Abstract
Background
Chromosome 7 has shown consistent evidence of linkage with a variety of phenotypes related to alcohol dependence in the Collaborative Study on the Genetics of Alcoholism (COGA) project. Using a sample of 262 densely affected families, a peak lod score for alcohol dependence of 2.9 was observed at D7S1799 (Wang et al., 2004, Hum Mol Genet). The lod score in the region increased to 4.1 when a subset of the sample was genotyped with the Illumina Linkage III panel for the Genetic Analysis Workshop 14 (GAW14; Dunn et al., 2005, BMC Genetics). To follow-up on this linkage region, we systematically screened SNPs across a 2 LOD support interval surrounding the alcohol dependence peak.
Methods
SNPs were selected from the HapMap Phase I CEPH data to tag linkage disequilibrium bins across the region. 1340 across the 18Mb region, genotyped by the Center for Inherited Disease Research (CIDR), were analyzed. Family-based association analyses were performed on a sample of 1172 individuals from 217 Caucasian families. Results: Eight SNPs showed association with alcohol dependence at p<0.01. Four of the eight most significant SNPs were located in or very near the ACN9 gene. We conducted additional genotyping across ACN9 and identified multiple variants with significant evidence of association with alcohol dependence.
Conclusions
These analyses suggest that ACN9 is involved in the predisposition to alcohol dependence. Data from yeast suggest that ACN9 is involved in gluconeogenesis and the assimilation of ethanol or acetate into carbohydrate.
Keywords: genetics, association, linkage disequilibrium, alcohol dependence, ACN9
Introduction
Alcohol dependence is a common complex disorder that affects millions of people worldwide, and causes considerable burden in terms of personal, interpersonal, and societal costs (1). Results from the National Comorbidity Study indicate that over 14% of adults in the United States have a lifetime history of alcohol dependence, making it one of the most prevalent adult psychiatric disorders (2). Family, twin, and adoption studies have convincingly demonstrated that genes play an important role in the development of alcohol dependence, with heritability estimates in the range of 50-60% for both men and women (3; 4). Efforts are now underway to identify specific genes involved in the development of the disorder.
The Collaborative Study on the Genetics of Alcoholism (COGA) is a multi-site collaboration aimed at identifying genes contributing to alcohol dependence. COGA ascertained families densely affected with alcohol dependence from treatment centers at multiple sites across the United States. Initially, an approximately 10 cM genome-wide microsatellite survey was conducted and linkage analyses were performed to detect chromosomal regions likely to harbor genes contributing to a variety of phenotypes related to alcohol dependence (5-7). In those regions with evidence of linkage, more extensive genotyping was performed, and association analyses were employed to identify specific genes involved in the predisposition to alcohol dependence and related phenotypes. COGA has also made use of electrophysiological endophenotypes (8-10), as a complement to clinical diagnoses in genetic analyses.
One region that has consistently emerged with significant evidence of linkage in the COGA project is chromosome 7q. In the initial COGA sample of 105 pedigrees, chromosome 7 provided the strongest evidence of linkage to alcohol dependence (11). Using the alcohol dependence criterion of meeting DSMIII-R alcohol dependence and Feighner definite alcoholism, the maximum multipoint lod score on chromosome 7 was 3.49 near the marker D7S1793. An independent sample consisting of an additional 157 extended families also showed modest, consistent evidence of linkage to chromosome 7, with a lod score of 1.3 (7). Additional microsatellite markers were genotyped on chromosome 7, and linkage analyses employing the full sample of 262 extended pedigrees yielded a peak LOD score of 2.9 at D7S1799 (12). Further genotyping was conducted as part of the Genetic Analysis Workshop 14 (GAW14) on a densely affected subset of the sample (N=143 pedigrees) using both the Affymetrix 10K Mapping SNPs and the Illumina Linkage Panel III (13). The LOD score at the peak increased to 4.1 using a reduced set of SNPs not in linkage disequilibrium with adjacent markers (14). In addition to the linkage observed in the COGA sample, linkage has been observed to this region of chromosome 7q in an Australian sample with P3 amplitude (15), a phenotype thought to index genetic vulnerability to alcohol dependence (9). In addition, there has been a recent report of linkage to this region with alcohol consumption phenotypes in the Nicotine Addiction Genetics project (16). Modest evidence of linkage to this region was also reported for an alcoholism phenotype using age and gender as covariates in an independent sample of multiplex families ascertained at Pittsburgh (17). Here, we report results from a systematic screen of SNPs across the chromosome 7 alcohol dependence linkage peak in the COGA sample in an effort to identify the gene(s) contributing to the observed linkage peak.
Methods
Sample
The Collaborative Study on the Genetics of Alcoholism (COGA) is a multi-site project, in which families were collected by six centers across the United States: Indiana University, State University of New York Health Science Center, University of Connecticut, University of Iowa, University of California/San Diego, and Washington University, St. Louis. Probands identified through inpatient or outpatient alcohol treatment programs by each of these six sites were invited to participate if they had a sufficiently large family (usually sibships > 3 with parents available) with two or more members in a COGA catchment area (6). Multiplex alcoholic families that had at least two biological first-degree relatives affected with alcohol dependence in addition to the proband were invited to participate in the more intensive stage of the study, which included obtaining blood for genetic analyses. Second and third degree relatives in the families were assessed when they were considered to be informative for the genetic linkage studies. The institutional review boards of all participating centers approved the study. Additional details about the study have been published previously (5; 6).
