Abstract
Ribonucleotide reductase (RNR) is an attractive target for anticancer agents given its central function in DNA synthesis, growth, metastasis, and drug resistance of cancer cells. The current clinically established RNR inhibitors have the shortcomings of short halflife, drug resistance, and iron chelation. Here we report the development of a novel class of effective RNR inhibitors addressing these issues. A novel ligand-binding pocket on the RNR small subunit (RRM2) near the C-terminal tail was proposed by computer modeling and verified by site-directed mutagenesis and NMR techniques. A compound targeting this pocket was identified by virtual screening of the NCI diverse small molecule database. By lead optimization we developed the novel RNR inhibitor COH29 which acted as a potent inhibitor of both recombinant and cellular human RNR enzymes. COH29 overcame hydroxyurea and gemcitabine resistance in cancer cells. It effectively inhibited proliferation of most cell lines in the NCI 60 human cancer panel, most notably ovarian cancer and leukemia, but exerted little effect on normal fibroblasts or endothelial cells. In mouse xenograft models of human cancer, COH29 treatment reduced tumor growth compared to vehicle. Site-directed mutagenesis, NMR and surface plasmon resonance biosensor studies confirmed COH29 binding to the proposed ligand-binding pocket and offered evidence for assembly blockade of the RRM1-RRM2 quaternary structure. Our findings offer preclinical validation of COH29 as a promising new class of RNR inhibitors with a new mechanism of inhibition, with broad potential for improved treatment of human cancer.
Keywords: ribonucleotide reductase, drug discovery, inhibition mechanism, structure activity relationship, drug resistance
INTRODUCTION
Cancer remains a leading cause of death worldwide. In the US alone, annual incidence exceeds one million with more than 500,000 deaths. There is still an unmet need for novel highly effective and selective antitumor agents with low toxicity that do not readily elicit tumor resistance. An attractive target is ribonucleotide reductase (RNR), the only enzyme responsible for the de novo conversion of ribonucleoside diphosphate (NDP) to deoxyribonucleoside diphosphate (dNDP).(1–3) RNR is the key regulator of intracellular dNTP supply.(4) Maintenance of a balanced dNTP pool is a fundamental cellular function because the consequences of imbalance in the substrates for DNA synthesis and repair include mutagenesis and cell death. RNR expression and activity is therefore tightly regulated both in the cell cycle and at the DNA damage checkpoints. (3, 5) Targeted inhibition of RNR depletes dNTPs, and could lead to aberrant replication forks, S-phase checkpoint activation and cell cycle arrest.(5)
Human RNR is composed of α subunits (RRM1) that contain the catalytic site and two binding sites for enzyme regulators, and β subunits (RRM2) with a binuclear iron co-factor that generates the stable tyrosyl radical necessary for catalysis.(6) Reduction of NDP to dNDP at the RRM1 catalytic center requires formation of the active quaternary structure, and transfer of radicals generated in the RRM2 subunit (~ 45 kDa) to the RRM1 subunit (~85 kDa) via a proposed 35 Å proton-coupled electron transfer (PCET) pathway.(7) Until recently the active quaternary structure of RNR holoenzyme as well as the PCET pathway was unclear.(8) However, the C-terminal tail of RRM2 is involved in the RRM1-RRM2 interface formation and radical transfer. (9)
Normal cells with a low proliferative status express very low levels of RNR whereas neoplastic cells overexpress RNR to manufacture dNTP pools to support DNA synthesis and proliferation. While both RRM1 and RRM2 are required for RNR holoenzyme activity, each subunit has differing significance in cancer. Overexpression of RRM2 promotes transformation and tumorigenic potential via its cooperation with several activated oncogenes.(10) Conversely, overexpression of RRM1 suppresses malignant potential in vivo.(11) In cancer cells in vitro, increased expression of RRM2 increased drug-resistance and invasive potential, whereas RRM2 suppression reversed drug resistance and decreased proliferation.(12–15) Clearly, RNR is directly involved in tumor growth, metastasis, and drug resistance. Therefore RNR, especially the RRM2 subunit, is an important target for anticancer agents. Strategies for RNR inhibition include free radical quenching (radical scavenger), dinuclear iron center disruption (iron chelator), interfering with catalysis and regulation at the RRM1 subunit by nucleoside analogs, perturbation of critical interactions between subunits, and inhibition of RRM1 or RRM2 expression.(3, 16, 17)
There is clinical experience for three RNR inhibitors; hydroxyurea (HU), 3-aminopyridine-2-carboxaldehyde thiosemicarbazone (3–AP, Triapine®), and GTI2040. HU has a 30 year history as a cancer therapeutic agent, and blocks DNA synthesis by reducing the RNR free radical.(18) However, resistance to HU is readily developed, limiting its usefulness. 3-AP, which is in human phase II clinical testing relies on iron chelation. Toxicities reported from the phase I trial were hypoxia, respiratory distress, and methemoglobulinemia, apparently due to iron chelation in the patients’ red blood cells (RBC).(19) We observed that 3-AP was associated with iron chelation in inhibition of the alternate β subunit, p53R2.(20) The RRM2 antisense oligonucleotide, GTI2040, showed strong antitumor activity in animal models, however, it had no significant benefit in human phase II trials.(21, 22) In summary, the current RNR inhibitors have drawbacks including short half-life, enzyme recovery, and strong iron chelation. Effective and specific RNR inhibitors for the clinic have yet to be developed. We present here the development using structure- and mechanism-based approaches of a novel potent RNR inhibitor, COH29, which binds a novel pocket located at the RRM1 and RRM2 interface. COH29 could overcome the known drawbacks of the existing RNR inhibitors and is a potentially attractive antineoplastic agent.
MATERIALS & METHODS
Protein Expression, Purification, Qualitative and Quantitative Analysis
Native hRRM1 and hRRM2 and mutant hRRM2 for in vitro activity assays were expressed in E. coli strain BL21 (DE3) (Stratagene, La Jolla, CA), purified by Ni-NTA (Novagen) affinity chromatography, and qualitatively and quantitatively analyzed as previously described.(23) Vector pET28a containing the hRRM2 cDNA (pET-hRRM2) was the template for site-directed mutagenesis, essentially as previously described.(23) Oligonucleotide primers synthesized using an Applied Biosystems DNA/RNA synthesizer (Model 392) were designed to generate the following mutations by PCR: Gly223Val, Ser263Lys, Gly267Val, Asp271Ala, Arg330Ala, Glu334Arg, Met350Gly, Val336Lys, Tyr369Phe, and Met372Phe. For NMR studies, perdeuterated proteins were prepared by growing cells in 2H2O-M9 medium containing [U-2H] glucose.(24) Details are provided in Supplemental Data.
Virtual-screening workflow
A working database prepared from known RRM2 inhibitors combined with 1441 compounds from the cleaned Developmental Therapeutics Program (DTP) NCI Diversity Set free-for-public access database of compounds representing the NCI library of 250,253 compounds (NCI2000) was virtually screened against pocket 5 using SYBYL FlexX docking tool (Tripos-Certara, Inc.). DTP compounds that successfully docked into pocket 5 were ranked using an embedded consensus docking score (25) and compared to known hRRM2 inhibitors. The 80 compounds with the highest docking scores and binding energies superior to the known inhibitors were compiled into a hit list, and obtained from NCI for in vitro validation. Details of the procedure have been described previously.(26)
In vitro Activity and screening Assay
The activities of recombinant hRRM2/hRRM1 were measured using a modified [3H] CDP reduction assay (5) as previously described.(23) Measurement of RNR activity in cell lysates was as previously described.(27, 28) Hit compounds (100 µM) were initially screened using an in vitro RNR activity.
hRRM2 Structure Model, Docking, and NMR Validation
We previously described the protein structure model.(23) See the Supplemental Data for details of docking protocols. The FlexX docking procedure was as previously described.(26) One-dimensional (1D) 1H NMR and saturation transfer difference (STD) NMR (29) experiments were performed and analyzed as detailed in the Supplemental Data
Surface Plasmon Resonance Analysis
The surface plasmon resonance measurements were performed using Biacore T1000 equipped with hRRM2 immobilized onto research grade CM4 sensor chips. See the Supplemental Data for further details.
