responsible governmental animal ethics committee (Regierungspraesidium Karlsruhe, Germany). For in vivo inoculation, a median laparotomy was performed under anaesthesia (isoflurane) prior to tumour cell injection. The caecum was exteriorized from the abdominal cavity and a mesocolic vein was prepared for injection. The CC531 cells (4 × 106) were injected slowly under microscopic control using a 28-gauge needle. Cells were injected for at least 60 sec., and then the puncture site was compressed between two cotton swabs for at least 1 min. to prevent bleeding. The caecum was then returned to the abdomen, and the musculature closed with a running suture (4–0 vicryl, Ethicon, Norderstedt, Germany), and the skin closed with metal clips [46]. Before tumour cell isolation, the rats were kept for 3, 6, 9, 13 and 21 days after tumour cell implantation. Then the abdominal cavity was opened and a 22 G cannula was inserted into the portal vein, through which the liver was perfused with Hank's balanced salt solution (HBSS) medium (20 ml/min., 37°C for 10 min.). This medium was replaced with pre-warmed perfusion medium [125 ml HBSS containing CaCl2 1M, 0.1% pronase, 100 mg collagenase Type IV (Serva), 37°C for the following 10 min.] to digest connective tissues. After getting the cells in suspension, they were filtered through a sterile filter (Cell strainer, 70 μm Nylon; BD, Heidelberg, Germany) and centrifuged (300 ×g for 10 min.). The resulting cell suspension of liver and tumour cells was transferred into 50 ml tubes and layered carefully onto a Ficoll gradient medium (Amersham pharmacia Biotech AB, Uppsala, Sweden). After centrifugation (15 min. at 500 ×g), the tumour cells were obtained from the top of the interface and resuspended in RPMI medium. To obtain a high purity of isolated tumour cells, CC531 cells were subsequently isolated by fluorescence-activated cell sorting technology using red fluorescent protein (RFP) as marker. Afterwards, the pure cells were pelleted at 3000 rpm for 5 min. and snap frozen at –80°C. An aliquot of the cells, which were isolated on day 21, was used for re-culturing CC531 cells in vitro. These cells were propagated every 3 days, but two time-points (14 and 22 days after tumour cell explantation) were chosen for subsequent microarray analysis, PCR and Western blot.
For the isolation of rat hepatocytes and Kupffer cells, the same perfusion method was performed as described above. However, to separate parenchymal from non-parenchymal cells, cell suspensions were gently pelleted (3 min. at 4°C, 70 ×g, maximal brake). The resulting pellet, containing mainly hepatocytes, was taken up in maintenance-medium without FCS. Trypan blue exclusion (1 part trypan blue: 2 parts cell suspension) was used for cell counting and assessing their viability by using a haemocytometer. A total of 4 × 107 hepatocytes with 95% viability were usually obtained from one rat liver. Afterwards, two methods were successfully used to isolate Kupffer cells as pure as possible. These included isolation using a 25%/50% two-step Percoll gradient or using a CD68 antibody coupled to magnetic beads. The anti-CD68 antibody was used for its specificity to macrophages including the Kupffer cell subpopulation (Supporting Information S1).
Co-culture/two compartment model
Briefly, this model is based on a two-compartment system in which hepatocytes or Kupffer cells, plated in the lower compartment, are co-cultured with CC531 tumour cells growing in the upper compartment, with the two cell types being separated by a porous membrane (0.4 μm pore size). This system, preventing a direct contact between the two compartments, allows the cells to be only indirectly influenced by molecules secreted from the cells in the other layer, respectively. This model was described in detail previously [47].
RNA-isolation and RT-PCR
The RNeasy mini-kit (Qiagen, Hilden, Germany) was used for RNA-isolation from CC531 cells, hepatocytes and Kupffer cells. The amount and purity of isolated RNA was measured in a spectrophotometer using the 260/280 ratio. After producing cDNA from the isolated RNA [47], amplicons of Cldn1, Cldn4 and γ-tubulin were generated with the respective primer sequences (Table S1). The PCR-reaction was performed in a thermal cycler (DNA engine, PTC200 Peltier; MJ Research, Waltham, MA, USA). After electrophoresis of PCR-amplicons on a polyacrylamide gel, it was incubated in ethidium bromide for 10 min. followed by visualization under UV-light (Gel Doc XR, BIO-RAD, Munich, Germany).
Real-time PCR
The LightCycler 480 Real-Time PCR system with the LC480 RNA Master Hydrolysis Probes and the human Universal ProbeLibrary kit (Roche, Mannheim, Germany) was used following the manufacturer’s protocol. The primer sequences used for CLDN1, CLDN4 and for GAPDH are shown in Table S1. One microlitre containing 50 ng cDNA per sample was pipetted in triplicate into 384-well plates and amplified at 60°C for 50 cycles. The cDNA input was normalized to the expression of the housekeeping gene GAPDH. As positive control, a mixture of 11 normal mucosa samples was used.
Microarray analysis
For RNA isolation from CC531cells, the RNeasy mini-kit (Qiagen) was used. RNA was eluted in water. The quality of total RNA was checked by gel analysis using the total RNA Nano chip assay on an Agilent 2100 Bioanalyzer (Agilent Technologies GmbH, Berlin, Germany). Only samples with RNA index values >8.5 were selected for expression profiling. RNA concentrations were determined using the NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) (Supporting Information S2).
Western blot analysis
Western blotting was performed as described previously [47]. Primary antibodies for CLDN1, CLDN4 and extracellular signal-regulated protein kinase (ERK)2 as well as their corresponding secondary antibodies are shown in Table S1.
Immunohistochemistry (IHC)
Formalin-fixed and paraffin-embedded tissue sections with a thickness of 4 μm were taken from 32 CRC and 8 liver metastasis specimens. These sections were deparaffinized in xylene and rehydrated in gradually decreasing concentrations of ethanol. The slides were washed in Tris-buffered saline pH 7.4 (10 mM Tris–HCl, 0.85% NaCl and 0.1% bovine serum albumin) and subjected to immunostaining. Consecutive tissue sections were boiled in 10 mM citrate buffer for 10 min. in a microwave oven for antigen retrieval. The sections were incubated with the same claudin antibodies used in Western blot, as well as with normal mouse IgG1 (DAKO Corporation, Glostrup, Denmark) as negative control. The slides were rinsed in washing buffer and incubated with anti-goat or anti-rabbit horseradish peroxidase (HRPO)-conjugated IgG (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) for 1 hr at room temperature. Thereafter, tissue sections were incubated with 100 μl streptavidin-phosphatase (KPL) for 35 min. at room temperature. The tissue sections were then washed in washing buffer and each section was subjected to 100 μl of the PhThaloRED activator- buffered substrate mixture (KPL) and counterstained with Mayer’s haematoxylin.