We analyzed a set of 217 Caucasian families here because (1) the marker selection strategy (detailed below) was based on patterns of LD among Caucasians, and allele frequencies often differ between races, and (2) the maximal lod score with alcohol dependence in the region (4.1) was based on a sample of Caucasian families. These 217 families contained a total of 1172 individuals with genotype and phenotype data: 554 females and 618 males. There were 855 affected individuals in the sample (288 females, 567 males). Mean age of affected females was 34.5 years (SD=9.7), unaffected females: 50.5 years (SD=15.9). Mean age of affected males was 39.4 years (SD=13.3), unaffected males: 50.4 years (SD=17.4). Thus, for both genders, unaffected individuals were significantly older than affected individuals and past the mean age of onset of dependence in this sample, suggesting that there is a low probability that they will convert to affected status. The average number of individuals per family with genotype and phenotype information was 5.4, with an average of 3.9 affected individuals per family. These individuals formed 921 sibling pairs, 20 half-sibs, 150 cousin pairs, 810 parent-child pairs, 82 grandparent-grandchild pairs and 509 avuncular pairs. All individuals in the genetic analysis sample were interviewed as adults (≥18 years of age) using the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA), a highly reliable, psychiatric interview (18; 19). The definition of alcohol dependence used in association analyses required individuals to meet criteria for DSMIII-R alcohol dependence and Feighner definite alcoholism (20). This was the phenotype that yielded the linkage peak from which the region was delineated for the SNP screen (14). Seventy-three percent of the sample used in genetic analyses was affected with alcohol dependence. There are also high rates of comorbid psychiatric disorders in the sample: 37% meet criteria for dependence on an illicit drug, 41% report a major depressive episode, 19% meet criteria for childhood conduct disorder, and 14% have a diagnosis of adult antisocial personality disorder. Additional information about comorbidity in the COGA sample has previously been reported elsewhere (21; 22). In the sample analyzed here, the highest level of educational attainment was less than a high school degree in 23%, a high school education in 31%, some college in 30%, a college degree in 11%, and a postgraduate degree in 5%. At the time of interview, 35% of the sample was unemployed, and 65% was currently employed. The modal current household gross income was $20 – 29,000/year.
SNP Selection Procedure
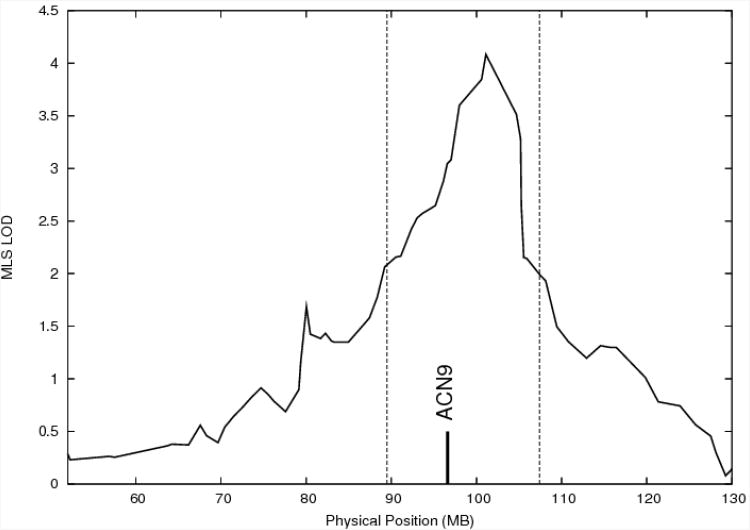
SNPs were selected to cover a 2-LOD support interval on either side of the linkage signal at 7q22, based on the peak observed in the GAW sample (14); that peak was narrower than the peak in the original ∼10 cM microsatellite linkage screen (12). The region was bounded by rs194506 (89.487 Mb) and rs441534 (107.423 Mb) (dbSNP 124/NCBI Human Build 35.1), and covered ∼18Mb (Figure 1). HapMap Phase I CEPH data (build 16c.1, June 2005) was used to select SNPs; only common SNPs (minor allele frequency ≥ 10%) were considered. A total of 4067 SNPs meeting this criteria were identified across the region. SNPs were grouped into linkage disequilibrium (LD) bins (23), based on the “greedy” algorithm (24; 25). Using this method, each bin had at least 1 SNP that satisfied r2≥ 0.8 with all other SNPs in the bin. Tag SNPs were selected for each bin in an 8:1 ratio (e.g., bins with 1-8 SNPs get 1 tag, 9-16 get 2, etc.). SNPs with the highest r2 with other SNPs in the bin were chosen to be tag SNPs. The tagSNP selection method allowed for a reduction in SNPs of 61%, yielding 1581 SNPs. An additional 55 nonsynonymous polymorphic HapMap SNPs were added to the set of SNPs (23/55 had MAF<10%). Selected SNPs were scored by the Center for Inherited Disease Research for expected performance on the Illumina platform. Failed tag SNPs were replaced by the next best tag. If there were no passing tags then the entire LD bin was selected for genotyping. The final list of 1536 SNPs covered 221 genes. It consisted of 883 intra-genic SNPs (654 in introns, 68 in exons, 128 in untranslated regions, and 33 SNPs within 2 kb of the first 5′ promoter and 500 bp of the 3′ end of the largest known transcript [known as “locus” SNPs in dbSNP]) and 653 inter-genic SNPs. Note that although dbSNP build 124/NCBI Human Build 35.1 was used for SNP selection, information about the location of SNPs presented in the paper tables is based on the updated dbSNP 126/NCBI 36.1 data.