Real-time Cell Proliferation, Viability Assays, and Cell Cycle Analysis
The human epidermal carcinoma cell line KB was purchased from American Type Culture Collection (ATCC). Cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and 1% Penicillin-Streptomycin (GibcoBRL). Development of the Gem-resistant clone (KBGem) and hydroxyurea resistant clone (KBHU) have been described previously.(12, 13) Human normal dermal fibroblasts (NHDF) and umbilical vein endothelial cells (HUVEC) were purchased from Clonetics, Lonza, San Diego, CA. Cultures were maintained at 37°C in a humidified atmosphere of 5% CO2.
Cell proliferation was monitored using a W200 real-time cell electronic sensing analyzer (RT-CES) 16X workstation (Acea Biosciences) (30). Each well of the RT-CES plate was seeded with 2,500 cells. Twenty-four hours later, the index and curve of cell proliferation of quadruplicate samples were monitored and plotted every half hour for 120 hours. Viability was assessed by MTS assays performed according to manufacturer's instructions (CellTiter 96 AQueous Assay reagent; Promega) on 10 replicates of 2.500 cells/well in a 96-well plate treated with test drugs for 72 hours. The IC50 was the concentration of compound that reduced viability to 50% of a DMSO control. Cell cycle distribution and apoptosis were determined using a Becton Dickinson FACScalibur flow cytometer to detect propidium iodide and Annexin V signal as previously described.(30)
Mouse Tumor Xenografts
Based on protocol 07050 approved by the Institutional Animal Care and Use Committee of City of Hope, Female NSG mice (NOD/SCID/IL2Rgamma null, from Jackson Laboratories, Bar Harbor, Maine) aged 8 – 10 weeks were supplied by ARC of City of Hope. Each mouse was injected with 5×106 Molt-4 or TOV-112D cells (obtained from the ATCC) subcutaneously in the right flank, and tumor volume was monitored (0.5×l×w2). After the tumors reached approximately 70 mm3, COH29 in 30% solutol was administered daily by gavage in a one or two dose schedule. Mice were sacrificed on the 28th day after cancer cell transplantation
COH29 HPLC-MS/MS Assay
COH29 in mouse plasma was measured using an HPLC-tandem mass spectrometric assay. All reagents were purchased from Fisher Scientific (Madison, WI). Instrumentation consisted of an Agilent 1100 Capillary LC system (Agilent Technologies, Palo Alto, CA) in line with a Micromass Quattro Ultima Triple Quadrupole Mass Spectrometer (Micromass, Beverly, MA). Further details are in the Supplemental Data.
RESULTS
Protein structure description
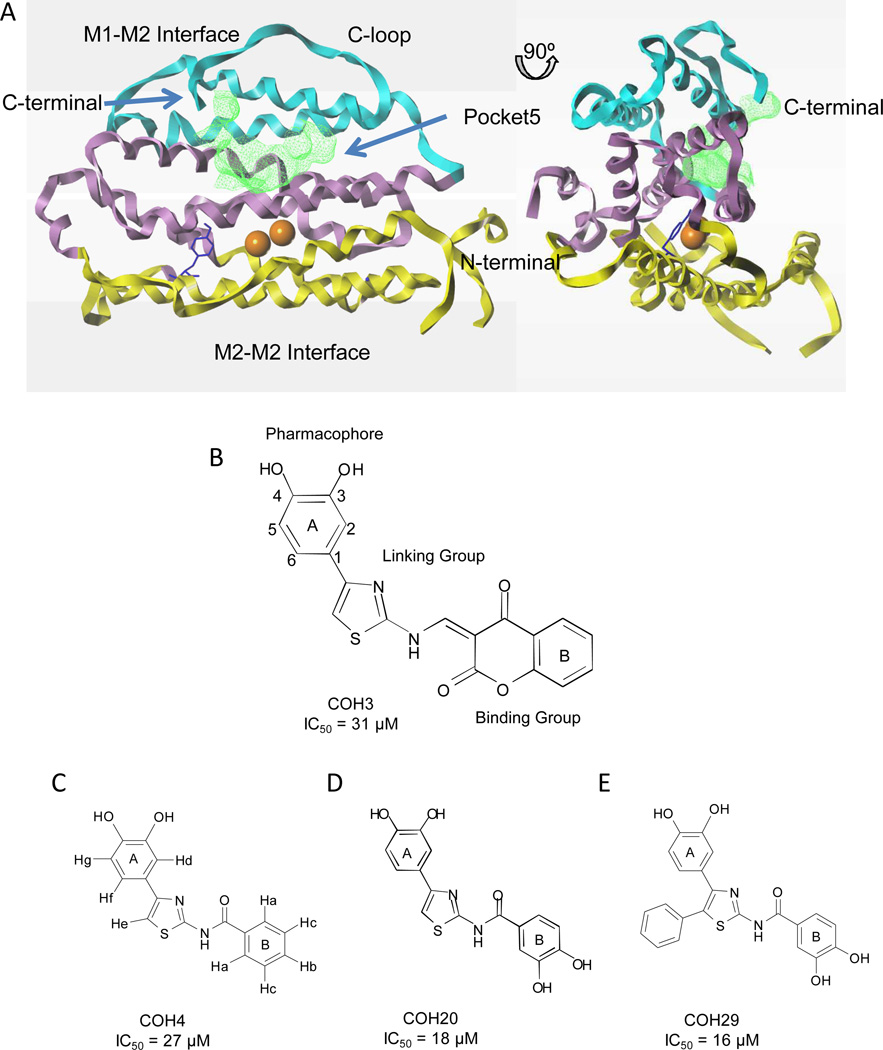
To design a novel class of specific RRM2 inhibitors, we used the only human RRM2 structure then available, PDB ID 2UW2. The main structural motif of hRRM2 is eight bundled long helices (αA to αH) with connecting shorter helices (α1 to α3) and loops. The unusual α-barrel structure has three layers of helices hosting a diiron-radical cofactor and a structurally flexible C-terminal tail (C-loop) (Fig. 1A) invisible by current biophysical techniques, but known to be at the RRM1-RRM2 interface and involved in the radical transfer (9). The exact active quaternary structure of the mammalian class Ia RNR holoenzyme is still unknown. The model of the R1R2 holoenzyme complex built based on the E. coli subunit structures, where the upper layer αH and the C-loop are close to the RRM1-RRM2 interface, remains the most accepted (31, 32).
Fig. 1. Identification of the hRRM2 ligand binding pocket, and the key compounds.
A) Two views of cartoon diagram of hRRM2 with the upper level (helices αG, αH; in cyan), middle level (helices αD, αE, αF; in violet), lower level (αA, αB, αC; in yellow), dinuclear irons (orange), radical harboring Y176 (blue), and pocket 5 colored green. C-loop, C-terminal, N-terminal, M1-M2 interface and M2-M2 interface are marked. The figure was drawn with Sybylx1.3. B-E) Structure and IC50 of the key compounds COH4, COH20, and COH29, which have substitutions in the binding and/or pharmacophore groups from COH3.
