Abstract
The luminescence of rac-[Ru(bpy)2(dppz)]2+ (bpy = 2,2′-bipyridine and dppz = dipyrido[3,2-a:2′,3′-c]phenazine) was explored in the presence of RNA oligonucleotides containing a single RNA mismatch (CA and GG) in order to develop a probe for RNA mismatches. While there is minimal luminescence of [Ru(bpy)2(dppz)]2+ in the presence of matched RNA due to weak binding, the luminescence is significantly enhanced in the presence of a single CA mismatch. The luminescence differential between CA mismatched and matched RNA is substantially higher compared to the DNA analogue, and therefore, [Ru(bpy)2(dppz)]2+ appears to be also a sensitive light switch probe for a CA mismatch in duplex RNA. Although the luminescence intensity is lower in the presence of RNA than DNA, Förster resonance energy transfer (FRET) between the donor ruthenium complex and FRET acceptor SYTO 61 is successfully exploited to amplify the luminescence in the presence of the mismatch. Luminescence and quenching studies with sodium iodide suggest that [Ru(bpy)2(dppz)]2+ binds to these mismatches via metalloinsertion from the minor groove. This work provides further evidence that metalloinsertion is a general binding mode of octahedral metal complexes to thermodynamically destabilized mismatches not only in DNA, but also in RNA.
INTRODUCTION
RNA has many important and complex roles in cellular processes and the development of luminescent probes for RNA could lead to a greater understanding of these roles. The study of protein processes has been facilitated by the family of green fluorescent proteins (GFP), however, there are no intrinsically fluorescent RNA motifs for studying RNA processes. Therefore, several approaches have been developed for tagging RNA with fluorescent probes.1 Recently, RNA mimics of green fluorescent protein have been reported where non-fluorescent dyes become fluorescent upon binding to a RNA aptamer selected by systematic evolution of ligands by exponential enrichment (SELEX).2 However, the rational design of a selective luminescent probe for RNA is made difficult by the complexity of RNA structures.
There is growing evidence that RNA base mismatches have an important biological function as they often occur in phylogenetically conserved regions.3,4 For example, a CA RNA mismatch is common in group I introns but not ribosomal RNAs in a database of RNA secondary structures.5 In addition, the role of RNA mismatches in the active sites of catalytic RNA, and in RNA-protein and RNA-RNA binding has been reported.4,6,7 A luminescent probe for RNA mismatches may provide greater insight into these cellular processes and the role of RNA therein. Here, we examine the luminescence of an octahedral metal complex in the presence of RNA mismatches.
Our laboratory has developed a series of octahedral metal complexes for targeting DNA base mismatches and explored their potential as cancer diagnostic and therapeutic agents.8–10 We have reported that rhodium complexes with sterically expansive ligands, such as [Rh(bpy)2(chrysi)]3+ (chrysi = chrysene-5,6-quinone diimine), bind specifically to DNA mismatches as the chrysi ligand is too wide to intercalate into well-matched DNA.11 Instead, the complexes bind via metalloinsertion, where the chrysi ligand inserts into the thermodynamically destabilized mismatch site from the minor groove and the mismatched base pair is ejected into the major groove.12 The binding affinity of the rhodium complex is correlated with the thermodynamic destabilization of the mismatch and the complex recognizes 80% of mismatched sites in all sequence contexts.11
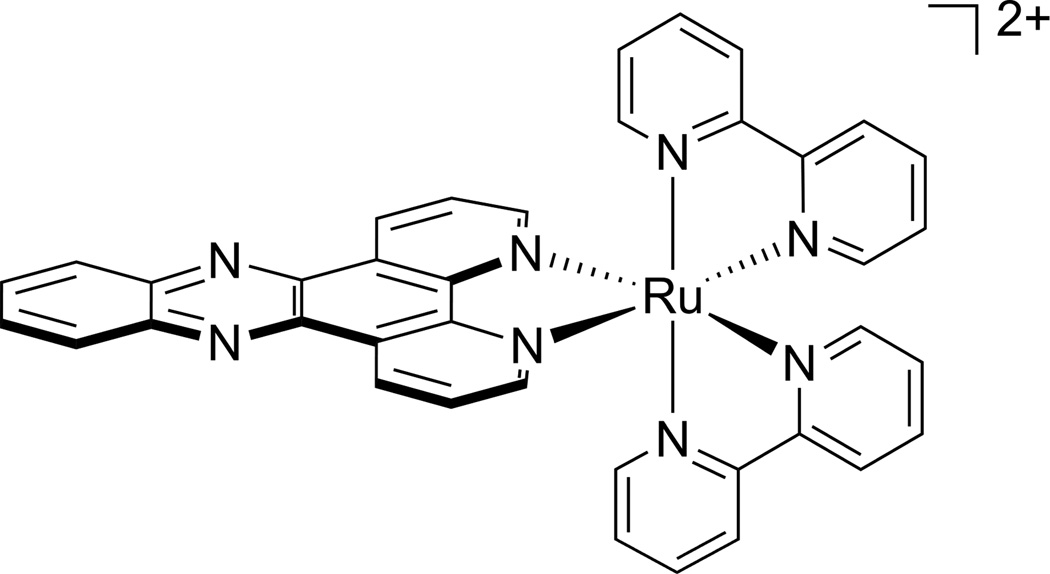
We have also been interested in developing luminescent analogues of the rhodium metalloinsertors and we have investigated ruthenium complexes as potential luminescent probes for DNA base mismatches. It is well known that [Ru(bpy)2(dppz)]2+ (Figure 1, dppz = dipyrido[3,2-a:2′,3′-c]phenazine) behaves as a light switch for well-matched DNA. Hydrogen bonding interactions between the phenazine nitrogen atoms of the dppz ligand and solvent quenches the luminescence, however, the phenazine nitrogens are protected from solvent upon intercalation of the dppz ligand into DNA switching on the luminescence.9 While crystal structures show intercalation between matched base pairs from the minor groove,13,14 solution studies by us are consistent with intercalation from the major groove,15 similar to that seen for other metallointercalators.16 Solution studies by others have proposed complex binding from the minor groove,17 and therefore, the energetic difference between binding from the major and minor groove must be small.
