Abstract
The great complexity of glycosylated biomolecules necessitates a set of powerful analytical methodologies to reveal functionally important structural features. Mass spectrometry (MS), with its different ionization techniques, mass analyzers, and detection strategies, has become the most important analytical method in glycomic and glycoproteomic investigations. In combination with MS, microscale separations (based on capillary chromatography and electrophoresis) and carbohydrate microchemistry, we feature here conceptually important applications of the recent years. This review focuses on methodological advances pertaining to disease biomarker research, immunology, developmental biology, and measurements of importance to biopharmaceuticals. High-sensitivity determinations and sample enrichment/preconcentration are particularly emphasized in glycomic and glycoproteomic profiling.
Introduction
The enormous structural diversity of glycoconjugates reflects their multilateral importance in biochemical recognition. Extensive glycosylation of proteins is featured inside different cells, on their surfaces, and the extracellular spaces of diverse organisms. While glycosylated structures were traditionally considered within the domain of multicellular eukaryotic systems, studies of the last decade have documented the presence of oligosaccharides (often with unusual monosaccharides) in the microbial world as well [1,2]. Although the methodologies for glycoanalysis have advanced substantially during the last several years [3,4], identifying and quantifying the glycome and glycoproteome still represents a daunting task for the current and future generations of glycoscientists. Many modern glycoconjugate analytical techniques rely on mass spectrometry (MS), which has gradually become the most prominent tool in the structural characterization of glycoproteins. Additionally, capillary-based separation methods coupled with MS enhance the positive identification of glycan isomers, describe the sites of glycosylation, and decipher their microheterogeneity. The structural complexity of the resulting glycomic and glycoproteomic data needs extensive use of bioinformatic tools [5] for structural interpretation. Yet different approaches to understanding glycan-protein interactions have been pursued through the technologies of glycan and lectin arrays pioneered a decade ago (see the review by L. Mahal, this issue), which appear complementary to MS-based systems.
Scope of Investigations
From microbes to the most sophisticated multicellular organisms, glycoconjugates are increasingly recognized as the key determinants in both extracellular and intracellular functions. Biological investigators with interests ranging from embryology and developmental biology, to evolutionary development and physiology, increasingly subscribe to the “glycobiology approach.” At different levels of experimental difficulties, there are now methodological options to tackle some of the most difficult problems of glycoprotein structural characterization. In the time-honored approach, some investigators isolate the glycoproteins of interest through affinity chromatography or gel electrophoresis. The isolated and purified glycoproteins can then be subjected to a controlled protease-based degradation, followed by a further chromatographic separation and measurement of glycopeptides (glycoproteomic approach), and additionally or alternatively, a sample aliquot can be deglycosylated, either enzymatically or chemically, to yield a series of oligosaccharides for further (glycomic) measurements. The glycoprotein amounts available through such isolations often determine the success of structural characterization. Fortunately, the sensitivity, mass resolution, and mass accuracy of today’s MS-related techniques enable extensive characterization of both the polypeptide and glycosylated parts of fairly complex biomolecules. This is seen in examples of identifying the microbial virulence factors [6,7]. In less frequent situations, sufficient quantities of isolated glycoconjugates permit the use of protein crystallography and NMR techniques to appreciate the most intimate details of the glycan interactions with their biologically relevant binding proteins [8].
While research activities in mammalian (and human) glycobiology continue, there has been increasing interest in microbial and parasitic systems during the recent years. Apparently, a number of surface-layer glycoproteins have now been described in a number of species, featuring both N- and O-linked glycans and some unusual monosaccharides in their glycan structures [6,7]. While there is less information available at this time [9] on other parasites, genomic studies point to the occurrence of glycosylation throughout the entire range of eukaryotic systems (worms, insects, plants, fish, etc.). Here, glycomic and glycoproteomic comparative measurements are likely to yield information of value to developmental biologists studying appropriate “model systems” (as an example, see Drosophila [10]). In particular, isotopic labeling, MS-techniques, and appropriate uses of knock-out technologies for glycosyltransferase genes can all provide valuable information. From the human health viewpoint, protozoans [9], parasitic helminthes [11] and ticks [12] have already been targets of glycomic studies.