Figure 1.
Linkage on chromosome 7 in the GAW sample (14) for the alcohol dependence phenotype, with the position of ACN9 annotated. The vertical dashed lines indicate the 2 lod support interval surrounding the peak that was screened with tagSNPs.
Genotyping and Analysis
Genotyping was conducted by the Center for Inherited Disease Research (CIDR) using the Illumina technology on a BeadLab station with GoldenGate chemistry. 1536 SNPs were attempted across the region, with 3,539,740 genotypes released for 1436 SNPs. CIDR cited the following reasons for dropping loci: poorly defined clusters; excessive replicate and/or Mendelian errors; more than 50% missing data; or all samples genotyping as heterozygous. An additional 96 SNPs were flagged with atypical clustering, and these SNPs were also omitted from analyses. Accordingly, 1340 SNPs passed all quality control checks and were used in analyses. The missing data rate was 0.056%. The program Pedcheck (26) was used by CIDR to check for Mendelian inconsistencies; the Mendelian consistency rate was 99.91%. Further checks were performed by COGA collaborators using the Prest program (27), and a small number of questionable relationships were removed, such that there were no Mendelian inconsistencies remaining in the data.
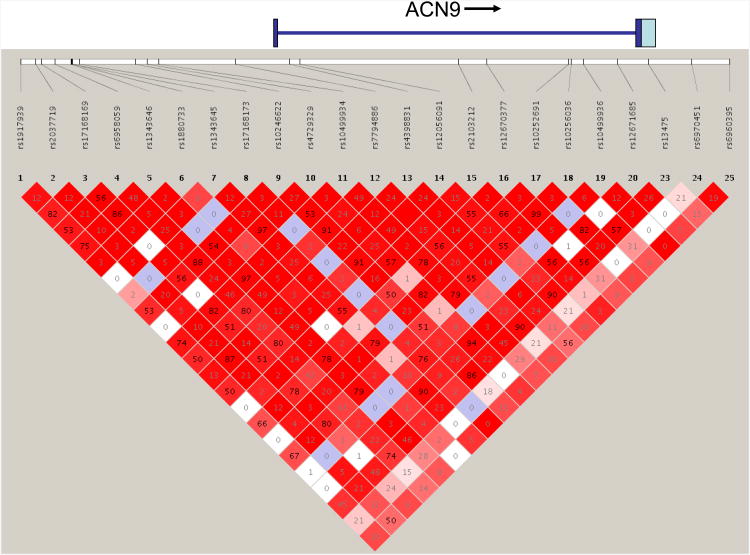
Findings for the most promising gene were followed up by conducting additional genotyping in order to more thoroughly evaluate the evidence for association. Publicly available databases, dbSNP (http://www.ncbi.nlm.nih.gov/SNP/) and HapMap (http://www.hapmap.org), were used to identify SNPs within and flanking the gene. SNPs with an r2 < 0.9 in HapMap with any of the associated CIDR SNPs were selected for genotyping. An additional 16 SNPs within and flanking ACN9 were genotyped using a modified single nucleotide extension reaction, with allele detection by mass spectroscopy [Sequenom MassArray system; Sequenom, San Diego, CA]. All genotypic data were checked for Mendelian inheritance of marker alleles with the USERM13 (28) option of the MENDEL linkage computer programs. Trio data from Caucasian individuals genotyped in the COGA dataset was entered into the program Haploview (29) to examine the linkage disequilibrium structure of the genotyped SNPs. Figure 2 shows the linkage disequilibrium (LD) structure across the region.
Figure 2.
Linkage disequilibrium across the SNPs genotyped in and around ACN9. D′ is illustrated by red shading, with darker shades indicating higher D′. r2 is indicated by the number inside the shaded block.
Hardy-Weinberg equilibrium was assessed for all SNPs. Among 601 unrelated individuals used in checks of the 1340 CIDR SNPs, 68 SNPs significantly deviated from Hardy-Weinberg at p<0.05, and 12 SNPs were significant at p<0.01. These numbers are very close to that expected by chance (67 and 13, respectively, based on 1340 SNPs). None of the SNPs most significantly associated with alcohol dependence (p<0.01) had significant deviations from Hardy-Weinberg equilibrium. Those SNPs with potential Hardy-Weinberg problems are indicated in Table 1 with asterisks, so that this information can be considered in interpretation of results. None of the additional SNPs genotyped by COGA showed significant deviation from Hardy-Weinberg.
