Abstract
In humans, mutations of the gene encoding for thyroid transcription factor-2 (TTF-2 or FOXE1) result in Bamforth syndrome. Bamforth syndrome is characterized by agenesis, cleft palate, spiky hair and choanal atresia. TTF-2 null mice (TTF-2−/−) also exhibit cleft palate, suggesting its involvement in the palatogenesis. However, the molecular pathology and genetic regulation by TTF2 remain largely unknown. In the present study, the recombinant expression vector pBROAD3-TTF-2 containing the promoter of the mouse ROSA26 gene was created to form the structural gene of mouse TTF-2 and was microinjected into the male pronuclei of fertilized ova. Sequence analysis confirmed that the TTF-2 transgenic mouse model was established successfully. The transgenic mice displayed a phenotype of cleft palate. In addition, we found that TTF-2 was highly expressed in the medial edge epithelium (MEE) from the embryonic day 12.5 (E12.5) to E14.5 in TTF-2 transgenic mice. These observations suggest that overexpression of TTF-2 during palatogenesis may contribute to formation of cleft palate.
Keywords: TTF-2, overexpression, transgenic mouse, cleft palate
Introduction
The formation of the secondary palate (palatogenesis) in mammals involves the orchestration of several processes to produce the correct separation of the oral and nasal cavities. Failure of palatogenesis results in cleft palate, one of the most common birth defects in humans [1].
Murine palatogenesis takes place between embryonic days 12.5 and 15.5 (E12.5-E15.5) [1]. Palatal shelves grow out bilaterally from maxillary prominences. Around E14.5, palatal shelves rapidly elevate to a horizontal position, and become adherent in the midline, before opposing palatal shelves finally fuse. During the initial stage of the fusion process, the epithelia covering the tip of each palatal shelf (MEE) adhere to form a midline epithelial seam (MES) [1]. The MES disappear through the combination of programmed cell death [2–4], epithelial-mesenchymal transformation (EMT) [5–7] and migration to the oral and nasal palatal epithelia [8]. Failure of any of these processes can result in isolated cleft palate.
Cleft palate can be caused by mutations in a variety of genes including transcription factors, growth factor receptors, extracellular matrix components, and cell surface adhesion molecules [9–17] and decreased malonylcarnitine [18]. Thyroid transcription factor-2 was first identified as a thyroid-specific DNA binding factor recognizing thyroglobulin (Tg) [19] and thyroperoxidase (TPO) [20] promoters. Zannini et al. [21] cloned the rat cDNA encoding TTF-2 and demonstrated that TTF-2 is a 42-kD forkhead-containing highly enriched protein in thyroid follicular cells. TTF-2, encoded by the TTF-2/foxe1 gene, is a promoter-specific transcriptional repressor [12] that displays both promoter and transcriptional activation domain specificity [22].
Forkhead domain-containing proteins are involved in embryonic pattern formation and regional specification. Forkhead genes have also been identified as factors that bind to regulatory elements in mammalian genes expressed in terminally differentiated cells [23]. Gene-targeting experiments have demonstrated that TTF-2/foxe1 null mice exhibit cleft palate [24]. In humans, mutations of the gene encoding for TTF-2 result in Bamforth syndrome, characterized by thyroid agenesis, cleft palate, spiky hair and choanal atresia [25].
The aim of this study was to investigate the phenotype of TTF-2 transgenic mice from E12.5 to E15.5. We demonstrated that overexpression of TTF-2 during the palatogenesis may contribute to cleft palate.
Materials and methods
Mouse breeding
C57BL/6J mice (Jackson Laboratory, Bar Harbor, MI, USA) were maintained at a temperature of 22°C on 12 hr light/12 hr dark cycles and were provided access to food and filtered water. C57BL/6J male and female mice (6 weeks old) were mated overnight. Vaginal plugs were observed the following morning which was considered day 0 of the embryo (E0).
Generation of plasmid vector
Recombinant plasmid expression vector pBROAD3-TTF-2 coding mouse TTF-2 protein was generated by our laboratory. It contained the ROSA26 gene promoter and the 1113 bp gene fragment whose codes start from the ATG initiation codon in the 593rd nt of the TTF-2 gene to the TGA termination codon at the 1705th nt. The plasmid pBROAD3- TTF-2 was linked by 3,-UTR and the poly(A) signal sequence of the human EF-1α gene. The full-length of pBROAD3-TTF-2 was 5.4 kb. The pBROAD3-TTF-2 was transfected into the bone marrow mesenchymal cells of C57BL/6J mice. Desired protein production was verified by western blotting analysis.
