Abstract
Autophagy is an intracellular process in which a cell digests its own constituents via lysosomal degradative pathway. Though autophagy has been shown in several cardiac diseases like heart failure, hypertrophy and ischaemic cardiomyopathy, the role and the regulation of autophagy is still largely unknown. Bcl-2-associated athanogene (BAG-1) is a multifunctional pro-survival molecule that binds with Hsp70/Hsc70. In this study, myocardial adaptation to ischaemia by repeated brief episodes of ischaemia and reperfusion (I/R) prior to lethal I/R enhanced the expression of autophagosomal membrane specific protein light chain 3 (LC3)-II, and Beclin-1, a molecule involved in autophagy and BAG-1. Autophagosomes structures were found in the adapted myocardium through electron microscopy. Co-immunoprecipitation and co-immunofluorescence analyses revealed that LC3-II was bound with BAG-1. Inhibition of autophagy by treating rats with Wortmannin (15 μg/kg; intraperitoneally) abolished the ischaemic adaptation-induced induction of LC3-II, Beclin-1, BAG-1 and cardioprotection. Intramyocardial injection of BAG-1 siRNA attenuated the induction of LC3-II, and abolished the cardioprotection achieved by adaptation. Furthermore, hypoxic adaptation in cardiac myoblast cells induced LC3-II and BAG-1. BAG-1 siRNA treatment attenuated hypoxic adaptation-induced LC3-II and BAG-1, and abolished improvement in cardiac cell survival and reduction of cell death. These results clearly indicate that myocardial protection elicited by adaptation is mediated at least in part via up-regulation of autophagy in association with BAG-1 protein.
Keywords: autophagy, ischaemia, reperfusion, cardiac adaptation, BAG-1
Introduction
Autophagy is an intracellular process in which cell digests its own constituents via the lysosomal degradative pathway [1]. Autophagy is a major pathway for degrading excess or damaged organelles and proteins [2–4]. It has been shown that autophagy is induced under various conditions like starvation [5], oxidative stress [4, 6] and developmental cues [7]. While proliferating cells can remove the ineffective mitochondria and lysosomes by cell division, cardiac myocytes cannot do so because they are terminally differentiated, and they maintain homeostasis only by activation of degrading pathways such as macroautophagy, microautophagy and chaperone-mediated autophagy [8]. Autophagy has been seen in myocardial aging [2] and in several cardiac diseases including heart failure [9], hypertrophy [10] and ischaemic cardiomyopathy [11]. Inefficient or lack of autophagy causes the development of age-related disorders and poor performance of the myocardium [12]. Recently, autophagy is shown as an adaptive response in failing hearts for protecting the myocardium from haemodynamic stress [13] and shown to render cell survival in the myocardium against ischaemia reperfusion (I/R) injury [14]. In another study, it is shown that cardiac ischaemia-induced autophagy is protective, but reperfusion-induced autophagy is detrimental causing cell death [15]. Nevertheless, the role of autophagy in the myocardium, i.e.whether it mediate cell survival or cell death is controversial [5, 12–16], and also the regulation and the underlying signalling mechanisms of autophagy are still poorly understood. Cardiac adaptation to ischaemic stress achieved via repeated brief periods of I/R is a well-known strategy for the protection of the myocardium against subsequent I/R injury [17]. Myocardial adaptation is known to induce redox signalling, which converts death signal into survival signal [17]. In the present study, in order to evaluate the role of autophagy in the myocardium, we evaluated the function of autophagy in cardiac ischaemic adaptation against I/R injury. Although I/R injury is shown to induce autophagy in the myocardium [14, 15], the role of autophagy during cardiac I/R remains controversial with respect to the crucial question whether autophagy contributes to cardiac cell survival or cell death. In order to address this question, we evaluated the function of autophagy during myocardial adaptation, which is a state-of-the-art technique to protect the myocardium against the subsequent I/R injury [17]. Cardiac adaptation to ischaemic stress induces the production of reactive oxygen species, which in turn potentiates the redox signalling and activates redox sensitive transcription factors and survival proteins [17].
Several stress conditions like heat shock, oxidative stress, etc. cause damage to a number of proteins, which are recognized by molecular chaperones, leading to the activation of either cellular refolding processes or the degradation machinery directed to the destruction of damaged proteins [18, 19]. However, molecular mechanisms underlying the cooperation between the molecular chaperones and the degradation machinery remain largely unknown. Bcl-2-associated athanogene (BAG-1) is a multifunctional protein, acts as a co-chaperone, and exerts many of its actions via binding with chaperone molecules heat shock protein 70 (Hsp70) and heat shock cognate protein (Hsc70) [20, 21]. BAG-1 is known to suppress apoptosis induced by various stimuli including serum withdrawal, heat shock and radiation – where BAG-1 binds with Bcl-2 and prevent the release of pro-apoptotic factors [22]. BAG-1 protects cardiac myocytes from simulated I/R injury [23]. In addition, BAG-1 directly binds with the components of ubiquitinylation machinery and helps proteasome-mediated degradation [24]. Recently, it has been shown that autophagy is being regulated by Bcl-2 binding molecules [25], and in another study, BAG-1 has been identified in the molecular chaperone complex at the lysosomal membrane required for protein degradation [26]. Very recently, it has been demonstrated that the autophagosomal membrane contains significantly higher amount of Hsc70 proteins [27]. Based on the above reports, in the present study, we sought to evaluate the possible role of BAG-1 in the regulation of autophagy.
When autophagy is induced, phagophore [otherwise known as isolation membrane] formation occurs via Beclin-1-dependent mechanism [28, 29]. In a separate pathway, processing of microtubule-associated protein light chain 3 (LC3) is initiated, where LC3 is cleaved to form LC3-I, which is further processed to form LC3-II [28, 29]. LC3-II is being incorporated into the isolation membrane, which surrounds the autophagic substrates like damaged proteins or organelles [28, 29]. The final structure is known as autophagosome. Autophagosomes then fuse with lysosomes for the digestion of autophagic substrates [28, 29]. The autophagosomal membrane bound LC3-II is being used as a specific marker to study autophagy [30, 31]. In mammalian cells, autophagy is shown to be activated by class III phosphatidylinositol-3 (PI-3) kinase complex containing Vps34 and Beclin-1 [28, 29]; Wortmannin, an inhibitor of class III PI-3 kinase has been used in several studies to suppress autophagy [32, 33]. In the present study, we injected rats with Wortmannin (15 μg/kg, intraperitoneally) 30 min. before isolation of heart in order to suppress the autophagy elicited by ischaemic adaptation.
Materials and methods
Animals
All animals used in this study received humane care in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals and experiments involving animals and adhered to principles stated in the Guide for the Care and Use of Laboratory Animals, NRC Publication, 1996 edition. Sprague Dawley male rats weighing between 250–300 gm were fed ad libitum regular rat chow with free access to water until the start of the experimental procedure. The rats were randomly assigned to one of the following three groups: normal control [N], I/R and ischaemic adaptation.
Isolated working heart preparation
Rats were anaesthetized with sodium pentobarbital (80 mg/kg, intraperitoneally), (Abbott Laboratories, North Chicago, IL, USA) and with the anticoagulant heparin sodium (500 IU/kg, i.v.) (Elkins-Sinn, Inc., Cherry Hill, NJ, USA). After ensuring sufficient depth of anaesthesia, thoracotomy was performed; hearts were isolated and perfused in the retrograde Langendorff mode at 37°C at a constant perfusion pressure of 100 cm of water (10 kPa) for a 5-min. washout period. The perfusion buffer used in this study consisted of a modified Krebs-Henseleit bicarbonate buffer (KHB) (in mM: sodium chloride 118, potassium chloride 4.7, calcium chloride 1.7, sodium bicarbonate 25, potassium biphosphate 0.36, magnesium sulphate 1.2 and glucose 10).