Table 1. SNPs from the CIDR screen on chromosome 7 yielding evidence of association at p < 0.050.
| SNP | Allele | HMMAF | P-value | Bin ID (size) | Position | Gene location | Function |
|---|---|---|---|---|---|---|---|
| rs2157745 | [A/C] | 0.108 | 0.0010 | 92,543,595 | . | ||
| rs10499934 | [A/G] | 0.267 | 0.0016 | 96,575,512 | [ACN9]# | . | |
| rs7794886 | [T/C] | 0.442 | 0.0033 | 76 (2) | 96,586,948 | ACN9 | INTRON |
| rs12056091 | [T/C] | 0.442 | 0.0033 | 76 (2) | 96,596,607 | ACN9 | INTRON |
| rs1917939 | [T/C] | 0.319 | 0.0035 | 96,554,800 | [ACN9]# | . | |
| rs10953089 | [T/C] | 0.158 | 0.0050 | 92,566,363 | SAMD9 | LOCUS | |
| rs13228694 | [T/C] | 0.142 | 0.0050 | 99,778,243 | PILRB | INTRON | |
| rs7778571 | [A/G] | 0.233 | 0.0071 | 99,502,753 | ZNF3 | INTRON | |
|
| |||||||
| rs1548457 | [A/G] | 0.375 | 0.0108 | 93,030,646 | CALCR | INTRON | |
| rs3802030* | [A/C] | 0.254 | 0.0110 | 90,322,514 | PFTK1 | INTRON | |
| rs484491 | [T/C] | 0.358 | 0.0116 | 105,164,306 | . | ||
| rs7785892 | [T/C] | 0.136 | 0.0146 | 106,721,411 | COG5 | INTRON | |
| rs460 | [T/C] | 0.142 | 0.0161 | 97,252,046 | . | ||
| rs5015755 | [A/C] | 0.211 | 0.0184 | 99,851,338 | ZCWPW1 | INTRON | |
| rs41835* | [G/C] | 0.317 | 0.0201 | 106,052,801 | . | ||
| rs1485004 | [A/G] | 0.325 | 0.0201 | 95,108,442 | . | ||
| rs176574 | [C/G] | 0.392 | 0.0204 | 105,606,231 | . | ||
| rs10250228* | [T/C] | 0.433 | 0.0205 | 43 (2) | 92,939,016 | CALCR | INTRON |
| rs10271241 | [T/C] | 0.183 | 0.0220 | 90,138,516 | . | ||
| rs2428161 | [A/G] | 0.283 | 0.0226 | 104,379,803 | . | ||
| rs6465475 | [T/A] | 0.242 | 0.0245 | 95,162,803 | . | ||
| rs10487140 | [A/G] | 0.158 | 0.0263 | 95,163,077 | . | ||
| rs8 | [T/C] | 0.150 | 0.0272 | 92,246,265 | CDK6 | INTRON | |
| rs17862241 | [A/G] | 0.467 | 0.0280 | 103 (2) | 90,059,284 | . | |
| rs7795063 | [G/C] | 0.450 | 0.0302 | 106,165,365 | . | ||
| rs10264271 | [T/C] | 0.184 | 0.0309 | 100,957,895 | EMID2 | INTRON | |
| rs12113318 | [A/C] | 0.103 | 0.0315 | 102,471,663 | FBXL13 | INTRON | |
| rs999885 | [T/C] | 0.458 | 0.0320 | 99,539,112 | AP4M1 | INTRON | |
| rs9655757 | [T/A] | 0.167 | 0.0327 | 96,278,455 | . | ||
| rs9648946 | [T/G] | 0.483 | 0.0346 | 106,564,404 | PRKAR2B | INTRON | |
| rs7796370 | [A/C] | 0.192 | 0.0359 | 91,739,652 | ANKIB1 | INTRON | |
| rs3814098 | [G/C] | 0.136 | 0.0361 | 90,676,805 | PFTK1 | mRNA-UTR | |
| rs1858823 | [T/C] | 0.300 | 0.0371 | 93,856,176 | . | ||
| rs4278097 | [A/G] | 0.183 | 0.0385 | 11 (4) | 90,093,442 | . | |
| rs885972 | [G/C] | 0.183 | 0.0385 | 33 (2) | 90,099,133 | . | |
| rs12704555 | [A/G] | 0.183 | 0.0385 | 33 (2) | 90,101,159 | . | |
| rs4730025 | [T/A] | 0.142 | 0.0387 | 104,014,555 | LHFPL3 | INTRON | |
| rs1721492* | [A/G] | 0.217 | 0.0388 | 105,013,416 | . | ||
| rs2253833 | [T/C] | 0.233 | 0.039 | 106,992,905 | DUS4L | INTRON | |
| rs1227519 | [T/A] | 0.125 | 0.0420 | 95,156,136 | . | ||
| rs3801944 | [A/G] | 0.267 | 0.0424 | 10 (2) | 107,042,784 | BCAP29 | INTRON |
| rs3823646 | [A/G] | 0.400 | 0.0441 | 131 (2) | 99,595,548 | GAL3ST4 | NONSYNON |
| rs2023772 | [A/C] | 0.433 | 0.0455 | 43 (2) | 92,969,098 | CALCR | INTRON |
| rs10273733 | [T/C] | 0.267 | 0.0494 | 10 (2) | 107,045,357 | BCAP29 | INTRON |
| rs1968201 | [A/G] | 0.133 | 0.0504 | 291 (2) | 102,594,671 | LOC646485 | LOCUS |
| rs3087615 | [A/C] | 0.142 | 0.0504 | 291 (2) | 102,739,359 | PMPCB | NONSYNON |
SNPs showing significant deviations from Hardy Weinberg
HMMAF=HapMap Minor Allele Frequency
Note: Position, Gene location, and function as listed in NCBI. SNPs without a Bin ID listed were the only SNP genotyped in that bin. Bin size refers to the number of SNPs in the bin.