Preparing of the gene element for microinjection
The pBROAD3-TTF-2 was digested with PacI. The reaction condition was in 100 μl (400 μg) plasmid, 10× Buffer 30 μl, PacI 6 μl, 0.1% BSA 30 μl, 140 μl of deionized water at 37°C or 2 hrs. Following electrophoresis with 0.8% low melting point agarose gel, the gel slice containing the 5.4 kb target gene fragment was excised to purify the DNA fragment by using the Agarose Gel DNA Extraction Kit [QIAGEN China (Shanghai) Co., Ltd., Pudong, Shanghai, China].
Microinjection and fertilized ovum transplantation
C57BL/6J female mice (8–10 weeks old) were given intraperitoneal injection of PMSG (5 IU; Sigma-Aldrich, USA) and HCG (5 IU; Sigma-Aldrich Corp., St. Louis, MO, USA). The purified DNA was diluted in sterile PBS buffer at the final concentration of 2–6 μg Pml (about 2000 copy PPl). The dissolved DNA was microinjected into the male pronucleus of the fertilized ovum. Next, the fertilized ova were cultured and the 2-cell-stage embryos were transplanted into a corner of the uterus of the pseudopregnant C57BL/6J mice.
Genotyping of the transgenic foetus
The genotype of the mice was determined by PCR using genomic DNA extracted from tail biopsies of the transgenic foetus. The 5′-primer sequence of TTF-2 gene was 5′-ACTCCCAGTTCAATTACAGCTGTTG-3′. The 3′-primer sequence was 5′-GGGCGGCGGTTGGTGTTACGTTTGG-3′. The amplification was obtained with 35 cycles at 95°C for 1 min., 58°C for 1 min. and 72°C for 30 sec. after the mixture was incubated at 95°C for 10 min., followed by a final extension of 10 min. at 72°C. The PCR product was analysed by electrophoresis with 1% agarose gel (Sigma-Aldrich).
Southern blotting identification of the transgenic foetus
The integration of the TTF-2 transgene into the genome was determined by Southern blotting analysis using mouse-tail DNA. The recombinant plasmid expression vector pBROAD3- TTF-2 was digested separately with Kpn I and EcoR I. This resulted in a 0.7 kb fragment as the template, and was radio-labelled for making probes. The DNA isolated from TTF-2 transgenic mice was digested with EcoR I and separated by electrophoresis with 2% agarose gel (Sigma-Aldrich). The DNA was denatured, neutralized and transformed to a Hybond nylon membrane, fixed, hybridized and exposed to detect the gene by using the standard Southern blotting analysis protocol.
RT-PCR detection of the mRNA expression of the transgenic mice
TTF-2 mRNA was measured by quantitative real-time RT-PCR and GAPDH gene used as an endogenous control. Relative quantification of TTF-2 mRNA was determined by using the comparative CT method (2−ΔΔCt). Data were normalized to GAPDH mRNA levels, and values were expressed as means ± S.E.M. (arbitrary units) of four independent groups (palatal developmental time series:E12.5-E15.5). Dissected palatal shelves were immediately frozen in liquid N2, disrupted in RLT buffer (Qiagen), and total RNAs were isolated by using Qiagen RNeasy Kit and Qiagen Omnisscript RT and random hexamers were used for RT reaction. Pyrobest™ DNA polymerase and the following primers were used for PCR: the 5′-primer sequence was 5′-ATTATGATTATTATTTATTATATTA′, and the 3′-primer sequence 5′-CGCGC CGCCGTCGCT GGCCGCGCCG′.
Western blotting detection of the TTF-2 expression of the palatal tissues
Western blotting assays were performed according to the standard procedures. The palatal proteins were transferred to a PVDF membrane (Millipore Corporation, Billerica, MA, USA). The membranes were probed with primary antibody at 1:500 dilution (Goat anti-mouse TTF-2 polyclonal antibody IgG; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) and visualized with the secondary antibody at 1:2000 dilution (AP-Goat Anti-Rabbit IgG; Santa Cruz) and the ECL kit (Bio-Rad, Benicia, CA, USA). The membranes were stripped and re-probed with β-actin to correct for differences in protein loading. The immunoreactive protein bands were visualized on X-ray film with an enhanced chemiluminescence light (ECL)-detection system (Bio-Rad). Relative grey values of the Western blotting were quantitatively analysed by using the Bio-Rad software (Bio-Rad). Data are presented as the mean ± S.E.M. (arbitrary units) of four independent groups.