Physical forces’ effect experiment
A total of 2 × 106 CC531 cells were seeded in 25 cm2 flat-bottom flasks or into round 50 ml glass bottles (Steiner GmbH, Siegen Eiserfeld, Germany), which were rotated on a roller (Stovall Life Science Incorporated, Greensboro, NC, USA) at a speed of 1 rpm, preventing the cells from adhesion to each other and onto the flask bottom. After 24 hrs, the cells in flat flasks and half of the cells in round bottles were harvested for PCR analysis; the other half of cells was seeded in flat flasks till the next day to investigate the influence of adhesion status on claudin expression and then harvested after determining their viability under the microscope. This procedure was done daily for 3 days after seeding the cells (Fig. 1).
Fig 1.
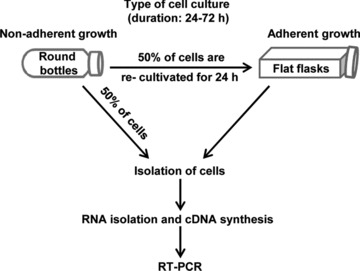
Scheme indicating the experimental procedure for assessing the physical force’s effects.
Small interfering RNA (siRNA) knockdown experiments
The siRNA duplexes designed against Cldn1 or Cldn4 and nonspecific siRNA used as negative control were purchased from Invitrogen (Table S1). CC531 cells cultured in 6-well plates were transfected with 100 nM siRNA or negative control using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. At 24, 48 and 72 hrs after treatment, the cells were harvested for PCR analysis.
Cell proliferation assay (MTT)
To assess the effect of siRNA transfection on the proliferation of CC531 cells, the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was used (Supporting Information S3).
Migration assay
This assay was performed to investigate the effect of Cldn1 or Cldn4 down-regulation on the migration of CC531 cells. The bottom layer in 24-well plates consisted of 50 μl FCS, which was gently over-layered with 200 μl semi-liquid RPMI medium (containing 0.4% methylcellulose and 20% FCS) resulting in the chemotaxis mixture. An incubation time of 24 hrs was needed to build the chemotaxis gradient. Then, 2 × 104 CC531 cells were seeded on 8 μm pore size polycarbonate membranes (Millicell; Millipore, Schwalbach, Germany), which were transferred onto the prepared wells. The next day, CC531 cells were transfected with nonsense or specific siRNA against Cldn1 or Cldn4 as described before. Migrating cells were counted under a microscope for 3 subsequent days. To that purpose, the membrane was removed with transfected, non-migrated cells and placed onto a fresh well with chemotaxis gradient.
Colony formation assay
To determine the effect of siRNA knockdown of Cldn1 or Cldn4 on the ability of CC531 cells to form colonies, the procedure previously detailed [48] was performed. Clusters of ≥30 cells were counted as colony, whereas clusters of ≥60 cells were considered as large colony.
Patients and tissue samples
For the immunohistochemical analyses of CLDN1 and CLDN4, 32 primary CRC tissue specimens with adjacent non-neoplastic tissue and 8 liver metastases were obtained from the Institute of Pathology, University of Heidelberg.
For real-time PCR analysis, 67 sporadic CRC patients, who were admitted and underwent surgery in the time between (January 1998–July 2001) at the Municipal Hospital in Nuernberg (Department of Abdominal-, Thorax- and Endocrine Surgery) were selected.
The samples included in this study (for IHC and real-time PCR, see Table 1) were used based on the patients informed consent and approved by the Ethics Committee of the Universities of Heidelberg and Erlangen.
Table 1.
General information on the two groups of CRC patients of this study
| Parameter | Nürnberg patients | Heidelberg patients |
|---|---|---|
| Analysis method | Real-time PCR | IHC |
| Number of patients | n= 67 | n= 32 |
| Age (average) | 67 years | 65 years |
| Gender | 42 male/25 female | 22 male/10 female |
| Location | Colon (n= 23) Rectum (n= 44) | Colon (n= 10) Rectum (n= 22) |
| Tumour stage* (UICC) | I (n= 11) II (n= 25) III (n= 20) IV (n= 11) | II (n= 24) IV (n= 8) |
| Tumour grade | G2 (n= 52) G3 (n= 15) | G2 (n= 25) G3 (n= 7) |
All lesions were diagnosed as adenocarcinoma.
Statistical analysis
For the calculations of gene/protein expression levels in semi-qRT-PCR/Western blot, the intensity of the target gene/protein was normalized to that of the housekeeping gene γ- tubulin/loading control ERK2. The densitometric analysis was performed with the Quantity One Program (Biorad Laboratories GmbH, Munich, Germany).The chi-square test (×2 test) was used to test the significance of colony formation inhibition or increased migration in the siRNA- versus nonsense-treated cells. For IHC, to determine the correlation between CLDN1/CLDN4 and tumour grade/stage and metastasis, the Wilcoxon–Mann-Whitney test was used. Calculations were carried out using the ADAM statistical software package (DKFZ, Heidelberg, Germany). For real-time PCR, correlation between CLDN1 and CLDN4 expressions as well as with patient’s age was assessed with Spearman’s rank correlation coefficient. The non-parametric Kruskal–Wallis test was used to test for an association between tumour stage and CLDN1/CLDN4 expression. The prognostic value of CLDN1 and CLDN4 expression on overall survival was determined with Cox’s proportional hazard regression model. Overall survival was calculated as time from surgery to time of death. P-values below 0.05 were considered statistically significant. Calculations were carried out with the statistical software R 11.0 and StatXact 6.
Results
Expression profile of Cldn1 and Cldn4 in re-isolated CC531 cells
cDNA microarray was used to analyse the alterations in expression profile of rat genes for discovering a possible correlation with liver metastasis formation of CC531 cells. To that purpose, CC531 cells had been intraportally injected into Wag-Rij rats and re-isolated from rat livers at different time-points (3, 6, 9, 14 and 21 days) (Figs S1 and S2). From 16 claudin group genes included in this array, 10 were not significantly modulated (<2-fold) and 6 were significantly down-regulated in the initial phase of metastasis formation. Of these, Cldn6 and Cldn12 were 2- to 5-fold down-regulated and Cldn1, 3, 4 and 9 were >5-fold down-regulated. For the experiments described in this article, we concentrated on Cldn1 and Cldn4 which showed ≥8-fold reduced gene expression in the re-isolated metastatic cells as compared to CC531 control cells (Fig. 2A). Interestingly, both genes were at first down-regulated with a nadir (8- or 11-fold down-regulation) on day 6, followed by gradual up-regulation within the observation period. These results were confirmed with RT-PCR (maximum down-regulation of >80% on day 6 for both genes; Fig. 2B and C) and Western blot (specific bands below detection limit, >90% inhibition on day 6; Fig. 2B and D). As shown in Figure 2B, there was not only a tight correlation between RNA and protein levels of Cldn1 or Cldn4, but also a nearly parallel modulation without a discernible delay in time between RNA and protein levels. It is noteworthy that the transcription repressor gene Snail showed an inverse modulation: an increased expression during the first week (up to 3.8-fold) with the peak of its expression corresponding to the nadir of Cldn1 and Cldn4 down-regulation (Fig. 2A).
