Abstract
Developing proteomic biomarkers is valuable for evaluating therapeutic effects of drugs and generating better treatment strategies. However, conventional protein analysis is often challenging due to inadequate sample size of clinical specimens, lack of assay reproducibility, accuracy and sensitivity. A novel capillary isoelectricfocusing (IEF) immunoassay system (NanoPro) was used to study the dynamic phosphorylation status of signaling molecules in non-small cell lung cancer (NSCLC) cells treated with EGFR tyrosine kinase and MEK inhibitors. NanoPro showed the same dynamic ERK phosphorylation as western blotting with good assay reproducibility using 1,000 times less protein. The IEF separation in NanoPro system enables multiple protein phosphorylation isoforms to be resolved and detected simultaneously. With NanoPro, we identified a specific on-target MEK response pattern to MEK inhibitor PD325901, which was not detectable by western blotting. We also revealed a MEK2 signal that may be associated with NSCLC cell sensitivity to EGFR inhibitor erlotinib, and distinguish erlotinib-sensitive from -intrinsic as well as -acquired resistant cells. Moreover, NanoPro could differentiate human ERK1 isoforms from the mouse isoforms based on their pI differences and demonstrated that erlotinib effectively inhibited ERK phosphorylation in targeted human xenograft cancer cells but not in surrounding mouse stromal cells. With 8 ug of tumor aspirates, we precisely quantified the response of 18 signaling molecules to erlotinib and MEK1 inhibitor treatments in a NSCLC patient.
NanoPro’s higher sensitivity, better resolution of protein phosphorylation status and reduced tissue requirement warrant NanoPro’s investigation for future drug development and evaluation of drug effects of targeted therapies.
Keywords: Lung cancer, Methodology for proteomics, Novel assay technology, Xenograft models, Cellular responses to anticancer drugs
Introduction
Cancer is a leading cause of death in the world (1). With increasing understanding of the biology of cancer, targeted treatments according to the molecular features of the tumor have emerged as the mainstream of anti-cancer therapies (2). Examples include several tyrosine kinase inhibitors (TKIs), such as imatinib for chronic myeloid leukemia carrying the Bcr-Abl fusion gene (3) and unresectable gastrointestinal stromal tumors carrying the mutant KIT gene (4), and epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors for lung adenocarcinoma harboring the mutant EGFR gene (5, 6). Because TKIs possess several “off-target” effects, which may be responsible for unwanted side effects, proteomic assessment of the drug effect in tumors may be useful in order to distinguish on-target from off-target effects (7).
The study of on-targets and off-targets of specific drugs requires sufficient tumor material, and this is at times challenging (e.g. when using diagnostic tools such as fine-needle aspirations). Larger biopsies and repeat biopsies (e.g. in cases where drug resistance has occurred), although important for treatment decisions, may be potentially hazardous and are not feasible in all patients (8, 9).
Whereas relatively small amounts of tumor cells may allow the study of genetic alterations (i.e. mutations, amplifications, fusion genes) and assessment of mRNA expression, profiling of proteins in tumors, in particular the dynamics of protein phosphorylation, is more complex. Mass spectrometry, conventional western blotting and immunohistochemical staining (IHC) have been used to evaluate the expression and activity (e.g. phosphorylated forms) of proteins. The quantity of proteins required for mass spectrometry is in the order of several milligrams, and micrograms per target for conventional western blotting, while fine-needle aspirations or core biopsies may only provide micrograms of protein material (10). Although IHC requires less material, IHC procedures are not standardized and interpretation is subjective and non-quantitative (11, 12).
Nano-fluidic proteomic immunoassay (NanoPro) is an automated capillary isoelectric focusing immunoassay system (13, 14) recently developed and commercialized by ProteinSimple. In NanoPro analysis, proteins and respective isoforms are separated by charge, followed by target specific antibody probing. NanoPro can simultaneously separate, detect and quantify multiple protein phosphorylation isoforms, thus allowing more sensitive and accurate dissection of cell signaling events. Because as little as 25 cells may be sufficient for each experiment (13), NanoPro could be a valuable assay to study the pharmacodynamics of targeted therapies. NanoPro has been applied to evaluate the dynamics of activities of oncoproteins in hematologic malignancies (14–17). In this report, using NanoPro, we studied oncoprotein activation status in lung cancer cell lines treated with tyrosine kinase and MEK inhibitors. Discrete MEK signal profiles were revealed by NanoPro but not conventional western, which correlated with specific cell responses to drug treatments. We further demonstrated the applications of NanoPro in evaluating human cancer specific signaling responses in xenograft tumors, and the feasibility of proteomic assessment in a biopsy of a lung cancer patient.
Materials and Methods
Drugs and cancer cell lines
Erlotinib was obtained from LC laboratories (Woburn, MA) and PD325901 (18) was purchased from EMD Millipore Chemicals (Darmstadt, Germany). Drugs were dissolved in DMSO at a concentration of 10 mM.
PC-9 was kindly provided by Dr. James Chih-Hsin Yang (National Taiwan University, Taiwan) and H4006 by Dr. Tetsuya Mitsudomi (Aichi cancer center, Japan); H322, H2122, and HCC827 were obtained from American Type Culture Collection (ATCC, Manassas, VA). The cell lines were not tested and authenticated by the authors. Cells were maintained in RPMI media containing 10% serum. Erlotinib acquired-resistant HCC827 and H4006 subclones were established by incubating the cells in escalating concentrations of erlotinib and were labeled as HCC827R and H4006R, respectively. Resistant cells were maintained in media containing 500 nM erlotinib.
Lung cancer specimens
A patient with lung adenocarcinoma underwent a core biopsy of a lung lesion, followed by treatment with a combination of AZD6244, a MEK1/2 inhibitor, and erlotinib (NCT01229150). The second day after treatment, a fine-needle aspiration of an axillary lymph node metastasis was performed. Pathologic report indicated that the aspirated material contained more than 50% of cancer cells and some necrotic tissue. Both pre- and post-treatment specimens were lysed and run on NanoPro system. Use of tumor specimens was approved by the institute review broad of the National Cancer Institute, National Institutes of Health.
Cell viability test
1,500 cells were plated onto 96-well plate one day before drug treatment. Cells were treated with various concentrations of erlotinib for 72 hours. The IC50 value was determined using CellTiter 96 AQueousOne Solution Cell Proliferation Assay (Promega, Fitchburg, WI).
Nano-fluidic proteomic immunoassay (NanoPro) analysis
Cell and tissue samples were lysed with M-Per buffer (Thermo Scientific Inc., Waltham, MA) containing phosphatase and protease inhibitors (EMD Millipore, Billerica, MA). Lysates were mixed with ampholyte premix and fluorescently labeled pI standards for analysis on the NanoPro1000 system (ProteinSimple, Santa Clara, CA) according to manufacturer’s instructions. In brief, a mixture of 30–50 ng cell lysate, 1x fluorescent pI standard ladder 3, and 1x premix 5–8 ampholyte (ProteinSimple) was loaded onto the capillaries. Capillary isoelectric focusing electrophoresis was performed at 21,000 microwatts for 40 minutes. The separated proteins were immobilized to the capillary wall by exposing to UV light for 100 seconds at instrument default setting. Immunoprobing was performed using primary antibodies obtained from Cell Signaling Technology (Danvers, MA) (anti-phospho-ERK, anti-AKT, and anti-pJNK T183/Y185), Millipore (Billerica, MA) (anti-ERK1/2, anti-pMEK1 pT292, and anti-MEK1), Abcam (Cambridge, MA) (anti-Alas1, anti-pMEK2 T394, and anti-pSTAT3 Y705), Epitomics (Burlingame, CA) (anti-MEK2 and anti-pMEK1 S218/S222), and Novus (Littleton, CO) (anti-MEK1 pT386), and then probed with HRP-conjugated, goat anti-rabbit or goat anti-mouse secondary antibodies (Jackson ImmunoResearch, West Grove, PA). All antibody incubation and wash steps were programmed and performed automatically in NanoPro system, the incubation time was 2 hours for primary antibodies, and 1 hour for secondly antibodies. A 1:1 Luminol and peroxide (ProteinSimple) mixture was then added to generate chemiluminescent light, which was captured by a charge-coupled device (CCD) camera. The digital image was analyzed and quantified with Compass software (ProteinSimple) according to the user guide provided by the manufacturer. In compass analysis, peak area corresponds with signal strength and was used for quantitation. Experiments were performed at least in triplicate.