Immunohistochemistry detection of the expression of TTF-2 of the palatal tissues
The palatal shelves of control mice and transgenic mice on E12.5 to E15.5 were fixed with 4% paraformaldehyde and embedded in paraffin. Immunohistochemistry staining was performed in accordance with the manufacturer's instructions [26] and visualized by diaminobenzidine (DAB) staining. The same primary antibody was used at 1:200 dilution. The paraffin-embedded ovary cancer using anti-TTF-2 IgG was used as a positive control. No staining was observed when omitting primary antibody or replacing the primary antibody with isotype serum as a negative control. Immunoreactivity was analysed through Image Pro Plus software (Media Cybernetics, Bethesda, MD, USA). For every section, the integral optical density of every visual field was calculated.
Statistical analysis
Statistical analysis for all data was carried out by using the statistical package for social science (version 11.0, SPSS Inc, Chicago, IL, USA). The numerical values are presented as mean ± S.D. The results of RT-PCR, Western blotting and immunohistochemical staining were analysed with the Student's t-test. The comparison of means between groups was carried out by using one-way anova.
Results
The results of microinjection and fertilized ova transplantation
In this study, 1,203 fertilized ova were obtained and 982 fertilized ova were microinjected with TTF-2 gene. A total of 580 live fertilized ova were transplanted after microinjection to the oviducts of 29 pseudopregnant female mice. In all, 68 embryos were obtained for analysis. As shown in Figure 1A and C, palatal shelves were fused on E18 in the control groups, but were not fused on E18 in the TTF-2 transgenic mice (Fig. 1B and D).
Fig 1.
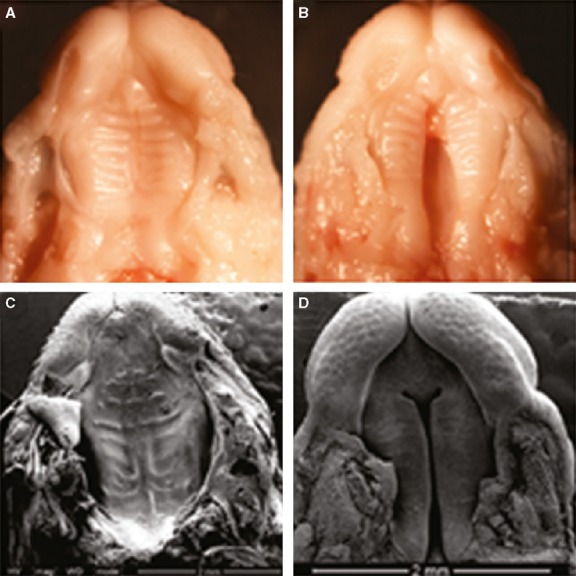
Micrographs and Scanning electron micrographs of the palatal shelves region of control mice and TTF-2 transgenic mice. (A) and (C) showed that the palatal shelves were fused on E18 in the control groups. However, the palatal shelves were not fused on E18 in the TTF-2 transgenic mice as shown in (B) and (D).
Six founder mice were bred with C57BL/6J mice to produce offspring (F1). The F1 mice with the integrated transgene were determined by PCR and Southern blotting analysis using tail DNA as templates. F1 mice mated brother-sister produced an F2 generation for further studies. One of founder mice failed to pass the transgene to offspring, probably due to the mosaic effect on the founder mouse. In total, 42 transgenic mice with cleft palate were born and died 24 hrs after their birth.
Gene expression determined by PCR and Southern blots
Among 42 transgenic mice and 68 embryos, a 784 bp positive band was seen in all samples by PCR detection (all the transgenic mice had a 784 bp positive band). Figure 2A shows the results of PCR using genomic DNA extracted from tail biopsies of the transgenic foetus. The DNA of the 72 positive TTF-2 PCR mice was then subjected to Southern blotting analysis. Figure 2B shows the results of Southern blots of the genome DNA of the positive PCR mice.
Fig 2.