At the end of 10 min., after the attainment of steady state cardiac function, baseline functional parameters were recorded. Ischaemic adaptation group hearts were subjected to four cyclic episodes of 5 min. ischaemia each followed by 10 min. of reperfusion. I/R and ischaemic adaptation group hearts were then subjected to 30 min. of global ischaemia followed by 2 hrs of reperfusion. The first 10 min. of reperfusion was in the retrograde mode to allow for post-ischaemic stabilization and there after in the antegrade working mode to allow for assessment of functional parameters, which were recorded at 30-, 60- and 120-min. reperfusion. Control hearts were collected after 2 hrs and 45 min. retrograde perfusion in Langendorff apparatus. Wortmannin (15 μg/kg) was injected into rats via intraperitoneal route 30 min. before isolation of heart.
Isolated non-working heart and drug treatment
After anaesthetizing and heparinizing rats as mentioned under working heart preparation, hearts were isolated and perfused in Langendorff mode for 15 min. I/R group hearts were subjected to 30 min. of ischaemia followed by 2 hrs of reperfusion. Ischaemic adaptation group hearts were subjected to four cyclic episodes of 5 min. of ischaemia and 10 min. of reperfusion, followed by 30 min. of ischaemia and 2 hrs of reperfusion. Normal control hearts were collected after 10 min. of retrograde perfusion in Langendorff apparatus. Wortmannin (15 μg/kg) was injected into rats via intraperitoneal route 30 min. before isolation of heart.
Cardiac function assessment
Aortic pressure was measured using a Gould P23XL pressure transducer (Gould Instrument Systems Inc., Valley View, OH, USA) connected to a side arm of the aortic cannula. The signal was amplified using a Gould 6600 series signal conditioner and monitored on a CORDAT II real-time data acquisition and analysis system (Triton Technologies, San Diego, CA, USA). Heart rate, left ventricular developed pressure (defined as the difference of the maximum systolic and diastolic aortic pressures), and dp/dt were all derived or calculated from the continuously obtained pressure signal. Aortic flow was measured using a calibrated flow meter (Gilmont Instrument Inc., Barrington, IL, USA) and coronary flow was measured by timed collection of the coronary effluent dripping from the heart.
Infarct size estimation
At the end of reperfusion, the left ventricle was cut into transverse slices. The slices were incubated in 1% triphenyl tetrazolium solution in phosphate buffer (Na2-HPO4- 88 mM, NaH2-PO4- 1.8 mM) for 20 min. at 37°C. This procedure distinguishes necrotic tissue from viable myocardium. The slices were stored for 48 hrs in 10% buffered formalin. The heart slices were photographed and the weights of the slices were monitored. Digital images of the slices were magnified, and the area of necrosis in each slice was quantified by computerized planimetry. The risk and infarct volumes in cubic centimetre of each slice were then calculated on the basis of slice weight to remove the introduction of any errors due to non-uniformity of heart slice thickness. The risk volumes and infarct volumes of each slice were summed to obtain the risk and infarct volumes for the whole heart. Infarct size was taken to be the percent infarct volume of risk volume for any one heart.
Transmission electron microscopy (TEM)
Small tissue samples about 1 mm3 were fixed by immersion in 4% glutaraldehyde, post-fixed for 1 hr in 1% OsO4 in 0.1 M cacodylate buffer dehydrated and embedded in Epon 812 at 60°C for 48 hrs, as previously described [34, 35]. Routine 60-nm ultra-thin sections were cut and mounted on coated grids, stained with 1% uranyl acetate and Reynold's lead citrate. The TEM examination was performed with Philips CM 12 at 60 kV and digital images were recorded with Morada CCD, iTEM (Olympus Soft Imaging Solutions, Munster, Germany).
In vivo BAG-1 siRNA treatment
Survival surgeries in animals were performed as mentioned in our previous studies [36]. Briefly, the rats were anaesthetized by intraperitoneal injection of ketamine [100 mg/kg] and xylazine (2 mg/kg). Additional doses of ketamine and xylazine were given during the protocol as needed to maintain anaesthesia. Intraperitoneal dosage of buprenorphine hydrochloride (0.1 mg/kg) was given for analgesia. Before starting the surgery, rats were also given subcutaneous gentamycin (1 mg/kg). Tracheal intubation was performed with a 14-gauge catheter, and animals were artificially ventilated with mixture of air and oxygen using a respirator (Harvard-rodent ventilator model 683, South Natick, MA, USA). Respiratory rates were maintained at 70/min. and tidal volume 10 ml/kg. For direct gene delivery, a small oblique thoracotomy is performed lateral to the midsternal line in the fourth intercostal space to expose the heart. A single injection of either control or BAG-1 siRNA (100 μl of 10 μM) was delivered with a 30-gauge needle into the anterior left ventricular wall 1 mm into the myocardium at a very shallow angle (10–20°) in a cranial direction approximately 3 mm below the auricle of the left atrium between the left anterior descending artery and the left atrial branch. The chest was closed in layers with 4–0 nylon suture and animal was allowed to recover. After the operation, intraperitoneal injection of buprenorphine hydrochloride (0.1 mg/kg) for analgesia and gentamycin (1 mg/kg) was added every 12 hrs for 3 days. Forty-eight hours after survival surgery animals’ hearts were isolated and subjected to working heart studies.
Cytosolic extract preparation
About 100 mg of left ventricular tissue was homogenized in 1 ml of buffer containing 25 mM Tris, 25 mM NaCl, 1 mM sodium orthovanadate, 10 mM sodium fluoride, 10 mM sodium pyrophosphate, 10 nM okadaic acid, 0.5 mM ethylenediaminetetraacetic acid (EDTA), 1 mM PMSF and protease inhibitors cocktail (leupeptin, aprotinin and pepstatin). Homogenates were centrifuged at 3000 rpm for 10 min. at 4°C, and the supernatant was again centrifuged at 10,000 rpm for 20 min. at 4°C. The resultant supernatant was saved as cytosolic extract. Total protein concentration in the cytosolic extract was determined using BCA Protein Assay Kit (Pierce, Rockford, IL, USA).
Immunoprecipitation
Cytosolic extract containing 500 μg of total protein was immunoprecipitated with BAG-1 (Santa Cruz Biotechnology, Santa Cruz, CA) and protein A Sepharose beads (Zymed, San Francisco, CA, USA) for overnight at 4°C. Pellets were collected by centrifuging at 10,000 ×g for 30 sec. at 4°C, and washed three times with ice-cold wash buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and 2mM EDTA). Pellets were resuspended in 1 × Laemmli sample buffer, and boiled at 95–100°C for 10 min. Western immunoblotting was done as described below.
Western blot analysis
Cytosolic proteins were separated in SDS-PAGE and transferred to nitrocellulose filters. Filters were blocked in 5% non-fat dry milk, and probed with a primary antibody for overnight. Primary antibodies such as BAG-1, LC3, Beclin-1, Hsp70, Hsc70 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were obtained from Santa Cruz Biotechnology and used at 1:500 dilutions, except in case of GAPDH (1:1000). Caspase-3 antibody obtained from Cell Signaling Technology, Inc. (Danvers, MA, USA) and used in 1:1000 dilution. Caspase-3 antibody detects endogenous levels of full length caspase-3 (35 kD) and the cleaved fragment (17 kD) of caspase-3 resulting from apoptosis. Protein bands were identified with horseradish peroxidase-conjugated secondary antibody (1:2000 dilution) and Western blotting Luminol reagent (Santa Cruz Biotechnology). The resulting blots were digitized, subjected to densitometric scanning using a standard NIH image program and normalized against loading control.
Immunofluorescence techniques and image analysis
Heart tissue samples collected at the end of experiments were fixed in 2% buffered paraformaldehyde (pH 7.4), embedded and frozen in O.C.T. compound, and subjected to cryosectioning. The 5-μm cryosections of the obtained specimens were processed for immunofluorescence analysis. The primary antibody against (i) Bag-1 [rabbit polyclonal IgG from Santa Cruz Biotechnology, Inc., http://www.scbt.com], (ii) LC3 (goat polyclonal IgG from Santa Cruz Biotechnology, Inc.), were used in 1:250 dilution. This was followed by incubation with secondary fluorochrome-conjugated antibody and counterstaining with Heochest 33342 (Molecular Probes, Inc., Eugene OR, USA) diluted 1:1000. The secondary antibodies used were (i) ALEXA 488 conjugated goat anti-rabbit IgG (Molecular Probes, Inc.) (ii) ALEXA 594 conjugated chicken anti-IgG (Rockland, Inc., Gilbertsville, PA, USA); negative controls for non-specific binding included normal goat serum without primary antibody or with secondary antibody alone. The labelled specimens were rinsed, mounted in Gelvatol (Monsanto Corp., St. Louis, MO, USA), and cover slipped for fluorescence microscopy.