Fig 2.
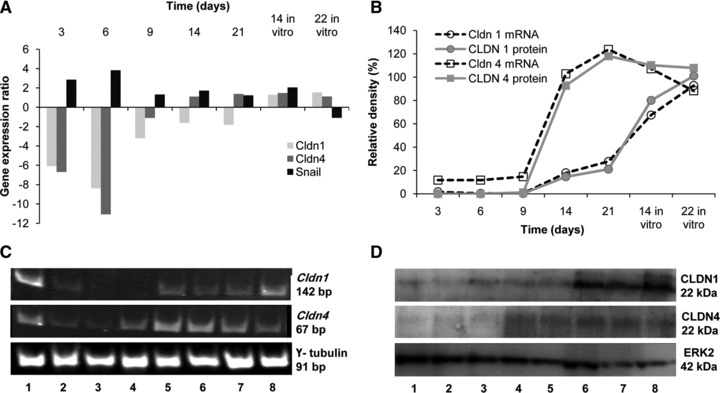
Down-regulation of Cldn1 and Cldn4 in CC531 cells homing into the liver. (A) Expression profile of claudins (1, 4) and Snail in CC531 cells as shown by microarray analysis. The values represent the gene expression in isolated metastasizing cells in comparison to the expression in cells growing in vitro. (B) The diagram represents the mRNA or protein expression levels in re-isolated CC531 cells in percentage of the expression detected in control cells (100%). Values were calculated using the pixel density of each PCR/or Western blot band normalized to the corresponding value of γ-tubulin or ERK2, respectively. (C) Expression of claudins (1, 4) in CC531 cells as shown by RT-PCR. Lane 1: control CC531 cells, lanes 2–6: CC531 cells isolated from the liver after 3, 6, 9, 14 and 21 days, respectively, lanes 7, 8: CC531 cells re-isolated after 21 days and cultured in vitro for 14 and 22 days, respectively. (D) Expression of CLDNs (1, 4) in CC531 cells as shown by Western blot. Lanes 1–5: CC531 cells isolated from the liver after 3, 6, 9, 14 and 21 days, respectively, lanes 6, 7: CC531 cells isolated after 21 days and cultured in vitro for 14 and 22 days, respectively, lane 8: control CC531 cells.
Effect of co-culture conditions on the expression of Cldn1 and Cldn4
In an attempt to explain the initial down-regulation of Cldn1 and Cldn4, CC531 cells were co-cultured with isolated rat hepatocytes and Kupffer cells.
No down-regulation effect on claudin expression was noticed, whereas both genes were up-regulated after co-culture with hepatocytes. As shown in Figure 3A, a substantial up-regulation (up to 4- and 3-fold) in Cldn1 expression was noticed in CC531 cells co-cultured with hepatocytes for 48 and 72 hrs, respectively. Similarly, the expression of Cldn4 in these cells was up-regulated to 1.6- and 1.8-fold, compared to control CC531 cells.
Fig 3.
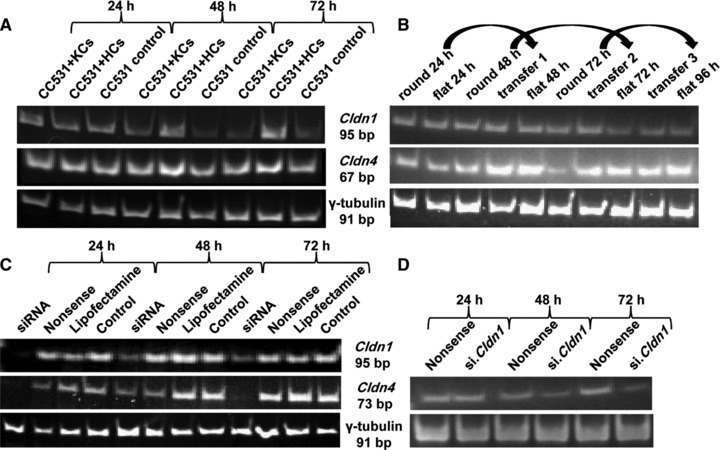
Modulation of Cldn1 and Cldn4 expression in vitro. (A) Expression of claudins (1, 4) in CC531 cells co-cultured for 24 to 72 hrs with Kupffer cells (KCs) and hepatocytes (HCs) in comparison to the housekeeping gene γ-tubulin as shown by RT-PCR. (B) Expression of claudins (1, 4) in CC531 cells harvested from round and flat flasks as shown by RT-PCR. Lanes 1, 3, 6: CC531 cells harvested from round bottles after 24, 48 and 72 hrs, respectively. Lanes 2, 5, 8, 10: CC531 cells harvested from flat flasks after 24, 48, 72 and 96 hrs, respectively. Lanes 4, 7, 9: CC531 cells harvested from flat flasks after being transferred from round bottles after 24, 48 and 72 hrs, respectively. (C) Down-regulation of claudins (1, 4) in CC531 cells after siRNA transfection as shown by RT-PCR. (D) Down-regulation of Cldn4 after 24–72 hrs in CC531si.Cldn1 cells (compared to CC531nonsense cells) as shown by RT-PCR.
Effect of altered adhesion on Cldn4 and Cldn1 expression
This experiment was performed to investigate the effect of physical forces (friction and shearing) on circulating tumour cells as a possible reason for the initial down-regulation of Cldn(1, 4) expression. As shown in Figure 3B, no change in Cldn1 expression was noticed either in CC531 cells growing continuously in flat flasks or in round bottles. On the contrary, Cldn4 mRNA expression was 2- and 1.8-fold higher in CC531 cells growing in flat flasks than their counterparts growing in round bottles at 48 and 72 hrs after seeding the cells, respectively (Fig. 3B). Furthermore, transferring tumour cells from a non-adhesive state in round bottles to growing in flat bottom flasks for 24 hrs caused >2.5-fold increased expression of Cldn4, whereas no effect on Cldn1 expression was noticed. Accordingly, the physical conditions and the adhesion status of the cells affect differently the expression of Cldn1 and Cldn4, suggesting a direct relationship with the latter, but not with the former.
Inhibition of Cldn1 and Cldn4 expression by siRNA
As shown by RT-PCR in Figure 3C, exposure to siRNA species directed against Cldn1 and Cldn4 caused reduced expression of mRNA to 24% and 15%, respectively.