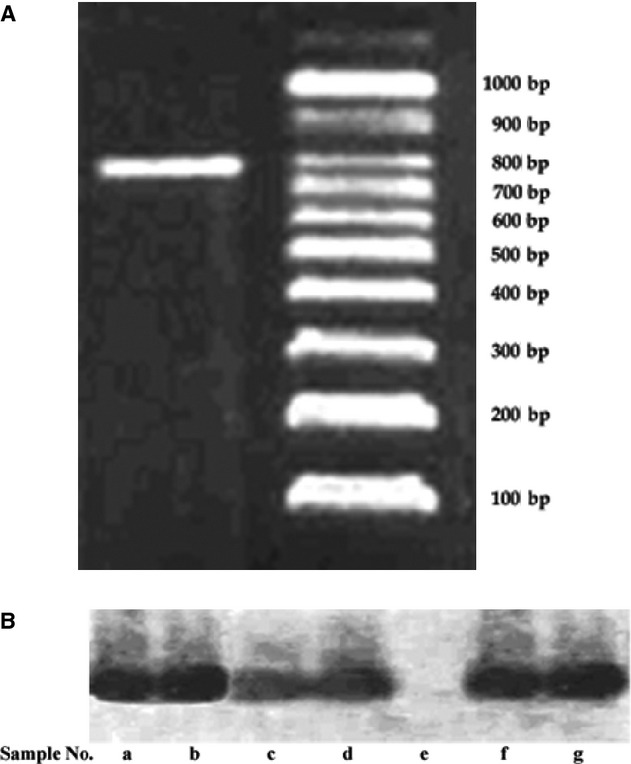
(A) Results of PCR using genomic DNA extracted from tail biopsies of the transgenic foetus. (B) Results of Southern blots detection of the genome DNA of the positive PCR transgenic mice.
TTF-2 mRNA expression of the palatal tissues
In the transgenic mice group, the 508 bp band was found on E12.5, 13.5, 14.5 and 15.5. In the control group, the 508 bp band was found on the E12.5, 13.5, 14.5. However, there was no 508 bp band on E15.5 in the control group (Fig. 3). The results of relative TTF-2 mRNA expression of the transgenic mice and control group mice are shown in Figure 4A. There was no statistical difference in the TTF-2 mRNA expression changes of the transgenic mice on E12.5, 13.5, 14.5 and 15.5(P > 0.05). The TTF-2 mRNA expression in the palatal tissues emerged on E12.5 in the control group. The expression level was the highest on E13.5 and began to decrease from E14.5. There was no detectable TTF-2 mRNA expression on E15.5(P < 0.05). TTF-2 mRNA expression was higher in the transgenic mice than in the control group on all other embryonic days(P < 0.05). As shown in Figure 4A, there was no apparent change in TTF-2 mRNA expression in the TTF-2 transgenic mice. However, there was a significant change in the control group mRNA expression, which reached its peak on E13.5 and started to decline thereafter. The TTF-2 mRNA expression was not detectable from E15.5.
Fig 3.
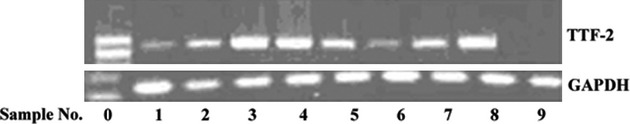
In the transgenic mice group, the 508 bp band was found on E12.5, 13.5, 14.5 and 15.5. In the control group, the 508 bp band was found on the E12.5, 13.5, 14.5 However, there was no 508 bp band found on E15.5 in the control group. 0:Marker;1: Positive control;2: E 12 Trs;3: E 13 Trs;4: E 14 Trs;5: E 15 Trs;6: E 12 Ctr;7: E 13 Ctr;8: E 14 Ctr;9: Ed15 Ctr (E: embryonic day;Trs: transgenic group;Ctr: control group).
Fig 4.
