Abstract
The autism susceptibility candidate 2 (AUTS2) gene is associated with multiple neurological diseases, including autism, and has been implicated as an important gene in human-specific evolution. Recent functional analysis of this gene has revealed a potential role in neuronal development. Here, we review the literature regarding AUTS2, including its discovery, expression, association with autism and other neurological and non-neurological traits, implication in human evolution, function, regulation, and genetic pathways. Through progress in clinical genomic analysis, the medical importance of this gene is becoming more apparent, as highlighted in this review, but more work needs to be done to discover the precise function and the genetic pathways associated with AUTS2.
Keywords: AUTS2, autism, neurodevelopment, human evolution
Neurodevelopmental disorders
Neurodevelopmental disorders are characterized by motor, speech, cognitive, and behavioral dysfunctions caused by impairment in growth and development of the central nervous system (CNS). Neurodevelopmental disorders encompass, but are not limited to, intellectual disability (ID), developmental delay (DD), and autism spectrum disorders (ASDs) [1]. ASDs are known as pervasive developmental disorders that are common (1/88 in the USA) [2] and highly heritable [3]. ASDs are characterized by variable deficits in social communication, language, and restrictive and repetitive behaviors, and present as a wide spectrum of phenotypes [4]. Other neurological abnormalities, including ID, DD, epilepsy, sensory and motor abnormalities, gastrointestinal phenotypes, developmental regression, sleep disturbance, mood disorders, conduct disorders, aggression, and attention deficit hyperactivity disorder (ADHD), are also frequently associated with ASD [4]. Despite the heritability of these disorders, no single gene has been identified as causative for ASD alone. Rather, several different genes have been implicated in these disorders containing either common variants with small effects or rare variants with larger consequences [5]. Over the years, studies examining individual patients, together with advances in sequencing technologies that have allowed the examination of a large number of individuals, have produced a myriad of new ASD, ID, and DD candidate genes, including AUTS2.
The discovery of AUTS2
AUTS2 was first identified in 2002 when it was found to be disrupted as a result of a balanced translocation in a pair of monozygotic (MZ) twins with ASD [6]. AUTS2 was mapped to 7q11, spans 1.2 Mb, and is approximately 340 kb upstream from the Williams–Beuren syndrome (WBS) critical region, a region that – when deleted – causes a neurodevelopmental disorder characterized by a distinctive ‘elfin’ facial appearance, a cheerful demeanor, developmental delay, strong language skills, and cardiovascular problems [7]. The AUTS2 protein sequence is highly conserved, with 62% amino acid conservation between humans and zebrafish [8]. It contains regions of homology to other proteins, such as the dwarfin family consensus sequence, human topoisomerase, and fibrosin (FBRS), a fibroblast growth factor [6]. In addition, the Drosophila gene tay has limited similarity to AUTS2. tay mutants have reduced walking speed and activity, thought to be associated with structural defects in the protocerebral bridge [9]. Sequence analysis of AUTS2 identified no membrane-spanning domains, but identified two proline-rich domains and a predicted PY (ProTyr) motif (PPPY) at amino acids 515–519 (Figure 1) [6]. The PY motif is a potential WW-domain-binding region that is involved in protein–protein interactions and is present in the activation domain of various transcription factors, suggesting that AUTS2 may be involved in transcriptional regulation [8]. Other predicted protein motifs include several cAMP and cGMP-dependent protein kinase phosphorylation sites, and putative N-glycosylation sites [6]. In addition, AUTS2 has eight CAC (His) repeats (Figure 1) [6], which have been shown to be associated with localization at nuclear speckles [10] – subnuclear structures where components of the RNA splicing machinery are stored and assembled [11]. Evidence of nuclear localization sequences as well as several predicted protein–protein interaction domains (SH2 and SH3) were also observed for this protein (Figure 1). No evidence was found for any signal peptide in AUTS2, indicating that it is not secreted or exposed to the cellular membrane [12]. No DNA-binding domains have been identified. Taken together, sequence analysis has revealed limited insight into the function of this gene.
Figure 1.
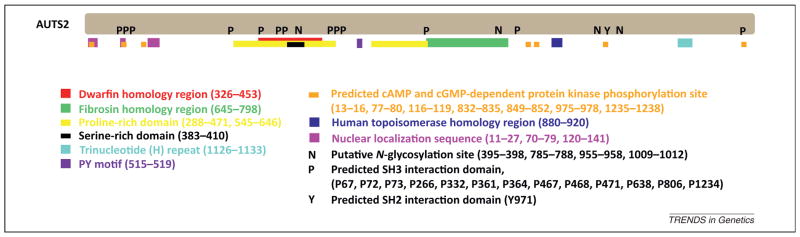
Schematic of the AUTS2 protein. AUTS2 (1259 amino acids) is shown as a gray bar (individual amino acids in single-letter code). The locations of predicted domains, motifs, regions of homology, and other characterized sequences are shown below and within the protein. Numbers in parenthesis represent the amino acid location. The figure is based on predicted features in [6,12].
AUTS2 is a nuclear protein that is expressed in the CNS
Multiple reports have characterized the expression of AUTS2 in different organisms, concluding that it is primarily expressed in the brain. Northern blot shows strong AUTS2 expression in human fetal brain in the frontal, parietal, and temporal regions, but not in the occipital lobe. Expression was also identified in the skeletal muscle and kidney, with lower expression in the placenta, lung, and leukocytes [6]. In human post-mortem fetal brain, AUTS2 mRNA expression was found in the telencephalon (uniformly), ganglionic eminence, cerebellum anlagen, and, more weakly, in the medulla oblongata at 8 weeks. AUTS2 was also found to be strongly expressed in the cortical plate and ventricular zone. Fetal (23 weeks) human brains showed AUTS2 expression in the dentate gyrus, CA1 and CA3 pyramidal cell subregions, the ganglionic eminence, caudate nucleus, and putamen nuclei [13]. AUTS2 was also shown to be expressed in the neocortex and prefrontal cortex up to the late mid-fetal stage [14]. Gene expression profiles from 10 human ocular tissues found AUTS2 to be the 20th highest expressed gene in the sclera [15]. Sequencing of total RNA from human brain and liver found a large fraction of reads (up to 40%) to be within introns [16]. The authors identified enrichment of intronic RNA in brain tissues, particularly for genes involved in axonal growth and synaptic transmission. AUTS2 was among the 10 genes with the highest intronic RNA score in fetal brain. Three of the top 10 genes – neurexin 1 (NRXN1), protocadherin 9 (PCDH9), and methionine sulfoxide reductase A (MSRA) – have also been implicated in autism. In addition, for long introns, including the first half of AUTS2, there is a 5′ to 3′ slope in read coverage, with significantly higher levels of RNA at the 5′ end. The authors reason that, in the fetal brain, intronic RNAs are subjected to brain-specific regulatory pathways that regulate alternative splicing programs to control neuronal development [16].