To further investigate a possible interdependence of these two genes, the expression of Cldn4 and Cldn1 was investigated in CC531si.Cldn1 cells and CC531si.Cldn4 cells, respectively. Expression of Cldn4 was down-regulated by 50% in tumour cells transfected with siRNA against Cldn1 (Fig. 3D), whereas inhibition of Cldn4 did not exert the same effect on Cldn1 expression (data not shown).
Effect of Cldn1 and Cldn4 knockdown on cell growth
The effect of Cldn1 or Cldn4 inhibition on the proliferation and growth of CC531 cells was studied by MTT test. A non significant reduction in proliferation of CC531 cells was noticed (Fig. 4A).
Fig 4.
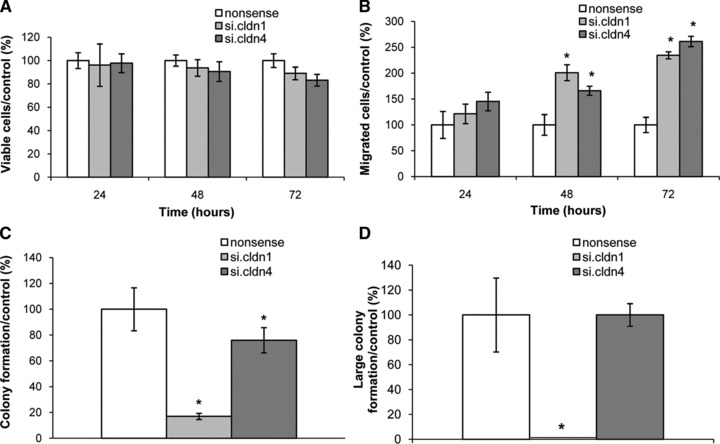
Knockdown effects of Cldn1 and Cldn4 on cellular functions of CC531 cells. (A) Proliferation of CC531 cells in response to si.Cldn1 or si.Cldn4. (B) Increased migration of CC531 cells in response to si.Cldn1 or si.Cldn4. (C) Inhibition of colony formation of CC531 cells in response to siRNA down-regulation of claudins (1, 4). (D) Inhibition of large colony formation of CC531 cells in response to siRNA down-regulation of Cldn1 or Cldn4. Data (n= 3) are shown as means ± S.D. in percentage of nonsense-transfected cells, an asterisk denotes a significant difference to control cells (P < 0.05).
Impact on cell migration caused by inhibition of Cldn1 or Cldn4
Figure 4B shows a 21% increase in tumour cell migration caused by inhibition of Cldn1 after 24 hrs, which further increased to 101% after 48 hrs and 135% after 72 hrs (P < 0.05). Similarly, inhibition of Cldn4 caused significantly (P < 0.05) increased tumour cell migration also after 48 and 72 hrs (66% and 161%, respectively; Fig. 4B).
Impact on colony formation after Cldn1 or Cldn4 knockdown
As shown in Figure 4C, the inhibition of colony formation differed following down-regulation of Cldn1 and Cldn4. The former caused a significant decrease in colony formation (by 83%, P < 0.05) and completely inhibited the formation of large colonies. Compared to this pronounced effect, down-regulation of Cldn4 caused minor (24%), but still significant (P < 0.05), inhibition of colony formation and did not affect the formation of large colonies (Fig. 4C and D).
Expression of CLDN1 and CLDN4 in neoplastic human CRC tissues
The expression of CLDN1 and CLDN4 was investigated in 32 human colorectal adenocarcinoma specimens (Table 2). The patients had a median age of 65 years and were classified into UICC stages II (n= 24) and IV (n= 8) and graded as G2 (n= 25) and G3 (n= 7) (Table 2). The histopathological analysis revealed that the expression of CLDN1 was high in 91% (n= 30) and that of CLDN4 in 85% (n= 28) of all tumour specimens. Comparing the CLDN expression related to UICC stages, CLDN1 and CLDN4 had significantly lower expression in stage IV than in stage II (P= 0.01 and P= 0.05, respectively). In line with this, liver metastases showed lower expression of CLDN1 and CLDN4 than in the corresponding primary carcinomas (Fig. 5). This difference was significant for CLDN1 (P < 0.05) but not for CLDN4
Table 2.
Pathological evaluation of immunohistochemical staining for the 32 CRC samples and 8 corresponding liver metastasis
| TNM Classification | CLDN1 | CLDN4 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Primary tumour | Liver metastasis | Primary tumour | Liver metastasis | ||||||||||
| pT3 pN2 pM1, G3 | ++ | (+) | n.i. | + | |||||||||
| pT4 pN2 pM1, G2 | ++ | (+/−) | ++ | +/− | |||||||||
| pT3 pN1 pM1, G2 | + | (+/−) | (+/−) | + | |||||||||
| pT4 pN1 pM1, G3 | + | + | ++ | +/− | |||||||||
| pT3 pN1 pM1, G3 | ++ | (+/−) | + | (+/−) | |||||||||
| pT3 pN0 pM1, G3 | ++ | (+/−) | + | (+/−) | |||||||||
| pT4 pN2 pM1, G3 | - | (+/−) | + | - | |||||||||
| pT4 pN2 pM1, G2 | (+/−) | +/− | +/− | +/− | |||||||||
| pT3 pN0, G2 | ++ | / | + | / | |||||||||
| pT3 pN0, G2 | ++ | / | ++ | / | |||||||||
| pT3 pN0, G2 | ++ | / | ++ | / | |||||||||
| pT3 pN0, G3 | ++ | / | ++ | / | |||||||||
| pT3 pN0, G2 | ++ | / | ++ | / | |||||||||
| pT3 pN0, G2 | ++ | / | ++ | / | |||||||||
| pT3 pN0, G2 | ++ | / | ++ | / | |||||||||
| pT3 pN0, G2 | ++ | / | ++ | / | |||||||||
| pT3 pN0, G2 | ++ | / | +/− | / | |||||||||
| pT3 pN0, G2 | ++ | / | ++ | / | |||||||||
| pT3 pN0, G3 | ++ | / | + | / | |||||||||
| pT3 pN0, G2 | ++ | / | ++ | / | |||||||||
| pT3 pN0, G2 | ++ | / | ++ | / | |||||||||
| pT3 pN0, G2 | ++ | / | ++ | / | |||||||||
| pT3 pN0, G2 | ++ | / | ++ | / | |||||||||
| pT3 pN0, G2 | ++ | / | ++ | / | |||||||||
| pT3 pN0, G2 | ++ | / | + | / | |||||||||
| pT3 pN0, G2 | ++ | / | ++ | / | |||||||||
| pT3 pN0, G2 | ++ | / | ++ | / | |||||||||
| pT3 pN0, G2 | ++ | / | + | / | |||||||||
| pT3 pN0, G2 | ++ | / | ++ | / | |||||||||
| pT3 pN0, G2 | ++ | / | + | / | |||||||||
| pT3 pN0, G2 | ++ | / | ++ | / | |||||||||
| pT3 pN0, G2 | ++ | / | ++ | / | |||||||||
++: strong positive; +: positive; (+): faint positive; +/−: focal positive; (+/−): focal faint positive; n.i.: not available.