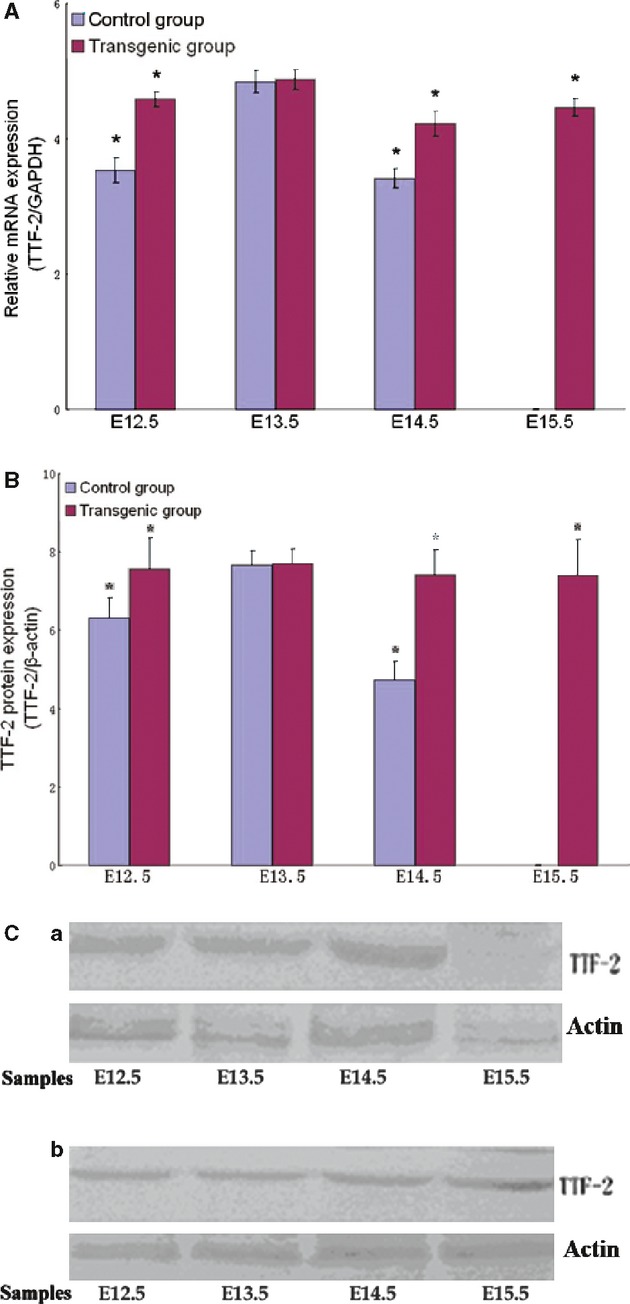
(A) Relative TTF-2 mRNA expression in the TTF-2 transgenic mice and the control group. As can be seen from (A), there was no obvious change on TTF-2 mRNA expression in the TTF-2 transgenic mice. However, there was an apparent change in the control group and reached its peak on E13.5 and started to decline later. The TTF-2 mRNA expression was not detected on E15.5. (B) TTF-2 protein expression in the TTF-2 transgenic mice and the control group. The change in tendency of the TTF-2 protein in the control group was basically consistent with the change in TTF-2 mRNA level. The tendency of the TTF-2 protein expression change of the TTF-2 transgenic mice was not obvious. (C) The Western blot results of the TTF-2 expression of the transgenic mice and control group on E12.5, 13.5, 14.5 and 15.5.
Western blotting analysis
The results of TTF-2 protein expression of the transgenic mice and control group mice are presented in Figure 4B and C. The pattern of the TTF-2 protein in the control group was basically consistent with that of the mRNA level. It has been suggested that the increase in TTF-2 transcription level is the main reason for the increased expression of TTF-2 protein. The tendency of TTF-2 protein expression of the TTF-2 transgenic mice was not visible. Figure 4C shows the Western Blot results of TTF-2 expression of the transgenic mice and control group on E12.5, 13.5, 14.5 and 15.5.
Immunohistochemistry detection of TTF-2 in palatal tissues
The murine palatal process protruded from the maxillary process on E12.5. In the control group, there were TTF-2 positive cells confirmed by immunohistochemical staining on the epithelial cells of the palatine process. The palatine process took place on a position of verticality on E13.5. TTF-2 was expressed in the epithelial cells on the surface of the palatine process and the staining was stronger on E13.5 than that on E12.5. The palatine process took place on a horizontal position on top of the tongue on E14.5. The surface of the palatine process was a single layer of epithelial cells. However, the TTF-2 staining was weaker than that of E13.5. The palatine processes on both sides became adherent on E15.5 (Fig. 5A–D).
Fig 5.
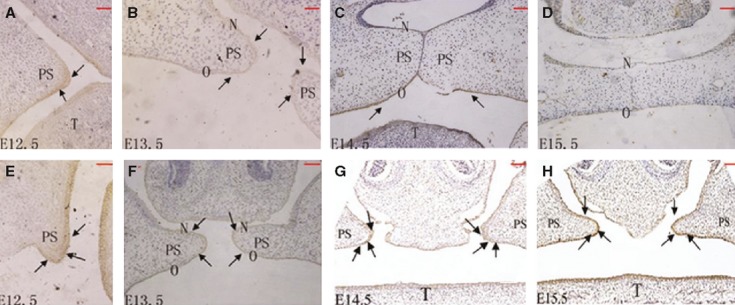
Immunohistochemical staining of TTF-2. TTF-2 expression in the palatal shelves on E12.5-15.5 in the control group mice (A–D) and in the transgenic mice (E–H).In the transgenic group, there was positive immunohistochemical staining of MEE cells, but no expression differences were found in the edge epithelium tissue of the palatine process from E 12.5 to E15.5. The positive immunohistochemical staining of MEE cells was increased in transgenic group compared with control group on the corresponding phase. N: nasal epithelium. O: oral epithelium. PS: palatal shelf. T: tongue. The positive immunohistochemical staining of MEE cells is indicated with arrows. Scale bars represent 50 μm.