A detailed analysis of Auts2 mRNA and protein expression in the developing mouse brain was published in 2010 [12]. The authors found that Auts2 is expressed in the developing cerebral cortex and cerebellum, and is located in the nuclei of neurons and some neuronal progenitors (Table 1). Auts2 expression was identified in numerous neuronal cell types, including glutamatergic neurons (cortex, olfactory bulb, hippocampus), GABAergic neurons (Purkinje cells), and tyrosine hydroxylase (TH)-positive dopaminergic neurons (substantia nigra and ventral tegmental area). Colocalization of Auts2 with only a subset of eomesodermin (Tbr2) and paired box 6 (Pax6)-positive cells was demonstrated in the ventricular and subventricular zones, suggesting that Auts2 might be expressed in the transition between radial glial and intermediate progenitors [12]. It was also suggested that Auts2 and T-box brain 1 (Tbr1) are coexpressed mostly in glutamatergic neuron populations in the forebrain, and other transcription factors likely influence expression of Auts2 in other regions. The report also notes that Auts2 could be expressed in a transient phase of neuronal maturation or differentiation in the cortex [12]. In zebrafish, using wholemount in situ hybridization, auts2 was shown to be expressed in the brain at 24, 48, 72 and 120 hours post-fertilization (hpf). At 48 hpf, auts2 is also expressed in the pectoral fin. From 24–130 hpf, auts2 is also weakly expressed in the eye [17]. In summary, AUTS2 has been shown to be a nuclear protein that is primarily expressed in the brain in various cell types as well as in regions implicated in ASD, such as the neocortex.
Table 1.
Auts2 expression in the developing mouse braina
| Timepointb | Auts2 expression |
|---|---|
| E11 | mRNA barely detectable. |
| E12–13 | Colocalization with Tbr1 in the cortical preplate. Tbr1 is a transcription factor specific for postmitotic projection neurons. |
| E12–14 | High expression in the developing cortex, thalamus, and cerebellum. There is continued expression in these regions throughout development, but levels fluctuate and are found in gradients. Different markers show Auts2 expression in multiple neuronal subtypes in the developing cortex. |
| E14 | Expression in the hippocampal primordium. Transient expression in the locus ceruleus and vestibular nuclei. |
| E16 | Expression in the cerebral cortex is now a gradient of high rostral to low caudal expression. |
| E19 | Highest expression in inferior and superior colliculi and the pretectum. |
| P0 | Auts2 expression becomes progressively more superficial in the frontal cortex. Coexpression with Tbr1 becomes rare as Tbr1 becomes more selective to layer 6. |
| E16–P21 | Auts2 is expressed mostly in the frontal cortex, hippocampus, and the cerebellum. In addition, high expression levels were detected in the developing dorsal thalamus, olfactory bulb, inferior colliculus and the substantia nigra. |
| P21 | Expression in developing thalamic areas, including the anterior thalamic nuclei and in ventrolateral/ventromedial nuclei. Auts2 is restricted to superficial layers in frontal cortex. Auts2 is expressed throughout the subgranular zone and the granule cell layer of the hippocampus. |
Summary based on [12].
E, embryonic day; P, postnatal day.
AUTS2 and ASD, ID, and DD
AUTS2 has been repeatedly implicated as an ASD candidate gene in recent years. Following the initial finding of an AUTS2 translocation in twins with autism [6], over 50 unrelated individuals with ASD, ID, or DD were identified with distinct structural variants disrupting the AUTS2 region in numerous different reports (Figure 2) [8,18–30]. Some of the structural variants are exclusively non-coding, suggesting that improper regulation and subsequent expression of AUTS2 could be involved in the progression of the disorder [17]. In addition to ASD, ID, and DD, many of these individuals also have other phenotypes, including epilepsy, brain malformations, or dysmorphic features. One group described an ‘AUTS2 syndrome’ in individuals with varying severity of growth and feeding problems, neurodevelopmental features, neurological disorders, dysmorphic features, skeletal abnormalities, and congenital malformations [26]. The spectrum of phenotypes observed in individuals with AUTS2 mutations is consistent with the wide range of ASD phenotypes. This suggests that AUTS2 is not associated with a specific subtype of ASD. It has also been noted that dysmorphic features were more pronounced in individuals with 3′ AUTS2 deletions, where most of the coding region resides [26]. However, copy-number variations (CNVs) at the AUTS2 locus have also been observed in unaffected individuals, indicating that structural rearrangements are tolerated in some cases [19,31]. This suggests that disruptions in AUTS2 may lead to neurodevelopmental disorders by being one of multiple genomic ‘hits’. The large number of independent publications implicating AUTS2 in ASD, ID, or DD provides strong evidence for its involvement in these disorders. It is worth noting, however, that no publication has shown single base-pair variants in the AUTS2 locus affiliated with ASD, despite numerous ASD-related exome sequencing studies [32–35].
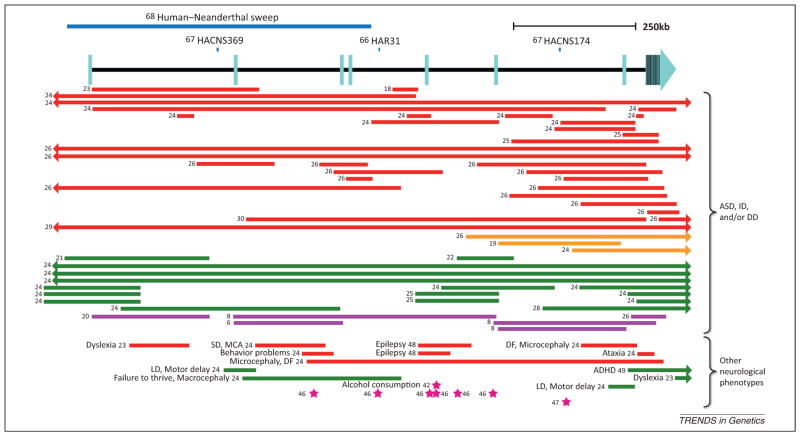
Figure 2.