Immunofluorescence and Nomarski Interference Contrast images were obtained using Olympus AX 80 microscope (Center Valley, PA, USA), fitted with a charge-coupled device (CCD) Hamamatsu Orca camera at magnification 40×. Each of the three colour images was scanned separately, by using Olympus 4’-6-Diamidino-2-phenylindole (DAPI), fluorescein isothiocyanate (FITC), and Rhodamine filter cubes. The background fluorescence was determined for BAG-1 and LC3-II from images specimens labelled with the secondary antibodies only. Higher magnification confocal immunofluorescence microscopic images for the group IA was conducted with a 100× 1.4 numerical aperture (zoom 2) objective lens on Nikon Eclipse E800/Bio-Rad Radiance 2100 confocal microscope (Hercules, CA, USA). The projections presented are individual XY confocal images from the respective stacks taken with 0.5-μm Z-steps. Inset ‘ZX’ presented in the panel H is ZX optical re-slicing of the region of interest (ROI) indicated in the panel with arrows (Note blue channel has been excluded). Spatial co-localization of BAG-1 (green channel) and LC3-II (red channel) in the panel H (ZX) appears in yellow colour as a result of interference between green and red colours. Pearson's correlation coefficient for signals of green and red channels in the indicated area is r= 0.29. Processing and analysis of digital images, including colocalization of immunostained BAG-1 and LC3 was conducted with SimplePCI High Performance Imaging software (Compix, Inc., Hamamatsu Co., http://www.cimaging.net) and Image J software. The results are representative images of three separate experiments.
Cell culture
Rat myocardium derived H9c2 cardiac myoblast cell line was purchased from the American Type Culture Collection (ATCC; Manasses, VA, USA). Cells were grown on Dulbecco's modified Eagle's medium (DMEM, Invitrogen, Grand Island, NY, USA) containing 4 mM l-glutamine, 4.5 g/l glucose, and 10% foetal bovine serum (Invitrogen) and incubated at 37°C in a humidified chamber with 5% CO2. To prevent the loss of differentiation potential, cells were not allowed to become confluent.
In vitro siRNA transfection
To each well of six-well tissue culture plate, 2 × 105 H9c2 cells were seeded with 2 ml DMEM growth medium containing antibiotics. Plates were incubated for 18–24 hrs till it reaches 70–80% confluency. BAG-1 and control siRNAs were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA), and used at the final concentration of 25 nM. siRNA transfection was done using TransPass R2 transfection reagent (New England BioLabs) as mentioned in the kit. Cells are grown in complete growth medium containing 10% serum for an additional period of 48 hrs. Follow-up experiments are performed at the end of 48 hrs incubation period.
Hypoxia-reoxygenation in cell culture
Normal or siRNA transfected H9c2 culture plates were subjected to either 30 min. of hypoxia or 30 min. of hypoxia followed by 1 hr of reoxygenation (H/R) or hypoxic adaptation, where the cells were subjected to three cyclic episodes of 5 min. hypoxia and 5 min. reoxygenation, followed by H/R [37]. Hypoxia was given by placing the cultured plates into an air-tight hypoxic chamber placed in a humidified 37°C CO2 incubator, and passing the mixture of 95% nitrogen and 5% CO2. Reoxygenation was given by placing back the cultured plates in a humidified 37°C CO2 incubator saturated with air.
Cell survival assay
At the end of experimentation, cells were washed with phosphate buffered saline (PBS). Viability of cells was studied using MTT Cell Proliferation Assay kit (Cayman Chemical Company, Ann Arbor, MI, USA) as mentioned.
Cell death assay
Cell death analysis was performed with the spent culture medium as substrate with lactate dehydrogenase (LDH) Cytotoxicity Assay kit (Cayman Chemical Company) as mentioned.
Immunofluorescence staining in H9c2 cells
Cells were washed with PBS and fixed using 4% paraformaldehyde solution for 20 min. After washing with PBS, the slides were blocked with Powerblock (BioGenex, San Ramon, CA, USA) for 10 min. Slides were again washed with PBS and incubated with either single or multiple primary antibodies (rabbit polyclonal BAG-1; goat polyclonal LC3) used at 1:20 dilution in PBS containing 1% BSA for 2 hrs at room temperature. Slides were washed and incubated with fluorescein-conjugated secondary antibodies (anti-rabbit Alexa Fluor 488; anti-goat Alexa Fluor 594) used at 1:500 dilution in dark for 45 min. at room temperature. Nuclear staining was done with To-Pro 1 iodide [Molecular Probes, Inc.] at 1:500 dilution for 45 min. in the dark. The slides were washed and covered with mounting medium, and examined under a fluorescence microscope. Confocal microscopic images were obtained using Zeiss LSM 510 (Thornwood, NY, USA) confocal laser scanning microscope with 40× 1.3 oil immersion objective by simultaneous recording in the 488, 530 and/or 560 \ channels as appropriate.
Cell lysate preparation
At the end of experimentation, cells were washed with PBS, and lysed in radio immuno precipitation assay (RIPA) buffer (Boston Bioproducts, Boston, MA, USA) supplemented with protease inhibitor cocktail (Sigma) and a phosphatase inhibitor (100 mM sodium orthovanadate) on ice for 1 hr. Lysate was centrifuged at 10,000 rpm for 10 min. at 4°C, and the supernatant was mixed with sample buffer and heated at 95°C for 5 min., and used for Western immunoblotting.
Statistical analysis
The values for myocardial functional parameters, total and infarct volumes and infarct sizes were all expressed as the mean ± standard error of mean (S.E.M.). Analysis of variance test followed by Bonferoni's correction was first carried out to test for any differences between the mean values of all groups. If differences between established, the values of the treated groups were compared with those of the control group by a modified t-test. The results were considered significant if P < 0.05.
Results
Myocardial adaptation to ischaemic stress enhances autophagy
In this study, when isolated rat hearts were subjected to 30 min. of global ischaemia followed by 2 hrs reperfusion, autophagic marker proteins such as LC3-II and Beclin-1 were slightly induced in the myocardium relative to control rat hearts (Fig. 1A). Moreover, cardiac adaptation by subjecting the hearts to four cyclic episodes of 5 min. of global ischaemia and 10 min. of reperfusion preceding I/R injury further induced both LC3-II and Beclin-1 (Fig. 1A). Quantitative analysis of Western immunoblotting of LC3 (Fig. 1B), Beclin-1 (Fig. 1C) and BAG-1 (Fig. 1D) show that LC3, Beclin-1 and BAG-1 were significantly up-regulated in the hearts after ischaemic adaptation. Furthermore, autophagosome formation in our experimental samples was confirmed by transmission electron microscopy. TEM images show that autophagosomes formed during I/R is selective containing mitochondria (Fig. 1E). However, several autophagosomes containing partially degraded cytoplasmic materials and mitochondria were found in the myocardium adapted for ischaemic stress (Fig. 1E). These results suggest that although autophagy is slightly induced during cardiac I/R, it is significantly induced when a survival signal is generated via ischaemic stress adaptation.
1.