In the transgenic group, TTF-2 positive MEE cells remained unchanged in the palatine process from E12.5 to E15.5. The number of positive cells was increased compared with that in the control group on the corresponding phase (Fig. 5E–H).
Discussion
It has been demonstrated that a variety of genes are involved in palatogenesis [27]. Mutations in genes are associated with regulating growth, differentiation, and EMT during the palatogenesis process, both in humans and in mouse models, all contributing to the formation of a cleft palate [28–30]. The pathogenesis of cleft palate, however, is still largely unknown, although genetic factors have been suggested.
Transgenic mice are now frequently used to create animal models to study the pathogenesis of human diseases. This system is likely to be the most sensitive assay for studying spatial and temporal regulation of a single gene product and its effect in the organism of a transgenic animal. The transgenic mouse is a particularly useful and popular model because its genome is very well characterized and can be manipulated with relative ease. However, animal models are relatively few in the field of human cleft palate. Transgenic models are valuable for studying the following points: (i) the identification of the tissue specificity of the expression directed by the promoter and (ii) the pathological effects linked to the expression of a single gene, enabling us to understand the pathogenesis of cleft palate.
The TTF-2, belonging to the forkhead/winged family, is a nucleoprotein. The pattern of TTF-2 expression during thyroid development has been extensively studied [21, 31]. Zannini et al. performed a detailed analysis of TTF-2 expression in thyroid precursors, to compare with the expression of TTF-1. TTF-2 is known to express in thyroid cell precursors at E 8.5 and could function as an inhibitor of differentiation in the early stages of thyroid development, perhaps by interfering with transcriptional activation of thyroid-specific gene expression by TTF-1 and, possibly, Pax-8 [21].
It was previously demonstrated that complete deletion or partial loss of its function may lead to cleft palate in both animal models (TTF-2−/− mouse) and human beings [28–30, 32]. The TTF-2 gene appears to play a role in normal palate development, as in situ expression studies [29] detect TTF-2 transcripts in developing palatal shelves. We carried out this experiment to understand the role of TTF-2 in the formation of palate. The overexpression of TTF-2 gene in the transgenic mice is useful for evaluating tissue specificity and temporo-spatial specificity of the TTF-2 gene. For this reason, we established the TTF-2 transgenic mice model for the first time, and analysed the expression patterns of the TTF-2 in the palate from E 12.5 to E 15.5.
The change in TTF-2 protein was less significant than that of mRNA, indicating that there are other regulatory mechanisms (post-transcription) involved, possibly via the phosphorylation of this protein. In the study, there was no significant difference in the TTF-2 mRNA and the TTF-2 protein in the palatal shelves on E13.5 between the transgenic and the control groups(P > 0.05). However, on the other embryonic days, the former expressions were higher than the control ones(P < 0.05)and there was no temporo-spatial change.
The expressions of TTF-2 in the MEE cells during palatogenesis in the control mice were found from E12.5 to E15.5 with a temporal-spatial change. The TTF-2 mainly expressed in the epithelium nucleolus, indicating its endogenous activation. The expression peak emerged in the later period of the growth of the palatine process. In the TTF-2 transgenic mice, it was indicated that TTF-2 had a high expression in MEE cells during the period of the palatal fusion. Previous studies revealed that the fate of the MEE, which is known to form the MES upon palatal shelf fusion, plays a very important role in palatal development. Three major mechanisms have been proposed for MEE degeneration: epithelial-mesenchymal transformation (EMT) [[6, 7, 33]]; MEE cell apoptosis (programmed cell death) [2]; and lateral migration of MEE cells [8]. Further studies will be needed to clarify how TTF-2 might affect MEE cellular functions. Does TTF-2 prevent apoptosis or epithelial to mesenchymal transformation?
The present study has demonstrated that TTF-2 was highly expressed in the palate at the key time point during palatogenesis in TTF-2 transgenic mice. To the best of our knowledge, this is the first report of the TTF-2 expression pattern and alterations during palatogenesis in TTF-2 transgenic mice.
The animal model established here may be useful for further studying the pathogenesis of cleft palate. In our study, we found that overexpression of TTF-2 caused the cleft palate. This might be a heredity mechanism for human cleft palate. Along with the identification of the TTF-2 downstream target genes, we can better understand the nature of the heredity mechanism by studying the abnormal TTF-2 expression. Our study indicates that TTF-2 may be involved in the palatogenesis and pathogenesis of cleft palate.