Ligand binding pocket identification
We identified five ligand binding pockets by mapping the hRRM2 structure. We discounted four pockets for either being too close to the iron center or too shallow. The largest cavity (pocket 5) had the greatest potential for RNR inhibition and was enclosed by 32 amino acid residues from the hRRM2 αE, αF, and αH helices, and the C-loop and is sufficiently far from the iron center to avoid ligand-induced iron chelation (Fig. 1A).
We tested these five pockets against known RRM2 ligands using the SYBYL FlexX built-in docking site search algorithm, which identifies the most fitted sites by mapping the entire protein (25). The majority of the test database compounds of about 100 known RR ligands including the inhibitors 3-AP, HU, and the HU derivative Schiff bases of hydroxysemicarbazide (SB-HSC), were docked into pocket 5 of hRRM2 by FlexX, consistent with the SiteID prediction. The most potent SB-HSC inhibitor of RRM2, compound HSC21 (20), docked at pocket 5 with numerous hydrogen bonds and charge interactions with the protein, indicating a reasonable shape and energetic fit, validating this pocket for further inhibitor screening
Inhibitor identification, SAR, and ligand-based optimization
We virtually screened the structure diversified NCI Diversity Set (NCI2000) for novel RRM2 inhibitors, then tested a hit list of 80 compounds using an in vitro RNR activity assay. Ten compounds inhibited the activity of the recombinant human RRM2/RRM1 complex by over 50%, and four by over 80%. Of these, compound NSC#659390 had the most favorable solubility and toxicity, and we used this formula to synthesize compound COH3 for further study.
COH3 is an aromatically substituted thiazole compound consisting of ring systems A and B flanking a linking group (Fig. 1B). We explored the pharmacophore features by structure-activity relationship (SAR) analysis of 20 COH3 analogues.(33) RNR inhibition was abolished by removing the 3, 4-dihydroxyl group from the phenyl ring, replacing it by 3, 5-dihydroxyl, or adding one more OH group, suggesting the dihydroxyl functional group at ortho position is mandatory for inhibition. Deleting the linking group, or replacing the binding group with bulkier or hydrophobic groups decreased RNR inhibition. Adding 3, 4-OH groups on the benzene (B) ring binding group (COH20, Fig. 1D) increased inhibition of recombinant RNR compared to COH3. We concluded that inhibition of RNR by the compound was likely due to radical quenching by the 3, 4-dihydroxyl group, which is proposed as the pharmacophore. The pharmacophore model that we generated for lead compound optimization is shown in Fig. S1.
Ligand binding mode by computer modeling
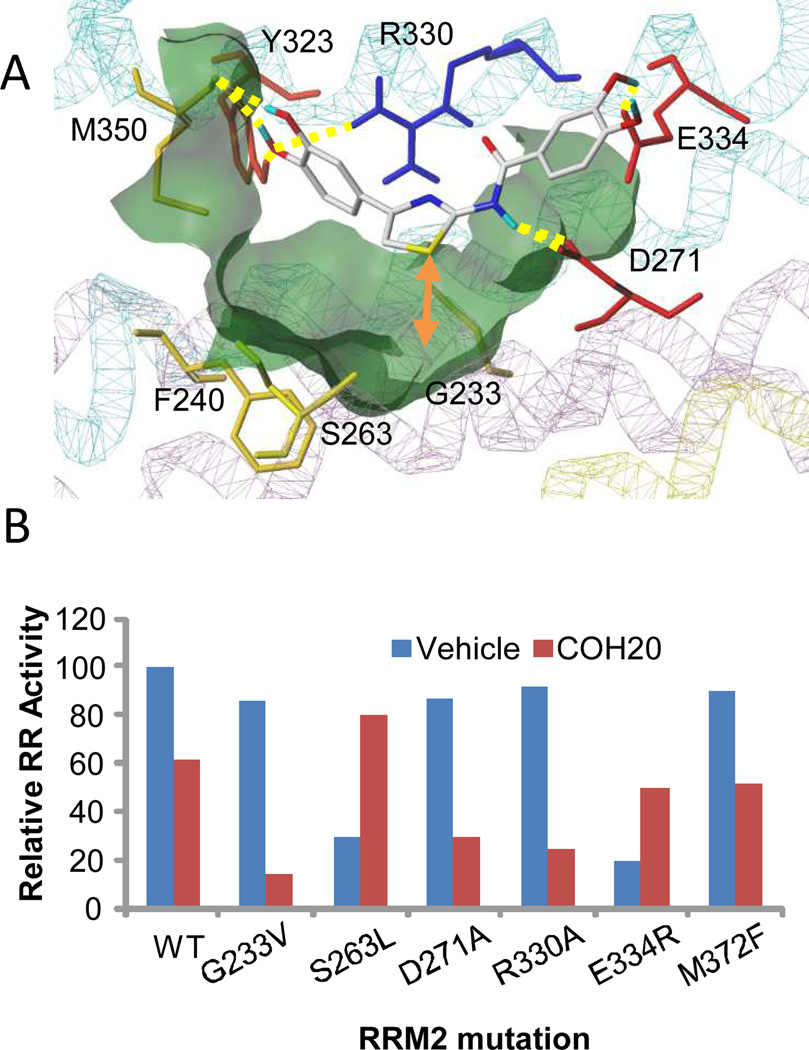
Pocket 5 is v-shaped with polar and charged residues at the surface opening and the tip touching the residues close to the interior diiron center (Figs. 1A, 2A). Docking studies of COH20 suggested several binding poses; however, the flat conformation had the best docking score and consistency with the experimental results. In this conformation, the pharmacophore group 3, 4-dihydroxyl of COH20 formed hydrogen bonds with Tyr323 and Met350, while the binding group extended outside the pocket, implying that structural hindrance between it, and the charged surface residues Asp271 and Glu334 prevented deep docking into the pocket. This is consistent with our SAR analysis showing that the bulkiness and hydrophobicity of the binding group attenuated inhibition. Other important features are that Tyr323 is the only pocket 5 tyrosine residue with a hydroxyl group of redox potential, and that the Arg330 at the edge of the pocket, between Tyr323 and Asp271, forms multiple hydrogen bonds with COH20 and caps the pocket.
Fig. 2. Validation of the ligand binding pocket.
A) The COH20 binding pose showing the intermolecular interaction with the key residues in the binding pocket. The surface of the binding pocket is colored green; protein structure shown with lines style; compound COH20 shown in stick rendering style and colored by atom types; hydrogen bondings are shown in yellow; the distance between COH20 and Gly233 is marked with orange arrow. B) Activity of recombinant RRM1/RRM2 proteins in the presence of vehicle (DMSO) or 10 µM COH20. Shown are results with wild-type RRM2 (WT), point mutations of five residues at the ligand binding pocket and one C-terminal residue
Site-directed mutagenesis study
We validated pocket 5 and the ligand binding mode by site-directed mutagenesis of twelve pocket 5 residues and two C-loop residues (Table 1). A non-pocket mutant E291A served as internal control. Six pocket residue mutations significantly altered or abolished RNR activity, indicating that they are critical to enzyme function and stability. We tested the effect in vitro of 10 µM COH20 on wild-type protein and six mutants; recombinant wild-type hRNR activity was inhibited by 40%, and RRM1/RRM2-R330A activity by over 75%, equating to a 40% increased sensitivity (Fig. 2B). We found similar results with G233V and D271A mutants, implicating these residues in ligand binding. In contrast, for both the S263L and E334R mutations inhibition was attenuated, indicating the positive roles for these residues in the inhibitor binding. The 40% increased COH20 inhibition by the C-terminal mutant M372F indicated that COH20 disturbed C-terminal function of the wild-type protein. One-dimensional (1D) 1H NMR and saturation transfer difference (STD) NMR validated these ligand-protein interactions and unambiguously confirmed Arg330 involvement in the ligand binding (Fig. S2, Table S1).