Figure 1.
Chemical structure of [Ru(bpy)2(dppz)]2+.
More recently, we reported that the luminescence of [Ru(bpy)2(dppz)]2+ is enhanced in the presence of a DNA mismatch and is correlated to the thermodynamic destabilization of the mismatch, suggesting metalloinsertion is the binding mode at the mismatch site.10 A recent X-ray crystal structure of Δ-[Ru(bpy)2(dppz)]2+ bound to an oligonucleotide containing two AA mismatches confirmed that the complex does bind to both mismatch sites by metalloinsertion;13 the complex inserts the dppz ligand from the minor groove and the mismatched AA base pairs are ejected. Therefore, metalloinsertion is a general binding mode for targeting DNA base mismatches.
While the luminescence of [Ru(bpy)2(dppz)]2+ is enhanced in the presence of a DNA mismatch, the luminescence differential between mismatched and matched DNA is only 1.5–3 fold, due to the strong intercalative binding of the complex to matched B-form DNA.10 In contrast, [Ru(bpy)2(dppz)]2+ only binds weakly to matched A-form RNA,9 and therefore, the luminescence differential for RNA mismatches may be improved compared to DNA mismatches.
EXPERIMENTAL
Materials
All reagents and solvents were purchased from commercial suppliers and used without further purification. Ultrapure DNase/RNase free distilled water (not treated with diethyl pyrocarbonate), RNaseZap and the SYTO Red Fluorescent Nucleic Acid Stains were purchased from Invitrogen. The RNA oligonucleotides 5′-GUC AXG AGA GCC UCA AAU CUC YUG AC -3′ [XY = CG, CA, GG] were purchased from Integrated DNA Technologies (IDT). The DNA oligonucleotides 5′-GTC AXG AGA GCC TCA AAT CTC YTG AC -3′ [XY = CG, CA] were synthesized on an ABI 3400 DNA synthesizer (Applied Biosystems) and purified as previously reported.18 [Ru(bpy)2(dppz)]Cl2 was synthesized according to literature procedures.9
Methods
RNase Free Conditions
To prevent degradation of the RNA oligonucleotides, RNase free conditions were maintained and solutions of the RNA oligonucleotides were stored at −80 °C when not in use. The aqueous buffer (5 mM Tris, pH 7.5, 50 mM NaCl) was made using DNase/RNase free distilled water. Glassware was decontaminated with RNaseZap then rinsed with DNase/RNase free distilled water and ethanol prior to use.
Oligonucleotide Preparation
The lyophilized oligonucleotides were re-suspended in aqueous buffer (5 mM Tris, pH 7.5, 50 mM NaCl) and the single-stranded concentration was quantified by measuring the absorbance at 260 nm using estimated extinction coefficients obtained from IDT. The oligonucleotides were annealed into the hairpin structure by heating at 90 °C for five minutes then cooling to room temperature over 1.5 hours.
Spectroscopy
UV-visible absorption spectra were recorded on a Beckman DU 7400 UV-visible spectrophotometer (Beckman Coulter). The concentrations of [Ru(bpy)2(dppz)]2+ (ε = 16,100 M−1 cm−1 at 444 nm) and ethidium bromide (ε = 5680 M−1 cm−1 at 476 nm) were determined by UV-visible absorption spectroscopy. Melting temperature experiments were carried out in triplicate on a Cary100 Bio UV-visible spectrophotometer by measuring the absorbance at 260 nm of 1.5 µM RNA in 5 mM Tris, pH 7.5, 50 mM NaCl. The melting temperatures were determined by fitting the data using the Boltzmann function in OriginPro 8.5. Errors are estimated to be ± 1 °C.
Circular dichroism spectra were recorded on an Aviv model 62A DS circular dichroism spectrometer in 5 mM Tris, pH 7.5, 50 mM NaCl. [RNA] = 1.5 µM, [Ru] = 0 µM or 1.5 µM. The blank was subtracted, a baseline correction was made by averaging the last five data points and the data was smoothed using the 2-point adjacent average smoothing function in OriginPro 8.5.