Schematic of the AUTS2 genomic region. Numbers to the left of the lines correspond to reference numbers. Human accelerated sequences are shown as blue lines above the gene [66–68]. Structural variants [6,8,18–26,28–30,48,49] are represented as colored lines (red, deletion; orange, inversion; green, duplication; purple, translocation). Single-nucleotide polymorphisms (SNPs) are shown as magenta stars. rs6943555 is associated with alcohol consumption [42]. SNPs in [46,47] are associated with bipolar disorder. SNPs in [46] are reported to be in strong linkage disequilibrium with each other. Arrows in bars signify that the structural variant extends past the gene in that direction. Exons are depicted as light-blue rectangles, as defined by the RefSeq genes track in the University of California, Santa Cruz (UCSC) Genome Browser. DD, developmental delay; DF, dysmorphic features; HACNS, human accelerated conserved non-coding sequence; HAR, human accelerated region; ID, intellectual disability; LD, language disability; MCA, multiple congenital anomalies; SD, seizure disorder. Figure adapted from [17].
The observation that AUTS2 variants are mostly CNVs may be due to the susceptibility of this region to chromosomal breakpoints. A 2011 report showed that the offspring of older male mice have an increased risk of de novo CNVs in specific locations, including the Auts2 locus [36]. Another report found that hydroxyurea, a ribonucleotide reductase inhibitor, as well as aphidicolin, a DNA polymerase inhibitor, induce a high frequency of de novo CNVs in cultured human cells, and found a clustering of CNVs in AUTS2 [37]. Aphidocolin also induced CNV formation in the Auts2 locus in non-homologous end-joining deficient mouse embryonic stem cells [38]. Because the AUTS2 locus is a hotspot for CNVs, and individuals with ASD generally carry more CNVs than their unaffected siblings [39], examining if these high numbers of ASD-associated CNVs around AUTS2 are consequential, and not merely a result of their susceptibility to CNVs, warrants investigation. There is also the possibility that these CNVs affect regulatory regions of other genes, including the nearby WBS critical region.
In 2013, a genome-wide analysis of DNA methylation was published on ASD discordant and concordant monozygotic twins. A region in the AUTS2 promoter (chr7: 68701907; hg18) was the 42nd most differentially methylated CpG site in the genome, suggesting that not only sequence variation but also epigenetic changes to the AUTS2 locus could be involved in the development of ASD-related traits [40]. Significant DNA methylation differences were often observed near other genes that have been previously implicated in ASD, including methyl-CpG binding domain protein 4 (MBD4) and microtubule-associated protein 2 (MAP2). The authors cautioned, however, that it is difficult to draw conclusions about the causality of the differentially methylated sites due to small sample size, lack of corresponding RNA expression data, the use of whole blood rather than brain tissue, and potential epigenetic effects due to medicine [40].
Combined, the evidence for a causative role of AUTS2 in DD and ID is convincing. However, for ASD the evidence presented so far suggests that disruptions in AUTS2 can play a causative role, but to demonstrate causality more research needs to done on cohorts of well-defined ASD patients and on the functional consequence of these disruptions.
AUTS2 and other neurological conditions
In addition to ASD, ID, and DD, AUTS2 has been implicated in other neurological disorders. Some of these disorders, such as epilepsy, have been shown to be linked to ASD. However, other AUTS2-associated phenotypes are ASD-independent. AUTS2 expression was found to have significant association with nicotine-dependence, cannabis-dependence, and antisocial personality disorder, although this study had a small number of cases and would need to be repeated with larger cohorts [41]. The study also suggested, although it did not reach significance, that AUTS2 expression is implicated in alcohol dependence [41]. In 2011 a genome-wide association meta-analysis found an AUTS2 non-coding single-nucleotide polymorphism (SNP), rs6943555, to be significantly associated with alcohol consumption [42]. The authors also reported increased AUTS2 expression in carriers of the minor A allele of rs6943555 compared with the T allele in 96 human prefrontal cortex samples. In addition, they identified significant differences in expression of Auts2 in whole-brain extracts of mice with differences in voluntary alcohol consumption. The authors also showed that downregulation of tay, which has sequence similarity to AUTS2, caused reduction in alcohol sensitivity in Drosophila [42]. Also implicating AUTS2 in drug dependence was a 2011 study showing that AUTS2 has a 3.01-fold change (downregulation) between 19 male heroin-dependent individuals and 20 controls in lymphoblastoid cell lines [43]. A follow-up study compared AUTS2 transcript levels of lymphoblastoid cell lines between 124 heroin-dependent and 116 control males using quantitative PCR – and found that average transcript levels of AUTS2 in the heroin-dependent group were significantly lower than in controls. They also found that AA homozygotes for rs6943555 were significantly over-represented in the heroin-dependent subjects [44]. Taken together, these reports show strong evidence for AUTS2 involvement in addiction and dependence.
In addition, the AUTS2 locus has been shown to be implicated or altered in individuals with schizoaffective disorder [45], bipolar disorder [46,47], epilepsy [48], ADHD [49], differential processing speed [50], suicidal tendencies under the influence of alcohol [51], and dyslexia [23], either through CNV or genome-wide association studies. A 2012 article sequenced balanced chromosomal abnormalities in patients with neurodevelopmental disorders, and found the AUTS2 locus to be perturbed in individuals with microcephaly, macrocephaly, ataxia, visual impairment, language disability, seizure disorder, dysmorphic features, behavioral problems, motor delay, or Rubinstein–Taybi syndrome [24]. It could be that the observation that most cases of AUTS2 structural variants are associated with ASD is attributed to more individuals with ASD being tested in this locus than patients with other neurological disorders – thereby leading to an underestimate in the link between AUTS2 and other neurological phenotypes. Taken together, these observations suggest that AUTS2 dysfunction is not restricted to ASD, DD, or ID, but instead AUTS2 dysfunction is involved in a wide range of neurological disorders. In addition, a few studies implicate AUTS2 in non-neurological disorders and traits (Box 1).
Box 1. AUTS2 and non-neurological disorders and traits.