These SNPs are not annotated as ACN9 in NCBI, but are very near the gene.
Multiplex families of alcoholics were used in tests of association between each of the SNPs and each of the phenotypes studied, using the Pedigree Disequilibrium Test (PDT) (30). The PDT uses all available trios in a family (two parents plus child genotyped) as well as discordant siblings. The PDT-ave statistic was computed, which averages the association statistic over all families (30). No corrections were made for multiple testing in the initial analyses of the 1340 SNP screen set, as these analyses were considered the first stage to consider genes for further follow-up. In the second stage of analysis, SNPs were tested across the ACN9 gene. A Nyholt correction was applied to the data to determine significance after taking into account multiple testing in the presence of correlated SNPs (31). This method uses information on the pairwise linkage disequilibrium between the genotyped SNPs to compute the number of “effectively independent” SNPs. Using the updated method of Li and Ji (32), the effective number of SNPs (Meff) based on the 23 SNPs genotyped in this study was 10. With Meff = 10, the Bonferroni corrected significance threshold required across the gene is p=0.005.
Results
Table 1 shows the SNPs from the CIDR screen yielding evidence of association at p<0.050, with p-values <0.01 differentiated with a horizontal rule on the table. SNPs are listed in order of ascending p-values, with the most significant SNPs at the top of the table. The results for all 1340 genotyped SNPs are available in the supplementary on-line material. Of the eight SNPs listed in Table 1 with p<0.01, four were located in, or very near, the gene ACN9: rs10499934, rs7794886, rs12056091, and rs19179391. Although rs7794886 and rs12056091 are in the same bin and display high LD (HapMap r2=.93), the other two SNPs are in separate LD bins and provide independent evidence for association. Accordingly, additional genotyping was undertaken to more thoroughly investigate this gene. The results from association analyses of all SNPs genotyped across ACN9, including those genotyped by CIDR, are shown in Table 2. Twelve of the 23 genotyped SNPs in ACN9 were significant at p<0.05. Eight SNPs surpassed the Bonferroni corrected significance level suggested by the Nyholt correction of p<0.005. The top two most significant SNPs, rs10246622 (p=0.000098) and rs13475 (p=0.00014), showed substantial LD (r2 = 0.88), making haplotype analyses uninformative, as only the 1 1 and 2 2 haplotypes were observed with considerable frequency. Figure 1 shows the location of the ACN9 gene with reference to the linkage peak observed in the GAW data from which the region for the SNP screen was selected. ACN9 is located centromeric of the linkage peak, with a lod score of 3.08. The peak lod score was 4.08 in the region; thus, ACN9 is located at a 1 lod distance from the peak.
Table 2.
P-values from family-based association tests of SNPs located across ACN9.
| Marker | SNP location | Alleles | Position | MAF | P-value |
|---|---|---|---|---|---|
| rs1917939* | upstream of the ACN9 gene | T/C | 96554800 | 0.26 | 0.0035 |
| rs2037719* | A/G | 96557076 | 0.30 | 0.8918 | |
| rs17168169 | A/C | 96557937 | 0.24 | 0.0016 | |
| rs6958059 | G/A | 96559977 | 0.35 | 0.0223 | |
| rs1343646 | A/C | 96562388 | 0.22 | 0.0246 | |
| rs1880733 | A/C | 96562442 | 0.08 | 0.8201 | |
| rs1343645 | T/C | 96562521 | 0.11 | 0.0585 | |
| rs17168173 | G/T | 96563649 | 0.01 | 0.0675 | |
| rs10246622 | G/A | 96571993 | 0.33 | 0.000098 | |
| rs4729329 | T/A | 96573695 | 0.11 | 0.0911 | |
| rs10499934* | A/G | 96575512 | 0.22 | 0.0016 | |
| rs7794886* | T/C | 96586948 | 0.35 | 0.0033 | |
|
| |||||
| rs4398831* | intron 1 | A/G | 96594993 | 0.33 | 0.8436 |
| rs12056091* | T/C | 96596607 | 0.35 | 0.0033 | |
| rs2103212 | A/T | 96620240 | 0.07 | 0.1048 | |
| rs12670377 | A/C | 96624561 | 0.24 | 0.0054 | |
| rs10252691 | C/A | 96636756 | 0.10 | 0.0681 | |
| rs10256036 | A/T | 96637118 | 0.24 | 0.0022 | |
| rs10499936 | G/T | 96638937 | 0.01 | 0.1195 | |
| rs12671685 | G/A | 96644065 | 0.12 | 0.0269 | |
|
| |||||
| rs13475 | 3′ UTR | G/A | 96648665 | 0.36 | 0.00014 |
|
| |||||
| rs6970451* | downstream of the ACN9 gene | G/C | 96655200 | 0.29 | 0.2257 |
| rs6960395 | G/A | 96660805 | 0.34 | 0.9284 | |
Note: asterisk indicates SNPs genotyped as part of the original CIDR panel
SNPs significant at p<.005 are indicated in bold
MAF=Minor Allele Frequency, based on COGA data
Discussion
In this paper, we undertook a systematic screen of SNPs covering a 2 LOD support interval around a linkage peak previously reported for alcohol dependence (12; 14). This screen led to the identification of a novel gene that appears to affect susceptibility to alcohol dependence, ACN9. ACN9 was originally identified in a collection of respiratory-competent yeast mutants that were unable to utilize acetate as a carbon source (33). ACN9 appears to be involved in gluconeogenesis and is required for the assimilation of ethanol or acetate into carbohydrate (34). Little is known about the human homolog of the gene. Accordingly, this gene likely would not have been prioritized for investigation were it not for the results from the systematic screen of the linked region.