Immunoblotting
For conventional western blots, cells were incubated in serum-free media for 24 hours, treated with various concentrations of drugs for one hour, and stimulated by 20 ng/ml EGF for 10 min. Cells were then harvested, trypsinized, lysed in RIPA buffer containing protease (Santa Cruz Biotechnology, Santa Cruz, CA) and phosphatase inhibitors (Thermo Scientific, Waltham, MA) for 30 min at 4°C, and then centrifuged at 13,000 rpm at 4°C for 10 min. Xenograft tumors were excised, resuspended in RIPA buffer containing protease and phosphatase inhibitors, homogenized for 30 sec, incubated on ice for one hour, and centrifuged at 13,000 rpm at 4°C for 40 min. Supernatant was collected.
Western blots were performed as previously described (19). Antibodies were obtained from Cell Signaling Technology (Danvers, MA) (EGFR, pERK1/2, ERK1/2, pJNK), Abcam (Cambridge, MA) (EGFR pY1068, MEK1/2 pS218/S222, MEK2 pT396, and MEK1/2), and Sigma Aldrich (St. Louis MI) (Actin). Quantitation of bands was performed using ImageJ software (NIH, Bethesda, MD).
Immunoblotting for anti-MEK2 siRNA knockdown samples were analyzed using the automated capillary-based Simon SimpleWestern™ System (ProteinSimple; Santa Clara, CA) as previously described (20). In brief, 20 ng of cell lysate was mixed with a master mix containing 1x sample buffer, 1x fluorescent molecular weight markers and 40 mM DTT (ProteinSimple, Santa Clara, CA), then heated at 95 °C for 5 minutes prior to loading to the plate for analysis. During capillary electrophoresis, proteins were separated on the basis of molecular weight through the stacking and separation matrices at 250 volts for 40 minutes and then immobilized on the capillary wall using UV light. Capillaries were next incubated with a blocking reagent (ProteinSimple), and then immunoprobed with anti-MEK2 antibody (Epitomics) and HRP-conjugated goat anti-rabbit secondary antibody (Jackson ImmunoResearch). Chemiluminescent signals were detected and analyzed as described in the NanoPro analysis.
Xenograft study
Three million HCC827 cells were injected subcutaneously into the flank of athymic nude mice. Once the tumor diameter reached 5 mm, or tumor volume reached ~60 mm3 using the V = 1/2(L × W2) formula, mice were treated with either one dose of water or 100 mg/kg erlotinib suspended in water by oral gavage. Animals were sacrificed 24 hours after treatment. Xenograft, skin, and lung tissue were collected and snap frozen. Specimens were stored at −80°C before further western or NanoPro analysis.
siRNA transfection
MEK2 siRNA and non-specific control siRNA were purchased from Dharmacon (Lafayette, CO). siRNA was transfected using Lipofetamine RNAiMAX transfection reagent (Invitrogen) following manufacturer’s recommendation. Cells were trypsinized 48 hours after transfection for further investigation.
Results
NanoPro is a sensitive method to study individual ERK phosphorylation isoforms in response to erlotinib treatment
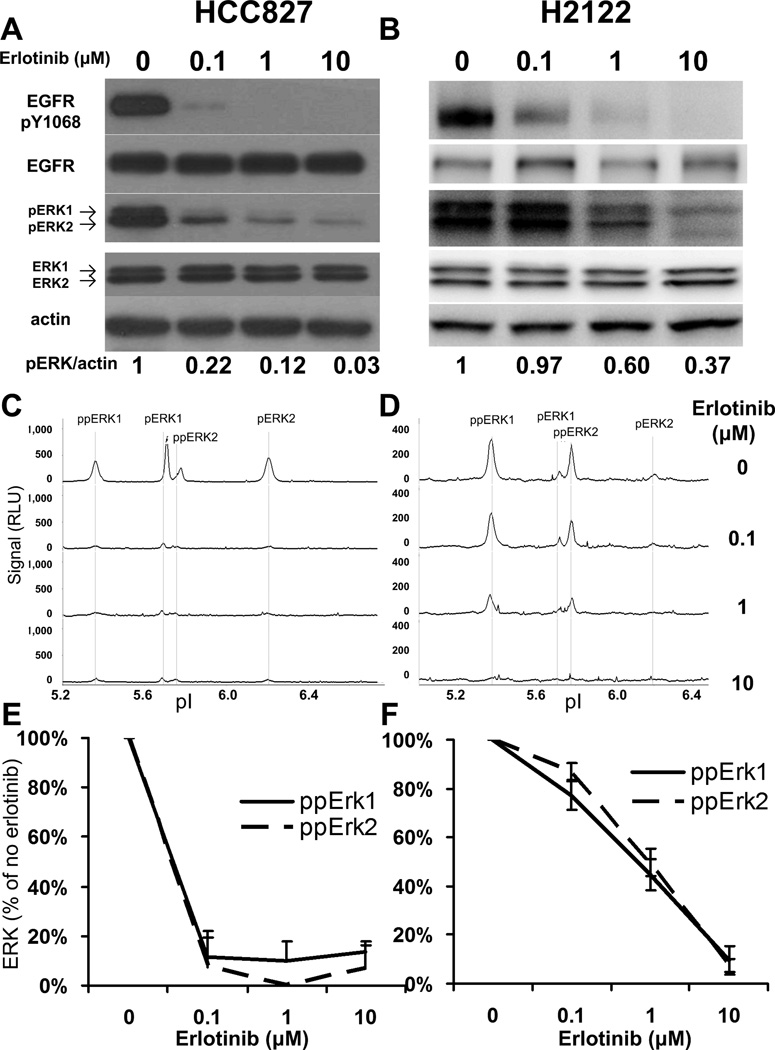
HCC827 cells, carrying amplified EGFR with E746-A750 deletion, are sensitive to erlotinib, and H2122 cells, carrying mutant KRAS gene, are resistant to erlotinib (21). Using 30 µg of cell lysates, we observed that erlotinib inhibited EGFR phosphorylation in both HCC827 and H2122 cell lines (Figure 1A and 1B) by western blotting. Erlotinib at 0.1 µM was sufficient to almost completely inhibit the downstream ERK1/2 phosphorylation in HCC827 cells, whereas 10 µM of erlotinib only partially inhibited ERK1/2 phosphorylation in H2122 cells (Figure 1A and 1B). Using 30 ng of cell lysate, we further analyzed above samples by NanoPro and compared results with western blot data. Since protein isoforms are separated by charge in the NanoPro system, it has been shown that NanoPro can distinguish and quantify non-phosphorylated, mono- and dual-phosphorylated ERK1/2 forms (designated later in the text as ERK1/2, pERK1/2, and ppERK1/2, respectively), which cannot be resolved by western blot analysis (13). On assaying phospho-ERK1/2, we observed decreases of phospho-ERK1/2 isoforms in both HCC827 and H2122 cells upon erlotinib treatment (Figure 1C and 1D). The signals of phospho-Erk isoforms were quantitated using Compass software and inhibition of Erk phosphorylation by erlotinib treatment were expressed relative to that of the no drug control (Figure 1E and 1F). The signals of the ppERK1 and ppERK2 decreased by more than 80% upon 0.1 µM erlotinib treatment in HCC827 cells and upon 10 µM erlotinib treatment in H2122 cells, consistent with western blot results. Samples were analyzed in triplicate. As shown in figure 1E and 1F, %CV of less than 15% was observed in samples with less than 60% signal decrease. Though higher %CV was observed in samples with more than 90% signal decrease (due to a reduction in the signal to noise ratio) less than 11% standard deviation was observed in those samples. This observation indicates good assay reproducibility of NanoPro analysis compared to conventional western blotting. In summary, NanoPro provided the same dynamics of phospho-ERK activities as western blot, while using 1,000 times less protein material. The NanoPro results were highly quantitative and reproducible and distinguished the activities of ERK1 from ERK2 and their respective phosphorylated isoforms.