A few reports have implicated AUTS2 in non-neurological disorders and traits. In 2004, 18 cases of childhood hyperdiploid acute lymphoblastic leukemia (ALL) were examined to identify the relationship between extra copies of chromosomes and increased gene expression. The authors identified multiple regions with increased expression that correlated poorly or not at all with the presence of extra copies of chromosomes, including 7q11.2. AUTS2 showed consistently higher expression levels in the cDNA samples of patients than in normal mononuclear cells, possibly implicating the gene in ALL [69]. In 2008 it was reported that paired box 5 (PAX5) can be rearranged with a variety of partners, including AUTS2 (one case) in pediatric ALL [70]. Two years later a second case of PAX5–AUTS2 fusion was identified in pediatric ALL [71]. In 2012, the third case of PAX5–AUTS2 fusion was identified in a patient with pediatric ALL, providing additional evidence that PAX5–AUTS2 is a recurring gene fusion in ALL [72]. Two of the three PAX5–AUTS2 cases had CNS diseases either at the time of diagnosis or relapse [72]. Individual reports, some of which identify single patients, have also implicated the AUTS2 locus in the aging of human skin [73], lung adenocarcinoma [74], lethal prostate cancer [75], the number of corpora lutea in pigs [76], early-onset androgenetic alopecia [77], and metastatic non-seminomatous testicular cancer [78]. Despite several reports suggesting a role for AUTS2 in non-neurological disorders and traits, disruption of AUTS2 is most often reported to be associated with neurological phenotypes.
The function and regulation of AUTS2
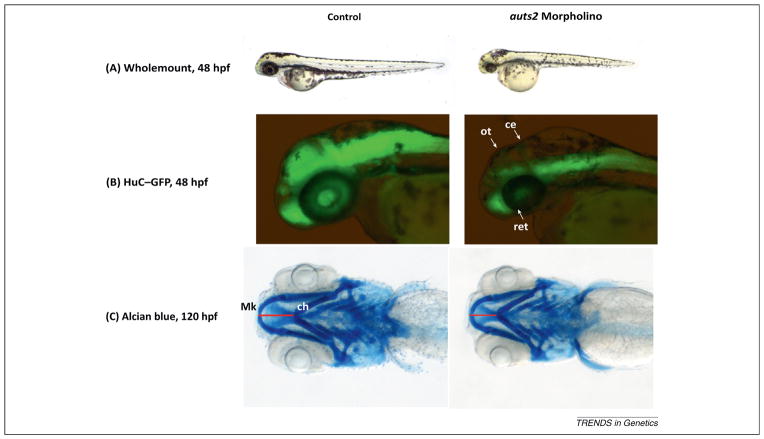
Despite the many articles linking AUTS2 to human disease and other traits, few papers have been published describing the function of the gene. In 2013, morpholino knockdowns of auts2 were performed in zebrafish by two different groups [17,26]. The observed phenotypes are summarized in Figure 3 and Table 2. Using HuC (Hu antigen C), a neuronal marker, both groups observed a decrease in neuronal cells in the brain (Figure 3B). Increased apoptosis and cell proliferation in the brain was reported, and it was noted that this observation could be a result of morphant cells failing to differentiate into mature neurons, which matches the HuC results [17]. Although increased cell proliferation was observed in one study [17], another study described decreased cell proliferation [26]. The differences in this phenotype could be due to differences in the stains used (proliferating cell nuclear antigen, PCNA, which marks cells in early G1- and S-phase versus phosphohistone-H3, a marker of cells in G2 and M phase). Both reports, however, found that auts2 knockdown cells show more replicating DNA, but fewer cells dividing into daughter cells. The craniofacial phenotype of the morphant fish was also characterized in one of the studies, finding that they have micrognathia (undersized jaw) and retrognathia (receded jaw) (Figure 3C) [26]. Given that migrating neural crest cells play an important role in craniofacial development [52], it is possible that this phenotype is a result of defects in neuronal cell development. In addition, less movement was reported in morphant fish, and this could be caused by fewer motor neuron cell bodies in the spinal cord, together with improperly angled and weaker projections, and/or fewer sensory neurons, both of which were observed in morphant fish [17]. Although one group observed overall stunted development [17] (Figure 3A), the other reported a phenotype restricted to the brain and jaw [26]. A potential cause for the difference in this phenotype, alongside the differences in cell proliferation phenotypes, could be due to the use of different morpholinos for these assays: an auts2 translational morpholino [17] versus splicing morpholinos [26]. Both groups were able to rescue the morphant phenotype by injecting full-length human AUTS2 mRNA together with the morpholino [17,26]. The morphant phenotype was also rescued by injecting the shorter C-terminal isoform of AUTS2, suggesting that the final nine exons of AUTS2 contain the crucial region of the gene, at least for the dysmorphic phenotype observed in knockdown fish. This is in line with the observation that dysmorphic features were more pronounced in individuals with 3′ AUTS2 deletions [26]. The zebrafish knockdown phenotypes appear to be an overall neurodevelopment defect, making it difficult to truly parse out the function of this gene. To understand AUTS2 function better, a conditional knockout mouse should be developed.
Figure 3.
auts2 zebrafish knockdown phenotype. (A) At 48 hours post-fertilization (hpf), fish injected with a 5 bp mismatch auts2 morpholino (MO) control have a similar morphology to wild type fish, whereas fish injected with a corresponding translational MO display a stunted developmental phenotype that includes a smaller head, eyes, body, and fins. (B) At 48 hpf, HuC–GFP fish injected with a 5 bp mismatch auts2 control MO display normal levels of developing neurons in the brain, whereas translational MO injected fish display less developing neurons in the cerebellum (ce), optic tectum (ot), and retina (ret). (C) At 120 hpf, fish injected with an auts2 splicing MO and stained with Alcian blue show a significant reduction in the distance between the Meckel (Mk) and ceratohyal cartilages (ch) (shown as a red line) compared to controls, indicating a reduced lower-jaw size. Panels (A, B) adapted from [17], (C) adapted from [26].