Acknowledgments
This research was supported by grants from the National Natural Science Foundation of China (Nos. 81000425 and 30530730) and Sichuan Key Technology R&D Program (No. 2010SZ0098).
Conflict of interest
The authors confirm that there are no conflicts of interest.
References
- 1.Ferguson MW. Palate development. Development. 1988;103:41–60. doi: 10.1242/dev.103.Supplement.41. [DOI] [PubMed] [Google Scholar]
- 2.Cuervo R, Covarrubias L. Death is the major fate of medial edge epithelial cells and the cause of basal lamina degradation during palatogenesis. Development. 2004;131:15–24. doi: 10.1242/dev.00907. [DOI] [PubMed] [Google Scholar]
- 3.Mori C, Nakamura N, Okamoto Y, et al. Cytochemical identification of programmed cell death in the fusing fetal mouse palate by specific labeling of DNA fragmentation. Anat Embryol. 1994;190:21–8. doi: 10.1007/BF00185843. [DOI] [PubMed] [Google Scholar]
- 4.Taniguchi K, Sato N, Uchiyama Y. Apoptosis and heterophagy of medial edge epithelial cells of the secondary palatine shelves during fusion. Arch Histol Cytol. 1995;58:191–203. doi: 10.1679/aohc.58.191. [DOI] [PubMed] [Google Scholar]
- 5.Fitchett JE, Hay ED. Medial edge epithelium transforms to mesenchyme after embryonic palatal shelves fuse. Dev Biol. 1989;131:455–74. doi: 10.1016/s0012-1606(89)80017-x. [DOI] [PubMed] [Google Scholar]
- 6.Griffith CM, Hay ED. Epithelial–mesenchymal transformation during palatal fusion: carboxyfluorescein traces cells at light and electronmicroscopic levels. Development. 1992;116:1087–99. doi: 10.1242/dev.116.4.1087. [DOI] [PubMed] [Google Scholar]
- 7.Shuler CF, Halpern DE, Guo Y, et al. Medial edge epithelium fate by cell lineage analysis during epithelial-mesenchymal transformation in vivo. Dev Biol. 1992;154:318–30. doi: 10.1016/0012-1606(92)90071-n. [DOI] [PubMed] [Google Scholar]
- 8.Carette MJ, Ferguson MW. The fate of medial epithelial cells during palatal fusion in vitro: an analysis by DiI labelling and confocal microscopy. Development. 1992;114:379–88. doi: 10.1242/dev.114.2.379. [DOI] [PubMed] [Google Scholar]
- 9.Wilkie AO, Morriss-Kay GM. Genetics of craniofacial development and malformation. Nat Rev Genet. 2001;2:458–68. doi: 10.1038/35076601. [DOI] [PubMed] [Google Scholar]
- 10.Pauws E, Stanier P. FGF signalling and SUMO modification: new players in the aetiology of cleft lip and/or palate. Trends Genetics. 2007;23:631–40. doi: 10.1016/j.tig.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 11.Jugessur A, Murray JC. Orofacial clefting: recent insights into a complex trait. Curr Opin Genet Dev. 2005;15:270–8. doi: 10.1016/j.gde.2005.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vieira AR, Avila JR, Daack-Hirsch S, et al. Medical sequencing of candidate genes for nonsyndromic cleft lip and palate. PLoS Genet. 2005;1:e64. doi: 10.1371/journal.pgen.0010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zucchero TM, Cooper ME, Maher BS, et al. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med. 2004;351:769–80. doi: 10.1056/NEJMoa032909. [DOI] [PubMed] [Google Scholar]
- 14.Riley BM, Mansilla MA, Ma J, et al. Impaired FGF signaling contributes to cleft lip and palate. Proc Natl Acad Sci USA. 2007;104:4512–7. doi: 10.1073/pnas.0607956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alkuraya FS, Saadi I, Lund JJ, Mass RL, et al. SUMO1 haploinsufficiency leads to cleft lip and palate. Science. 2006;313:1751. doi: 10.1126/science.1128406. [DOI] [PubMed] [Google Scholar]
- 16.Andreou AM, Pauws E, Jones MC, et al. TBX22 missense mutations found in patients with X-linked cleft palate affect DNA binding, sumoylation, and transcriptional repression. Am J Hum Genet. 2007;81:700–12. doi: 10.1086/521033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gritli-Linde A. Molecular control of secondary palate development. Dev Biol. 2007;301:309–26. doi: 10.1016/j.ydbio.2006.07.042. [DOI] [PubMed] [Google Scholar]