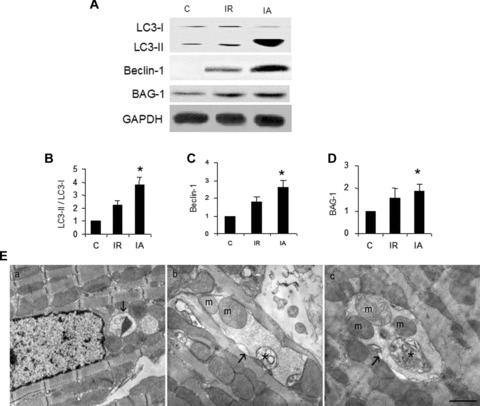
Ischaemic adaptation induces autophagy. (A) – Cytoplasmic extract from left ventricular tissue was isolated, proteins were separated by SDS-PAGE and analysed by Western immunoblotting using specific antibodies. (B-D) – Quantitative analysis of Western immunoblotting of LC3 (B), Beclin-1 (C) and BAG-1 (D); the results are represented as fold change in comparison to control heart samples. Quantitation of LC3 is calculated by analysing the expression ratio of LC3-II / LC3-I. The expression of Beclin-1 and BAG-1 is normalized with the expression of GAPDH. *P < 0.05 versus control. (E) – Transmission electron micrographs of left ventricular tissue sections showing autophagosomes, indicated by arrows in control (a) ischaemia and reperfusion (b) and ischaemic adaptation followed by I/R (c). Autophagosomes contain mitochondria (m) and partially degraded material, which is indicated by * in (b) and (c). Scale bar represents 2 μ. Results are representative images of three to five separate samples. C – Control, I/R – ischaemia and reperfusion, IA – Ischaemic adaptation followed by I/R.
Ischaemic adaptation enhanced BAG-1 in the myocardium
I/R injury slightly induced the expression of BAG-1 protein, but it was significantly elevated after ischaemic adaptation preceding I/R injury (Fig. 1A and D). These results show a positive correlation between the induction of autophagy and BAG-1 protein.
Autophagosomal membrane protein LC3 binds with BAG-1
Cytoplasmic protein extract was immunoprecipitated with BAG-1 specific antibody and the immunoprecipitate was analysed for autophagosomal membrane protein LC3-II by Western immunoblotting. We found that LC3-II protein was present in the BAG-1 immunoprecipitate, where significantly higher amount of LC3-II was bound in the BAG-1 immunoprecipitate obtained from the hearts subjected to adaptation relative to I/R (Fig. 2). Further, in order to confirm the binding of LC3 with BAG-1, we performed co-immunofluorescence analysis with BAG-1 and LC3 specific antibody using myocardial tissue sections. Figure 3 shows the staining of BAG-1 in green channel (B and F), and the staining of LC3 in red channel (C and G). The merged picture (D and H) shows the co-localization LC3 and BAG-1 in yellow. Taken together, our results indicate that BAG-1 binds with autophagosomal membrane specific protein LC3 in the cytoplasmic region.
2.
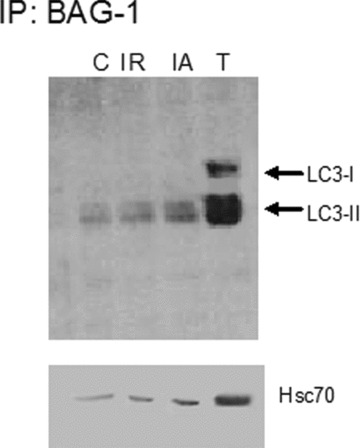
LC3-II interacts with BAG-1. Cytoplasmic extracts of the left ventricular tissue is immunoprecipitated with BAG-1 specific antibodies. BAG-1 immunoprecipitates were separated by SDS-PAGE, and Western immunoblotting was performed with specific antibodies. Results are representative images of three separate experiments. C – Control, I/R – Ischaemia and reperfusion, IA – Ischaemic adaptation followed by I/R, T – cytosolic extract from IA sample.
3.
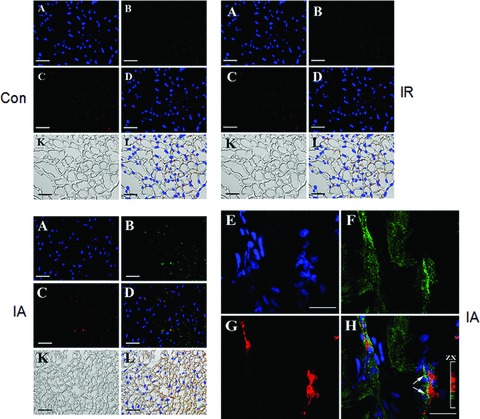
Colocalization of BAG-1 and LC3. Immunofluorescence imaging of the expression of BAG-1 (B) and LC3 (C) in tissue sections obtained from hearts subjected to ischaemia and reperfusion (I/R), ischaemic adaptation followed by ischaemia and reperfusion (IA), and from control (Con) rats. (A), (E) – counterstaining of nuclei with Hoechst 33342 (blue channel); (B), (F) – immunofluorescence staining of BAG-1 (Alexa 488, green channel); (C), (G) – immunofluorescence staining of LC3-II (Alexa 594, red channel); (D), (H) – overlay of blue, green and red channels; (K) – Nomarski interference contrast images of the specimens; L – overlay of images presented in the (D) and (K). Confocal images showing the colocalization of BAG-1 and LC3 in IA heart samples (E-H). Bars in the images (A), (B), (C), (D), (K) and (L) are representing 100 μm. Bars in the images (E)–(H) are representing 10 μm.
Inhibition of autophagy inhibits BAG-1 expression
Treatment with Wortmannin significantly attenuated adaptation-induced improvement in cardiac functional parameters such as left ventricular developed pressure and aortic flow (Fig. 4). However, heart rate and coronary flow were not significantly affected by Wortmannin treatment (Fig. 4). Also, Wortmannin treatment partially abolished the adaptation-induced reduction of infarct size (Fig. 5A and B). Western blot analyses show that treatment with Wortmannin attenuated cardiac adaptation-induced LC3-II, Beclin-1 and BAG-1 (Fig. 5C–F).
4.
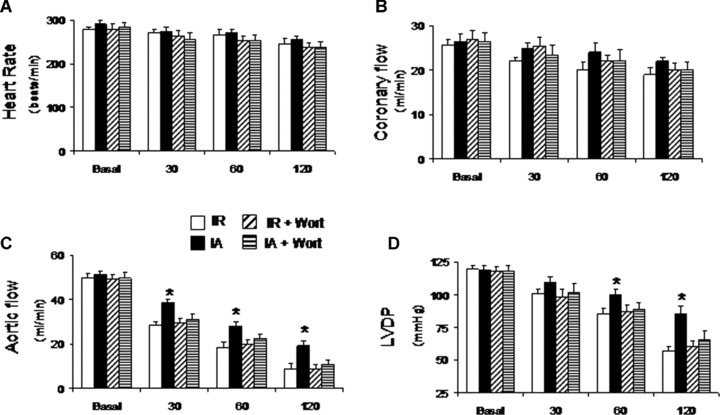
Myocardial function after treatment with Wortmannin, an inhibitor of autophagy. Rats were injected with Wortmannin (15 μg/kg, intraperitoneally) 30 min. before isolation of heart. Hearts were perfused in Langendorff working heart apparatus in working mode. Myocardial functional parameters like heart rate (A), coronary flow (B), aortic flow (C) and left ventricular developed pressure (D) were recorded. Results are representative images of six animals ± S.E.M. *P< 0.05 versus IA + Wort.
5.
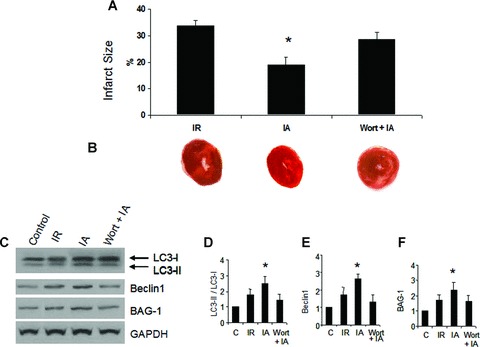
Inhibition of autophagy inhibits IA-induced reduction of myocardial infarct size and BAG-1. Rats were injected with Wortmannin (15 μg/kg, intraperitoneally) 30 min. before isolation of heart. Hearts were perfused in Langendorff mode, and at the end of reperfusion, hearts were collected and cut into several sections. Heart sections were stained with 1% triphenyltetrazolium chloride solution. Infarct size analysis (A) was carried out using Scion Image software. (B) – Representative cardiac tissue sections showing infarct zone (white area). *P< 0.05 versus I/R, and Wort + IA. (C) – Hearts were isolated from Wortmannin treated rats, and perfused in Langendorff mode as mentioned in methods. Cytoplasmic extract from left ventricular tissue was isolated, and the proteins were separated by SDS-PAGE and analysed by Western immunoblotting with specific antibodies. (D–F) – Quantitative analysis of Western immunoblotting of LC3 (D), Beclin-1 (E) and BAG-1 (F); the results are represented as fold change in comparison to control heart samples. Quantitation of LC3 is calculated by analysing the expression ratio of LC3-II / LC3-I. The expression of Beclin-1 and BAG-1 is normalized with the expression of GAPDH. *P< 0.05 versus control. Results are representative images of three to five separate samples. C – Control, I/R – Ischaemia and reperfusion, IA – Ischaemic adaptation followed by I/R, Wort + IA, Wortmannin treatment followed by IA.