Table 2.
auts2 morpholino knockdown phenotypes
| Assay following morpholino injectiona | Developmental phenotype | Refs |
|---|---|---|
| Wholemount | Overall stunted development, including smaller head and eyes (Figure 3A). Less movement when prodded. | [17] |
| Microcephaly with no overall developmental delay. | [26] | |
| Alcian blue staining | Micrognathia (undersized jaw) and retrognathia (receded jaw) (Figure 3C). | [26] |
| HuC–GFP zebrafish line | Fewer developing neurons in the dorsal region of the midbrain, including the optic tectum, the midbrain- hindbrain boundary (including the cerebellum), the hindbrain and the retina [17] (Figure 3B). | [17] |
| HuC/D staining | Reduction in HuC/D-positive postmitotic neurons as well as a loss of bilateral symmetry. | [26] |
| TUNEL staining | Increased apoptosis in the midbrain. | [17] |
| PCNA staining | Increased cell proliferation in the forebrain, midbrain and hindbrain. | [17] |
| Phosphohistone H3 | Decreased cell proliferation in the brain. | [26] |
| Tg(mnx1:GFP) zebrafish line | Fewer motor neuron cell bodies in the spinal cord and weaker, improperly angled projections. | [17] |
| HNK-1 staining | Fewer sensory neurons in the spinal cord. | [17] |
HNK-1, neural cell adhesion molecule 1/Ncam1 (CD57); HuC/D, Hu antigen C/D [ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3/4]; mnx1, motor neuron and pancreas homeobox 1; PCNA, proliferating cell nuclear antigen; Tg, transgenic; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end-labeling.
Given the observation that non-coding regions within AUTS2 have been implicated in human evolution (Box 2) and disease, the regulatory landscape around AUTS2 was investigated [17]. Twenty-three enhancers were identified in zebrafish, 10 of which are active in the brain. Three mouse brain enhancers were found to overlap a purely non-coding ASD-associated deletion, and four different mouse enhancers (two of which were positive in the brain) were found to reside in regions implicated in human evolution, supporting the idea that this gene is tightly regulated, and that enhancers for this gene are important for health and evolution [17]. The enhancers described are potentially only a subset of the AUTS2 regulatory landscape – and it is possible that some of these enhancers regulate other genes, including those in the WBS critical region. Although the precise function of AUTS2 remains to be elucidated, current reports show it to be a crucial and tightly regulated gene involved in neurodevelopment.
Box 2. AUTS2 and human evolution.
In 2006 a comparative genomics approach was used to search the human genome for regions that have significantly changed in humans in the past 5 million years, since the divergence from chimpanzees, but are highly conserved in other species [66,79]. They identified 202 such regions which they termed human accelerated regions (HARs). These HARs are strong candidates for sequences responsible for the evolution of human-specific traits. An intronic region in AUTS2 (Figure 2) ranked as the 31st most accelerated region in their study. Similarly, in 2006 a different group combed the genome for conserved non-coding sequences in the human lineage that displayed accelerated evolution [67]. The authors identified 902 human accelerated conserved non-coding sequences (HACNSs). HACNSs 174 and 369 both lay within introns of AUTS2 (Figure 2). With the publication of the draft sequence of the Neanderthal genome in 2011, it was found that the first half of AUTS2 displayed the strongest statistical signal in a genomic screen differentiating modern humans from Neanderthals (Figure 2) [68]. This region contains 293 consecutive SNPs where only ancestral alleles were observed in the Neanderthals, only two of which are coding variants [a G to C non-synonymous substitution at chr7:68,702,743 (hg18) only in the Han Chinese and a C to T synonymous change at chr7:68,702,866 (hg18) within the Yoruba and Melanesian populations]. Other regions that were found to have the most significant human-Neanderthal changes also include genes that are involved in cognition and social interaction, including dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A (DYRK1A), neuregulin 3 (NRG3) and Ca2+-dependent secretion activator 2 (CADPS2) [68]. The authors conclude that multiple genes involved in cognitive development were positively selected during the evolution of modern humans [68]. Taken together, these studies suggest that significant changes in AUTS2 occurred specifically in modern humans and it is conceivable, based on the neurological role that this gene plays, that these changes could lead to cognitive traits specific to humans.
AUTS2 gene pathways
A 2010 study used radiation hybrid genotyping data to test for interaction of 99% of all possible gene pairs across the mammalian genome [53]. AUTS2 was the known gene with the greatest number of edges, or connectivity [53]. Despite that finding, little is known about the genetic pathways in which AUTS2 is involved. However, a few articles have provided evidence linking AUTS2 to other proteins and pathways.
One potential pathway was revealed by examining genes that can oscillate expression during somitogenesis. Two papers found that the expression of AUTS2 oscillates in phase with other notch pathway genes, suggesting that it is a component of the notch signaling pathway [54,55]. Notch signaling has been shown to be involved in neuronal migration through its interaction with Reelin, a gene implicated in ASD and a target of Tbr1 [56,57].
Although not reaching significance, a group found that Auts2 has a 1.33-fold change in cerebellar gene expression in methyl CpG binding protein 2 (Mecp2)-null mice. Loss of MECP2 function can cause neurodevelopmental disorders including Rett syndrome and autism [58]. The authors also compared their data with data generated from other gene expression studies. They found that Auts2 is consistently altered in both their datasets, as well as in post-mortem Rett syndrome patient brain, and is mutated in fibroblasts and lymphocytes [58].
Starting at mouse embryonic (E) day 12, Auts2 mRNA is expressed in the cortical preplate, where it colocalizes with Tbr1, a transcription factor that exerts positive and negative control of regional and laminar identity in postmitotic neurons [12,59]. Using Tbr1 antibodies for chromatin immunoprecipitation (ChIP) of E14.5 cortex, it was shown that the Auts2 promoter is a direct transcriptional target of Tbr1 in the developing neocortex and is involved in frontal identity [59].
SATB homeobox 2 (Satb2) is one of four genes (including Tbr1) that regulates projection identity within the layers of the mammalian cortex. In 2012 a report showed that, in mice, Tbr1 expression is dually regulated by Satb2 and B cell CLL/lymphoma 11B (Ctip2) in cortical layers 2–5. The authors also demonstrated that Satb2 regulates Auts2. They showed that, similarly to Tbr1, Auts2 is expressed in the deep and upper layers of the cortex. They investigated whether the loss of Tbr1 expression in the upper layer neurons in Satb2 mutants coincides with changes in Auts2 expression. They observed that there was a significant loss of Auts2 expression in the upper layers of Satb2 mutants, similar to the loss of Tbr1 in Satb2 mutants. The authors did not observe any changes in Auts2 expression in layers 5 or 6. Their results suggest that Satb2 regulates the expression of Tbr1, which in turn regulates Auts2 expression in callosal projection neurons [60].