Table 1.
Relative enzyme activity of wild type and mutant hRRM2
| Protein | Relative Activity (%) | |
|---|---|---|
| Control | WT | 100 ± 5.6 |
| E291A | 99.8 ± 5.4 | |
| Ligand Binding |
G233V | 85.7 ± 3.5 |
| F236A | 9.2 ± 3.4 | |
| F240A | 3.1 ± 1.5 | |
| S263L | 29.8 ± 1.4 | |
| G267V | 0.0 ± 0.0 | |
| D271A | 86.9 ± 4.6 | |
| Y323F | 57.8 ± 0.1 | |
| F326A | 64.3 ± 4.9 | |
| R330A | 91.6 ± 3.5 | |
| E334R | 18.8 ± 2.0 | |
| N345A | 70.8 ± 0.9 | |
| M350G | 2.0 ± 2.8 | |
| C-loop | Y369F | 0.0 ± 0.0 |
| M372F | 89.6 ± 2.8 |
Protein activity expressed as percentage of dCDP formation (see Materials and Methods). Enzyme activity was measured in the presence of 0.5 µM hRRM1 and 2 µM hRRM2 in 100 µl total sample volume. Each value is the average of at least four determinations with deviations < 0.3.
Structure-based optimization of the lead compounds
We conducted another round of lead optimization based on structural features of the ligand binding pocket and the proposed flat conformation ligand binding mode, in which there is a space between COH20 and Gly233-hRRM2 of the αE-helix deep in the pocket (Fig. 2A). First we tested whether replacement of Gly233 with valine, which is longer and bulkier, could provide extra protein-ligand interactions. The G223V mutant retained most of the enzyme activity (80% of wild-type, Table 1 and Fig. S3B), however, 10 µM COH20 markedly decreased the in vitro relative activity to 10% for G233V vs 60% for wild-type protein, Fig. 2B). The point mutation contributed 50% activity loss, indicating Gly233 is involved in the inhibition and Val233 significantly contributes to this as predicted. By NMR analysis, G233V caused significant line broadening of COH20 (Fig. S2E), demonstrating the mutation changed the ligand-protein interaction exchange rates or binding kinetics. The sum of the results suggested that the inhibitory activity of COH20 could be optimized by addition of a side-chain. We synthesized and tested compounds with a variety of side chains.(33) The most effective was COH29 (Fig. 1E), N-(4–(3,4-dihydroxyphenyl)-5-phenylthiazol-2-yl)-3,4-dihydroxybenzamide, which had an extra phenyl ring structure from the linking aminothiazole ring of COH20 (Fig. 1D). COH29 inhibited recombinant RRM1/RRM2 activity in vitro with an IC50 of 16 µM. Docking of COH29 to hRRM2 showed a similar binding mode in pocket 5 as COH20 with the extra phenyl ring touching Gly233 as predicted (Fig. S3A). Mutagenesis studies confirmed residues Asp271, Tyr323, Phe326, Val327, and Met350 are involved in COH29 binding.
COH29 binds RRM2, interfering with RRM1-RRM2 interactions
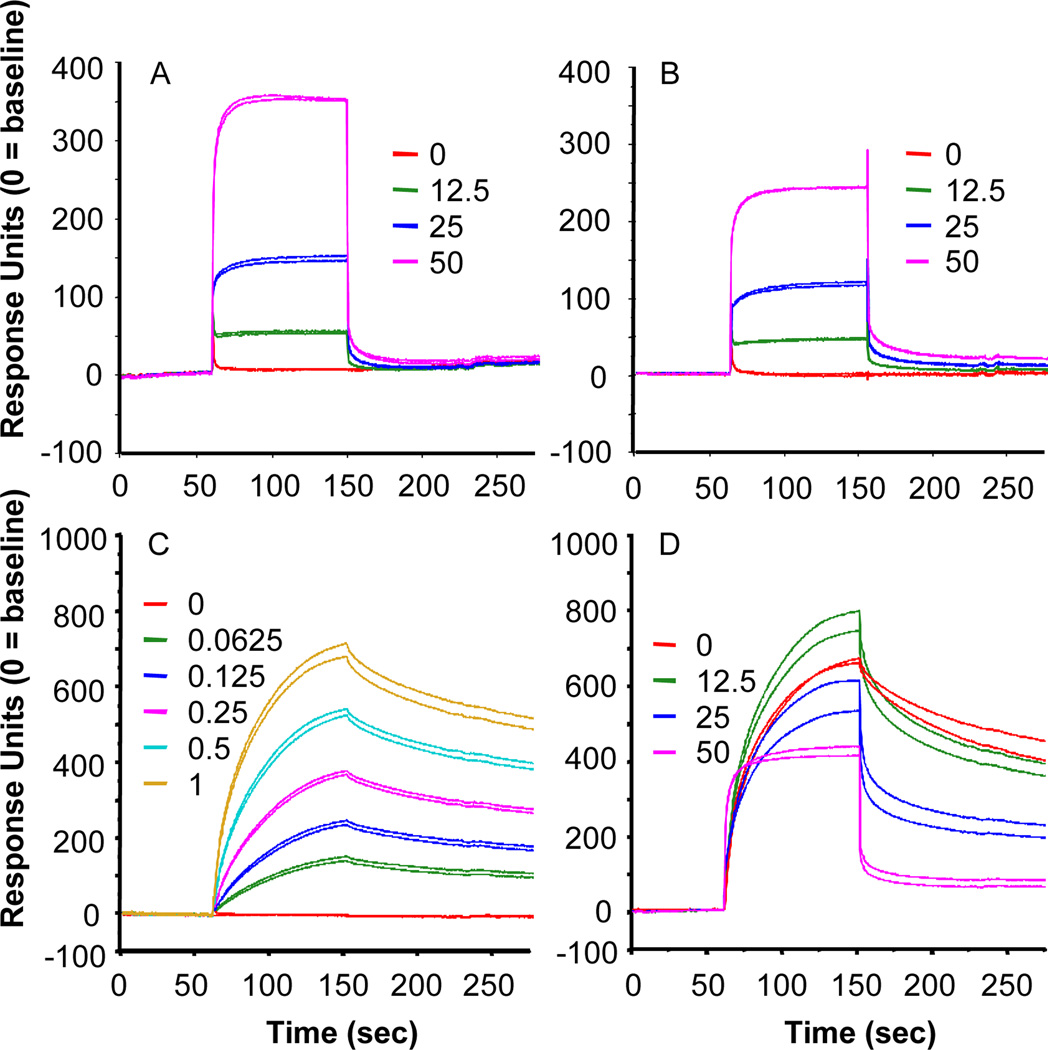
We used surface plasmon resonance biosensor experiments to study real-time COH29 – hRRM2 interaction. There was a dose-dependent interaction between immobilized full-length hRRM2 and COH29, reflected in the initial rising portion of the sensorgram (Fig. 3A). Removal of 41 C-terminal amino acids from hRRM2 greatly decreased COH29 binding (Fig. 3B), showing the C-terminal tail is important. Next, binding of RRM1 to immobilized RRM2 was confirmed over a range of concentrations of RRM1 (0 – 1 µM) (Fig. 3C). When a fixed concentration of RRM1 (0.5 µM) in the presence of escalating COH29 concentrations (0 – 50 µM) was tested with immobilized RRM2, COH29 interfered with RRM1-RRM2 binding in a dose-dependent manner (Fig. 3D). COH29 enhanced RRM1-RRM2 interaction at low dose (12.5 µM) but disrupted the complex at higher dose (> 25µM).