In vivo treatment with BAG-1 siRNA attenuated autophagy in the myocardium
We found that BAG-1 protein was simultaneously induced when autophagy was induced. In order to further demonstrate the role of BAG-1 in autophagy, we studied the effect of BAG-1 inhibition in autophagy. We employed RNA interference technique to suppress the expression of BAG-1 protein through direct injection of BAG-1 siRNA into the myocardium of rats. Forty-eight hours after siRNA injection, the hearts were collected and subjected to ischaemic adaptation followed by I/R. Cardiac function like aortic flow, left ventricular developed pressure and the first derivative of left ventricular developed pressure (dp/dt) were markedly reduced by BAG-1 siRNA treatment when compared to control siRNA treated hearts (Fig. 6A–C). Western immunoblot analysis shows that BAG-1 siRNA injection suppressed BAG-1 protein expression when compared to control siRNA treated hearts (Fig. 6D).
6.
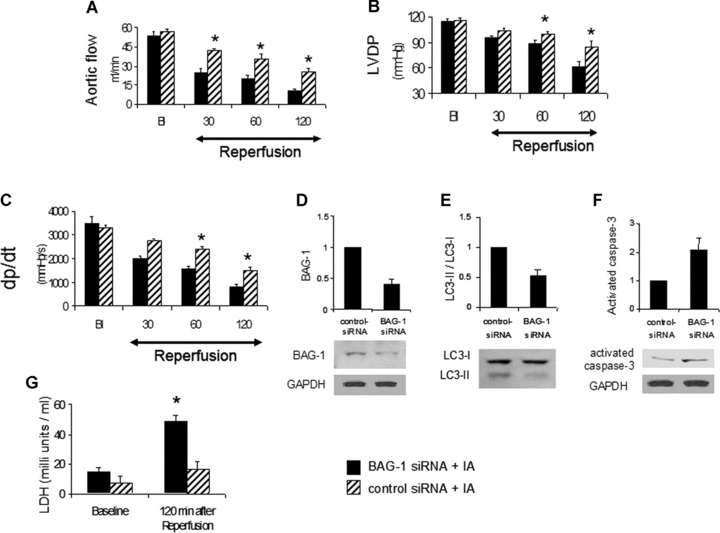
In vivo treatment with BAG-1 siRNA attenuates the effects of ischaemic adaptation. BAG-1 and control siRNA was directly injected into the myocardium of rats as mentioned in methods. 48 hrs after intramyocardial injection, hearts were isolated and subjected for ischaemic adaptation followed by I/R (IA) as mentioned in methods. Cardiac functional parameters like aortic flow (A), left ventricular developed pressure (B), and the first derivative of left ventricular developed pressure (C, dp/dt) were measured in working mode during reperfusion at different time periods like 30, 60 and 120 min. B, Baseline. (D–F) – Quantitative analysis of Western blotting expression of BAG-1 (D), LC3 (E) and activated caspase-3 (F). The representative Western blot images are shown at the bottom. The results are represented as fold change in comparison to control siRNA treated heart samples. Quantitation of LC3 is calculated by analysing the expression ratio of LC3-II / LC3-I. The expression of BAG-1 and activated caspase-3 is normalized with the expression of GAPDH. (C) – Lactate dehydrogenase activity assay was performed with the coronary effluent collected at 2 hrs reperfusion period. *P< 0.05 versus control-siRNA of the same time period. Figures are representative images of three different samples.
Moreover, BAG-1 siRNA treatment attenuated adaptation-induced up-regulation of autophagic marker protein LC3-II relative to control siRNA treated hearts (Fig. 6E). Cardiac cell death by apoptosis was evaluated by examining the protein expression of cleaved or activated caspase-3. Figure 6F shows that BAG-1 siRNA treatment markedly up-regulated the expression of activated caspase-3 when compared to control siRNA treated hearts. Furthermore, LDH activity, a marker for cell death, was significantly increased in the coronary effluent collected from BAG-1 siRNA treated hearts during reperfusion period relative to control siRNA treated hearts (Fig. 6G). These results clearly indicate that BAG-1 siRNA treatment attenuated autophagy and at the same time up-regulated cell damage via necrosis and apoptosis, which is evident from increased LDH activity in the coronary effluent of BAG-1 siRNA treated hearts.
Hypoxic adaptation in cardiac myoblast cells induces autophagy and BAG-1
Our in vivo study results show that cardiac adaptation induced autophagy in association with BAG-1 protein. These results were further tested in vitro using H9c2 cardiac myoblast cells. H9c2 cells were subjected to 30 min. of hypoxia followed by 60 min. of reoxygenation (H/R). The hypoxic adaptation was induced by three repeated cycles of 5 min. of hypoxia and 5 min. of reoxygenation preceding H/R. H/R in cardiac myoblast cells induced the expression of LC3-II, and BAG-1 proteins (Fig. 7A–C). However, when H9c2 cells were subjected to hypoxic adaptation preceding the H/R, LC3-II and BAG-1 proteins were significantly enhanced as shown by Western immunoblotting in Fig. 7A–C. In accordance with the results shown in Fig. 3, our immunofluorescence analyses of H9c2 cells showed that the expression of LC3 and BAG-1 was increased in cells subjected to hypoxic adaptation followed by H/R. Representative immunofluorescence images (Fig. 7D and E) showing the staining of BAG-1 and LC3, respectively, in cells subjected hypoxic adaptation. Figure 7F shows the co-localization BAG-1 and LC3 in H9c2 cells subjected to hypoxic adaptation. This indicates that the results obtained in cardiac myoblast cells are in consistent with the results obtained in rat myocardium.
7.
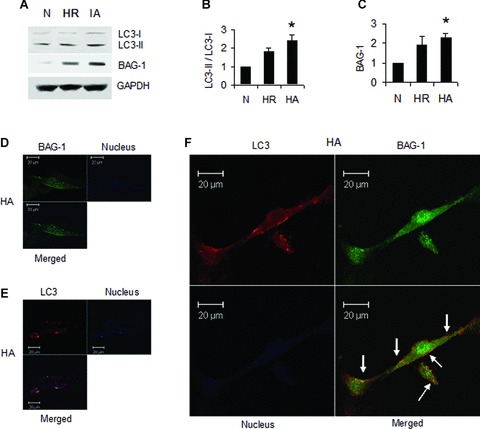
Hypoxic adaptation in H9c2 cardiac myoblast cells induces autophagy. H9c2 cardiac myoblast cells were subjected to hypoxia followed by reoxygenation (H/R) or hypoxic adaptation (HA) as mentioned in methods section. (A)-Representative Western blot images showing the expression of LC3-I, LC3-II and BAG-1 in total cell lysate. (B–C) – Quantitative analysis of Western immunoblotting of LC3 (B), and BAG-1 (C); the results are represented as fold change in comparison to control cells. Quantitation of LC3 is calculated by analysing the expression ratio of LC3-II/LC3-I. The expression of BAG-1 is normalized with the expression of GAPDH. *P< 0.05 versus control. Results are representative images of three to five separate samples. N-normal, H/R-hypoxia and reperfusion, HA- hypoxic adaptation followed by H/R. (D–E) – Confocal microscopic images showing the expression of BAG-1 (D) and LC3 (E) in HA samples. (F) – Confocal microscopic images showing the colocalization of BAG-1 and LC3 in HA samples. Colocalization is indicated by arrows. Counterstaining of nucleus in images D-F was done with Topro-1-iodide (Invitrogen). Figures are representative images of three different samples.