GTF2I repeat domain containing 1 (GTF2IRD1) is one of 26 genes deleted in WBS, and encodes a putative transcription factor expressed throughout the brain during development. Gtf2ird1 knockout mice display reduced innate fear and increased sociability, phenotypes consistent with WBS [61]. Microarray screens were used to find transcriptional targets of Gtf2ird1 in brain tissue from Gtf2ird1 knockout mice at two timepoints – E15.5 and birth [postnatal (P) day 0] – versus wild type littermates. Auts2 was one of only two genes identified in both (E15.5 and P0) microarray experiments to be altered compared to controls. In P0 mouse brains of knockout mice, Auts2 was increased by 1.3-fold, whereas in E15.5 embryos it was decreased by 1.5-fold [62]. It is unclear if Auts2 is a target of Gtf2ird1 or if this observation reflects the proximity of the two genes.
Zinc finger matrin-type 3 (Zmat3, also known as Wig1), a transcription factor regulated by p53, plays an important role in RNA protection and stabilization and, as part of the p53 pathway, is a casual factor in neurodegenerative diseases. Wig1 downregulation by antisense oligonucleotide treatment led to a significant reduction in Auts2 mRNA levels in the brains of BACHD (bacterial artificial chromosome – HD) mice, a mouse model for Huntington’s disease (HD). The authors also reported a trend in reduction of Auts2 mRNA levels in the livers of BALB/c mice but no reduction in Auts2 levels in FVB (background strain of BACHD) mouse brains [63]. These results suggest a role for Wig1 in the regulation of Auts2 expression and further links Auts2 with pathways involved in the CNS.
Polycomb repressive complex 1 (PRC1) is a polycomb group (PcG) gene which acts as a developmental regulator through transcriptional repression. It is crucial for many biological processes in mammals, including differentiation. There are six major groups of PRC1 complexes, each containing a distinct polycomb group ring finger 1 (PCGF) subunit (PCGF1–6), a RING1 A/B ubiquitin ligase, and unique associated polypeptides. Using tandem affinity purification of PCGF3 and PCGF5, AUTS2 was recovered, implying a role for AUTS2 in transcriptional repression during development [64].
In 2013, the regulatory pathway for SEMA5A (semaphorin 5A), an autism candidate gene, was mapped in silico using expression quantitative trait locus (eQTL) mapping. The authors found that the SEMA5A regulatory network significantly overlaps with rare CNVs around ASD-associated genes, including AUTS2. Given the extensive trans-regulatory network associated with SEMA5A, the authors also investigated the possibility that there are several upstream master regulators that control this network. Performing eQTL mapping for expression levels of the eQTL-associated genes within the network (eQTLs of the eQTLs of SEMA5A), the authors identified 12 regions associated with the expression of 10 or more primary SEMA5A eQTL genes, including AUTS2. This study suggests that AUTS2 is involved, and may be a master regulator in ASD-related pathways [65].
Concluding remarks
As we identify the genes involved in ASD, DD, and ID, our ability to genetically diagnose these disorders improves, and future screens should assess AUTS2 for potential causative CNVs. However, before we are able to use AUTS2 as a diagnostic tool we must determine what makes a CNV in or around AUTS2 causative or benign and for what disorders (e.g., ID, DD, ASD, ASD with ID/DD, etc.). This includes a deeper investigation of the regulatory network of this gene. Although not in immediate sight, a major step in developing future ASD and ASD-related phenotype treatments relies on a solid understanding of the pathways involved and how they interact. Multiple reports have implicated AUTS2 in addiction and other neurological phenotypes, but the mechanism and certainty of these involvements remain unclear, highlighting the need for deeper investigations into the function of this gene and its role in development and disease. Future work using an Auts2 mouse knockout should reveal greater detail of the function of this gene. In addition, genomic studies such as RNA-seq following the knockdown of this gene and chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) could identify the various gene pathways and regions of the genome with which this gene interacts. Obtaining a better understanding of the pathways associated with AUTS2 will allow us to comprehend better the biological systems that can be perturbed when the function of this gene is disrupted, as well as how nucleotide changes within the gene might have led to human-specific traits. In summary, we can presume that this gene is involved in neurodevelopment, and may play a role in ASD and ASD-related phenotypes. There are also significant data suggesting that AUTS2 has human-specific variants that could possibly contribute to human cognition. It is important to differentiate the evolution and phenotypic data surrounding this gene. The data suggests that genes involved in human specific cognition may also play a role in human-specific disorders of the brain.
Acknowledgments
We would like to thank Christelle Golzio, Nicholas Katsanis, and Erik A. Sistermans for sharing their work on auts2 including their morpholino results used in Figure 3C. We would also like to thank members of the Ahituv lab for helpful comments. N.A. and N.O. received support for this research from the Simons Foundation (SFARI grant 256769 to N.A.), National Human Genome Research Institute (NHGRI) grant number R01HG005058, National Institute of Child Health and Human Development (NICHD) grant number R01HD059862, and National Institute of Neurological Disorders and Stroke (NINDS) grant number R01NS079231. N.O. is also supported in part by a Dennis Weatherstone pre-doctoral fellowship from Autism Speaks.