Luminescence spectra were recorded on an ISS-K2 spectrofluorometer in 5 mM Tris, pH 7.5, 50 mM NaCl at ambient temperature in aerated solutions. Samples containing [Ru(bpy)2(dppz)]2+ (0.5 µM or 1 µM) were excited at 440 nm and the emission intensity was integrated from 560 to 800 nm. Samples containing ethidium bromide (100 nM) were excited at 512 nm and the emission intensity was integrated from 540 to 800 nm. Experiments were performed at least three times and the standard deviations were calculated.
RESULTS
RNA Hairpins
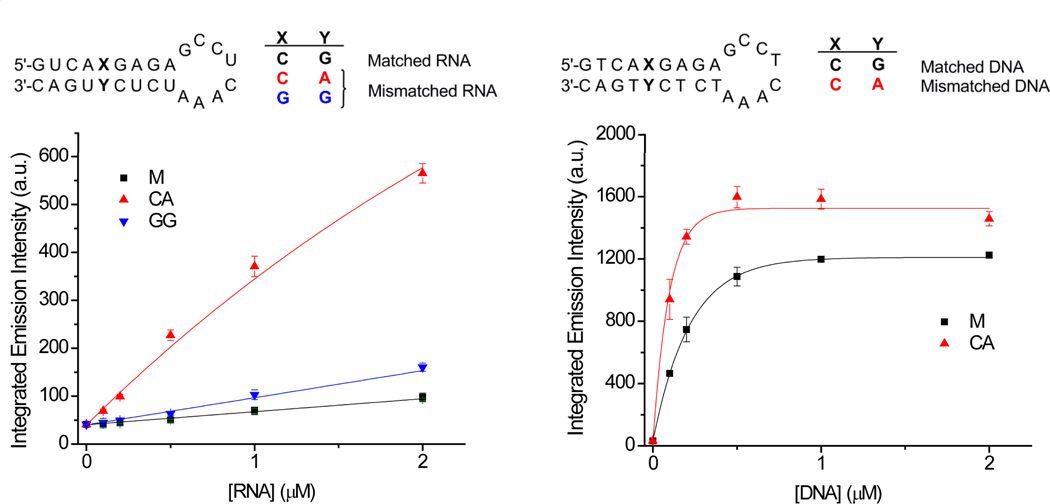
The oligonucleotide sequences used to study the binding of [Ru(bpy)2(dppz)]2+ to RNA are shown in Figure 2. The 26-mer oligonucleotides are designed to fold into a hairpin structure with a single base mismatch at the center of the duplex. Previously we have reported that the luminescence intensity of [Ru(bpy)2(dppz)]2+ bound to DNA mismatches is correlated to the relative thermodynamic stability of the mismatch.10 To investigate whether this is also the case for RNA mismatches, two mismatches (CA and GG) were first examined, as they were expected to have varying thermodynamic stabilities. Circular dichroism (CD) experiments were carried out to ensure that all of the RNA hairpins were in the same A-RNA conformation, and the spectra were seen to be very similar (Figure S1).
Figure 2.
Plots of integrated emission intensities (λex 440 nm) of rac-[Ru(bpy)2(dppz)]2+ (0.5 µM) in the presence of increasing concentrations of hairpin oligonucleotides (RNA: left, DNA: right) containing mismatches (0–2 µM) in 5 mM Tris, pH 7.5, 50 mM NaCl. The sequences of the RNA (left) and DNA (right) hairpins are shown above. Error bars indicate standard deviations for at least three replicates.
Melting temperature experiments revealed that the hairpins containing a single mismatch are thermodynamically destabilized compared to the well-matched hairpin; the mismatches cause a 15–16 °C decrease in duplex melting temperature (Table 1, Figure S2). The two mismatched hairpins have similar melting temperatures within experimental error. While the melting temperature gives an indication of the thermodynamic stability of the duplex, consideration of other thermodynamic parameters, such as Gibbs free energy, is useful for determining the thermodynamic destabilization of the two mismatches. Based on Gibbs free energies determined for similar RNA duplexes with mismatches,5,19 we predict the stability of the mismatches is GG > CA.
Table 1.
Melting temperatures of RNA hairpinsa
| RNA Hairpin | Tm (°C) |
|---|---|
| M | 73 |
| CA | 58 |
| GG | 57 |
1.5 µM RNA (5 mM Tris, pH 7.5, 50 mM NaCl). The matched (M) and mismatched (CA, GG) oligonucleotides shown in Figure 2 were used. Errors are estimated to be ± 1°C.
Steady State Luminescence of [Ru(bpy)2(dppz)]2+ Bound to RNA Mismatches
The steady state luminescence of 0.5 µM rac-[Ru(bpy)2(dppz)]2+ was measured in the presence of increasing concentrations of matched and mismatched RNA (0–2 µM) in aqueous buffer. The luminescence of [Ru(bpy)2(dppz)]2+ in the presence of matched RNA is very low; little binding in the hairpin loop is apparent. However, the luminescence is enhanced in the presence of the CA and GG mismatches (Figure 2).20
At 2 µM RNA, the luminescence differential between matched and mismatched RNA is 5.8 and 1.6 fold for the CA and GG mismatches respectively. The luminescence differential for the CA RNA mismatch is significant given that the ruthenium luminescence is typically only enhanced 1.5–3 fold in the presence of DNA mismatches.10 To investigate this further, analogous experiments were carried out with matched and CA mismatched DNA hairpins with identical sequences to the RNA hairpins (Figure 2).21 In contrast to RNA, the luminescence differential between matched and CA mismatched DNA is less than 1.5 fold at 2 µM. In addition, the emission intensity for both DNA hairpins is significantly higher and binding saturation occurs at a much lower concentration than for the analogous RNA hairpins.