BAG-1 siRNA treatment in cardiac myoblast cells attenuated hypoxic adaptation-induced autophagy
When cardiac myoblast cells are treated with either control or BAG-1 siRNA, the hypoxic adaptation-induced expression of BAG-1 protein was decreased in BAG-1 siRNA treated cells compared to control siRNA treated cells (Fig. 8A and B). At the same time, the hypoxic adaptation-induced expression of LC3-II was also attenuated in BAG-1 siRNA treated cells compared to control siRNA treated cells (Fig. 8A and C).
8.
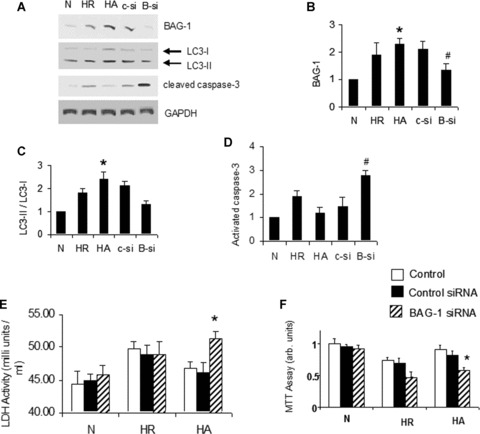
In vitro BAG-1 siRNA treatment in H9c2 cells attenuate the effects of hypoxic adaptation H9c2 cardiac myoblast cells were subjected to 30 min. of hypoxia and 60 min. reoxygenation (H/R). Hypoxic adaptation (HA) was given by subjecting the cells for three cyclic episodes of 5 min. of hypoxia and 5 min. of reoxygenation, followed by H/R. A group of cells were treated with either control or BAG-1 siRNA as mentioned in methods. (A)-Representative Western blot images showing the expression of BAG-1, LC3-I, LC3-II, and cleaved caspase-3 in total cell lysate. (B–D) – Quantitative analysis of Western blotting expression of BAG-1 (B), LC3 (C) and activated caspase-3 (D). The results are represented as fold change in comparison to normal cells. Quantitation of LC3 is calculated by analysing the expression ratio of LC3-II/LC3-I. The expression of BAG-1 and activated caspase-3 is normalized with the expression of GAPDH. *P< 0.05 versus N, #P< 0.05 versus HA. (E) – Lactate dehydrogenase activity was measured using the spent culture medium collected at the end of the experiment. (C) – MTT assay was performed with the cultured cells as mentioned in methods. N, Normal cells; C-si, cells treated with control siRNA followed by HA; B-si, cells treated with BAG-1 siRNA followed by HA. * in (E) and (F) indicates P< 0.05 versus HA-control.
Further, cell death by apoptosis was evaluated by examining the protein expression of activated caspase-3. Figure 8A and D shows that in vitro BAG-1 siRNA treatment preceding HA in H9c2 cardiac cells significantly enhanced the expression of activated caspase-3 when compared to cells treated with control siRNA. The release of LDH into the spent culture medium show that H/R in cardiac myoblast cells significantly induced LDH release (Fig. 8E). H/R-induced LDH release is significantly attenuated by hypoxic adaptation in myoblast cells (Fig. 8E). Hypoxic adaptation-induced attenuation of LDH release was blocked when the cells are treated with BAG-1 siRNA compared to control siRNA (Fig. 8E). Moreover, cell survival assay using 3’(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT) in H9c2 cells indicated that the hypoxic adaptation-induced improvement in cell survival probability was attenuated by treating cells with BAG-1 siRNA relative to control siRNA (Fig. 8F). Altogether these results clearly indicate that autophagic marker LC3-II and cellular survivability in cardiac myoblast cells were decreased when the expression of BAG-1 was diminished via treatment with BAG-1 siRNA.
Altogether, our results show that ischaemic adaptation of the myocardium enhances autophagy and BAG-1, and the autophagosomal membrane specific protein microtubule-associated protein LC3 interacts with BAG-1. Moreover, the essential role of BAG-1 in the induction of autophagy has been proved by BAG-1 siRNA treatment in vivo and in vitro. These results indicate that ischaemic adaptation-induced cardiac protection is mediated at least in part via up-regulation of autophagy, and BAG-1 appears to play an important role in the induction of autophagy.
Discussion
A lot of work has been done on the mechanism of ischaemic adaptation-induced cardioprotection [17], and recently, the protective effect of autophagy has been demonstrated under anoxia-reoxygenation in isolated cardiomyocytes [37].
In this study, our results reveal the following novel findings: (i) ischaemic adaptation of the myocardium enhances autophagy and BAG-1, (ii) autophagosomal membrane specific protein LC3-II interacts with BAG-1, (iii) inhibition of autophagy by treating rats with Wortmannin attenuates BAG-1 protein expression and (iv) suppression of BAG-1 protein expression by in vivo BAG-1 siRNA treatment into the myocardium attenuated adaptation-induced autophagy. Further, treatment with BAG-1 siRNA in H9c2 cardiac myoblast cells attenuated hypoxic adaptation-induced LC3-II and cell survival.
Is autophagy in the myocardium protective or detrimental?
Autophagy acts as cellular defence mechanism against bacterial toxins [33]. Autophagy is shown to be induced in several cardiac diseases like heart failure [9], cardiac hypertrophy [10] and ischaemic cardiomyopathy [11]. Hamacher-Brady et al. have demonstrated that enhancing autophagy protects against I/R injury in cardiac myocytes [14]. Overexpression of Bnip3, a pro-apoptotic molecule present in the mitochondrial membrane, triggered up-regulation of autophagy and protected cardiac myocytes from I/R injury-related apoptosis [38]. A recent study demonstrates that autophagy elicited by hypoxia or ischaemia via AMP activated protein kinase-dependent mechanism may provide protective role against cardiac injury; but autophagy elicited by ischaemia followed by reperfusion occurring via Beclin-1-dependent mechanism may play a detrimental role in the myocardium [15]. In another study, reduction of autophagy by RNA interference of Beclin-1, a key molecule involved in mediating autophagy, enhances cardiac cell survival [39]. In order to address the controversial role of autophagy in the myocardium, in the present study we examined the function of autophagy during cardiac adaptation to ischaemic stress. Our study, in accordance with previous results [14], shows that autophagy was slightly up-regulated by I/R injury (Fig. 1). In addition, our results indicate that cardiac adaptation to ischaemic stress further enhanced autophagy as demonstrated by the up-regulation of autophagosomal membrane specific protein LC3-II, and Beclin-1 (Fig. 1A–C), and by the presence of autophagosomes (Fig. 1E). Further, hypoxic adaptation in cardiac myoblast cells induced the expression of LC3-II as shown by Western immunoblotting and immunoflorescence analyses (Fig. 7). These results suggest that cardiac adaptation, a state-of-the-art technique to protect the myocardium against I/R injury, may trigger cell survival at least in part via the up-regulation of autophagy.
BAG-1, a co-chaperone, binds with autophagosomal membrane protein LC3
BAG-1 is a multifunctional pro-survival molecule that binds with Bcl-2 and protects cells from apoptosis [21, 22]. BAG-1 functions as a nucleotide exchange factor of mammalian cytosolic Hsc70, thereby triggering substrate unloading from the chaperone [40]. BAG-1 is shown to promote the association of Hsp70/Hsc70 with proteasome in vitro[41]. The ubiquitin domain protein BAG-1 and the carboxyl terminus of the Hsc70-interacting protein (CHIP) ubiquitin ligase can cooperate to shift the activity of the Hsc70/Hsp70 chaperone system from protein folding to degradation [42]. BAG-1 has been shown to protect cardiac myocytes from simulated I/R injury-induced apoptosis in association with Hsp70 and Hsc70 chaperone molecules [23]. This BAG-1-mediated car-dioprotection was shown to be critical upon cytoplasmic localization but independent of its association with proteasome as shown that BAG-1 induced cardioprotection was not affected by a BAG-1 deletion mutant lacking N-terminal ubiquitin-like domain, which mediates interaction with proteasome [23]. These results suggest that BAG-1 may provide a novel cardioprotective role, which may be independent of its association with proteasome [23].