References
- 1.Fleischhacker WW, Brooks DJ. Neurodevelopmental Disorders. Springer; 2006. [Google Scholar]
- 2.Baio J, et al. Prevalence of autism spectrum disorders – autism and developmental disabilities monitoring network, 14 sites, United States, 2008. MMWR Surveill Summ. 2012;61:1–19. [PubMed] [Google Scholar]
- 3.Risch N, et al. A genomic screen of autism: evidence for a multilocus etiology. Am J Hum Genet. 1999;65:493–507. doi: 10.1086/302497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geschwind DH. Advances in autism. Annu Rev Med. 2009;60:367–380. doi: 10.1146/annurev.med.60.053107.121225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sultana R, et al. Identification of a novel gene on chromosome 7q11.2 interrupted by a translocation breakpoint in a pair of autistic twins. Genomics. 2002;80:129–134. doi: 10.1006/geno.2002.6810. [DOI] [PubMed] [Google Scholar]
- 7.Martens MA, et al. Research review: Williams syndrome: a critical review of the cognitive, behavioral, and neuroanatomical phenotype. J Child Psychol Psychiatry. 2008;49:576–608. doi: 10.1111/j.1469-7610.2008.01887.x. [DOI] [PubMed] [Google Scholar]
- 8.Kalscheuer VM, et al. Mutations in autism susceptibility candidate 2 (AUTS2) in patients with mental retardation. Hum Genet. 2007;121:501–509. doi: 10.1007/s00439-006-0284-0. [DOI] [PubMed] [Google Scholar]
- 9.Poeck B, et al. Locomotor control by the central complex in Drosophila – an analysis of the tay bridge mutant. Dev Neurobiol. 2008;68:1046–1058. doi: 10.1002/dneu.20643. [DOI] [PubMed] [Google Scholar]
- 10.Salichs E, et al. Genome-wide analysis of histidine repeats reveals their role in the localization of human proteins to the nuclear speckles compartment. PLoS Genet. 2009;5:e1000397. doi: 10.1371/journal.pgen.1000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 12.Bedogni F, et al. Autism susceptibility candidate 2 (Auts2) encodes a nuclear protein expressed in developing brain regions implicated in autism neuropathology. Gene Expr Patterns. 2010;10:9–15. doi: 10.1016/j.gep.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lepagnol-Bestel AM, et al. SLC25A12 expression is associated with neurite outgrowth and is upregulated in the prefrontal cortex of autistic subjects. Mol Psychiatry. 2008;13:385–397. doi: 10.1038/sj.mp.4002120. [DOI] [PubMed] [Google Scholar]
- 14.Zhang YE, et al. Accelerated recruitment of new brain development genes into the human genome. PLoS Biol. 2011;9:e1001179. doi: 10.1371/journal.pbio.1001179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner AH, et al. Exon-level expression profiling of ocular tissues. Exp Eye Res. 2013;111:105–111. doi: 10.1016/j.exer.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ameur A, et al. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat Struct Mol Biol. 2011;18:1435–1440. doi: 10.1038/nsmb.2143. [DOI] [PubMed] [Google Scholar]
- 17.Oksenberg N, et al. Function and regulation of AUTS2, a gene implicated in autism and human evolution. PLoS Genet. 2013;9:e1003221. doi: 10.1371/journal.pgen.1003221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinto D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bakkaloglu B, et al. molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet. 2008;82:165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang XL, et al. A de novo balanced translocation breakpoint truncating the autism susceptibility candidate 2 (AUTS2) gene in a patient with autism. Am J Med Genet A. 2010;152A:2112–2114. doi: 10.1002/ajmg.a.33497. [DOI] [PubMed] [Google Scholar]
- 21.Glessner JT, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ben-David E, et al. Identification of a functional rare variant in autism using genome-wide screen for monoallelic expression. Hum Mol Genet. 2011;20:3632–3641. doi: 10.1093/hmg/ddr283. [DOI] [PubMed] [Google Scholar]
- 23.Girirajan S, et al. Relative burden of large CNVs on a range of neurodevelopmental phenotypes. PLoS Genet. 2011;7:e1002334. doi: 10.1371/journal.pgen.1002334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Talkowski ME, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagamani SCS, et al. Detection of copy-number variation in AUTS2 gene by targeted exonic array CGH in patients with developmental delay and autistic spectrum disorders. Eur J Hum Genet. 2013;21:1–4. doi: 10.1038/ejhg.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beunders G, et al. Exonic deletions in AUTS2 cause a syndromic form of intellectual disability and suggest a critical role for the C Terminus. Am J Hum Genet. 2013;92:210–220. doi: 10.1016/j.ajhg.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Girirajan S, et al. Global increases in both common and rare copy number load associated with autism. Hum Mol Genet. 2013;22:2870–2880. doi: 10.1093/hmg/ddt136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuscó I, et al. Autism-specific copy number variants further implicate the phosphatidylinositol signaling pathway and the glutamatergic synapse in the etiology of the disorder. Hum Mol Genet. 2009;18:1795–1804. doi: 10.1093/hmg/ddp092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tropeano M, et al. Male-biased autosomal effect of 16p13.11 copy number variation in neurodevelopmental disorders. PLoS ONE. 2013;8:e61365. doi: 10.1371/journal.pone.0061365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jolley A, et al. De novo intragenic deletion of the autism susceptibility candidate 2 (AUTS2) gene in a patient with developmental delay: a case report and literature review. Am J Med Genet A. 2013;161:1508–1512. doi: 10.1002/ajmg.a.35922. [DOI] [PubMed] [Google Scholar]
- 31.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Roak BJ, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chahrour MH, et al. Whole-exome sequencing and homozygosity analysis implicate depolarization-regulated neuronal genes in autism. PLoS Genet. 2012;8:e1002635. doi: 10.1371/journal.pgen.1002635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flatscher-Bader T, et al. Increased de novo copy number variants in the offspring of older males. Transl Psychiatry. 2011;1:e34. doi: 10.1038/tp.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arlt M, Ozdemir A. Hydroxyurea induces de novo copy number variants in human cells. Proc Natl Acad Sci USA. 2011;108:17360–17365. doi: 10.1073/pnas.1109272108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arlt MF, et al. De novo CNV formation in mouse embryonic stem cells occurs in the absence of Xrcc4-dependent nonhomologous end joining. PLoS Genet. 2012;8:e1002981. doi: 10.1371/journal.pgen.1002981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong C, et al. Methylomic analysis of monozygotic twins discordant for autism spectrum disorder and related behavioural traits. Mol Psychiatry. 2013 doi: 10.1038/mp.2013.41. http://dx.doi.org/10.1038/mp.2013.41. [DOI] [PMC free article] [PubMed]
- 41.Philibert RA, et al. Transcriptional profiling of subjects from the Iowa adoption studies. Am J Med Genet B: Neuropsychiatr Genet. 2007;144B:683–690. doi: 10.1002/ajmg.b.30512. [DOI] [PubMed] [Google Scholar]