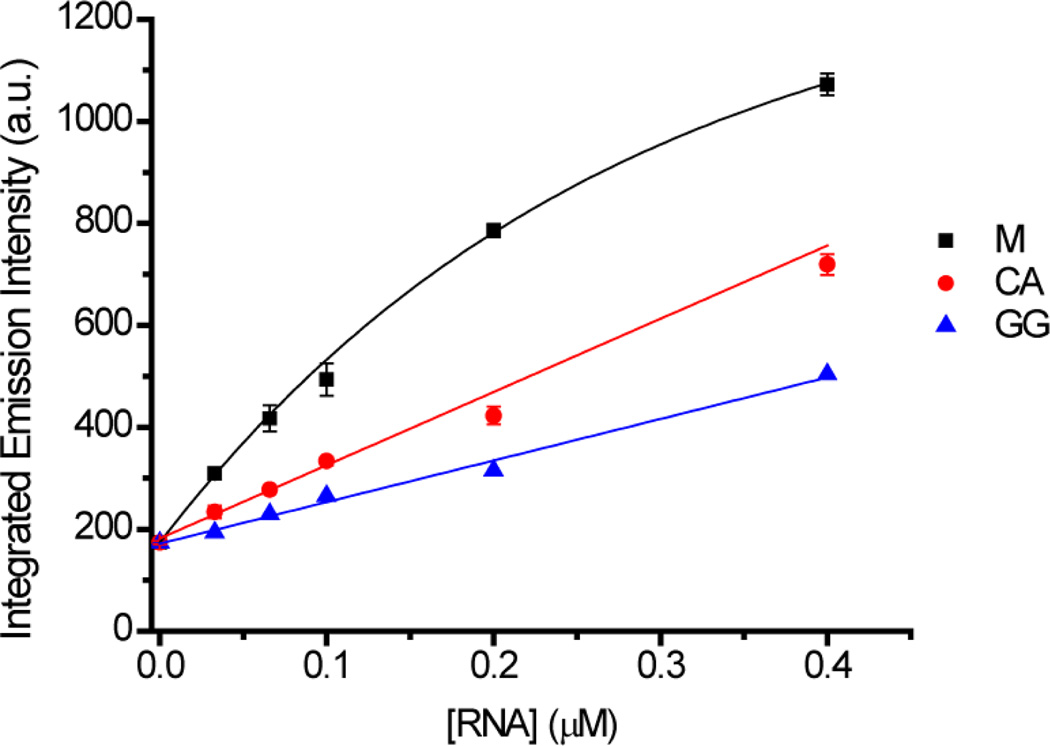
As [Ru(bpy)2(dppz)]2+ is a “light switch” molecule, luminescence is only observed when the dppz ligand is protected within the base stack. To investigate whether intercalation or metalloinsertion is the cause of the enhanced luminescence with a RNA mismatch, fluorescence studies with the known intercalator ethidium bromide were carried out (Figure 3). The fluorescence of ethidium bromide (0.1 µM) increases over 5 fold in the presence of 2 µM matched RNA. However, the fluorescence is greatly reduced in the presence of a mismatch, suggesting that ethidium bromide binds to these hairpins with lower affinity than the matched analogue. This contrasts previous results with DNA where ethidium bromide binds to matched and mismatched DNA with similar affinity.10 Nevertheless, it is clear that ethidium bromide does not report the presence of RNA mismatches by enhanced luminescence like [Ru(bpy)2(dppz)]2+.
Figure 3.
Plots of integrated emission intensities (λex 512 nm) of ethidium bromide (0.1 µM) in the presence of increasing concentrations of RNA hairpins containing mismatches (0–0.4 µM) in 5 mM Tris, pH 7.5, 50 mM NaCl. Hairpins: M (black), CA (red), GG (blue). Error bars indicate standard deviations in the measurements calculated for three replicates.
Based on these results and the very weak intercalative binding of ruthenium to the matched RNA hairpin, it is unlikely that the enhanced luminescence of [Ru(bpy)2(dppz)]2+ in the presence of a RNA mismatch results from intercalation of the complex. A RNA conformational change upon ruthenium binding to the mismatched RNA hairpins has also been discounted since the CD spectrum of the CA mismatch is unchanged in the presence of [Ru(bpy)2(dppz)]2+ (Figure S1). Instead, we propose that [Ru(bpy)2(dppz)]2+ binds to the CA and GG mismatched RNA hairpins via metalloinsertion, and this binding mode is the source of the enhanced luminescence in the presence of RNA mismatches. Further evidence for metalloinsertion as the binding mode is that the relative luminescence of the CA and GG mismatches is consistent with the predicted thermodynamic destabilization of the mismatch; the highest luminescence intensity is observed with the more thermodynamically destabilized CA mismatch. This correlation has been previously attributed to the binding of [Ru(bpy)2(dppz)]2+ and [Rh(bpy)2(chrysi)]3+ to DNA mismatches via metalloinsertion from the minor groove.10,11 Since A-form RNA has a narrower major groove and wider minor groove compared to B-form DNA, presumably, [Ru(bpy)2(dppz)]2+ also inserts into RNA mismatches from the minor groove rather than the major groove.