Figure 1. Profiles of ERK1/2 phosphorylation in erlotinib treated HCC827 and H2122 cells.
Cells were treated with indicated concentrations of erlotinib, followed by 20 ng/ml EGF stimulation. (A) and (B) Western blot showing phospho-EGFR and phospho-ERK response to erlotinib dosing in HCC827 and H2122 cells, respectively. 30 µg of protein lysate were loaded for each sample. Semi-quantitation of phospho-ERK1/2 intensities, calibrated by actin intensity, was provided at bottom of images. (C) and (D) NanoPro showing the profiles of phospho-ERK1/2 isoforms and their response to erlotinib dosing in HCC827 and H2122 cells, respectively. 30 ng of protein lysate were loaded for each sample. (E) and (F) Quantitation of dual-phospho ERK1/2 signal intensities as percentage of no drug treatment control in (C) and (D), respectively. Error bars are standard deviations from triplicate analysis.
NanoPro can distinguish ERK1/2 phosphorylation in human xenograft tumors from surrounding mouse stromal cells in response to erlotinib treatment
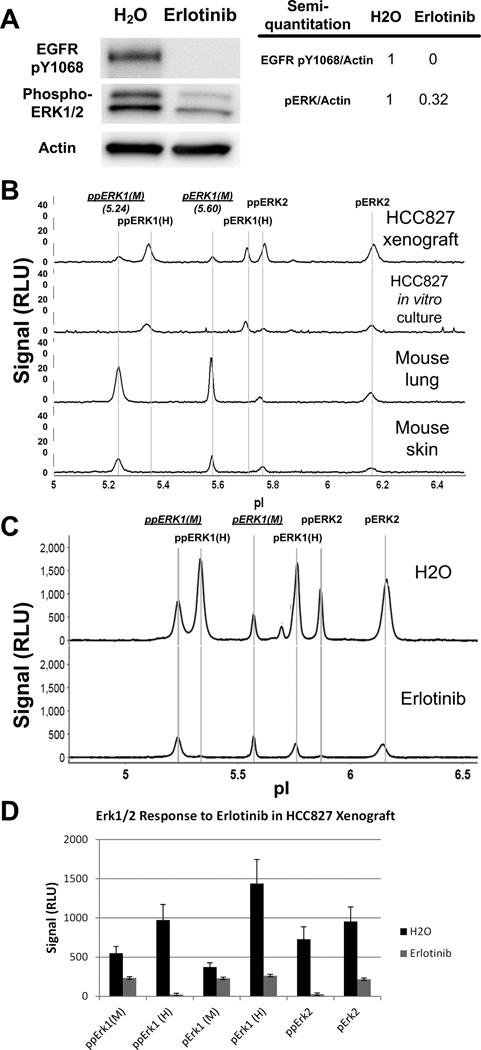
HCC827 xenograft mice were treated with one dose of either water or erlotinib, and xenograft samples were analyzed. Western blot analysis indicated that EGFR Y1068 was completely dephosphorylated upon erlotinib treatment, whereas ERK1/2 was only partly dephosphorylated (Figure 2A). On assaying ERK phosphorylation with NanoPro, we observed two additional signals, pI values of 5.24 and 5.60, in HCC827 xenograft samples that were not present in HCC827 in vitro cultured cells (Figure 2B). These two peaks have lower pI than the ppERK1 and pERK1 peaks observed in HCC827 cells. Since the theoretical pI value of mouse ERK1 is lower than that of human ERK1 (6.15 and 6.28, respectively, for non-phospho ERK1), we predicted that these two peaks are mouse ERK1 isoforms. Further analysis of mouse lung and skin samples confirmed the identity of the pI 5.24 and pI 5.60 peaks to be mouse ppERK1 and pERK1, respectively (Figure 2B). We also observed that, in erlotinib treated mouse xenografts, the human phospho-ERK1 signals decreased dramatically, whereas the mouse phospho-ERK1 signals decreased only modestly (Figure 2C and 2D). Further analysis of the lung and skin tissue samples from mice treated with erlotinib showed no significant decrease in mouse lung or only modest decrease of ERK phosphorylation in mouse skin, when compared to tissue samples from mice treated with water only (Figure S1A and S1B). NanoPro analysis data indicate that the residual phospho-ERK activities observed in western blot were derived from mouse stromal cells in the xenograft rather than from human cancer cells. These data demonstrate that NanoPro technology is able to distinguish human cancer cell-specific signals and their response to drug treatment from interfering mouse stromal cells in xenografts, and clearly revealed that erlotinib effectively inhibited down-stream Erk phosphorylation in targeted tumor cells but not surrounding stromal cells.
Figure 2. Profile of ERK1/2 phosphorylation in HCC827 xenografts.
HCC827 xenograft mice were treated with one dose of water or 100 mg/kg erlotinib, and sacrificed 24 hours after treatment. (A) Western blot analysis of EGFR and ERK phosphorylation in xenograft samples treated with or without erlotinib and semi-quantitation of phospho-EGFR and phospho-ERK1/2 intensities, calibrated by actin intensity. (B) NanoPro profiles of phospho-ERK1/2 isoforms in HCC827 xenograft, HCC827 in vitro culture, nude mouse lung, and nude mouse skin. (C) Representative profile of NanoPro analysis of ERK phosphorylation in xenograft samples treated with water or with erlotinib. pERK1 and ppERK1 of mouse origin are shown in underline with M in parenthesis. 30 ng of protein lysate were loaded for each sample in NanoPro analysis, except mouse lung samples that 50 ng protein was loaded. (D) Quantitation of ERK isoforms in response to erlotinib treatment in xenograft samples.
Specific target response pattern detected by NanoPro in response to MEK inhibitor treatment
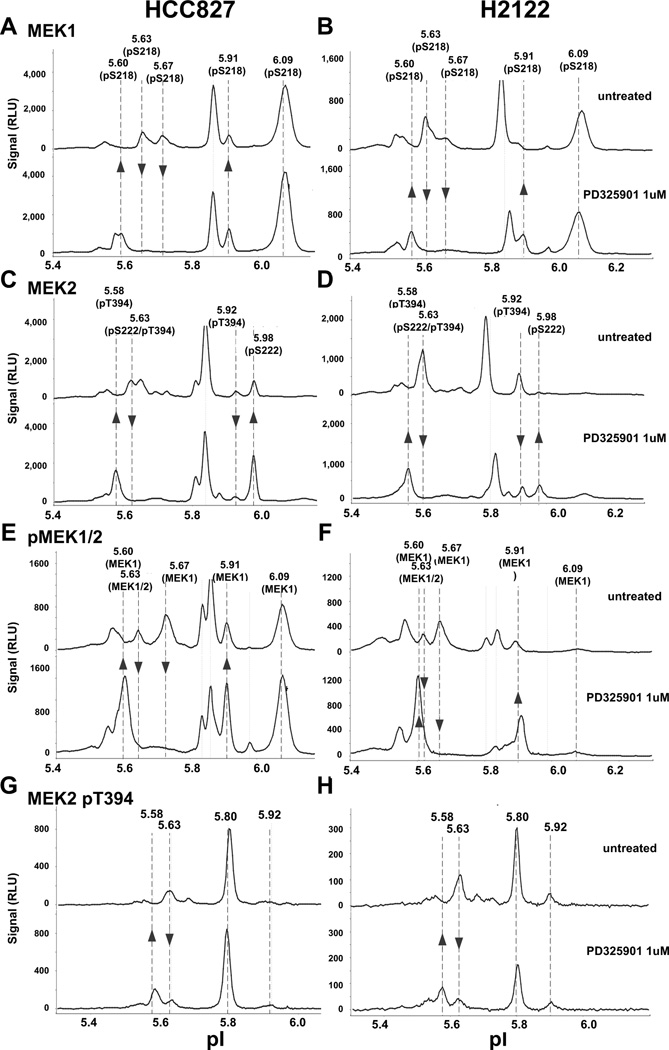
Drug treatment of NSCLC cells with PD325901, an allosteric MEK1/2 inhibitor, resulted in dephosphorylation of ERK1/2, up-regulation of MEK1/2 pS218/S222 in HCC827 cells, and slight down-regulation of MEK2 pT394 in H2122 cells as observed in western analysis (Figure S2A). Using NanoPro, we confirmed the drug inhibition on the phosphorylation of ERK isoforms (Figure S2B).