Fig 5.
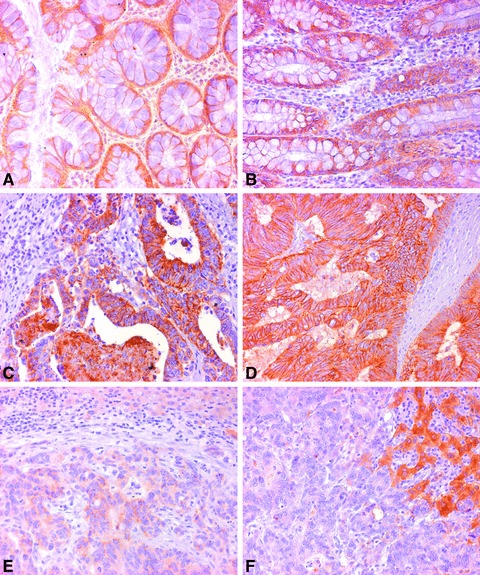
Expression of CLDN1 and CLDN4 proteins in human CRC and liver metastasis tissues compared to normal mucosa as shown by IHC. (A), (C) and (E) Expression of CLDN1 in normal mucosa, cancerous tissue and liver metastasis, respectively. (B), (D) and (F) expression of CLDN4 in normal mucosa, cancerous tissue and liver metastasis, respectively. Magnification ×64.
Correlation of CLDN4 or CLDN1 expression with prognosis in CRC patients
The 67 CRC patients (42 men and 25 women) had an average age of 67 years. All lesions were adenocarcinomas and were classified into four UICC stages (I, n= 11; II, n= 25; III, n= 20; IV, n= 11). The expression levels of CLDN1 and CLDN4 were significantly correlated (P < 0.05; Fig. 6A). No correlation between CLDN1 expression and age (P= 0.19), tumour stage (P= 0.88) or overall survival (P= 0.2) was seen. With respect to CLDN4, also no correlation with age (P= 0.69) or tumour stage (P= 0.38) was noticed. However, the overall survival of CRC patients with high or low risk in relation to the median split of CLDN4 expression levels differed significantly according to the log-rank test (P= 0.018; Fig. 6B). Similarly, using a univariate Cox model, there was an almost significant difference (P= 0.07) between the low- and high-risk groups taking all stages together, whereas in patients with tumour stages I–III, an elevated CLDN4 level was clearly associated with a less favourable prognosis (P= 0.05; Fig. 6C).
Fig 6.
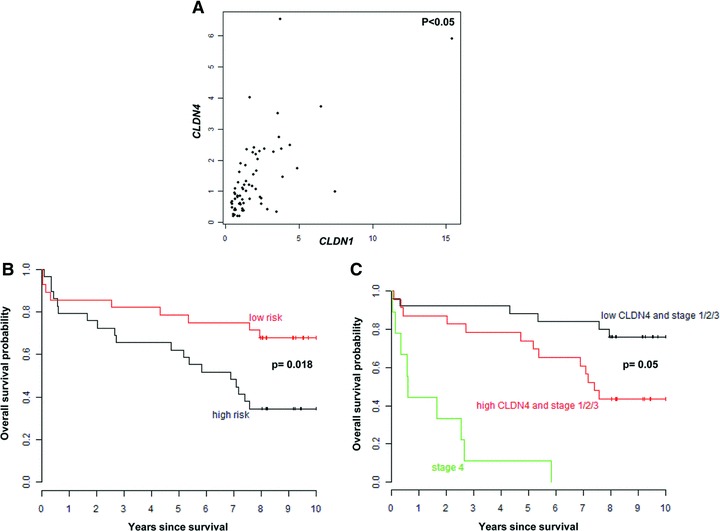
Correlation of CLDN1 or CLDN4 expression levels with prognosis in 67 CRC patients and with each other. (A) Scatterplot of the correlation between CLDN1 and CLDN4 expression levels assessed with the non-parametric correlation coefficient from Spearman. (B) The Kaplan–Meier plot represents the overall survival probability of CRC patients with high or low risk according to CLDN4 Median Split for overall survival. The log-rank test shows a significant difference (P= 0.018) between the two groups. (C) The Kaplan–Meier plot demonstrates the overall survival probability of CRC patients with high or low CLDN4 expression, dichotomized into high- and low-risk groups by the CLDN4 Median Split for overall survival and after separating the stages into I–III and IV stages. The Cox model shows a significant association between CLDN4 elevated levels and reduced overall survival (P= 0.05).
Discussion
Liver metastasis is a salient problem for patients with CRC. A comparison of the histological aspects of metastases with their primary carcinoma shows a high degree of similarity, as does the genetic signatures of tumour suppressors and oncogenes between the respective tumour origin and spread [5]. We reasoned that certain features that enable CRC cells to metastasize are activated for a relatively short period during invasion of the liver. Therefore, we investigated temporal changes in gene expression when CRC cells home to the liver. These investigations found a transient down-regulation of genes known to be essential for cell–cell contacts. In the first few days of metastasis establishment, a considerable fraction (25%) of the claudin family of genes were significantly down-regulated (>5-fold), followed by return to original levels over the next days. When exploring the causes of this down-regulation, neither co-culture with hepatocytes nor with Kupffer cells, which compose the vast majority of cells in the liver, was able to elicit down-regulation of claudins. However, exposure of tumour cells to the physical forces involved in haematogenic spread of the primary tumour to the liver showed a direct effect on the expression of Cldn4, but not of Cldn1. This notwithstanding, Cldn1 and Cldn4 share a common transcription repressor, Snail [41, 49], with the initial up-regulation of Snail in freshly metastatic CC531 cells consistent with a reduction in the expression of Cldn1 and Cldn4. In an attempt to model down-regulation of claudin in vitro, we used siRNA silencing to investigate phenotypic effects on cellular proliferation, migration and the ability to achieve colony formation. Although cellular proliferation was not affected, migration was increased and colony formation significantly decreased.