Recently, it has been shown that autophagy is being regulated by Bcl-2 binding Beclin-1 [25]. BAG-1 is shown to present in the molecular chaperone complex at the lysosomal membrane along with Hsp70 and Hsc70 [26]. In another study, it has been demonstrated that autophagosomal membrane contains significantly higher amount of Hsc70 proteins [27]. Since BAG-1 can bind with both Bcl-2 and Hsc70 molecules, we studied the possible role of BAG-1 protein in the regulation of autophagy. In the present study, we found that I/R injury slightly up-regulated BAG-1 protein expression, but upon ischaemic adaptation the BAG-1 protein expression was further enhanced in the myocardium. Moreover, hypoxic adaptation against hypoxia and reoxygenation in cardiac myoblast cells also induced BAG-1 protein. Our in vivo and in vitro studies show that BAG-1 protein expression is positively correlated with the expression of autophagosomal marker proteins. Further, our immunoprecipitation experiment with the cytosolic extract of the myocardium show that BAG-1 binds with LC3-II, and the binding was enhanced during ischaemic adaptation (Fig. 2). In addition, our co-immunofluorescence analysis shows that LC3 is co-localized with BAG-1 in the myocardium (Fig. 3). However, it is not known that whether BAG-1 directly interacts LC3-II or indi-rectly through other molecules. BAG-1 is shown to exert many of its functions via interaction of heat shock protein Hsc70 [22]. Recently, it has been shown that Hsc70 is present in significantly higher level in autophagosomal membrane [27]. LC3-II is a specific molecule found in the autophagosomal membrane [29]. In our study, we found that Hsc70 co-immunoprecipitate with BAG-1. Based on the above evidence, we speculate that BAG-1 may interact with LC3 via Hsc70.
Inhibition of autophagy abolished the cardioprotective abilities of ischaemic adaptation and attenuated BAG-1 protein
In mammalian cells, autophagy is shown to be activated by class III PI3 kinase complex containing Vps34 and Beclin-1 [28, 29], and it is suppressed by the treatment with PI3 kinase inhibitor Wortmannin [33, 36]. In the present study, we treated rats with Wortmannin in order to suppress the autophagy found during ischaemic adaptation. Our results show that treatment of rats with Wortmannin attenuated both LC3-II as well as BAG-1 protein expression induced after cardiac adaptation against ischaemic stress. Wortmannin treatment further attenuated the cardiac adaptation-induced improvement in cardiac functional parameters (Fig. 4) and in myocardial infarct size (Fig. 5). Our results are in accordance with other studies [43, 44], where Wortmannin has been shown to abrogate the protective effects in the myocardium.
Though Wortmannin has been used as an inhibitor of autophagy in several studies [33, 36], one cannot rule out the possibilities of other effects of Wortmannin in the myocardium.
Role of BAG-1 in the induction of autophagy
In order to demonstrate the role of BAG-1 in the regulation of autophagy, we adopted RNA interference technique to suppress the expression of BAG-1 in vivo and in vitro by treating with BAG-1 siRNA. In vivo myocardial siRNA treatment was performed by direct injection of either BAG-1 or control siRNA into the rat myocardium. Two days after siRNA treatment, the hearts were isolated and subjected to ischaemic adaptation followed by I/R injury. BAG-1 siRNA treatment attenuated cardiac adaptation-induced improvement in cardiac functional parameters like left ventricular developed pressure and aortic flow, and markedly reduced the protein expression of BAG-1 and LC3-II relative to control siRNA treated hearts (Fig. 6). Furthermore, BAG-1 siRNA treatment enhanced myocardial damage demonstrated by elevated LDH activity observed in the coronary effluent collected form BAG-1 siRNA treated hearts with respect to control siRNA treated hearts (Fig. 6). Moreover, BAG-1 siRNA treatment in cardiac myoblast cells resulted in attenuation of hypoxic adaptation-induced (i) protein expression of BAG-1 and LC3-II, (ii) reduction of LDH release from cells and (iii) improvement in cell survival (Fig. 8). These results clearly indicate that treatment with BAG-1 siRNA in both in vivo and in vitro attenuated ischaemic/hypoxic adaptation-induced improvement in cardiac cell survival and autophagy. However, further studies with overexpression of BAG-1 in the myocardium may support the direct role of BAG-1 in the induction of autophagy. Though the cardioprotective role of multifunc-tional protein BAG-1 has been ascertained in the recent years [20–23], the present study indicates yet another role for BAG-1 conjugated with autophagy for the protection of myocardium against I/R injury.
In summary, cardioprotective adaptive response to I/R or hypoxia reoxygenation injury enhanced autophagic marker proteins and BAG-1. The binding between BAG-1 and LC3 have been demonstrated. Autophagic inhibition attenuated BAG-1 and cardioprotective adaptive responses. Suppression of BAG-1 with BAG-1 siRNA treatment attenuated adaptation-induced autophagic marker proteins cardiac improvements. These results indicate that cardioprotective abilities of ischaemic adaptation are mediated at least in part via up-regulation of autophagy, in association with BAG-1 protein in the myocardium.
Acknowledgments
This study was supported in part by NIH HL 34360, HL22559 and HL 33889. The views, opinions, and/or findings contained herein are those of the authors and should not be construed as an official Department of the Army position, policy or decision. The authors have no conflicting financial interests.
References
- 1.Cuervo AM. Autophagy: many paths to the same end. Mol Cell Biochem. 2004;263:55–72. doi: 10.1023/B:MCBI.0000041848.57020.57. [DOI] [PubMed] [Google Scholar]
- 2.Kundu M, Thompson CB. Macroautophagy versus mitochondrial autophagy: a question of fate? Cell Death Differ. 2005;12:1484–9. doi: 10.1038/sj.cdd.4401780. [DOI] [PubMed] [Google Scholar]
- 3.Iwata J, Ezaki J, Komatsu M, Yokota S, Ueno T, Tanida I, Chiba T, Tanaka K, Kominami E. Excess peroxisomes are degraded by autophagic machinery in mammals. J Biol Chem. 2006;281:4035–41. doi: 10.1074/jbc.M512283200. [DOI] [PubMed] [Google Scholar]
- 4.Kiffin R, Bandyopadhyay U, Cuervo AM. Oxidative stress and autophagy. Antioxid Redox Signal. 2006;8:152–62. doi: 10.1089/ars.2006.8.152. [DOI] [PubMed] [Google Scholar]
- 5.Codogno P, Meijer AJ. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 2005;12:1509–18. doi: 10.1038/sj.cdd.4401751. [DOI] [PubMed] [Google Scholar]
- 6.Moore MN. Autophagy as a second level protective process in conferring resistance to environmentally-induced oxidative stress. Autophagy. 2008;4:254–6. doi: 10.4161/auto.5528. [DOI] [PubMed] [Google Scholar]
- 7.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 8.Terman A, Brunk UT. Oxidative stress, accumulation of biological ‘garbage’, and aging. Antioxid Redox Signal. 2006;8:197–204. doi: 10.1089/ars.2006.8.197. [DOI] [PubMed] [Google Scholar]
- 9.Takemura G, Miyata S, Kawase Y, Okada H, Maruyama R, Fujiwara H. Autophagic degeneration and death of cardiomyocytes in heart failure. Autophagy. 2006;2:212–4. doi: 10.4161/auto.2608. [DOI] [PubMed] [Google Scholar]
- 10.Hein S, Arnon E, Kostin S, Schonburg M, Elsasser A, Polyakova V, Bauer EP, Klovekorn WP, Schaper J. Progression from compensated hypertrophy to failure in the pressure-overloaded human heart: structural deterioration and compensatory mechanisms. Circulation. 2003;107:984–91. doi: 10.1161/01.cir.0000051865.66123.b7. [DOI] [PubMed] [Google Scholar]