While HCC827 and H2122 cells exhibited different MEK1/2 peak profiles in un-treated baseline samples, a similar drug response signature was shared by both cell lines when treated with PD325901. For example, in comparison with H2122 cells, untreated HCC827 cells presented relatively higher MEK1 pS218 signal at pI 6.09 (compare Figure 3A & 3E with 3B & 3F), and MEK2 pS222 signal at pI 5.98 (compare Figure 3C & 3E with 3D & 3F) and a relatively lower MEK2 pT394 signal at pI 5.92 (compare Figure 3C & 3G with 3D & 3H). However with PD325901 treatment, as highlighted by arrows in Figure 3, specific MEK response patterns appeared in both cell lines. The pattern was composed of an increase of MEK1 pS218/pS222 (pI 5.60, 5.91, and 5.98 peaks) and MEK2 (pT394 pI 5.58) peaks; and a decrease of MEK1/2 pS218/S222 (pI 5.63) peak, MEK1 pS218 (pI 5.67) peak, and MEK2 pT394 (pI 5.63 and pI 5.92) peaks. These peaks were detected by the pan-reactive MEK1 (Figure 3A & 3B) and MEK2 antibodies (Figure 3C & 3D), as well as phospho-specific MEK1/2 pS218/222 (Figure 3E & 3F) and MEK2 pT394 antibodies (Figure 3G & 3H). Quantitation of the PD325901 responding MEK signals were shown in Figure S3.
Figure 3. Profiles of MEK signal response in NSCLC cells treated with MEK inhibitor PD325901.
HCC827 and H2122 cells were treated with or without 1 µM of PD325901, followed by 20 ng/ml EGF stimulation. NanoPro analysis of MEK1 in HCC827 (A) and H2122 (B), MEK2 in HCC827 (C) and H2122 (D), MEK1/2 pS218/S222 in HCC827 (E) and H2122 (F), MEK2 pT394 in HCC827 (G) and H2122 cells (H). Arrows indicate the responding MEK isoforms upon PD325901 treatment.
Peak assignments were established using isoform-specific and phospho-specific antibodies as shown in Figure S4. For example, overlaying MEK1/2 pS218/222 signals with MEK2 (Figure S4A and S4C) and MEK1 (Figure S4B and S4D) signals, indicated that the pI 5.67, 5.86, 5.91, and 6.09 peaks are MEK1 isoforms, the pI 5.80 and 5.98 are MEK2 isoforms. Overlaying MEK2 with MEK2 pS222 (Figure S4A and S4C) or MEK2 pT394 (Figure S4E) indicated that the pI 5.58, and 5.92 peaks are phosphorylated at pT394 but not pS222. pI 5.63 peak contains both MEK1 and MEK2 isoforms, and pI 5.63 and 5.80 peaks contain both MEK2 pS222 and pT394 isoforms. Comparison of MEK1 pS218 signals in PD325901 untreated (Figure S4B) and treated (Figure S4D) cells indicated that the pI 5.60 peak is a specific MEK1 pS218 isoform that responded to PD325901 treatment.
In summary, in both HCC827 and H2122 cells, NanoPro detected a specific MEK signal pattern in response to PD325901 treatment, which was not detectable using conventional western blot. The pattern clearly demonstrated the on-target effect of the MEK inhibitor PD325901.
A MEK2 signal associated with erlotinib sensitivity in NSCLC cells
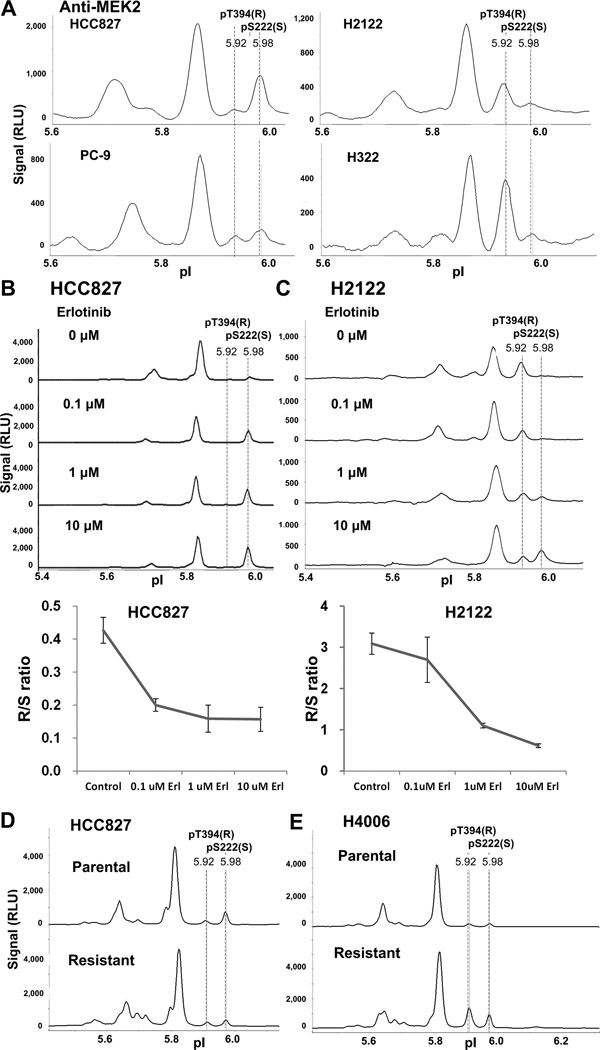
Erlotinib sensitive (HCC827 and PC-9) and resistant (H2122 and H322) NSCLC cells (Figure S5A), as well as acquired erlotinib resistant cells (HCC827R and H4006R) (Figure S5B), were analyzed by NanoPro, and peak profiles were compared. The analysis detected two MEK2 peaks, at pI 5.92 and pI 5.98,which correlated with NSCLC cell sensitivities to erlotinib. We observed that the pI 5.92 signals were always higher than the pI 5.98 signals in erlotinib-resistant H2122 and H322 cells with the pI 5.92 / pI 5.98 signal ratio of 4.16 and 4.78, respectively, whereas the pI 5.92 signals were lower than pI 5.98 signals in erlotinib-sensitive PC-9 and HCC827 cells with the pI 5.92 / pI5.98 signal ratio of 0.73 and 0.41 respectively, although the pattern was not as prominent in PC-9 as in HCC827 cells (Figure 4A and Table 1). The pI 5.92 and pI 5.98 peaks were hereafter designated as R (resistant) and S (sensitive), respectively. As demonstrated in the previous section, blotting with isoform-specific and phospho-specific antibodies identified the R and S peaks as MEK2 pT394 and MEK2 pS222/226, respectively. We observed an erlotinib dose-dependent decrease of the R signal and an increase in the S signal in both HCC827 cells (Figure 4B) and H2122 cells (Figure 4C). Plotting the R/S signal ratio with erlotinib concentration generated a curve corresponding with cell response to erlotinib (Figure 4B and 4C bottom), that is similar to the ERK phosphorylation response curve shown in figure 1E and 1F. In HCC827 cells, R/S ratio reached a low-end plateau at 0.1 µM erlotinib, a concentration that erlotinib efficiently inhibited downstream ERK phosphorylation; whereas, in H2122 cells, 10 µM of erlotinib was needed to decrease the R/S ratio to a comparable level. These data suggest that these MEK2 R and S peaks are related with cell response to erlotinib treatment.
Figure 4. MEK2 peaks indicate erlotinib sensitivity.