Down-regulation of claudins and the resulting effects may be related to initial events in the metastasis process: prior to dissemination, tumour cells need to overcome the restriction on movement resulting from cell–cell contacts and to leave the tumour of origin; these events are mirrored by the reduction in Cldn1 and Cldn4 and in an increased migration capacity. Furthermore, during dissemination, colony formation is not required until metastatic cells have initiated growth within their target organ. In line with these experimental data, a decrease in CLDN1 and CLDN4 expression was seen in the tumours of stage IV CRC patients, which is defined by overt metastasis. Liver metastasis had the lowest claudin expression of all CRC stages investigated. Additionally, elevated CLDN4 levels were associated with less favourable prognosis in stage I–III patients, suggesting that intensive expression of CLDN4 is a potential risk factor of progression in CRC. We propose the following hypothesis to explain early and late stages of cancer progression. Initially, high expression of Cldn1 and Cldn4 is a molecular prerequisite for increased TJs, which results in an increase in the strength of interaction between cells. This conclusion is supported by the observation that CLDN1-transfected-T84 cells aggregate and form multilayers to a greater extent than the parent T84 cells [36]. It remains unclear why an increase in association between cancer cells promotes aggressiveness as implied by a reduction in the overall survival of the stage I–III patients expressing high levels of CLDN1 and CLDN4. This could be due to increased clonogenicity resulting from elevated claudin (1, 4) expression as silencing of Cldn1 ([37] and this article) and Cldn4 (this article) results in reduced colony formation. In addition, enhanced Cldn1 expression may impact on pathways to enhance cancer growth. For example, activation of the β-catenin-T cell-specific transcription factor/lymphoid enhancer-binding factor (Tcf/LEF) transcriptional complex, known to be frequently disrupted in CRC, was responsible for Cldn1 up-regulation in the primary tumour [40]. Increased Cldn1, in turn, inhibits the tumour suppressor activity of membrane-localized E-cadherin, leading to activation of the β-catenin-Tcf/LEF pathway. In late stages of cancer progression, a reduction in intercellular interactions becomes essential for tumour cell dissemination. Correspondingly, reduced, claudin expression promotes detachment, migration and metastasis formation. In agreement with this model, a transient loss of the intestinal homeobox transcription factor CDX2 was observed in invasive CRC cells at the tumour host interface [50]. CDX2 was found to regulate directly the expression of intestinal claudins (CLDN3 and CLDN4) [51].
With respect to a possible crosstalk between Cldn1 and Cldn4, we show that siRNA-mediated inhibition of Cldn1 induced parallel inhibition of Cldn4, but not vice versa, suggesting cross-regulation between these two claudins in CRC cells. In view of the clear effect of si.Cldn4 on migration, it is unclear whether the effect of si.Cldn1 should be attributed to a modulation of Cldn1 or Cldn4, or both. However, colony formation was far less inhibited by si.cldn4 and therefore, a combination effect of si.cldn1 and si.cldn4 can be excluded.
Our results suggest that Cldn4 is the major player between Cldn1 and Cldn4. For example, in CC531 cells homing into the rat liver, Cldn4 changed to a greater extent than Cldn1. Furthermore, only Cldn4 responded to cell culture conditions favouring adhesion in our experiments. Nevertheless, there was a significant correlation between the two claudins in primary CRC samples. As described by other reports, altered CLDN1 and CLDN4 levels induce different effects: silencing of CLDN1 in SW620 metastatic human CRC cells resulted in a significantly decreased invasive capacity of SW620siRNA cells using the Boyden chamber invasion assay [37]. However, siRNA-mediated CLDN4 knockdown in SW480 CRC cells increased their invasive activity using a modified two-chamber invasion assay [34].
Intriguingly, the occurrence of CLDN1 and CLDN4 in human CRC tumours, as determined by IHC and RT-PCR, is in partial concordance with the results obtained from the rat in vivo model; CLDN1 and CLDN4 were up-regulated in primary tumour tissues but under-expressed in liver metastasis samples (as shown by IHC). Liver metastases have a tendency to colonize the whole organ. To facilitate their detaching, it seems plausible that their claudin levels are partly reduced. These changes in expression were conceivably pronounced in our rat model, because all cells were forced to adapt to the liver environment after inoculation. This encourages us to suggest our rat model as a helpful tool for studying the gene expression profile of claudins in tumour cells between their homing into the liver and the occurrence of clinical liver metastasis.
Based on the results obtained from this study, hepatocytes and Kupffer cells can be excluded as the main cause of the initial down-regulation of Cldn1 and Cldn4 during early metastatic events. Physical forces could be suggested as a reason for down-regulation of Cldn4, but not of Cldn1. However, two possibilities remain. First, whether or not immunological factors may play a role in this effect and second, if interaction with other liver cell types, such as stellate cells or endothelial cells, could influence claudin levels. Despite these open questions, Cldn1 and Cldn4 seem to play a crucial role within the early metastatic process and therefore deserve more attention. Further studies are required to address how the transient down-regulation of Cldn1 and Cldn4 can be prevented in patients and the influence this would have on the occurrence of liver metastasis. Such an experiment could be based on CC531 cells into which a miRNA against Cldn1 and/or Cldn4 has been transfected stably under the control of a system allowing their conditional silencing. This model would allow a final proof of the investigated proteins’ importance and is planned in future experiments.
In summary, we show that the initial phase of rat CRC cells homing to liver involves a transient down-regulation of Cldn1 and in particular of Cldn4. The transcription repressor Snail, which regulates both claudins, was concomitantly up-regulated during the early stages of metastasis before returning to normal expression levels. Silencing of Cldn1 and Cldn4 by siRNA increased migration and reduced colony formation, with these phenotypes consistent with metastatic homing. These model results were paralleled in human CRC tumour samples, which show increased CLDN1 and CLDN4 expression in UICC stages I–III, and significantly reduced expression in stage IV and in liver metastasis. The results obtained with human specimens give first evidence of a modulated claudin expression similar to those in the rat model. However, a prospective study is needed to corroborate these results, taking into account separately the entities, colonic and rectal cancers. That research could be driven by our hypothesis that primary CRC tumours have an initial growth advantage from increased claudin expression, whereas metastasizing cells require a transient reduction in claudin expression to be liberated from the primary tumour and to then initiate metastatic growth in the liver.
Acknowledgments
R.G. was supported by a scholarship from Al-Baath University (Syria). We thank Dr. George Reid for language revision and Dr. Bernhard Korn for microarray analysis.
Conflict of interest
The authors confirm that there are no conflicts of interest.
Supporting Information
(A--E) Photographs of rat liver taken at3, 6, 9, 14 and 21 days after inoculation of CC531 cells, beforere-isolation of the metastatic tumour cells.
Fig. S2 The density plot illustrates the CC531 population which was obtained by FACS sorting. The marker protein RFP was used for isolating CC531 cells without contaminating liver cells.