Quenching Studies with NaI
We have proposed that [Ru(bpy)2(dppz)]2+ binds to the CA RNA hairpin via metalloinsertion from the minor groove. To investigate this further, quenching studies with the anionic luminescent quencher iodide were carried out. The ability of iodide to quench the luminescence of [Ru(bpy)2(dppz)]2+ is dependent on accessibility to the complex in the presence of the polyanion RNA.22 In contrast to DNA, the wider and shallower minor groove of RNA is expected to be more accessible than the major groove and therefore, greater iodide quenching might be expected of [Ru(bpy)2(dppz)]2+ bound from the minor groove. In order to account for the increased ionic strength in the presence of the quencher, a constant ionic strength was maintained by addition of the non-quencher KCl.
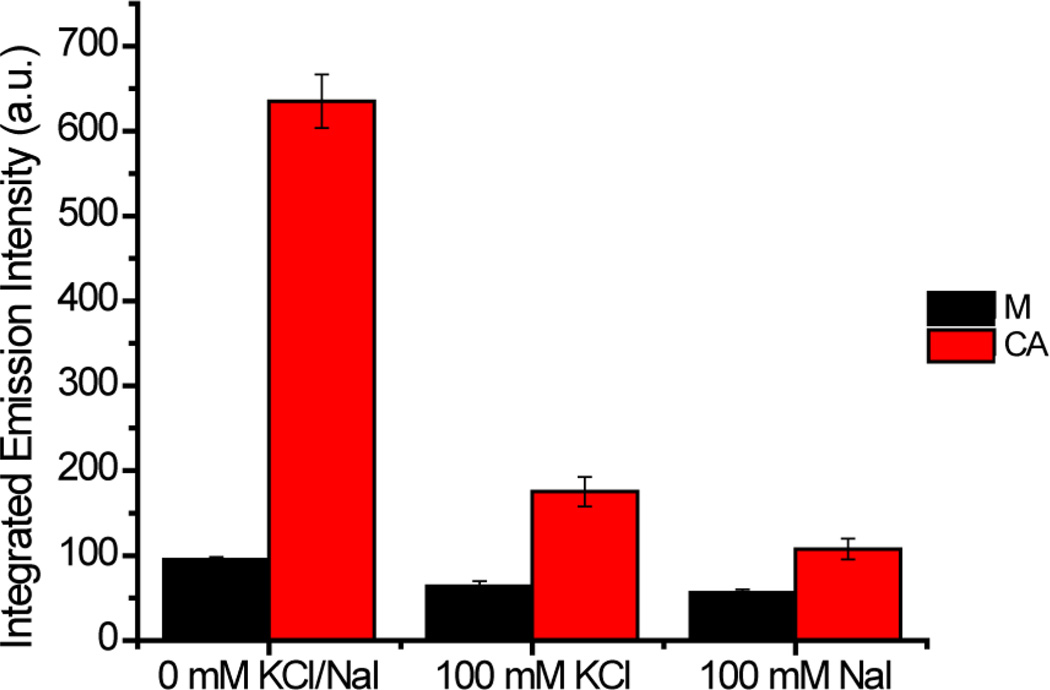
The luminescence of [Ru(bpy)2(dppz)]2+ bound to the CA RNA hairpin significantly decreases upon addition of 100 mM KCl as the increased ionic strength inhibits complex binding. Therefore, the concentration of both the complex and RNA was increased to 1 µM to obtain measurable luminescence in the presence of KCl (Figure 4). The luminescence decreases even further in the presence of the quencher (100 mM NaI), suggesting that [Ru(bpy)2(dppz)]2+ is binding to the mismatch from the more accessible minor groove. As a control, analogous experiments were carried out with the matched RNA hairpin. Consistent with earlier results, the luminescence of [Ru(bpy)2(dppz)]2+ in the presence of the matched RNA hairpin is very low, even at the higher ruthenium and RNA concentrations, and addition of 100 mM KCl leads to the expected luminescence decrease. However, it is difficult to draw definitive conclusions about the effect of the quencher iodide as the emission is very low at the higher ionic strength. Based on these results, quenching experiments with the GG mismatched hairpin were not carried out as similarly low levels of luminescence were expected.
Figure 4.
Plots of integrated emission intensities (λex 440 nm) of rac-[Ru(bpy)2(dppz)]2+ (1 µM) with matched and CA mismatched RNA (1 µM) in the presence and absence of KCl or NaI (100 mM) in 5 mM Tris, pH 7.5, 50 mM NaCl. Error bars indicate standard deviations for at least three replicates.