(A) Comparison of MEK2 pT394 (pI 5.92, R in parenthesis denotes resistant signal) and MEK2 pS222 (pI 5.98, S in parenthesis denotes sensitive signal) signals among HCC827, PC-9, H322, and H2122 cells. (B and C) NanoPro profile of MEK2 signals in HCC827 (B) and H2122 (C) cells treated with indicated concentrations of erlotinib, followed by EGF stimulation. (B and C bottom) Quantitation of pT394 and pS222 signals in HCC827 and H2122 cells treated with indicated concentrations of erlotinib. Ratio of pT394(R)/pS222(S) is plotted versus erlotinib concentration. Error bars are standard deviations from triplicate analysis. (D and E) Comparison of MEK2 pT394(R) and pS222(S) signals between parental and erlotinib-resistant HCC827 cells (D) and between parental and erlotinib-resistant H4006 cells (E). 40 ng of protein lysate were loaded for each sample.
Table 1.
The MEK2 R/S ratio in erlotinib sensitive, intrinsic erlotinib resistant, and acquired erlotinib resistant lung adenocarcinoma cell lines.
| R/S | IC50 (µM erlotinib) |
|
|---|---|---|
| Sensitive | ||
| PC-9 | 0.73 ± 0.11 | 0.1 |
| HCC827 | 0.41 ± 0.01 | 0.03 |
| H4006 | 1.21 ± 0.02 | 0.05 |
| Intrinsic resistant | ||
| H322 | 4.78 ± 0.44 | > 3.0 |
| H2122 | 4.16 ± 0.98 | > 3.0 |
| Acquired resistant | ||
| HCC827R | 0.77 ± 0.03 | >3.0 |
| H4006R | 1.87 ± 0.03 | >3.0 |
We further quantitated and compared the R and S signals in erlotinib-sensitive HCC827 and H4006 cells to their corresponding acquired erlotinib-resistant HCC827R and H4006R cells (Figure S5B). The acquired resistant subclones showed a higher R/S ratio than their parental sensitive cells (Figure 4D and 4E, and Table 1). Our observation suggests that the MEK2 R/S ratio may be associated with erlotinib sensitivity in vitro.
To further verify that the R and S signals are MEK2-specific, cells were transfected with MEK2 specific siRNA: knockdown of MEK2 eliminated R and S signals confirming that the R and S signals are MEK2 specific (Figure S6).
Pharmacodynamic effects assessed by NanoPro in a lung cancer patient
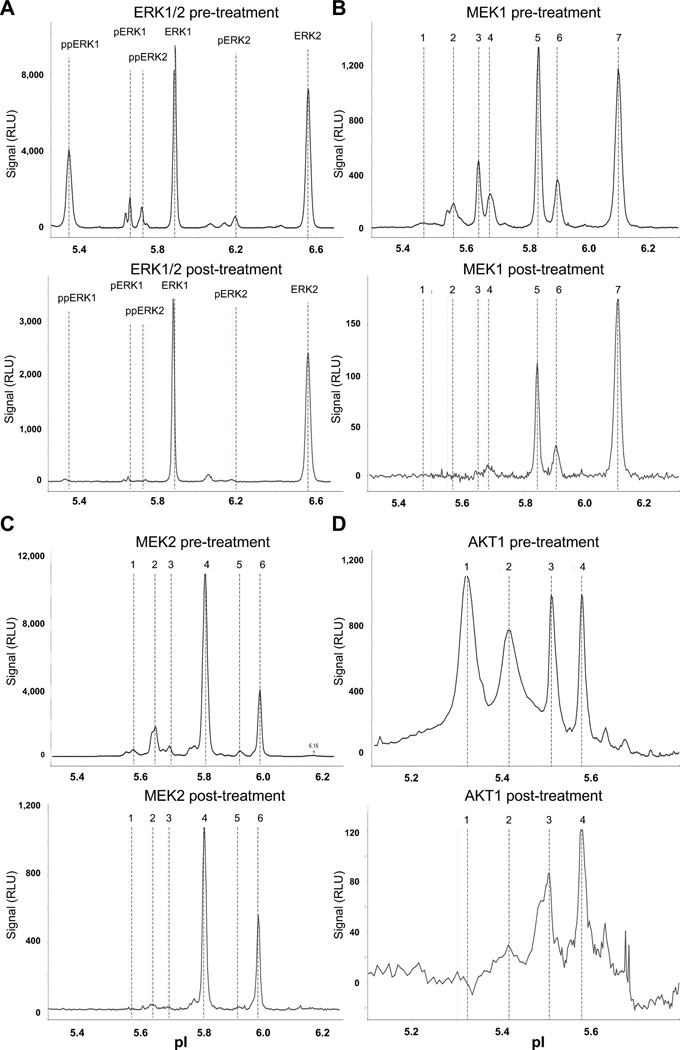
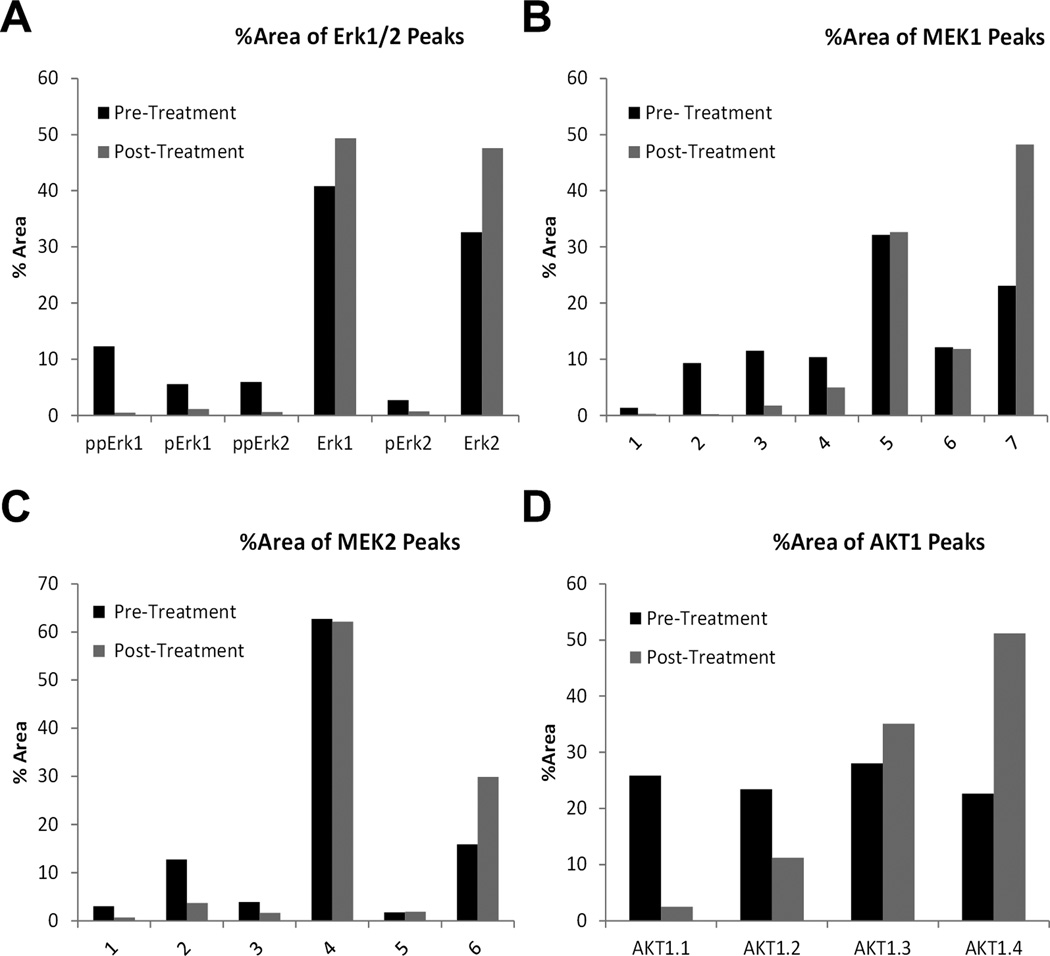
Tumor specimens were acquired from an advanced lung cancer patient before and after treatment with a combination of erlotinib and AZD6244 (Selumetinib, a MEK1/2 allosteric inhibitor; NCT01229150). Using 8 µg of each sample lysate, we profiled 18 protein targets, including phospho-ERK, total ERK, MEK1/2, phospho-MEK1 and phospho-MEK2, AKT1, phospho-AKT, phospho-JNK, phospho-STAT3 isoforms and loading controls. Analysis profiles of ERK1/2, MEK1, MEK2 and AKT1 are presented in Figure 5, and the quantitation of the ERK1/2, MEK1, MEK2 and AKT1 isoforms are shown in Figure 6. The signals of all tested targets, including the loading control Alas1 (Figure S7A), were smaller in the post-treatment specimen even though equal amount of protein lysates were loaded, probably due to presence of necrotic tissue in the aspirated specimen (Figure 5 and S7). As shown in Figure 5, by using pan-reactive antibodies, NanoPro revealed the distribution of different phosphorylated and non-phosphorylated isoforms. For the MEK1, MEK2 and AKT 1 profiles, more acidic, lower pI signals, representing more phosphorylated isoforms, were designated with lower numeric peak labels. More basic, higher pIs signals, representing less phosphorylated MEK1, MEK2 and AKT isoforms, were designated with higher numeric labels. Although signals were low in post-treatment samples, both phospho and non-phospho ERK, MEK and AKT signals were clearly detectable in the post-treatment samples (Figure 5). These peaks most likely were derived from the remaining viable cells in the post-treatment samples, as some peaks with shifted pI would be observed in the NanoPro assay system, if the signals were derived from degraded proteins in dead cells. As shown in Figure 5 (bottom panels), no Erk1, Erk2, MEK1, MEK2 and AKT1 peak shifting was detected.