The sequences of human and rat claudin1/claudin4 primers and siRNAs as well as primary and secondary antibodies used for detection of CLDN1, CLDN4 and ERK2 proteins
References
- 1.Mathers C, Fat DM, et al. WHO. The global burden of desease: 2004 update. Geneva: World Health Organization; 2008. pp. 7–22. [Google Scholar]
- 2.Hess KR, Varadhachary GR, SH Taylor, et al. Metastatic patterns in adenocarcinoma. Cancer J. 2006;106:1624–33. doi: 10.1002/cncr.21778. [DOI] [PubMed] [Google Scholar]
- 3.Garden OJ, Rees M, Poston GJ, et al. Guidelines for resection of colorectal cancer liver metastases. Gut. 2006;55:iii1–8. doi: 10.1136/gut.2006.098053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 5.Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9:274–84. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 6.Ferrara N. Molecular and biological properties of vascular endothelial growth factor. J Mol Med. 1999;77:527–43. doi: 10.1007/s001099900019. [DOI] [PubMed] [Google Scholar]
- 7.de Castro-Carpeno J, Belda-Iniesta C, Casado Saenz E, et al. EGFR and colon cancer: a clinical view. Clin Transl Oncol. 2008;10:6–13. doi: 10.1007/s12094-008-0147-3. [DOI] [PubMed] [Google Scholar]
- 8.Zeng ZS, Shu WP, Cohen AM, et al. Matrix metalloproteinase-7 expression in colorectal cancer liver metastases: evidence for involvement of MMP-7 activation in human cancer metastases. Clin Cancer Res. 2002;8:144–8. [PubMed] [Google Scholar]
- 9.Soler AP, Miller RD, Laughlin KV, et al. Increased tight junctional permeability is associated with the development of colon cancer. Carcinogenesis. 1999;20:1425–31. doi: 10.1093/carcin/20.8.1425. [DOI] [PubMed] [Google Scholar]
- 10.Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol. 2001;2:285–93. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- 11.Van Itallie CM, Anderson JM. Claudins and epithelial paracellular transport. Annu Rev Physiol. 2006;68:403–29. doi: 10.1146/annurev.physiol.68.040104.131404. [DOI] [PubMed] [Google Scholar]
- 12.Mitic LL, Anderson JM. Molecular architecture of tight junctions. Annu Rev Physiol. 1998;60:121–42. doi: 10.1146/annurev.physiol.60.1.121. [DOI] [PubMed] [Google Scholar]
- 13.Furuse M, Sasaki H, Tsukita S. Manner of interaction of heterogeneous claudin species within and between tight junction strands. J Cell Biol. 1999;147:891–903. doi: 10.1083/jcb.147.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartsock A, Nelson WJ. Adherens and tight junctions: structure, function and connections to the actin cytoskeleton. Biochim Biophys Acta. 2008;1778:660–9. doi: 10.1016/j.bbamem.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angelow S, Ahlstrom R, Yu AS. Biology of claudins. Am J Physiol-Renal Physiol. 2008;295:F867–76. doi: 10.1152/ajprenal.90264.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furuse M, Tsukita S. Claudins in occluding junctions of humans and flies. Trends Cell Biol. 2006;16:181–8. doi: 10.1016/j.tcb.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Niessen CM. Tight junctions/adherens junctions: basic structure and function. J Invest Dermatol. 2007;127:2525–32. doi: 10.1038/sj.jid.5700865. [DOI] [PubMed] [Google Scholar]
- 18.Morin PJ. Claudin proteins in human cancer: promising new targets for diagnosis and therapy. Cancer Res. 2005;65:9603–6. doi: 10.1158/0008-5472.CAN-05-2782. [DOI] [PubMed] [Google Scholar]
- 19.Swisshelm K, Macek R, Kubbies M. Role of claudins in tumorigenesis. Adv Drug Deliv Rev. 2005;57:919–28. doi: 10.1016/j.addr.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Hough CD, Sherman-Baust CA, Pizer ES, et al. Large-scale serial analysis of gene expression reveals genes differentially expressed in ovarian cancer. Cancer Res. 2000;60:6281–7. [PubMed] [Google Scholar]
- 21.Michl P, Buchholz M, Rolke M, et al. Claudin-4: a new target for pancreatic cancer treatment using Clostridium perfringens enterotoxin. Gastroenterology. 2001;121:678–84. doi: 10.1053/gast.2001.27124. [DOI] [PubMed] [Google Scholar]
- 22.Long H, Crean CD, Lee WH, et al. Expression of Clostridium perfringens enterotoxin receptors claudin-3 and claudin-4 in prostate cancer epithelium. Cancer Res. 2001;61:7878–81. [PubMed] [Google Scholar]
- 23.Sanada Y, Oue N, Mitani Y, et al. Down-regulation of the claudin-18 gene, identified through serial analysis of gene expression data analysis, in gastric cancer with an intestinal phenotype. J Pathol. 2006;208:633–42. doi: 10.1002/path.1922. [DOI] [PubMed] [Google Scholar]
- 24.Cheung ST, Leung KL, Ip YC, et al. Claudin-10 expression level is associated with recurrence of primary hepatocellular carcinoma. Clin Cancer Res. 2005;11:551–6. [PubMed] [Google Scholar]
- 25.Kominsky SL, Argani P, Korz D, et al. Loss of the tight junction protein claudin-7 correlates with histological grade in both ductal carcinoma in situ and invasive ductal carcinoma of the breast. Oncogene. 2003;22:2021–33. doi: 10.1038/sj.onc.1206199. [DOI] [PubMed] [Google Scholar]
- 26.Johnson AH, Frierson HF, Zaika A, et al. Expression of tight-junction protein claudin-7 is an early event in gastric tumorigenesis. Am J Pathol. 2005;167:577–84. doi: 10.1016/S0002-9440(10)62999-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinugasa T, Huo Q, Higashi D, et al. Selective up-regulation of claudin-1 and claudin-2 in colorectal cancer. Anticancer Res. 2007;27:3729–34. [PubMed] [Google Scholar]
- 28.Usami Y, Chiba H, Nakayama F, et al. Reduced expression of claudin-7 correlates with invasion and metastasis in squamous cell carcinoma of the esophagus. Hum Pathol. 2006;37:569–77. doi: 10.1016/j.humpath.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 29.Kominsky SL, Argani P, Korz D, et al. Loss of the tight junction protein claudin-7 correlates with histological grade in both ductal carcinoma in situ and invasive ductal carcinoma of the breast. Oncogene. 2003;22:2021–33. doi: 10.1038/sj.onc.1206199. [DOI] [PubMed] [Google Scholar]