Luminescence Amplification Through Förster Resonance Energy Transfer
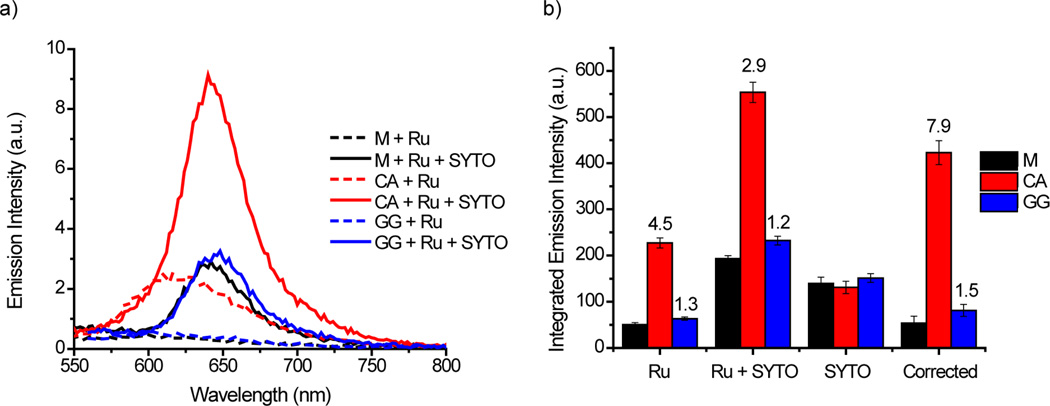
While [Ru(bpy)2(dppz)]2+ has improved luminescence differential in the presence of the CA RNA mismatch compared with the DNA analogue, the luminescence intensity is very low due to weaker binding to RNA. However, the luminescence can be amplified through Förster resonance energy transfer (FRET) between the donor [Ru(bpy)2(dppz)]2+ and a suitable acceptor fluorophore with an excitation wavelength of around 600 nm, the emission wavelength of [Ru(bpy)2(dppz)]2+ upon excitation at 440 nm. SYTO 61 from the nucleic acid stain SYTO® Red series was chosen as the acceptor fluorophore. FRET between the ruthenium donor (0.5 µM) and SYTO acceptor (1 µM) is observed upon excitation at 440 nm in the presence of the CA RNA mismatch (0.5 µM, Figure 5a). The emission maximum is red-shifted and the intensity increases approximately 3 fold, features consistent with the ruthenium donor emission being replaced by SYTO 61 emission. For comparison, a RNA concentration of approximately 2 µM would be required to achieve similar levels of luminescence in the absence of SYTO 61.
Figure 5.
a) Emission spectra and b) integrated emission intensities (λex 440 nm) of rac-[Ru(bpy)2(dppz)]2+ (0.5 µM) with matched and mismatched RNA (0.5 µM) in the presence and absence of SYTO 61 (1 µM) in 5 mM Tris, pH 7.5, 50 mM NaCl. Corrected = (Ru + RNA + SYTO) − (RNA + SYTO). The numbers above the bars indicate the luminescence differential between mismatched and matched RNA. Luminescence differential = (Mismatched Integrated Emission Intensity)/(Matched Integrated Emission Intensity). Error bars indicate standard deviations for at least three replicates.
Luminescence amplification through FRET using SYTO 61 was also investigated with the other hairpins (Figure 5). While relatively low levels of luminescence are observed with the matched and GG RNA hairpins in the absence of SYTO 61, the luminescence is significantly enhanced in the presence of SYTO 61 and the emission maximum is also red-shifted as with the CA RNA hairpin (Figure 5a). However, control experiments with the RNA hairpins and SYTO 61 in the absence of [Ru(bpy)2(dppz)]2+ reveal that the luminescence of SYTO itself upon excitation at 440 nm contributes to the luminescence amplification with the matched and GG RNA hairpins. This increases the background luminescence signal and as a result, the luminescence differential between the CA mismatch and matched RNA decreases from 4.5 to less than 3 fold in the presence of SYTO 61. However, the contribution from SYTO only excitation can be corrected by subtraction and the luminescence for the matched RNA hairpin in the presence of SYTO 61 is reduced to similar levels observed in the absence of SYTO 61. As a result of this correction, the luminescence differential for the CA RNA hairpin is significantly improved to almost 8 fold and the differential for the GG RNA hairpin is also improved slightly to 1.5 fold.
DISCUSSION
While the DNA “light switch” behavior of [Ru(bpy)2(dppz)]2+ is well established, studies with RNA have been limited due the weaker binding and consequently lower luminescence of the complex.9 In this work, we investigated the luminescence of [Ru(bpy)2(dppz)]2+ in the presence of a series of RNA hairpins containing a single mismatch (CA and GG) at the center of the duplex. These studies reveal that [Ru(bpy)2(dppz)]2+ has minimal luminescence in the presence of matched RNA but there is a significant luminescent enhancement in the presence of a RNA mismatch. In comparison, [Ru(bpy)2(dppz)]2+ binds to both matched and mismatched DNA and therefore, [Ru(bpy)2(dppz)]2+ is a better luminescent probe for RNA rather than DNA mismatches.