Figure 5. Signaling molecule responses in a lung carcinoma patient following erlotinib and AZD6244 treatment.
(A) Profile of ERK1/2 signals. (B) Profile of MEK1 signals. (C) Profile of MEK2 signals. (D) Profile of AKT1 signals. 50 ng of protein lysate were loaded for each sample. See Figure 6 for isoform signal quantitation.
Figure 6. Quantitation of signaling molecule responses in the lung carcinoma patient following erlotinib and AZD6244 treatment.
(A) Quantitation of ERK1/2 signals. (B) Isoform distribution of MEK1 signal. (C) Isoform distribution of MEK2 isoforms. (D) Isoform distribution of AKT1 signal. Column values are percentage of the signal peak area (corresponding to signal strength) of respective isoform over the summation of peak areas for all isoforms. 50 ng of protein lysate were loaded for each sample.
Besides the overall decrease in signals, NanoPro data showed that drug treatment also dramatically inhibited the phosphorylation of all the major responding signaling molecules, i.e. ERK1/2, MEK1, MEK2 and AKT1. The distribution of phosphorylation isoforms was quantified as percentage of the signal peak area (obtained from Compass analysis, that corresponds with signal strength) of the respective isoform over the summation of peak areas for all isoforms. As shown in Figures 5 and 6, in the post-treatment specimen the percentage of the more phosphorylated ERK1/2, MEK1, MEK2 and AKT1 isoforms (lower pI signals designated with lower numeric labels) decreased and the percentage of the less phosphorylated ERK1/2, MEK1, MEK2 and AKT1 isoforms (higher pI signals designated with higher numeric labels) increased. The inhibition of phosphorylation was further confirmed upon assaying phospho-ERK1/2, MEK1 pT292, MEK1 pT386, JNK pT183/Y185, and STAT3 pY705 in the post-treatment specimen (Figure S7).
Discussion
Using NanoPro, an automated capillary isoelectric focusing immunoassay system, we studied dynamic oncoprotein phosphorylation in non-small cell lung cancer cells and drug responses to targeted therapies. Using high resolution isoelectric focusing, followed by target specific immunoprobing, NanoPro is able to separate different protein phosphorylation isoforms by charge differences, and detect them using pan reactive antibodies. While western blot has been the most commonly used assay to study protein phosphorylation, our study demonstrates that NanoPro data provides more detailed information on the activation status of each individual phosphorylated isoform, information which is not accessible using conventional immunoassay technologies. In our study, a complex MEK response profile was revealed by NanoPro, showing the on-target effect of MEK inhibitor PD325901. We also identified a MEK2 signal associated with erlotinib sensitivity in NSCLC cell lines. NanoPro was able to separate human from mouse ERK1 isoforms in xenograft tumor samples, thus allowing differentiation of human cancer cell specific signals from mouse stroma interference. NanoPro is 1000 times more sensitive, in terms of the amount of protein loaded, than conventional western blotting and detected the dynamics of ERK1/2 isoform activities upon erlotinib treatment with good assay reproducibility. We also showed the feasibility of evaluating the dynamics of target protein activities using a minute tumor specimen (total 8 µg) from a lung cancer patient, obtained by fine needle aspiration.
In the era of molecular targeted therapy, it has become important in drug development to be able to ascertain that a drug hits its claimed target in tumor tissue. In addition, the maximum tolerated dose (MTD), which has been used to develop cytotoxic agents in the past, may not be necessary to achieve maximal treatment effect of targeted therapies (22, 23). In addition, kinase inhibitors have been reported to have limited success in cancer treatment due to kinome reprogramming in response to drug treatment, thus it is desirable to monitor the dynamic and system changes of signals in multiple key pathways (24). Serial tumor biopsies have been advocated to study the direct effect of drugs on their targets (25). Serial biopsies have also been useful to study mechanisms of drug resistance and to potentially offer alternative treatments (26). However, this has not been broadly accepted because of the intrinsic risk involved in performing serial biopsies in patients with metastatic solid tumors. Our data demonstrate the feasibility of using fine needle aspirate samples for proteomics study of pharmacodynamics in molecular targeted therapy, and this could potentially be of great value for clinical development of molecular targeted therapies.
Whereas conventional western blotting resolves proteins according to their molecular weights, NanoPro resolves protein isoforms by isoelectric focusing points (13). Although NanoPro is not a substitute for size-based western blots, NanoPro provides more information regarding the activation status of target proteins than western blots, in addition to its higher sensitivity and better assay reproducibility. As an example, in erlotinib treated NSCLC cells, the signal detected by western blotting (Figure 1A & B) only provided general ERK1/2 de-phosphorylation in response to erlotinib treatment and phospho-specific antibody is required to detect the Erk1/2 activation. Whereas, using a pan-reactive antibody, Nanopro is able to measure the distribution of different ERK1/2 isoforms and their individual responses to drug treatment (Figure 1C &D), thus offers the opportunity for a more detailed and in-depth study of signaling molecule activation. The anti-phosho-ERK1/2 antibody we used for both western blot and NanoPro was produced by immunizing animals with a synthetic phospho-peptide corresponding to residues surrounding T202/Y204 of human ERK1 and recognized dual-phospho-ERK1/2 and mono-phospho-ERK1 pT202 (http://www.cellsignal.com/products/4370.html). Both dual-phospho-ERK1/2 and mono-phospho-ERK1/2 responded to erlotinib treatment to the same degree (Figure 1). Interestingly, we observed relatively high mono-phospho-ERK1 and mono-phospho-ERK2 activities in HCC827 cells (Figure 1C). Although mono-phospho-ERK1 pY204 is 500 fold less active than the fully activated dual-phospho-ERK1, the crystal structure indicates that T202 phosphorylation may have a significant role in the conformational change of the activation-loop of the ERK1 (27).
MEK1/2 phosphorylations were profiled in PD325901 treated NSCLC cells by both western blotting and NanoPro analysis, and compared. Conventional western blotting showed that drug treatment induced an up-regulation of MEK1/2 pS218/S222 in HCC827 cells and a slight down-regulation of MEK2 pT394 in H2122 cells (Figure S2A). Besides the up-regulation of total MEK1/2 pS218/222 signals in HCC827 (Figure 3E) and slight down-regulation of MEK2 pT394 signals in H2122 (Figure 3H) as observed in western blots, NanoPro also revealed that in both HCC827 cells and H2122 cells, upon PD325901 treatment, a specific drug response or MEK phosphorylation pattern was formed containing different MEK1 pS218, MEK2 pS222, and MEK2 pT394 isoforms (Figure 3A–3H). Although overall peak profiles are different between the HCC827 and H2122 cells (see a more detailed explanation in the Result section), the drug response patterns are similar in both cell lines (highlighted by arrows in Figure 3A–3H), indicating specific on-target effect of the MEK inhibitor.