The most significant SNP in the CIDR screen, rs2157745, is located in a region with no genes listed in NCBI. It is ∼20kb past the 3′ end of SAMD9, making it unlikely that it would be involved in the function or expression of SAMD9. There is a large gene upstream (CDK6, a cyclin-dependent kinase), whose promoter might be in this region. One of the other top eight SNPs is located in SAMD9. This gene is believed to be involved in the regulation of extraosseous calcification, and has been associated with familial tumoral calcinosis (FTC), a rare autosomal recessive disorder characterized by the progressive deposition of calcified masses in cutaneous and subcutaneous tissues, resulting in ulcerative lesions and severe skin and bone infections (35). There is little evidence for expression of SAMD9 in the brain (35); thus, it was not considered a high priority for further follow-up for potential association with alcohol dependence.
We have previously reported association between alcohol dependence and related phenotypes and several genes located elsewhere on chromosome 7, including a muscarinic cholinergic receptor gene, CHRM2(12), and two taste receptor genes, TAS2R16(36) and TAS2R38(37). Those genes were selected based on proximity to a more distal linkage peak with an electrophysiological endophenotype (38), and none are located within the 2-lod interval of the linkage peak for alcohol dependence studied here. Finally, we note that at the time when the SNP selection took place for this study, only the Phase I build 16c.1 data were available from HapMap. Hapmap Phase II build 21 has added many additional SNPs, nearly doubling the number of nonsynonymous polymorphisms and creating many additional LD bins. In addition, we limited our SNP selection to SNPs with a minor allele frequency of >10%. This was because the SNP selection algorithm that was utilized takes advantage of LD patterns observed in the HapMap Caucasian population, and the statistics used to estimate LD are sensitive to the allele frequency. Accordingly, rarer SNPs would not have been detected using our screening strategy. Thus, the screening panel genotyped here did not exhaustively cover all genetic variation across the linkage peak, but was focused on common alleles that might contribute to risk for this common disease. We detected evidence of association with a novel gene ACN9 that appears to be involved in the predisposition to human alcohol dependence. Although little is known about the function of ACN9, data from yeast show that the yeast homologue to ACN9 is involved in acetate utilization suggesting that this gene may alter risk for alcohol dependence via alcohol metabolic pathways. Other genes in the metabolic pathway from alcohol to acetaldehyde to acetate are known to affect risk for alcoholism (39). Furthermore, comparison of the physical position of the SNPs associated with alcohol dependence with multiple alignments across all 28 vertebrate species (UCSC Genome Browser Human assembly hg18) indicates that 4 of the SNPs (rs17168169, rs1343646, rs12670377, and rs12671685) are located in evolutionally conserved regions spanning 34kb upstream of the gene, further suggesting its functional role during evolution. We hope that our finding of association with alcohol dependence will stimulate further research on the function of ACN9. In addition, we continue to follow-up other association signals with additional genotyping.
Supplementary Material
Acknowledgments
The Collaborative Study on the Genetics of Alcoholism (COGA) (Co-Principal Investigators: L. Bierut, H. Edenberg, V. Hesselbrock, B. Porjesz) includes nine different centers where data collection, analysis, and storage take place. The nine sites and Principal Investigators and Co-Investigators are: University of Connecticut (V. Hesselbrock); Indiana University (H. Edenberg, J. Nurnberger Jr., P.M. Conneally, T. Foroud); University of Iowa (S. Kuperman, R. Crowe); SUNY HSCB (B. Porjesz, H. Begleiter); Washington University in St. Louis (L. Bierut, A. Goate, J. Rice); University of California at San Diego (M. Schuckit); Howard University (R. Taylor); Rutgers University (J. Tischfield); Southwest Foundation (L. Almasy). Zhaoxia Ren serves as the NIAAA Staff Collaborator. This national collaborative study is supported by the NIH Grant U10AA08401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA). In memory of Henri Begleiter, Ph.D. and Theodore Reich, M.D. Principal and Co-Principal Investigators of COGA, we acknowledge their immeasurable and fundamental scientific contributions to COGA and the field. SS was supported by American Cancer Society grant IRG-58-010-50. NLS was supported by NIDA grant K01DA015129. Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, Contract Number N01-HG-65403.