Fig. 3. COH29 interferes with the RRM1/RRM2 interaction.
Representative sensorgrams of the interaction of 0.0 – 50.0 µM COH29 with (A) immobilized full-length hRRM2, and (B) C-terminal truncated hRRM2, (C) 0.0 – 1.0 µM hRRM1 with immobilized hRRM2 (KD = 0.07844 µM), and (D) COH 29 (0.0 – 50.0 µM) and 0.5 µM hRRM1 interaction with immobilized hRRM2.
COH29 inhibits intracellular hRNR activity
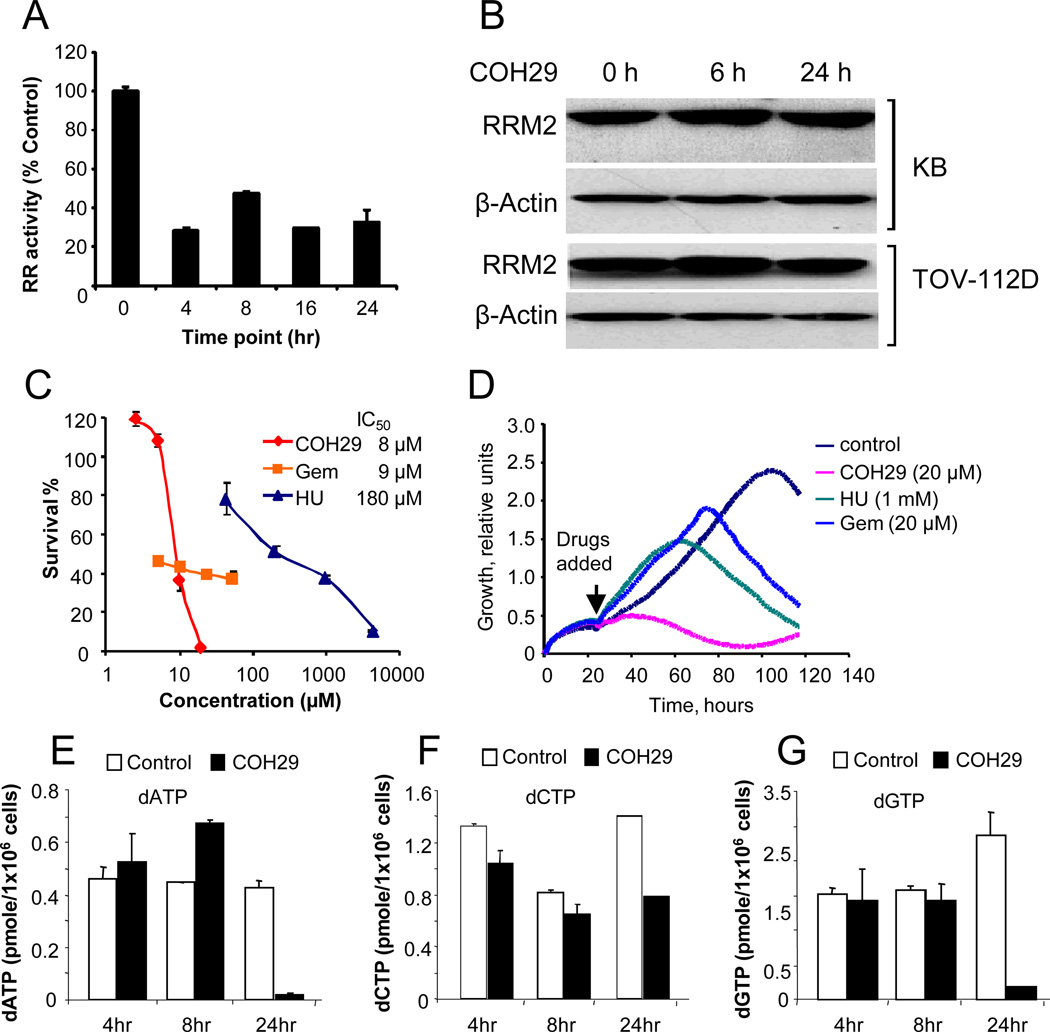
We showed that in cultured KB human cancer cells endogenously expressed RNR activity was approximately 50% decreased after 24 h incubation with 10 µM COH29 (Fig. 4A), whereas hRRM2 protein level was unaffected in both KB and TOV-112D cells (Fig. 4B). We compared the efficacy of COH29 to HU and gemcitabine (Gem) in KB cells. The IC50 of COH29 is 8 µM, similar to that of Gem (9 µM) but dramatically lower than that of HU (180 µM) in cell proliferation assays (Fig. 4C). Consistent with this, real-time KB cell growth was essentially halted by addition of 20 µM COH29 at 24 h, whereas cell growth continued after addition of either 20 µM Gem or 1 mM HU for approximately 60-70 hours (Fig. 4D).
Fig. 4. Effect of COH29 on RNR activity and cell growth.
A) Time course of RNR activity in the presence of 20 µM COH29 in KB cells in culture. The assay was performed in triplicate, shown are the mean ± standard deviation. B) Western blot of lysates of KB and TOV112D cell lines treated with 20 µM COH29 for the times indicated. Loading control is β-actin. C) 72-hr cell viability assay for KB cells in the presence of COH29, Gem, or HU. Shown as an insert in the graph are the estimated IC50 for each compound. D) Real-time proliferation curves of KB cells over 96 hr after the addition of RNR inhibitors as indicated. Each trace was an average of 4 replicates. E-G) dNTP Pools in Molt-4 cells at different time points in the presence and absence (control) of 10 µM COH29
Depletion of dNTP pools is a consequence of RNR inhibition. We assayed the levels of dATP, dCTP and dGTP at 4, 8, and 24 hours of exposure of MOLT-4 cells to 10 µM COH29. The dNTP levels were differentially affected, but by 24 h all pools were greatly decreased (Fig. 4E-G). Unbalanced dNTP pools should activate the replication checkpoint and result in S phase-arrest. Indeed, KB cells were arrested at S-phase after 48 h of COH29 treatment (Fig. S4, bottom), and underwent apoptosis (Fig. S4, top and middle).
COH29 and COH20 cytotoxicity in human cancer cells
The cytotoxicity of COH20 and COH29 was examined with the NCI broad panel of 60 human cancer cell lines. Impressive inhibition of most of the cancer cell lines was observed in in vitro MTT cytotoxicity assays, with ovarian cancer and leukemia cells being the most sensitive to 10 µM COH29 and to COH20 (Fig. S5). COH29 is more effective than COH20 in most of the cell lines. Importantly, COH29 was 10-fold less toxic to human normal fibroblast cells (NHDF) and human umbilical vein endothelial (HUVEC) cells (Fig. 5A, 5B) than to the KB cancer cell line (IC50 of 82 µM in NHDF).
Fig. 5. Activity of COH20 and COH29 in cancer and normal cell lines.
A) Dose-response 72 h viability curve for NHDF and (B) HUVEC cells. C-D) Activity of COH29 in drug-resistant clones; C) Comparison of 72 h viability of HU-resistant cells (KBHURs) to COH29 or HU, and D) 72 h viability of gemcitabine-resistant cells (KB-Gem) to COH29 or gemcitabine
COH29 overcomes resistance to Gem and HU
We used clones of KB resistant to HU (KB-HURs), and Gem (KB-Gem) as model systems to test COH29 in drug-resistant cells. COH29 retained activity in HU and Gem resistant cells in 72 h toxicity assays, with an IC50 of 8 µM in KB-HURs and KB-Gem (Fig. 5C, 5D). In contrast, HU was far less cytotoxic in the KBHURs line than in the parental line (IC50 320 µM vs 180 µM, respectively). As expected, gemcitabine was far less active in the resistant clone than in the KB parental line, as shown by the IC50 increasing from 9 µM to 1000 µM. The real-time growth curves for KBHURs in the presence of HU (2–50 mM) and COH29 (2–250 µM) clearly showed that the HU-resistant cells remained sensitive to COH29 (Fig. S6A). Real-time growth curves for KB-Gem in the presence of Gem (20–500 µM) and COH29 (2–250 µM) similarly showed that COH29 cytotoxic activity is strong and is unaffected by Gem resistance (Fig. S6B).