- 30.Sauer T, Pedersen MK, Ebeltoft K, et al. Reduced expression of Claudin-7 in fine needle aspirates from breast carcinomas correlate with grading and metastatic disease. Cytopathology. 2005;16:193–8. doi: 10.1111/j.1365-2303.2005.00257.x. [DOI] [PubMed] [Google Scholar]
- 31.Michl P, Barth C, Buchholz M, et al. Claudin-4 expression decreases invasiveness and metastatic potential of pancreatic cancer. Cancer Res. 2003;63:6265–71. [PubMed] [Google Scholar]
- 32.Rangel LB, Agarwal R, D’Souza T, et al. Tight junction proteins claudin-3 and claudin-4 are frequently overexpressed in ovarian cancer but not in ovarian cystadenomas. Clin Cancer Res. 2003;9:2567–75. [PubMed] [Google Scholar]
- 33.Soini Y. Expression of claudins 1, 2, 3, 4, 5 and 7 in various types of tumours. Histopathology. 2005;46:551–60. doi: 10.1111/j.1365-2559.2005.02127.x. [DOI] [PubMed] [Google Scholar]
- 34.Ueda J, Semba S, Chiba H, et al. Heterogeneous expression of claudin-4 in human colorectal cancer: decreased claudin-4 expression at the invasive front correlates cancer invasion and metastasis. Pathobiology. 2007;74:32–41. doi: 10.1159/000101049. [DOI] [PubMed] [Google Scholar]
- 35.de Oliveira SS, de Oliveira IM, De Souza W, et al. Claudins upregulation in human colorectal cancer. FEBS Lett. 2005;579:6179–85. doi: 10.1016/j.febslet.2005.09.091. [DOI] [PubMed] [Google Scholar]
- 36.Huo Q, Kinugasa T, Wang L, et al. Claudin-1 protein is a major factor involved in the tumorigenesis of colorectal cancer. Anticancer Res. 2009;29:851–7. [PubMed] [Google Scholar]
- 37.Dhawan P, Singh AB, Deane NG, et al. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J Clin Invest. 2005;115:1765–76. doi: 10.1172/JCI24543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takehara M, Nishimura T, Mima S, et al. Effect of claudin expression on paracellular permeability, migration and invasion of colonic cancer cells. Biol Pharm Bull. 2009;32:825–31. doi: 10.1248/bpb.32.825. [DOI] [PubMed] [Google Scholar]
- 39.Jakab C, Rusvai M, Galfi P, et al. Expression of claudin-1, -3, -4, -5 and -7 proteins in low grade colorectal carcinoma of canines. Histol Histopathol. 2010;25:55–62. doi: 10.14670/HH-25.55. [DOI] [PubMed] [Google Scholar]
- 40.Miwa N, Furuse M, Tsukita S, et al. Involvement of claudin-1 in the beta-catenin/Tcf signaling pathway and its frequent upregulation in human colorectal cancers. Oncol Res. 2001;12:469–76. doi: 10.3727/096504001108747477. [DOI] [PubMed] [Google Scholar]
- 41.Ikenouchi J, Matsuda M, Furuse M, et al. Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. J Cell Sci. 2003;116:1959–67. doi: 10.1242/jcs.00389. [DOI] [PubMed] [Google Scholar]
- 42.Escaffit F, Boudreau F, Beaulieu JF. Differential expression of claudin-2 along the human intestine: implication of GATA-4 in the maintenance of claudin-2 in differentiating cells. J Cell Physiol. 2005;203:15–26. doi: 10.1002/jcp.20189. [DOI] [PubMed] [Google Scholar]
- 43.Sakaguchi T, Gu X, Golden HM, et al. Cloning of the human claudin-2 5’-flanking region revealed a TATA-less promoter with conserved binding sites in mouse and human for caudal-related homeodomain proteins and hepatocyte nuclear factor-1alpha. J Biol Chem. 2002;277:21361–70. doi: 10.1074/jbc.M110261200. [DOI] [PubMed] [Google Scholar]
- 44.Marquet RL, Westbroek DL, Jeekel J. Interferon treatment of a transplantable rat colon adenocarcinoma: importance of tumour site. Int J Cancer. 1984;33:689–92. doi: 10.1002/ijc.2910330521. [DOI] [PubMed] [Google Scholar]
- 45.Wittmer A, Khazaie K, Berger MR. Quantitative detection of lac-Z-transfected CC531 colon carcinoma cells in an orthotopic rat liver metastasis model. Clin Exp Metastasis. 1999;17:369–76. doi: 10.1023/a:1006643831825. [DOI] [PubMed] [Google Scholar]
- 46.Eyol E, Boleij A, Taylor RR, et al. Chemoembolisation of rat colorectal liver metastases with drug eluting beads loaded with irinotecan or doxorubicin. Clin Exp Metastasis. 2008;25:273–82. doi: 10.1007/s10585-008-9142-x. [DOI] [PubMed] [Google Scholar]
- 47.Georges R, Adwan H, Zhivkova M, et al. Regulation of osteopontin and related proteins in rat CC531 colorectal cancer cells. Int J Oncol. 2010;37:249–56. [PubMed] [Google Scholar]
- 48.Adwan H, Bauerle TJ, Berger MR. Downregulation of osteopontin and bone sialoprotein II is related to reduced colony formation and metastasis formation of MDA-MB-231 human breast cancer cells. Cancer Gene Ther. 2004;11:109–20. doi: 10.1038/sj.cgt.7700659. [DOI] [PubMed] [Google Scholar]
- 49.Martinez-Estrada OM, Culleres A, Soriano FX, et al. The transcription factors Slug and Snail act as repressors of Claudin-1 expression in epithelial cells. Biochem J. 2006;394:449–57. doi: 10.1042/BJ20050591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brabletz T, Spaderna S, Kolb J, et al. Down-regulation of the homeodomain factor Cdx2 in colorectal cancer by collagen type I: an active role for the tumour environment in malignant tumour progression. Cancer Res. 2004;64:6973–7. doi: 10.1158/0008-5472.CAN-04-1132. [DOI] [PubMed] [Google Scholar]
- 51.Satake S, Semba S, Matsuda Y, et al. Cdx2 transcription factor regulates claudin-3 and claudin-4 expression during intestinal differentiation of gastric carcinoma. Pathol Int. 2008;58:156–63. doi: 10.1111/j.1440-1827.2007.02204.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A--E) Photographs of rat liver taken at3, 6, 9, 14 and 21 days after inoculation of CC531 cells, beforere-isolation of the metastatic tumour cells.
Fig. S2 The density plot illustrates the CC531 population which was obtained by FACS sorting. The marker protein RFP was used for isolating CC531 cells without contaminating liver cells.
The sequences of human and rat claudin1/claudin4 primers and siRNAs as well as primary and secondary antibodies used for detection of CLDN1, CLDN4 and ERK2 proteins