Footnotes
Only rs7794886 and rs12056091 are listed as in ACN9 in Table 1 because these are the notations as listed in NCBI.
Financial Disclosures: All authors were provided with the Biological Psychiatry guidelines from the web for Disclosure of Biomedical Financial Interests and Potential Conflicts of Interest. Dr. Raymond Crowe reported that he has consulted with a law firm that is defending the Pfizer company in lawsuits against its product Zoloft. All other authors reported no interests to disclose related to this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Harwood H. Updating estimates of the economic costs of alcohol abuse in the United States: Estimates, update methods, and data. Report prepared by The Lewin Group for the National Institute on Alcohol Abuse and Alcoholism 2000 [Google Scholar]
- 2.Kessler RC, McGonagle KA, Zhao S, et al. Lifetime and 12-month prevalence of DSM-III-R psychiatric disorders in the United States. Results from the National Comorbidity Survey. Archives of General Psychiatry. 1994;51:8–19. doi: 10.1001/archpsyc.1994.03950010008002. [DOI] [PubMed] [Google Scholar]
- 3.McGue M. The behavioral genetics of alcoholism. Current Directions in Psychological Science. 1999;8:109–115. [Google Scholar]
- 4.Heath AC, Bucholz KK, Madden PA, et al. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychological Medicine. 1997;27:1381–1396. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- 5.Begleiter H, Reich T, Hesselbrock V, et al. The Collaborative Study on the Genetics of Alcoholism. Alcohol Health & Research World. 1995;19:228–236. [PMC free article] [PubMed] [Google Scholar]
- 6.Reich T. A genomic survey of alcohol dependence and related phenotypes: Results from the Collaborative Study on the Genetics of Alcoholism (COGA) Alcoholism: Clinical and Experimental Research. 1996;20:133A–137A. doi: 10.1111/j.1530-0277.1996.tb01763.x. [DOI] [PubMed] [Google Scholar]
- 7.Foroud T, Edenberg HJ, Goate A, et al. Alcoholism susceptibility loci: Confirmation studies in a replicate sample and further mapping. Alcoholism: Clinical and Experimental Research. 2000;24:933–945. [PubMed] [Google Scholar]
- 8.Begleiter H, Porjesz B. What is inherited in the predisposition toward alcoholism? A proposed model. Alcoholism: Clinical and Experimental Research. 1999;23:1125–1135. doi: 10.1111/j.1530-0277.1999.tb04269.x. [DOI] [PubMed] [Google Scholar]
- 9.Porjesz B, Rangaswamy M, Kamarajan C, Jones K, Padmanabhapillai A, Begleiter H. The utility of neurophysiological markers in the study of alcoholism. Clinical Neurophysiology. 2005;116:993–1018. doi: 10.1016/j.clinph.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 10.Dick DM, Jones K, Saccone N, et al. Endophenotypes Successfully Lead to Gene Identification: Results from the Collaborative Study on the Genetics of Alcoholism. Behavior Genetics. 2006;36:77–86. doi: 10.1007/s10519-005-9001-3. [DOI] [PubMed] [Google Scholar]
- 11.Reich T, Edenberg HJ, Goate A, et al. Genome-wide search for genes affecting the risk for alcohol dependence. American Journal of Medical Genetics. 1998;81:207–215. [PubMed] [Google Scholar]
- 12.Wang JC, Hinrichs AL, Stock H, et al. Evidence of common and specific genetic effects: Association of the muscarinic acetylcholine receptor M2 (CHRM2) gene with alcohol dependence and major depressive syndrome. Human Molecular Genetics. 2004;13:1903–1911. doi: 10.1093/hmg/ddh194. [DOI] [PubMed] [Google Scholar]
- 13.Bailey-Wilson JE, Almasy L, Edenberg HJ, et al. Genetic analysis workshop 14: Introduction to workshop summaries. Genetic Epidemiology. 2005;29(Supplement 1):S1–S6. [Google Scholar]
- 14.Dunn G, Hinrichs AL, Bertelsen S, et al. Microsatellites versus single-nucleotide polymorphisms in linkage analysis for quantitative and qualitative measures. BMC Genetics. 2005;6(1):S122. doi: 10.1186/1471-2156-6-S1-S122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wright MJ, Luciano M, Hansell NK, Montgomery GW, Geffen GM, Martin NG. QTLs identified for P3 amplitude in a non-clinical sample: Importance of neurodevelopmental and neurotransmitter genes. Biological Psychiatry. 2007 Oct 17; doi: 10.1016/j.biopsych.2007.09.002. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 16.Agrawal A, Heath AC, Saccone SF, et al. Poster 242. Alcoholism: Clinical and Experimental Research. 2007;39:69A. [Google Scholar]