- 18.Hozyasz KK, Oltarzewski M, Dudkiewicz Z. Malonylcarnitine in newborns with non-syndromic cleft lip with or without cleft palate. Int J Oral Sci. 2010;2:136–41. doi: 10.4248/IJOS10047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Civitareale D, Lonigro R, Sinclair AJ, et al. A thyroid-specific nuclear protein essential for tissue-specific expression of the thyroglobulin promoter. EMBO J. 1989;8:2537–42. doi: 10.1002/j.1460-2075.1989.tb08391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Francis-Lang H, Price M, Polycarpou-Schwarz M, et al. Cell-type-specific expression of the rat thyroperoxidase promoter indicates common mechanisms for thyroid-specific gene expression. Mol Cell Biol. 1992;12:576–88. doi: 10.1128/mcb.12.2.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zannini M, Avantaggiato V, Biffali E, et al. TTF-2, a new forkhead protein, shows a temporal expression in the developing thyroid which is consistent with a role in controlling the onset of differentiation. EMBO J. 1997;16:3185–97. doi: 10.1093/emboj/16.11.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perrone L, Pasca di Magliano M, Zannini M, et al. The thyroid transcription factor 2(TTF-2) is a promoter-specific DNA-binding independent transcriptional repressor. Biochem Biophys Res Commun. 2000;275:203–8. doi: 10.1006/bbrc.2000.3232. [DOI] [PubMed] [Google Scholar]
- 23.Kaestner KH, Lee KH, Schlondorff J, et al. Six members of the mouforkhead gene family are developmentally regulated. Proc Natl Acad Sci USA. 1993;90:7628–31. doi: 10.1073/pnas.90.16.7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Felice M, Ovitt C, Biffali E, et al. A mouse model for hereditary thyroid dysgenesis and cleft palate. Nat Genet. 1998;19:395–8. doi: 10.1038/1289. [DOI] [PubMed] [Google Scholar]
- 25.Clifton-Bligh RJ, Wentworth JM, Heinz P. Mutations of the gene encoding human TTF-2 associated with thyroid agenesis, cleft palate and choanal atresia. Nat Genet. 1998;19:399–401. doi: 10.1038/1294. [DOI] [PubMed] [Google Scholar]
- 26.Harlow E, Lane D. Antibodies: a laboratory manual. New York: CSH Press; 1998. [Google Scholar]
- 27.Stanier P, Moore GE. Genetics of cleft lip and palate: syndromic genes contribute to the incidence of non-syndromic clefts. Hum Mol Genet. 2004;13:R73–81. doi: 10.1093/hmg/ddh052. [DOI] [PubMed] [Google Scholar]
- 28.Kaartinen V, Voncken JM, Shuler C, et al. Abnormal lung development and cleft palate in mice lacking TGF-beta3 indicates defects of epithelial–mesenchymal interaction. Nat Genet. 1995;11:415–21. doi: 10.1038/ng1295-415. [DOI] [PubMed] [Google Scholar]
- 29.Proetzel G, Pawlowski SA, Wiles MV, et al. Transforming growth factor-β3 is required for secondary palate fusion. Nat Genet. 1995;11:409–14. doi: 10.1038/ng1295-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lidral AC, Romitti PA, Basart AM, et al. Association of MSX1 and TGFβ3 with nonsyndromic clefting in humans. Am J Hum Genet. 1998;63:557–68. doi: 10.1086/301956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Damante G, Tell G, Di Lauro R. A unique combination of transcription factors controls differentiation of thyroid cells. Prog Nucleic Acid Res Mol Biol. 2001;66:307–56. doi: 10.1016/s0079-6603(00)66033-6. [DOI] [PubMed] [Google Scholar]
- 32.Mitchell LE, Murray JC, O'Brien S, et al. Evaluation of two putative susceptibility loci for oral clefts in the Danish population. Am J Epidemiol. 2001;153:1007–15. doi: 10.1093/aje/153.10.1007. [DOI] [PubMed] [Google Scholar]
- 33.Nawshad A, LaGamba D, Hay ED. Transforming growth factor beta (TGFbeta) signalling in palatal growth, apoptosis and epithelial mesenchymal transformation (EMT) Arch Oral Biol. 2004;49:675–89. doi: 10.1016/j.archoralbio.2004.05.007. [DOI] [PubMed] [Google Scholar]