We also used NanoPro to explore proteomic signatures related to drug sensitivity. EGFR mutations are important predictors of response to EGFR-TKI therapy in NSCLC (28, 29). Su et al. demonstrated that pretreatment presence of EGFR T790M mutation is a predictor for poor response to EGFR-TKI treatment of lung adenocarcinoma carrying common sensitizing EGFR mutations (30). A deletional BIM polymorphism in Asian lung cancer patients predicted poorer progression-free survival on erlotinib therapy (31). Using mass spectrometry, Zhang et al. demonstrated that three phosphorylation sites on the EGFR protein (Y1110, Y1172, and Y1197) are associated with erlotinib sensitivity in lung cancer cell lines (10). Here, using NanoPro assay, we identified a MEK2 signature, pI 5.92 (R signal = MEK2 pT394) and pI 5.98 (S signal = MEK2 pS222) peaks, that was associated to both intrinsic and acquired erlotinib resistance. Increased R/S signal ratios were observed in resistant NSCLC cells, and decreased R/S signal ratios were detected in cells responding to erlotinib treatment. The R and S peaks were identified to correspond to a MEK2 pT394 isoform and a MEK2 pS222 isoform, respectively. MEK2 T394, a homolog of MEK1 T386, is phosphorylated by kinases, including ERK1 (32, 33). Higher R signal (one of MEK2 pT394 specific signals) in the cells might indicate intrinsic kinase activities independent of EGFR signals, and hence the cells may not respond well to EGFR inhibitors. Even though the R signal was identified as containing the MEK2 phosphorylation at T394 and the S signal contains MEK2 phosphorylation at pS222, conventional western blotting using the phospho-specific antibodies was not able to detect and distinguish the R and S signals to evaluate their correspondence with cells sensitivity to erlotinib. According to the MEK2, MEKpS218/222 and MEK2 pT394 profiles in HCC827 and H2122 cells (Figure 3 and 4), the pI 5.92 MEK2 pT394 and pI 5.98 MEK2 pS222 isoform signals are less than 10% of total MEKpS218/222 or MEK2pT394 signal. Thus conventional western is not able to distinguish and detect this kind of isoform changes, as it measures total protein signal changes. These results warrant the evaluation of the clinical application of the MEK2 signature in lung adenocarcinomas. Though increased R/S ratios were observed in both the intrinsic and acquired resistant cells compared with the sensitive cells, the R/S ratios in the acquired resistant HCC827R and H4006R cells did not seem to correlate with the IC 50 of these cells. An explanation of this observation is that the parental sensitive cells and intrinsic resistant cells were grown in regular cell culture media, while the acquired resistant cells needed to be maintained in 500 nM erlotinib. To establish a cut-off R/S ratio for clinical application, it will require a careful evaluation with more patient samples.The complex peak profiles revealed by NanoPro may provide novel and critical information about the activation status of signaling molecules and how they respond to stimulus and drug treatments. As shown in this study, the response pattern to a MEK inhibitor is valuable in the evaluation of on-target effects of drug treatment; furthermore, the MEK2 peak correlation with cell sensitivity to erlotinib may be helpful formulate proper treatment strategy. Fan et al reported that a small presence of mono-phospho Erk2 (2.6% of total Erk) correlated with response to imatinib treatment in chronic myelogeneous leukemia (CML) patients (14). In the future, it might be of interest to explore the components and dynamics of individual phospho-protein isoforms in detail, in conjunction with other technologies e.g. mass spectrometry.
Subcutaneous xenografts in immunodeficient mice are still the most commonly used preclinical models to explore potential clinical activities of new anticancer drugs (34), however drugs active in xenografts have often failed during clinical development (35). Mouse stroma surrounding xenografts may confound interpretation of drug activities. Different signaling activation profiles were observed between tumor epithelium and stroma (16), thus it could be critical to access protein phosphorylation status in cancer cells for accurate interpretation of xenograft study results. As human and mouse have identical sequences around the ERK1 T202/Y204 area, anti-phospho-ERK1/2 antibodies bind to phospho-ERK1/2 of human origin as well as of mouse origin in the xenograft. Thus conventional immunoassays, e.g. western blotting or ELISA, would not be able to distinguish specific drug effect on human tumor cells from surrounding mouse tissue except that one has species-specific antibodies. However NanoPro could distinguish mouse from human mono-phospho-ERK1 and dual-phospho-ERK1 by their charge difference, and clearly demonstrated that erlotinib completely diminished ERK phosphorylation in human cancer cells, but only partially inhibited ERK phosphorylation in mouse stroma (Figure 2B and S 1). As EGFR-TKIs bind more tightly to mutant EGFR than to wild-type EGFR (36), including EGFR of mouse stromal cells, and ERK activities in the mouse tissue may depend not solely on EGFR signaling, it is not surprising to observe residual ERK phosphorylation of mouse origin in erlotinib treated xenograft as shown in Figure 2A.
In conclusion, we applied the automated capillary isoelectric focusing immunoaasy system to study signaling molecule activation status in tumor cells in vitro, in vivo and in human biopsy samples. During NanoPro analysis, different protein phosphorylation isoforms can be simultaneously separated, detected and quantified. Our study demonstrated that the NanoPro system is able to reveal the dynamics of individual isoform responses to treatment that are not detectable by western blotting, and offers the opportunity for novel biomarker discovery and therapeutic target identification. Credentialed with its higher sensitivity, better reproducibility and reduced tissue requirement compared to western blotting, NanoPro is appealing for drug development and proteomic assessment of drug effects in targeted therapies.
Supplementary Material
Acknowledgments
Financial support: Dr. Giuseppe Giaccone was supported by the Intramural Research Program, Center for Cancer Research, National Cancer Institute, National Institutes of Health.
Grant Support:
All authors in this manuscript were supported by the Intramural Research Program, National Institutes of Health, Center for Cancer Research, National Cancer Institute.