- 17.Hill SY, Shen S, Zezza N, Hoffman EK, Perlin M, Allan W. A genome wide search for alcoholism susceptibility genes. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2004;128B:102–113. doi: 10.1002/ajmg.b.30013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bucholz KK, Cadoret R, Cloninger CR, et al. A new, semi-structured psychiatric interview for use in genetic linkage studies: A report on the reliability of the SSAGA. Journal of Studies on Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- 19.Hesselbrock M, Easton C, Bucholz KK, Schuckit M, Hesselbrock V. A validity study of the SSAGA--A comparison with the SCAN. Addiction. 1999;94:1361–1370. doi: 10.1046/j.1360-0443.1999.94913618.x. [DOI] [PubMed] [Google Scholar]
- 20.Feighner JP, Robins E, Guze SB, Woodruff RA, Winokur G, Munoz R. Diagnostic criteria for use in psychiatric research. Archives of General Psychiatry. 1972;26:57–63. doi: 10.1001/archpsyc.1972.01750190059011. [DOI] [PubMed] [Google Scholar]
- 21.Dick DM, Agrawal A, Wang JC, et al. Alcohol Dependence with Comorbid Drug Dependence:Genetic and Phenotypic Associations Suggest A More Severe Form of the Disorder With Stronger Genetic Contribution to Risk. Addiction. 2007;102:1131–9. doi: 10.1111/j.1360-0443.2007.01871.x. [DOI] [PubMed] [Google Scholar]
- 22.Nurnberger JI, Jr, Weigand R, Bucholz KK, et al. A family study of alcohol dependence: coaggregation of multiple disorders in relatives of alcohol-dependent probands. Archives of General Psychiatry. 2004;61:1246–56. doi: 10.1001/archpsyc.61.12.1246. [DOI] [PubMed] [Google Scholar]
- 23.Saccone SF, Pergadia ML, Loukola A, et al. Genetic linkage to chromosome 22q12 for a heavy-smoking quantitative trait in two independent samples. American Journal of Human Genetics. 2007;80:856–866. doi: 10.1086/513703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. American Journal of Human Genetics. 2004;74 doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinds DA, Stuve LL, Nilsen GB, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–9. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 26.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. American Journal of Human Genetics. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McPeek MS, Sun L. Statistical tests for detection of misspecified relationships by use of genome-screen data. American Journal of Human Genetics. 2000;66:1076–1094. doi: 10.1086/302800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boehnke M. Allele frequency estimation from pedigree data. American Journal of Human Genetics. 1991;48:22–25. [PMC free article] [PubMed] [Google Scholar]
- 29.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 30.Martin ER, Monks SA, Warren LL, Kaplan NL. A test for linkage and association in general pedigrees: The Pedigree Disequilibrium Test. American Journal of Human Genetics. 2000;67:146–154. doi: 10.1086/302957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. American Journal of Human Genetics. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005 doi: 10.1038/sj.hdy.6800717. advance on-line publication. [DOI] [PubMed] [Google Scholar]
- 33.Dennis RA, Rhodey M, McCammon MT. Yeast mutants of glucose metabolism with defects in the coordinate regulation of carbon assimilation. Archives of Biochemistry and Biophysics. 1999;365:279–288. doi: 10.1006/abbi.1999.1163. [DOI] [PubMed] [Google Scholar]
- 34.Dennis RA, McCammon MT. Acn9 is a novel protein of gluconeogenesis that is located in the mitochondrial intermembrane space. European Journal of Biochemistry. 1999;261:236–243. doi: 10.1046/j.1432-1327.1999.00267.x. [DOI] [PubMed] [Google Scholar]
- 35.Topaz O, Indelman M, Chefetz I, et al. A deleterious mutation in SAMD9 causes normophosphatemic familial tumoral calcinosis. American Journal of Human Genetics. 2006;79:759–764. doi: 10.1086/508069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hinrichs AL, Wang JC, Bufe B, et al. Functional variant in a bitter taste receptor (hTAS2R16) influences risk for alcohol dependence. American Journal of Human Genetics. 2006;78:103–111. doi: 10.1086/499253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang JC, Hinrichs AL, Stock H, et al. Functional variants in TAS2R38 and TAS2R16 influence alcohol consumption in high-risk families of African-American origin. Alcoholism: Clinical and Experimental Research. 2007;31 doi: 10.1111/j.1530-0277.2006.00297.x. [DOI] [PubMed] [Google Scholar]
- 38.Jones KA, Porjesz B, Almasy L, et al. Linkage and linkage disequilibrium of evoked EEG oscillations with CHRM2 receptor gene polymorphisms: Implications for human brain dynamics and cognition. International Journal of Psychophysiology. 2004;53:75–90. doi: 10.1016/j.ijpsycho.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Edenberg HJ, Xuei X, Chen HJ, et al. Association of alcohol dehydrogenase genes with alcohol dependence: A comprehensive analysis. Human Molecular Genetics. 2006;15:1539–1549. doi: 10.1093/hmg/ddl073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.