In vivo tumor growth inhibition and activity
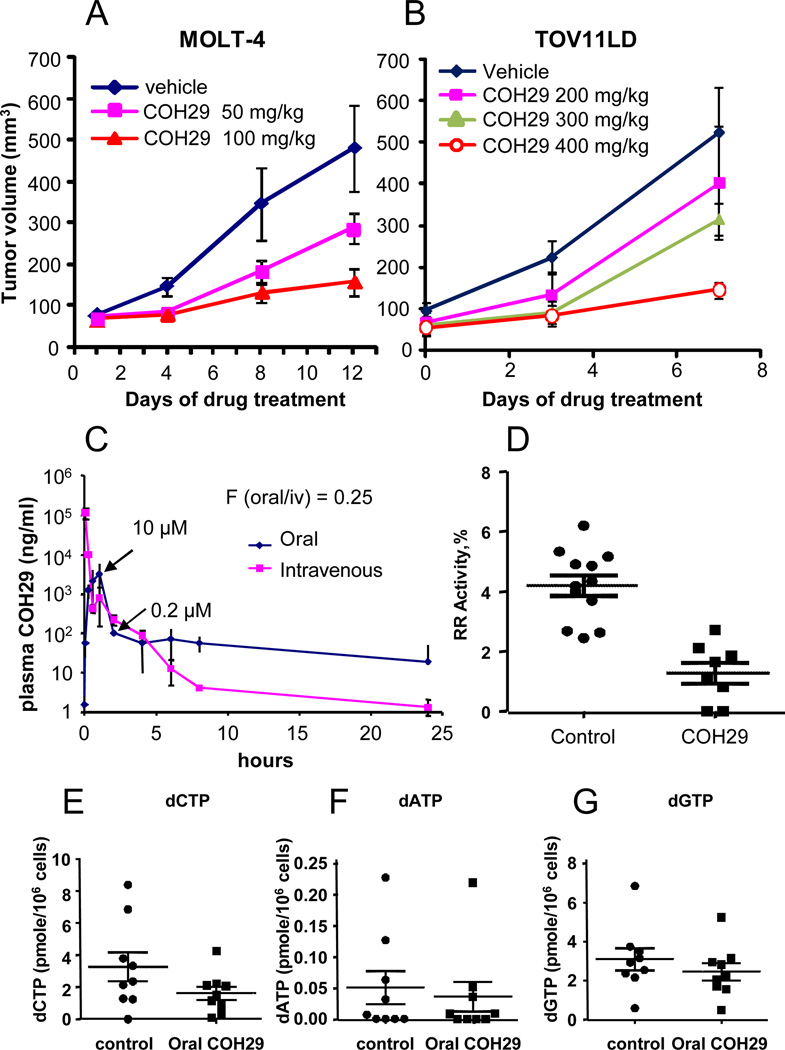
We investigated the antitumor activity of COH29 using murine tumor xenograft models of MOLT-4 leukemia cells or TOV11LD ovarian cancer cells, which were implanted and allowed to grow until the tumor was measurable at the subcutaneous site before oral administration of COH29 was begun. COH29 resulted in a dose-dependent inhibition of MOLT-4 tumor xenograft growth with twice-daily oral dosing at 50 mg/kg and 100 mg/kg, which was pronounced by Day 12 of treatment (Fig. 6A). Similarly, 7 days of treatment of mice bearing TOV11D xenografts with 200, 300, or 400 mg/kg/day COH29 resulted in a dose-dependent inhibition of tumor xenograft growth (Fig. 6B). Tumor growth was significantly inhibited compared with the control group.
Fig. 6. Activity of COH29 in mouse xenografts.
Subcutaneous xenograft growth curves for (A) MOLT-4 and (B) TOV112D in mice treated with oral COH29 daily as indicated. Shown are the average ± standard deviations for 4 animals/group. C) Pharmacokinetic analysis of 800 mg/kg COH29 administered orally or intravenously in mice. D) Activity of RNR in MOLT-4 tumor xenografts from mice treated with vehicle (solutol-15) or 100 mg/kg COH29. E-F) Intratumoral dNTP pools from MOLT-4 tumor xenografts.
Preliminary investigation of the pharmacokinetics (PK) of intravenous vs oral formulations of COH29 showed that following PO administration in female Balb/c nude mice at a dose 800 mg/kg in 30% solutol, COH29 was quickly absorbed with a mean Tmax of 0.5 h, and was slowly eliminated with a mean terminal elimination t1/2 of 10h (Fig. 6C). The COH29 peak plasma concentration was 10 µM. COH29 decreased to approximately 0.2 µM by 24 h. Compared to the intravenous route of administration, the oral bioavailability of COH29 was determined to be 25%. COH29 was also well tolerated in mice, with no appreciable side effects observed up to a daily dose of 500 mg/kg.
RNR activity in extracts of the MOLT-4 tumor xenografts shown in Fig. 6A was dramatically lower in the tumors from mice that had been treated with COH29 for 12 days (Fig. 6D), confirming that COH29 inhibits its target in vivo. Consistent with decreased RNR activity, intratumoral dNTP pools were also decreased in xenografts from mice treated with COH29 vs. control (Fig. 6E-G).
DISCUSSION
Using a structure- and mechanism-based approach we designed and developed a novel class of RNR inhibitors with potential clinical utility, having better drug resistance and cytotoxicity profiles than the clinically established agents. COH29 inhibits RNR activity in vitro in the micromolar range and is active in tissue culture and human tumor xenografts in mice. Using the NCI panel of 60 cancer cell lines we established that COH29 was broadly active, with ovarian cancer and leukemic cell lines being particularly susceptible. In contrast, cultured non-malignant human cells - dermal fibroblasts and HUVECs – were 10 times less sensitive to COH29 in viability assays, an encouraging indication that this compound would be relatively non-toxic in vivo where normal cell replication is lower than in cell culture. COH29 did not affect the expression of RRM2 in vitro, and we conclude that in vitro inhibition of RNR is via a direct effect on the enzyme activity and not by regulation of protein expression. Further, in cell cultures and in tumor xenografts we consistently detected the primary biological consequence of RNR inhibition - perturbation of dNTP pools. Consistent with inhibition of RNR and its established role of catalyzing the rate-limiting step in dNTP synthesis and therefore DNA synthesis (1, 2) we observed S-phase arrest in cell cultures treated with COH29.
Our successful development of COH29 is based on the novel binding pocket we identified (33), which is located in a position close to the R1-R2 interface that makes it potentially capable of multiple functional and biologically relevant effects. Notably, at the pocket apex the helix E residues Gly233 and Phe236 are close to the interior diiron center; near the surface residues Asp271 R330 and Glu334 form a charged cluster opposite the hydrophobic moiety capable of specific aromatic interactions formed by residues Phe236, Phe240, Tyr323, and Phe326. The C-loop of hRRM2 encloses the pocket close to the RRM2-RRM1 interface. Potential inhibitory ligand modes of action include: interference with RRM1-RRM2 holoenzyme assembly and radical transfer, disturbance of the iron center, and blocking the oxygen passage channel. The surface plasmon resonance biosensor experiments confirmed that COH29 binds hRRM2, involving the hRRM2 C-terminal tail; more importantly, COH29 interferes with RRM1-RRM2 interactions, therefore disrupting holoenzyme complex formation. This study suggested a new RNR inhibition mechanism acting via interference with the RRM2 side of the RRM1-RRM2 interface.