Abbreviations
- IEF
Isoelectricfocusing
- NSCLC
non-small cell lung cancer
- EGFR
Epidermal growth factor receptor
- TKI
Tyrosine kinase inhibitor
- IHC
immunohistochemical staining
- DMSO
Dimethyl sulfoxide
Footnotes
The authors disclose no potential conflicts of interest.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Martini M, Vecchione L, Siena S, Tejpar S, Bardelli A. Targeted therapies: how personal should we go? Nat Rev Clin Oncol. 2012;9:87–97. doi: 10.1038/nrclinonc.2011.164. [DOI] [PubMed] [Google Scholar]
- 3.O'Brien SG, Guilhot F, Larson RA, Gathmann I, Baccarani M, Cervantes F, et al. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2003;348:994–1004. doi: 10.1056/NEJMoa022457. [DOI] [PubMed] [Google Scholar]
- 4.Verweij J, Casali PG, Zalcberg J, LeCesne A, Reichardt P, Blay JY, et al. Progression-free survival in gastrointestinal stromal tumours with high-dose imatinib: randomised trial. Lancet. 2004;364:1127–1134. doi: 10.1016/S0140-6736(04)17098-0. [DOI] [PubMed] [Google Scholar]
- 5.Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 6.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 7.Wistuba II, Gelovani JG, Jacoby JJ, Davis SE, Herbst RS. Methodological and practical challenges for personalized cancer therapies. Nat Rev Clin Oncol. 2011;8:135–141. doi: 10.1038/nrclinonc.2011.2. [DOI] [PubMed] [Google Scholar]
- 8.Felip E, Rojo F, Reck M, Heller A, Klughammer B, Sala G, et al. A phase II pharmacodynamic study of erlotinib in patients with advanced non-small cell lung cancer previously treated with platinum-based chemotherapy. Clin Cancer Res. 2008;14:3867–3874. doi: 10.1158/1078-0432.CCR-07-5186. [DOI] [PubMed] [Google Scholar]
- 9.Kim ES, Herbst RS, Wistuba II, Lee JJ, Blumenschein GR, Jr, Tsao A, et al. The BATTLE trial: personalizing therapy for lung cancer. Cancer Discov. 2011;1:44–53. doi: 10.1158/2159-8274.CD-10-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang G, Fang B, Liu RZ, Lin H, Kinose F, Bai Y, et al. Mass spectrometry mapping of epidermal growth factor receptor phosphorylation related to oncogenic mutations and tyrosine kinase inhibitor sensitivity. J Proteome Res. 2011;10:305–319. doi: 10.1021/pr1006203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seidal T, Balaton AJ, Battifora H. Interpretation and quantification of immunostains. Am J Surg Pathol. 2001;25:1204–1207. doi: 10.1097/00000478-200109000-00013. [DOI] [PubMed] [Google Scholar]
- 12.Press MF, Sauter G, Bernstein L, Villalobos IE, Mirlacher M, Zhou JY, et al. Diagnostic evaluation of HER-2 as a molecular target: an assessment of accuracy and reproducibility of laboratory testing in large, prospective, randomized clinical trials. Clin Cancer Res. 2005;11:6598–6607. doi: 10.1158/1078-0432.CCR-05-0636. [DOI] [PubMed] [Google Scholar]
- 13.O'Neill RA, Bhamidipati A, Bi X, Deb-Basu D, Cahill L, Ferrante J, et al. Isoelectric focusing technology quantifies protein signaling in 25 cells. Proc Natl Acad Sci U S A. 2006;103:16153–16158. doi: 10.1073/pnas.0607973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan AC, Deb-Basu D, Orban MW, Gotlib JR, Natkunam Y, O'Neill R, et al. Nanofluidic proteomic assay for serial analysis of oncoprotein activation in clinical specimens. Nat Med. 2009;15:566–571. doi: 10.1038/nm.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seetharam M, Fan AC, Tran M, Xu L, Renschler JP, Felsher DW, et al. Treatment of higher risk myelodysplastic syndrome patients unresponsive to hypomethylating agents with ON 01910.Na. Leuk Res. 2012;36:98–103. doi: 10.1016/j.leukres.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maiso P, Liu Y, Morgan B, Azab AK, Ren P, Martin MB, et al. Defining the role of TORC1/2 in multiple myeloma. Blood. 2011;118:6860–6870. doi: 10.1182/blood-2011-03-342394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kentsis A, Reed C, Rice KL, Sanda T, Rodig SJ, Tholouli E, et al. Autocrine activation of the MET receptor tyrosine kinase in acute myeloid leukemia. Nat Med. 2012;18:1118–1122. doi: 10.1038/nm.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrett SD, Bridges AJ, Dudley DT, Saltiel AR, Fergus JH, Flamme CM, et al. The discovery of the benzhydroxamate MEK inhibitors CI-1040 and PD 0325901. Bioorg Med Chem Lett. 2008;18:6501–6504. doi: 10.1016/j.bmcl.2008.10.054. [DOI] [PubMed] [Google Scholar]
- 19.Lee JH, Giovannetti E, Hwang JH, Petrini I, Wang Q, Voortman J, et al. Loss of 18q22.3 involving the carboxypeptidase of glutamate-like gene is associated with poor prognosis in resected pancreatic cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:524–533. doi: 10.1158/1078-0432.CCR-11-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohn EA, Yang YA, Du Z, Nagano Y, Van Schyndle CM, Herrmann MA, et al. Biological responses to TGF-beta in the mammary epithelium show a complex dependency on Smad3 gene dosage with important implications for tumor progression. Mol Cancer Res. 2012 doi: 10.1158/1541-7786.MCR-12-0136-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gandhi J, Zhang J, Xie Y, Soh J, Shigematsu H, Zhang W, et al. Alterations in genes of the EGFR signaling pathway and their relationship to EGFR tyrosine kinase inhibitor sensitivity in lung cancer cell lines. PLoS ONE. 2009;4:e4576. doi: 10.1371/journal.pone.0004576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Postel-Vinay S, Arkenau HT, Olmos D, Ang J, Barriuso J, Ashley S, et al. Clinical benefit in Phase-I trials of novel molecularly targeted agents: does dose matter? British journal of cancer. 2009;100:1373–1378. doi: 10.1038/sj.bjc.6605030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain RK, Lee JJ, Hong D, Markman M, Gong J, Naing A, et al. Phase I oncology studies: evidence that in the era of targeted therapies patients on lower doses do not fare worse. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16:1289–1297. doi: 10.1158/1078-0432.CCR-09-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JH, Giovannetti E, Hwang JH, Petrini I, Wang Q, Voortman J, et al. Loss of 18q22.3 involving the carboxypeptidase of glutamate-like gene is associated with poor prognosis in resected pancreatic cancer. Clin Cancer Res. 2012;18:524–533. doi: 10.1158/1078-0432.CCR-11-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kummar S, Kinders R, Gutierrez ME, Rubinstein L, Parchment RE, Phillips LR, et al. Phase 0 clinical trial of the poly (ADP-ribose) polymerase inhibitor ABT-888 in patients with advanced malignancies. J Clin Oncol. 2009;27:2705–2711. doi: 10.1200/JCO.2008.19.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinoshita T, Yoshida I, Nakae S, Okita K, Gouda M, Matsubara M, et al. Crystal structure of human mono-phosphorylated ERK1 at Tyr204. Biochem Biophys Res Commun. 2008;377:1123–1127. doi: 10.1016/j.bbrc.2008.10.127. [DOI] [PubMed] [Google Scholar]
- 28.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. The New England journal of medicine. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 29.Yang CH, Yu CJ, Shih JY, Chang YC, Hu FC, Tsai MC, et al. Specific EGFR mutations predict treatment outcome of stage IIIB/IV patients with chemotherapy-naive non-small-cell lung cancer receiving first-line gefitinib monotherapy. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26:2745–2753. doi: 10.1200/JCO.2007.15.6695. [DOI] [PubMed] [Google Scholar]
- 30.Su KY, Chen HY, Li KC, Kuo ML, Yang JC, Chan WK, et al. Pretreatment epidermal growth factor receptor (EGFR) T790M mutation predicts shorter EGFR tyrosine kinase inhibitor response duration in patients with non-small-cell lung cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:433–440. doi: 10.1200/JCO.2011.38.3224. [DOI] [PubMed] [Google Scholar]
- 31.Ng KP, Hillmer AM, Chuah CT, Juan WC, Ko TK, Teo AS, et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nature medicine. 2012;18:521–528. doi: 10.1038/nm.2713. [DOI] [PubMed] [Google Scholar]
- 32.Gardner AM, Vaillancourt RR, Lange-Carter CA, Johnson GL. MEK-1 phosphorylation by MEK kinase, Raf, and mitogen-activated protein kinase: analysis of phosphopeptides and regulation of activity. Mol Biol Cell. 1994;5:193–201. doi: 10.1091/mbc.5.2.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cobb MH, Xu S, Cheng M, Ebert D, Robbins D, Goldsmith E, et al. Structural analysis of the MAP kinase ERK2 and studies of MAP kinase regulatory pathways. Adv Pharmacol. 1996;36:49–65. doi: 10.1016/s1054-3589(08)60576-1. [DOI] [PubMed] [Google Scholar]
- 34.Troiani T, Schettino C, Martinelli E, Morgillo F, Tortora G, Ciardiello F. The use of xenograft models for the selection of cancer treatments with the EGFR as an example. Crit Rev Oncol Hematol. 2008;65:200–211. doi: 10.1016/j.critrevonc.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Voskoglou-Nomikos T, Pater JL, Seymour L. Clinical predictive value of the in vitro cell line, human xenograft, and mouse allograft preclinical cancer models. Clinical cancer research : an official journal of the American Association for Cancer Research. 2003;9:4227–4239. [PubMed] [Google Scholar]
- 36.Yun CH, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, et al. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.