- 11.Yan L, Vatner DE, Kim SJ, Ge H, Masurekar M, Massover WH, Yang G, Matsui Y, Sadoshima J, Vatner SF. Autophagy in chronically ischemic myocardium. Proc Natl Acad Sci USA. 2005;102:13807–12. doi: 10.1073/pnas.0506843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Terman A, Brunk UT. Autophagy in cardiac myocyte homeostasis, aging, and pathology. Cardiovasc Res. 2005;68:355–65. doi: 10.1016/j.cardiores.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 13.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Mizote I, Matsumura Y, Asahi M, Nishida K, Hori M, Mizushima N, Otsu K. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–24. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 14.Hamacher-Brady A, Brady NR, Gottlieb RA. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. J Biol Chem. 2006;281:29776–87. doi: 10.1074/jbc.M603783200. [DOI] [PubMed] [Google Scholar]
- 15.Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100:914–22. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 16.Takagi H, Matsui Y, Sadoshima J. The role of autophagy in mediating cell survival and death during ischemia and reperfusion in the heart. Antioxid Redox Signal. 2007;9:1373–81. doi: 10.1089/ars.2007.1689. [DOI] [PubMed] [Google Scholar]
- 17.Das DK, Maulik N. Preconditioning potentiates redox signaling and converts death signal into survival signal. Arch Biochem Biophys. 2003;420:305–11. doi: 10.1016/j.abb.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 18.Kiffin R, Christian C, Knecht E, Cuervo AM. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15:4829–40. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poppek D, Grune T. Proteasomal defense of oxidative protein modifications. Antioxid Redox Signal. 2006;8:173–84. doi: 10.1089/ars.2006.8.173. [DOI] [PubMed] [Google Scholar]
- 20.Takayama S, Reed JC. Molecular chaper-one targeting and regulation by BAG family proteins. Nat Cell Biol. 2001;3:E237–41. doi: 10.1038/ncb1001-e237. [DOI] [PubMed] [Google Scholar]
- 21.Townsend PA, Cutress RI, Sharp A, Brimmell M, Packham G. BAG-1: a multifunctional regulator of cell growth and survival. Biochim Biophys Acta. 2003;1603:83–98. doi: 10.1016/s0304-419x(03)00002-7. [DOI] [PubMed] [Google Scholar]
- 22.Townsend PA, Stephanou A, Packham G, Latchman DS. BAG-1: a multi-functional pro-survival molecule. Int J Biochem Cell Biol. 2005;37:251–9. doi: 10.1016/j.biocel.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 23.Townsend PA, Cutress RI, Carroll CJ, Lawrence KM, Scarabelli TM, Packham G, Stephanou A, Latchman DS. BAG-1 proteins protect cardiac myocytes from simulated ischemia/reperfusion-induced apoptosis via an alternate mechanism of cell survival independent of the proteasome. J Biol Chem. 2004;279:20723–8. doi: 10.1074/jbc.M400399200. [DOI] [PubMed] [Google Scholar]
- 24.Alberti S, Demand J, Esser C, Emmerich N, Schild H, Hohfeld J. Ubiquitylation of BAG-1 suggests a novel regulatory mechanism during the sorting of chaperone substrates to the proteasome. J Biol Chem. 2002;277:45920–7. doi: 10.1074/jbc.M204196200. [DOI] [PubMed] [Google Scholar]
- 25.Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, Tsujimoto Y. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6:1221–8. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- 26.Agarraberes FA, Dice JF. A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J Cell Sci. 2001;114:2491–9. doi: 10.1242/jcs.114.13.2491. [DOI] [PubMed] [Google Scholar]
- 27.Overbye A, Fengsrud M, Seglen PO. Proteomic analysis of membrane-associated proteins from rat liver autophagosomes. Autophagy. 2007;3:300–22. doi: 10.4161/auto.3910. [DOI] [PubMed] [Google Scholar]
- 28.Martinet W, Knaapen MW, Kockx MM, De Meyer GR. Autophagy in cardiovascular disease. Trends Mol Med. 2007;13:482–91. doi: 10.1016/j.molmed.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 29.Hamacher-Brady A, Brady NR, Gottlieb RA. The interplay between pro-death and pro-survival signaling pathways in myocardial ischemia/reperfusion injury: apoptosis meets autophagy. Cardiovasc Drugs Ther. 2006;20:445–62. doi: 10.1007/s10557-006-0583-7. [DOI] [PubMed] [Google Scholar]
- 30.Asanuma K, Tanida I, Shirato I, Ueno T, Takahara H, Nishitani T, Kominami E, Tomino Y. MAP-LC3, a promising autophagosomal marker, is processed during the differentiation and recovery of podocytes from PAN nephrosis. FASEB J. 2003;17:1165–7. doi: 10.1096/fj.02-0580fje. [DOI] [PubMed] [Google Scholar]
- 31.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–11. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3’-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–8. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 33.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo Ml, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–66. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 34.Gherghiceanu M, Popescu LM. Electron microscope tomography: further demonstration of nanocontacts between caveolae and smooth muscle sarcoplasmic reticulum. J Cell Mol Med. 2007;11:1416–8. doi: 10.1111/j.1582-4934.2007.00166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mandache E, Popescu LM, Gherghiceanu M. Myocardial interstitial Cajal-like cells (ICLC) and their nanostructural relationships with intercalated discs: shed vesicles as intermediates. J Cell Mol Med. 2007;11:1175–84. doi: 10.1111/j.1582-4934.2007.00117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gurusamy N, Malik G, Gorbunov NV, Das DK. Redox activation of Ref-1 potentiates cell survival following myocardial ischemia reperfusion injury. Free Radic Biol Med. 2007;43:397–407. doi: 10.1016/j.freeradbiomed.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 37.Dosenko VE, Nagibin VS, Tumanovska LV, Moibenko AA. Protective effect of autophagy in anoxia-reoxygenation of isolated cardiomyocyte? Autophagy. 2006;2:305–6. doi: 10.4161/auto.2946. [DOI] [PubMed] [Google Scholar]
- 38.Hamacher-Brady A, Brady NR, Gottlieb RA, Gustafsson AB. Autophagy as a protective response to Bnip3-mediated apop-totic signaling in the heart. Autophagy. 2006;2:307–9. doi: 10.4161/auto.2947. [DOI] [PubMed] [Google Scholar]
- 39.Valentim L, Laurence KM, Townsend PA, Carroll CJ, Soond S, Scarabelli TM, Knight RA, Latchman DS, Stephanou A. Urocortin inhibits Beclin-1-mediated autophagic cell death in cardiac myocytes exposed to ischaemia/reperfusion injury. J Mol Cell Cardiol. 2006;40:846–52. doi: 10.1016/j.yjmcc.2006.03.428. [DOI] [PubMed] [Google Scholar]
- 40.Alberti S, Esser C, Hohfeld J. BAG-1 – a nucleotide exchange factor of Hsc70 with multiple cellular functions. Cell Stress Chaperones. 2003;8:225–31. doi: 10.1379/1466-1268(2003)008<0225:bnefoh>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Townsend PA, Cutress RI, Sharp A, Brimmell M, Packham G. BAG-1 prevents stress-induced long-term growth inhibition in breast cancer cells via a chaperone-dependent pathway. Cancer Res. 2003;63:4150–7. [PubMed] [Google Scholar]
- 42.Demand J, Alberti S, Patterson C, Hohfeld J. Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr Biol. 2001;11:1569–77. doi: 10.1016/s0960-9822(01)00487-0. [DOI] [PubMed] [Google Scholar]
- 43.Hua F, Ha T, Ma J, Li Y, Kelley J, Gao X, Browder IW, Kao RL, Williams DL, Li C. Protection against myocardial ischemia/ reperfusion injury in TLR4-deficient mice is mediated through a phosphoinositide 3-kinase-dependent mechanism. J Immunol. 2007;178:7317–24. doi: 10.4049/jimmunol.178.11.7317. [DOI] [PubMed] [Google Scholar]
- 44.Strande JL, Hsu A, Su J, Fu X, Gross GJ, Baker JE. Inhibiting protease-activated receptor 4 limits myocardial ischemia/ reperfusion injury in rat hearts by unmasking adenosine signaling. J Pharmacol Exp Ther. 2008;324:1045–54. doi: 10.1124/jpet.107.133595. [DOI] [PMC free article] [PubMed] [Google Scholar]