Currently many groups are investigating several proposed mechanisms for RNR inhibition (34–36). Recently Gräslund’s group proposed a binding pocket in a similar position to our pocket 5 in a molecular docking study of metal complexes of Triapine (3–AP) using a mouse RRM2 model (36). They found two binding pockets because of the smaller size of their ligand, which also docked deeper into the iron site. A mechanism of labilization of the RRM2 diferric center by 3-AP and subsequent 3-AP action as an iron chelator was proposed. In line with their finding, we noted that mutations around the tip of pocket 5 significantly reduced the enzyme activity (F236A, G267V; Table 1, Fig. S3B), supporting disturbance of the iron center by 3-AP. We note that clinically 3-AP produced iron-chelation-related toxicities (19) that we predict COH29 will avoid as our models indicate that it does not bind as deep into the iron site.
COH29 is a potential radical quencher due to the 3, 4-dihydroxybenzene group, which has a reduction potential lower than tyrosine (37). In a similar compound, 3, 4-dihydroxyphenylalanine (DOPA), the 3, 4-dihydroxyl group was readily oxidized and served as a radical trap (37). Substitution of the C-terminal residue in the PCET pathway, Tyr356 in E. coli RRM2 (Tyr369 in hRRM2), with DOPA resulted in the formation of a DOPA radical intercepting the radical transfer pathway and diminishing RNR activity (38, 39). COH29 binds close to the C-terminal loop and helix, and it is possible that it inhibited the enzyme activity by ligand-radical interception. Therefore, the ligands of the novel binding pocket could have multiple modes of inhibition of RNR activity, which makes the pocket an important and effective target for therapeutic agent design. The multiple functional features of the pocket and the structural features of COH29 may allow for multiple modalities of interference with RRM2 functions, in turn resulting in the high efficacy of the inhibitor.
We also compared the activity of COH29 to the clinically established RNR inhibitors HU and gemcitabine, neither of which are specific RNR inhibitors (17), in tumor cell cultures. In addition to off-target effects, acquired resistance to these drugs has limited their clinical effectiveness. For nearly two decades we have used the KB epithelioid cell line to study RNR and have derived HU and gemcitabine-resistant clones (12, 13). In addition to showing that in KB cells COH29 has similar antiproliferative activity to Gem, and is over 20-fold more active than HU, we showed that COH29 activity was unaffected in the drug resistant KBHURs and KB-Gem clones. This indicates that COH29 bypasses the mechanisms by which resistance to HU and gemcitabine develop. This is most likely due to the differences in the mechanisms of action. Unlike COH29, gemcitabine binds the RRM1 subunit, while HU interaction with the RRM2 subunit is less specific and does not involve pocket 5. (17) Resistance to gemcitabine and HU in the KB clones is at least partially mediated by overexpression of RRM2. (12, 13) Interestingly, we observed no increase in RRM2 expression in response to COH29. Similar to our results with COH29, others have found that 3-AP is active in KBHURs.(40) Taken together with preliminary evidence that COH29 is orally bioavailable and is well tolerated in mice all evidence indicates that COH29 is an attractive antineoplastic agent for further development.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NCI grant CA 127541-01 for each author. Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under grant number P30CA033572.
We would like to thank Dr. Frank Un of City of Hope for helpful discussion.
Footnotes
No potential conflicts of interest were disclosed.
REFERENCES
- 1.Jordan A, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 1998;67:71–98. doi: 10.1146/annurev.biochem.67.1.71. [DOI] [PubMed] [Google Scholar]
- 2.Kolberg M, Strand KR, Graff P, Andersson KK. Structure, function, and mechanism of ribonucleotide reductases. Biochim Biophys Acta. 2004;1699:1–34. doi: 10.1016/j.bbapap.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Cerqueira NM, Fernandes PA, Ramos MJ. Ribonucleotide reductase: a critical enzyme for cancer chemotherapy and antiviral agents. Recent Pat Anticancer Drug Discov. 2007;2:11–29. doi: 10.2174/157489207779561408. [DOI] [PubMed] [Google Scholar]
- 4.Julias JG, Pathak VK. Deoxyribonucleoside triphosphate pool imbalances in vivo are associated with an increased retroviral mutation rate. J Virol. 1998;72:7941–7949. doi: 10.1128/jvi.72.10.7941-7949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, et al. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 6.Larsson A, Sjoberg BM. Identification of the stable free radical tyrosine residue in ribonucleotide reductase. EMBO J. 1986;5:2037–2040. doi: 10.1002/j.1460-2075.1986.tb04461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ekberg M, Potsch S, Sandin E, Thunnissen M, Nordlund P, Sahlin M, et al. Preserved catalytic activity in an engineered ribonucleotide reductase R2 protein with a nonphysiological radical transfer pathway. The importance of hydrogen bond connections between the participating residues. J Biol Chem. 1998;273:21003–21008. doi: 10.1074/jbc.273.33.21003. [DOI] [PubMed] [Google Scholar]
- 8.Uppsten M, Farnegardh M, Domkin V, Uhlin U. The first holocomplex structure of ribonucleotide reductase gives new insight into its mechanism of action. J Mol Biol. 2006;359:365–377. doi: 10.1016/j.jmb.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 9.Stubbe J, Nocera DG, Yee CS, Chang MC. Radical initiation in the class I ribonucleotide reductase: long-range proton-coupled electron transfer? Chem Rev. 2003;103:2167–2201. doi: 10.1021/cr020421u. [DOI] [PubMed] [Google Scholar]
- 10.Fan H, Villegas C, Huang A, Wright JA. The mammalian ribonucleotide reductase R2 component cooperates with a variety of oncogenes in mechanisms of cellular transformation. Cancer Res. 1998;58:1650–1653. [PubMed] [Google Scholar]
- 11.Fan H, Huang A, Villegas C, Wright JA. The R1 component of mammalian ribonucleotide reductase has malignancy-suppressing activity as demonstrated by gene transfer experiments. Proc Natl Acad Sci U S A. 1997;94:13181–13186. doi: 10.1073/pnas.94.24.13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou BS, Hsu NY, Pan BC, Doroshow JH, Yen Y. Overexpression of ribonucleotide reductase in transfected human KB cells increases their resistance to hydroxyurea: M2 but not M1 is sufficient to increase resistance to hydroxyurea in transfected cells. Cancer Res. 1995;55:1328–1333. [PubMed] [Google Scholar]
- 13.Goan YG, Zhou B, Hu E, Mi S, Yen Y. Overexpression of ribonucleotide reductase as a mechanism of resistance to 2,2-difluorodeoxycytidine in the human KB cancer cell line. Cancer Res. 1999;59:4204–4207. [PubMed] [Google Scholar]