It is interesting to compare the binding of [Ru(bpy)2(dppz)]2+ to RNA with that of DNA. Due to the “light switch” properties of [Ru(bpy)2(dppz)]2+, binding to both RNA and DNA is only observed by luminescence when the dppz ligand is protected from solvent in the base stack. However, DNA and RNA duplexes exist in different forms, leading to different binding preferences for the complex (major versus minor groove binding, intercalation versus metalloinsertion). This is most obvious when comparing the binding of [Ru(bpy)2(dppz)]2+ to matched RNA and DNA. The complex binds strongly to B-form duplex DNA via intercalation from the major groove,10 whereas it only binds weakly to A-form well-matched duplex RNA.9 This results in the higher emission intensities and lower binding saturation concentrations for DNA compared with RNA. Intercalative binding of [Ru(bpy)2(dppz)]2+ from the major groove to matched RNA is less favorable since the major groove of A-form RNA is narrower and deeper than B-form DNA. While the RNA minor groove is wider and shallower, perceivably more conducive for ruthenium binding, the low luminescence of [Ru(bpy)2(dppz)]2+ in the presence of matched RNA suggests that intercalative binding from either groove is very weak.
Ruthenium binding to DNA and RNA mismatches appears to be more similar as the ruthenium luminescence is enhanced in both cases compared to the matched analogues. A recent crystal structure of [Ru(bpy)2(dppz)]2+ bound to an oligonucleotide containing two AA DNA mismatches demonstrated that, like the rhodium metalloinsertors,11 [Ru(bpy)2(dppz)]2+ binds via metalloinsertion from the minor groove at the mismatched sites with a conserved binding geometry.13 Furthermore, the ruthenium luminescence is correlated to the thermodynamic stability of the mismatch.10 In this work, higher ruthenium luminescence is observed with the more thermodynamically destabilized CA RNA mismatch compared with the GG mismatch and therefore, we propose that [Ru(bpy)2(dppz)]2+ also binds to these RNA mismatches via metalloinsertion. It is likely that metalloinsertion is from the minor groove, like with DNA mismatches, based on quenching studies with iodide and the narrow width of the RNA major groove. Despite the structural differences between B-form DNA and A-form RNA, this work suggests that metalloinsertion is a general binding mode for targeting mismatches not only in DNA, but also in RNA.
The luminescence differential between CA mismatched and matched RNA is significantly improved compared with the DNA analogues, since [Ru(bpy)2(dppz)]2+ does not bind to well-matched RNA. While the luminescence intensity is low due to the weak binding of the complex to RNA even in the presence of a mismatch, this can be amplified approximately 3 fold by exploiting FRET with SYTO 61. Furthermore, the luminescence differential increases to almost 8 fold after correcting for the emission of SYTO 61 bound to the CA RNA hairpin in the absence of ruthenium. Therefore, [Ru(bpy)2(dppz)]2+ is a sensitive “light switch” probe for a CA RNA mismatch. Interestingly, the CA RNA mismatch is a prevalent RNA mismatch in a database of RNA secondary structures containing ribosomal RNAs and group I introns.5 Thus, a CA RNA mismatch could be exploited in a biological setting, providing an RNA tag for targeting Ru luminescence.
CONCLUSIONS
[Ru(bpy)2(dppz)]2+ appears to be a more effective luminescent probe for RNA mismatches than for DNA mismatches. The luminescence of [Ru(bpy)2(dppz)]2+ is enhanced in the presence of CA and GG RNA mismatches, while the complex binds very weakly to matched RNA. In contrast, the complex binds strongly to both matched and mismatched DNA. Hence the differential luminescence associated with binding RNA mismatches is enhanced. This work furthermore suggests that [Ru(bpy)2(dppz)]2+ binds to RNA mismatches via metalloinsertion from the minor groove. Therefore, metalloinsertion is a general binding mode of octahedral metal complexes to thermodynamically destabilized mismatches not only in DNA, but also in RNA.
Supplementary Material
Acknowledgments
We thank the NIH (GM33309 to J. K. B.) for their financial support, the Lindemann Trust for a postdoctoral fellowship to A. J. M., the Tobacco-Related Disease Research Program (TRDRP) for a Dissertation Research Award to H. S. We are grateful to the Beckman Institute Laser Resource Center (BILRC) at Caltech for facilities.
Footnotes
Supporting Information.