- 42.Schumann G, et al. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc Natl Acad Sci USA. 2011;108:7119–7124. doi: 10.1073/pnas.1017288108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao D, et al. Comparative gene expression profiling analysis of lymphoblastoid cells reveals neuron-specific enolase gene (ENO2) as a susceptibility gene of heroin dependence. Addict Biol. 2011 doi: 10.1111/j.1369-1600.2011.00390.x. http://dx.doi.org/10.1111/j.1369-1600.2011.00390.x. [DOI] [PubMed]
- 44.Chen YH, et al. Genetic analysis of AUTS2 as a susceptibility gene of heroin dependence. Drug Alcohol Depend. 2013;128:238–242. doi: 10.1016/j.drugalcdep.2012.08.029. [DOI] [PubMed] [Google Scholar]
- 45.Hamshere ML, et al. Genetic utility of broadly defined bipolar schizoaffective disorder as a diagnostic concept. Br J Psychiatry. 2009;195:23–29. doi: 10.1192/bjp.bp.108.061424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hattori E, et al. Preliminary genome-wide association study of bipolar disorder in the Japanese population. Am J Med Genet B: Neuropsychiatr Genet. 2009;150B:1110–1117. doi: 10.1002/ajmg.b.30941. [DOI] [PubMed] [Google Scholar]
- 47.Lee H, et al. A genome-wide association study of seasonal pattern mania identifies NF1A as a possible susceptibility gene for bipolar disorder. J Affect Disord. 2012;145:200–207. doi: 10.1016/j.jad.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mefford HC, et al. Genome-wide copy number variation in epilepsy: novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genet. 2010;6:e1000962. doi: 10.1371/journal.pgen.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elia J, et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol Psychiatry. 2010;15:637–646. doi: 10.1038/mp.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luciano M, et al. Whole genome association scan for genetic polymorphisms influencing information processing speed. Biol Psychol. 2011;86:193–202. doi: 10.1016/j.biopsycho.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chojnicka I, et al. Possible association between suicide committed under influence of ethanol and a variant in the AUTS2 gene. PLoS ONE. 2013;8:e57199. doi: 10.1371/journal.pone.0057199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gilbert SF. Developmental Biology. 6. Sinauer Associates; 2000. [Google Scholar]
- 53.Lin A, et al. A genome-wide map of human genetic interactions inferred from radiation hybrid genotypes. Genome Res. 2010;20:1122–1132. doi: 10.1101/gr.104216.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.William D, et al. Identification of oscillatory genes in somitogenesis from functional genomic analysis of a human mesenchymal stem cell model. Dev Biol. 2007;305:172–186. doi: 10.1016/j.ydbio.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dequéant ML, et al. A complex oscillating network of signaling genes underlies the mouse segmentation clock. Science. 2006;314:1595–1598. doi: 10.1126/science.1133141. [DOI] [PubMed] [Google Scholar]
- 56.Hashimoto-Torii K, et al. Interaction between Reelin and Notch signaling regulates neuronal migration in the cerebral cortex. Neuron. 2008;60:273–284. doi: 10.1016/j.neuron.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang GS, et al. Transcriptional modification by a CASK-interacting nucleosome assembly protein. Neuron. 2004;42:113–128. doi: 10.1016/s0896-6273(04)00139-4. [DOI] [PubMed] [Google Scholar]
- 58.Ben-Shachar S, et al. Mouse models of MeCP2 disorders share gene expression changes in the cerebellum and hypothalamus. Hum Mol Genet. 2009;18:2431–2442. doi: 10.1093/hmg/ddp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bedogni F, et al. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proc Natl Acad Sci USA. 2010;107:13129–13134. doi: 10.1073/pnas.1002285107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Srinivasan K, et al. A network of genetic repression and derepression specifies projection fates in the developing neocortex. Proc Natl Acad Sci USA. 2012;109:19071–19078. doi: 10.1073/pnas.1216793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Young EJ, et al. Reduced fear and aggression and altered serotonin metabolism in Gtf2ird1-targeted mice. Genes Brain Behav. 2008;7:224–234. doi: 10.1111/j.1601-183X.2007.00343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O’Leary J, Osborne LR. Global analysis of gene expression in the developing brain of Gtf2ird1 knockout mice. PLoS ONE. 2011;6:e23868. doi: 10.1371/journal.pone.0023868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sedaghat Y, et al. Genomic analysis of wig-1 pathways. PLoS ONE. 2012;7:e29429. doi: 10.1371/journal.pone.0029429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gao Z, et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell. 2012;45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheng Y, et al. An eQTL mapping approach reveals that rare variants in the SEMA5A regulatory network impact autism risk. Hum Mol Genet. 2013;22:2960–2972. doi: 10.1093/hmg/ddt150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pollard KS, et al. Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2006;2:e168. doi: 10.1371/journal.pgen.0020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prabhakar S, et al. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- 68.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gruszka-Westwood AM, et al. Comparative expressed sequence hybridization studies of high-hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2004;41:191–202. doi: 10.1002/gcc.20085. [DOI] [PubMed] [Google Scholar]
- 70.Kawamata N, et al. Cloning of genes involved in chromosomal translocations by high-resolution single nucleotide polymorphism genomic microarray. Proc Natl Acad Sci USA. 2008;105:11921–11926. doi: 10.1073/pnas.0711039105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Coyaud E, et al. PAX5–AUTS2 fusion resulting from t(7;9)(q11.2;p13.2) can now be classified as recurrent in B cell acute lymphoblastic leukemia. Leuk Res. 2010;34:e323–e325. doi: 10.1016/j.leukres.2010.07.035. [DOI] [PubMed] [Google Scholar]
- 72.Denk D, et al. PAX5-AUTS2: a recurrent fusion gene in childhood B-cell precursor acute lymphoblastic leukemia. Leuk Res. 2012;36:e178–e181. doi: 10.1016/j.leukres.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lener T, et al. Expression profiling of aging in the human skin. Exp Gerontol. 2006;41:387–397. doi: 10.1016/j.exger.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 74.Weir B, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Penney KL, et al. Genome-wide association study of prostate cancer mortality. Cancer Epidemiol Biomarkers Prev. 2010;19:2869–2876. doi: 10.1158/1055-9965.EPI-10-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sato S, et al. Characterization of porcine autism susceptibility candidate 2 as a candidate gene for the number of corpora lutea in pigs. Anim Reprod Sci. 2011;126:211–220. doi: 10.1016/j.anireprosci.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 77.Li R, et al. Six novel susceptibility loci for early-onset androgenetic alopecia and their unexpected association with common diseases. PLoS Genet. 2012;8:e1002746. doi: 10.1371/journal.pgen.1002746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stadler ZK, et al. Rare de novo germline copy-number variation in testicular cancer. Am J Hum Genet. 2012;91:379–383. doi: 10.1016/j.ajhg.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pollard KS, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]