- 14.Zhou BS, Tsai P, Ker R, Tsai J, Ho R, Yu J, et al. Overexpression of transfected human ribonucleotide reductase M2 subunit in human cancer cells enhances their invasive potential. Clin Exp Metastasis. 1998;16:43–49. doi: 10.1023/a:1006559901771. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Zhou B, Xue L, Yen F, Chu P, Un F, et al. Ribonucleotide reductase subunits M2 and p53R2 are potential biomarkers for metastasis of colon cancer. Clin Colorectal Cancer. 2007;6:374–381. doi: 10.3816/CCC.2007.n.007. [DOI] [PubMed] [Google Scholar]
- 16.Nocentini G. Ribonucleotide reductase inhibitors: new strategies for cancer chemotherapy. Crit Rev Oncol Hematol. 1996;22:89–126. doi: 10.1016/1040-8428(95)00187-5. [DOI] [PubMed] [Google Scholar]
- 17.Shao J, Zhou B, Chu B, Yen Y. Ribonucleotide reductase inhibitors and future drug design. Curr Cancer Drug Targets. 2006;6:409–431. doi: 10.2174/156800906777723949. [DOI] [PubMed] [Google Scholar]
- 18.Nyholm S, Thelander L, Graslund A. Reduction and loss of the iron center in the reaction of the small subunit of mouse ribonucleotide reductase with hydroxyurea. Biochemistry. 1993;32:11569–11574. doi: 10.1021/bi00094a013. [DOI] [PubMed] [Google Scholar]
- 19.Yen Y, Margolin K, Doroshow J, Fishman M, Johnson B, Clairmont C, et al. A phase I trial of 3- aminopyridine-2-carboxaldehyde thiosemicarbazone in combination with gemcitabine for patients with advanced cancer. Cancer Chemother Pharmacol. 2004;54:331–342. doi: 10.1007/s00280-004-0821-2. [DOI] [PubMed] [Google Scholar]
- 20.Shao J, Zhou B, Zhu L, Bilio AJ, Su L, Yuan YC, et al. Determination of the potency and subunitselectivity of ribonucleotide reductase inhibitors with a recombinant-holoenzyme-based in vitro assay. Biochem Pharmacol. 2005;69:627–634. doi: 10.1016/j.bcp.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 21.Stadler WM, Desai AA, Quinn DI, Bukowski R, Poiesz B, Kardinal CG, et al. A Phase I/II study of GTI-2040 and capecitabine in patients with renal cell carcinoma. Cancer Chemother Pharmacol. 2008;61:689–694. doi: 10.1007/s00280-007-0524-6. [DOI] [PubMed] [Google Scholar]
- 22.Leighl NB, Laurie SA, Chen XE, Ellis P, Shepherd FA, Knox JJ, et al. A phase I/II study of GTI-2040 plus docetaxel as second-line treatment in advanced non-small cell lung cancer: a study of the PMH phase II consortium. J Thorac Oncol. 2009;4:1163–1169. doi: 10.1097/JTO.0b013e3181a949b2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou B, Su L, Yuan YC, Un F, Wang N, Patel M, et al. Structural basis on the dityrosyl-diiron radical cluster and the functional differences of human ribonucleotide reductase small subunits hp53R2 and hRRM2. Mol Cancer Ther. 2010;9:1669–1679. doi: 10.1158/1535-7163.MCT-10-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hajduk PJ, Mack JC, Olejniczak ET, Park C, Dandliker PJ, Beutel BA. SOS-NMR: a saturation transfer NMR-based method for determining the structures of protein-ligand complexes. J Am Chem Soc. 2004;126:2390–2398. doi: 10.1021/ja039480v. [DOI] [PubMed] [Google Scholar]
- 25.Rarey M, Kramer B, Lengauer T, Klebe G. A fast flexible docking method using an incremental construction algorithm. J Mol Biol. 1996;261:470–489. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- 26.Suetsugi M, Su L, Karlsberg K, Yuan YC, Chen S. Flavone and isoflavone phytoestrogens are agonists of estrogen-related receptors. Mol Cancer Res. 2003;1:981–991. [PubMed] [Google Scholar]
- 27.Steeper JR, Steuart CD. A rapid assay for CDP reductase activity in mammalian cell extracts. Anal Biochem. 1970;34:123–130. doi: 10.1016/0003-2697(70)90092-8. [DOI] [PubMed] [Google Scholar]
- 28.Zhou BS, Ker R, Ho R, Yu J, Zhao YR, Shih J, et al. Determination of deoxyribonucleoside triphosphate pool sizes in ribonucleotide reductase cDNA transfected human KB cells. Biochem Pharmacol. 1998;55:1657–1665. doi: 10.1016/s0006-2952(98)00042-2. [DOI] [PubMed] [Google Scholar]
- 29.Mayer M, Meyer B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J Am Chem Soc. 2001;123:6108–6117. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- 30.Zhang K, Wu J, Wu X, Wang X, Wang Y, Zhou N, et al. p53R2 inhibits the proliferation of human cancer cells in association with cell-cycle arrest. Mol Cancer Ther. 2011;10:269–278. doi: 10.1158/1535-7163.MCT-10-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uhlin U, Eklund H. Structure of ribonucleotide reductase protein R1. Nature. 1994;370:533–539. doi: 10.1038/370533a0. [DOI] [PubMed] [Google Scholar]
- 32.Ando N, Brignole EJ, Zimanyi CM, Funk MA, Yokoyama K, Asturias FJ, et al. Structural interconversions modulate activity of Escherichia coli ribonucleotide reductase. Proc Natl Acad Sci U S A. 2011;108:21046–21051. doi: 10.1073/pnas.1112715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yen Y, Horne D, Yuan Y-C, Zhou B-S, Perkin Harki AL, Su L. City of Hope, assignee. Ribonucleotide Reductatse Inhibitors and Methods of Use. patent 7956076. United States. 2011 inventors.
- 34.Aye Y, Stubbe J. Clofarabine 5'-di and -triphosphates inhibit human ribonucleotide reductase by altering the quaternary structure of its large subunit. Proc Natl Acad Sci U S A. 2011;108:9815–9820. doi: 10.1073/pnas.1013274108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Popovic-Bijelic A, Kowol CR, Lind ME, Luo J, Himo F, Enyedy EA, et al. Ribonucleotide reductase inhibition by metal complexes of Triapine (3-aminopyridine-2-carboxaldehyde thiosemicarbazone): a combined experimental and theoretical study. J Inorg Biochem. 2011;105:1422–1431. doi: 10.1016/j.jinorgbio.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo J, Graslund A. Ribonucleotide reductase inhibition by p-alkoxyphenols studied by molecular docking and molecular dynamics simulations. Arch Biochem Biophys. 2011;516:29–34. doi: 10.1016/j.abb.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 37.Jovanovic SV, Steenken S, Tosic M, Marjanovic B, Simic MG. Flavonoids as Antioxidants. Journal of the American Chemical Society. 1994;116:4846–4851. [Google Scholar]
- 38.Seyedsayamdost MR, Yee CS, Reece SY, Nocera DG, Stubbe J. pH Rate profiles of FnY356-R2s (n = 2, 3, 4) in Escherichia coli ribonucleotide reductase: evidence that Y356 is a redox-active amino acid along the radical propagation pathway. J Am Chem Soc. 2006;128:1562–1568. doi: 10.1021/ja055927j. [DOI] [PubMed] [Google Scholar]
- 39.Seyedsayamdost MR, Stubbe J. Site-specific replacement of Y356 with 3,4- dihydroxyphenylalanine in the beta2 subunit of E. coli ribonucleotide reductase. J Am Chem Soc. 2006;128:2522–2533. doi: 10.1021/ja057776q. [DOI] [PubMed] [Google Scholar]
- 40.Finch RA, Liu M, Grill SP, Rose WC, Loomis R, Vasquez KM, et al. Triapine (3-aminopyridine-2- carboxaldehyde- thiosemicarbazone): A potent inhibitor of ribonucleotide reductase activity with broad spectrum antitumor activity. Biochem Pharmacol. 2000;59:983–991. doi: 10.1016/s0006-2952(99)00419-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.