CD spectra and melting temperature curves of the RNA hairpins. This material is available for free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.(a) Rodriguez AJ, Condeelis J, Singer RH, Dictenberg JB. Semin. Cell Dev. Biol. 2007;18:202–208. doi: 10.1016/j.semcdb.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Tyagi S. Nature Methods. 2009;6:331–338. doi: 10.1038/nmeth.1321. [DOI] [PubMed] [Google Scholar]
- 2.Paige JS, Wu KY, Jaffrey SR. Science. 2011;333:642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battiste JL, Tan R, Frankel AD, Williamson JR. Biochemistry. 1994;33:2741–2747. doi: 10.1021/bi00176a001. [DOI] [PubMed] [Google Scholar]
- 4.Strobel SA, Ortoleva-Donnelly L, Ryder SP, Cate JH, Moncoeur E. Nat. Struct. Mol. Biol. 1998;5:60–66. doi: 10.1038/nsb0198-60. [DOI] [PubMed] [Google Scholar]
- 5.Kierzek R, Burkard ME, Turner DH. Biochemistry. 1999;38:14214–14223. doi: 10.1021/bi991186l. [DOI] [PubMed] [Google Scholar]
- 6.Cai Z, Tinoco I. Biochemistry. 1996;35:6026–6036. doi: 10.1021/bi952985g. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP, Zapp ML, Green MR, Szostak JW. Cell. 1991;67:529–536. doi: 10.1016/0092-8674(91)90527-6. [DOI] [PubMed] [Google Scholar]
- 8.(a) Zeglis BM, Pierre VC, Barton JK. Chem. Commun. 2007;44:4565–4579. doi: 10.1039/b710949k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Komor AC, Barton JK. Chem. Commun. 2013;49:3617–3630. doi: 10.1039/c3cc00177f. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Ernst RJ, Komor AC, Barton JK. Biochemistry. 2011;50:10919–10928. doi: 10.1021/bi2015822. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Komor AC, Schneider CJ, Weidmann AG, Barton JK. J. Am. Chem. Soc. 2012;134:19223–19233. doi: 10.1021/ja3090687. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) McConnell AJ, Lim MH, Olmon ED, Song H, Dervan EE, Barton JK. Inorg. Chem. 2012;51:12511–12520. doi: 10.1021/ic3019524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Friedman AE, Chambron JC, Sauvage JP, Turro NJ, Barton JK. J. Am. Chem. Soc. 1990;112:4960–4962. [Google Scholar]; (b) Jenkins Y, Friedman AE, Turro NJ, Barton JK. Biochemistry. 1992;31:10809–10816. doi: 10.1021/bi00159a023. [DOI] [PubMed] [Google Scholar]; (c) Hartshorn RM, Barton JK. J. Am. Chem. Soc. 1992;114:5919–5925. [Google Scholar]
- 10.Lim MH, Song H, Olmon ED, Dervan EE, Barton JK. Inorg. Chem. 2009;48:5392–5397. doi: 10.1021/ic900407n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Jackson BA, Barton JK. J. Am. Chem. Soc. 1997;119:12986–12987. [Google Scholar]; (b) Jackson BA, Alekseyev VY, Barton JK. Biochemistry. 1999;38:4655–4662. doi: 10.1021/bi990255t. [DOI] [PubMed] [Google Scholar]; (c) Jackson BA, Barton JK. Biochemistry. 2000;39:6176–6182. doi: 10.1021/bi9927033. [DOI] [PubMed] [Google Scholar]
- 12.(a) Pierre VC, Kaiser JT, Barton JK. Proc. Natl. Acad. Sci. USA. 2007;104:429–434. doi: 10.1073/pnas.0610170104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Cordier C, Pierre VC, Barton JK. J. Am. Chem. Soc. 2007;129:12287–12295. doi: 10.1021/ja0739436. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Zeglis BM, Pierre VC, Kaiser JT, Barton JK. Biochemistry. 2009;48:4247–4253. doi: 10.1021/bi900194e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song H, Kaiser JT, Barton JK. Nature. Chem. 2012;4:615–620. doi: 10.1038/nchem.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niyazi H, Hall JP, O’Sullivan K, Winter G, Sorensen T, Kelly JM, Cardin CJ. Nature Chem. 2012;4:621–628. doi: 10.1038/nchem.1397. [DOI] [PubMed] [Google Scholar]
- 15.(a) Dupureur CM, Barton JK. J. Am. Chem. Soc. 1994;116:10286–10287. [Google Scholar]; (b) Dupureur CM, Barton JK. Inorg. Chem. 1997;36:33–43. [Google Scholar]
- 16.(a) Kielkopf CL, Erkkila KE, Hudson BP, Barton JK, Rees DC. Nat. Struct. Biol. 2000;7:117–121. doi: 10.1038/72385. [DOI] [PubMed] [Google Scholar]; (b) Hudson BP, Dupureur CM, Barton JK. J. Am. Chem. Soc. 1995;117:9379–9380. [Google Scholar]
- 17.(a) Greguric A, Greguric ID, Hambley TW, Aldrich-Wright JR, Collins JG. J. Chem. Soc., Dalton Trans. 2002:849–855. [Google Scholar]; (b) Andersson J, Fornander LH, Abrahamsson M, Tuite E, Nordell P, Lincoln P. Inorg. Chem. 2013;52:1151–1159. doi: 10.1021/ic302626d. [DOI] [PubMed] [Google Scholar]; (c) Tuite E, Lincoln P, Nordén B. J. Am. Chem. Soc. 1997;119:239–240. [Google Scholar]
- 18.Brunner J, Barton JK. Biochemistry. 2006;45:12295–12302. doi: 10.1021/bi061198o. [DOI] [PubMed] [Google Scholar]
- 19.Davis AR, Znosko BM. Biochemistry. 2010;49:8669–8679. doi: 10.1021/bi100146z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Preliminary experiments were also carried out with a CC RNA hairpin. Titration experiments showed only a small increase in luminescence, not unlike that seen with the fully matched RNA. While the oligonucleotide sequences were designed to fold into a hairpin structure, it is possible that the CC sequence adopts a different, perhaps slipped secondary structure. Given the lack of luminescence enhancement with this sequence, further studies were carried out with the GG and CA RNA hairpins only.
- 21.Experiments with the GG DNA hairpin were not carried out as the luminescence is expected to be very similar to the matched DNA hairpin based on previous studies (see reference 10).
- 22.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3rd ed. New York: Springer; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.