The biotechnology industry has been producing a number of glycoprotein-based drugs, most notably monoclonal antibodies, as therapeutics against cancer and inflammatory diseases. In turn, the use of recombinant antibodies and the more recently introduced “biosimilars” necessitate very stringent analytical control of their chemical compositions and physicochemical properties. The glycans incorporated into the biopharmaceuticals are known to influence important properties, such as in vivo circulatory half-life and possible antibody-dependent cytotoxicity [13,14]. Additionally, antigenic epitopes could possibly be introduced during the manufacturing process involving non-human cell lines [15]. Thus, the immunogenicity of unusual glycans must be closely examined in the selection of different eukaryotic expression systems. Tracking “correct glycosylation” in recombinant products has resulted in the design of different analytical platforms and methodologies, some based on MS and the others on fluorescence labeling together with HPLC and CE. Several analytical strategies concerning IgG glycosylation have been reviewed [16]. The current needs for better standards for biological functions and structural attributes of antibody-based pharmaceuticals is likely to drive further methodological developments.
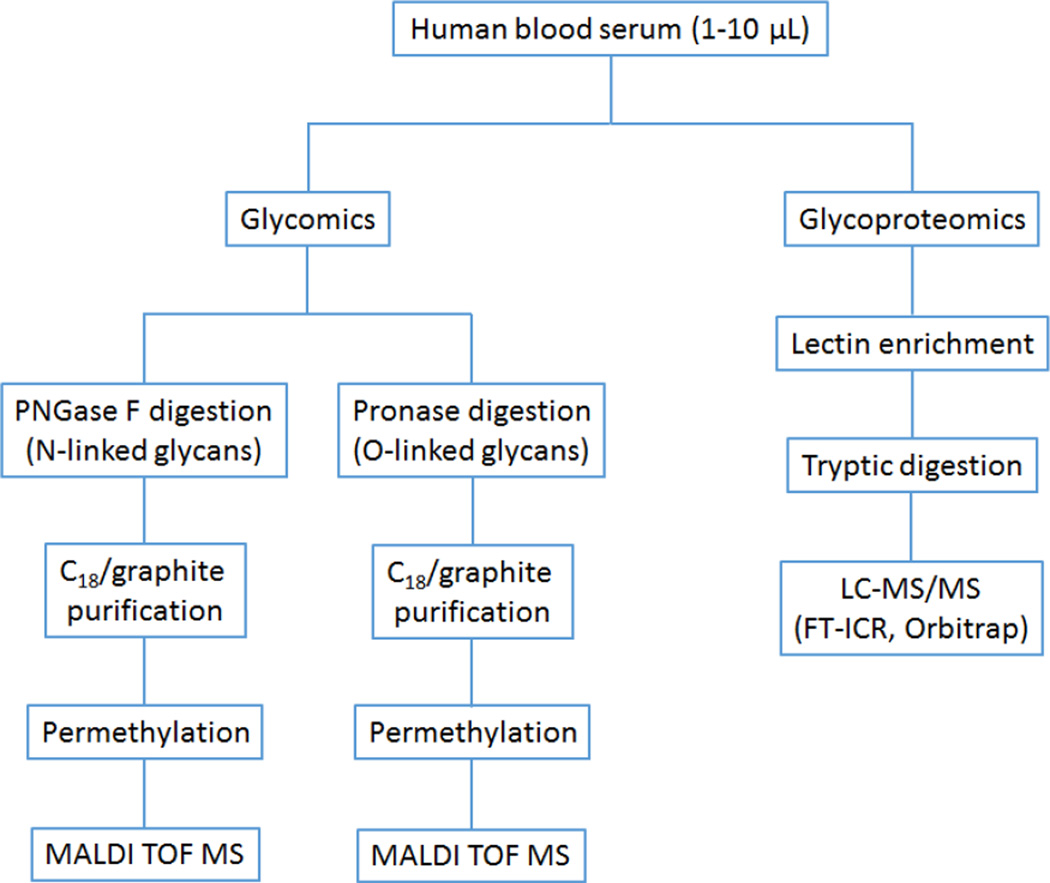
Among the most challenging areas of contemporary glycobiology are high-sensitivity, multicomponent analyses of complex biological mixtures, such as physiological fluids and tissue extracts (as an example, see Figure 1). This area has largely been driven by the search for biomarkers of human diseases and the many connections that glycobiology has to health-related issues. Many known human diseases and metabolic disorders have been tentatively linked to aberrantly glycosylated proteins for a number of years [17], but only recently has it become possible, through methodological improvements in analytical glycobiology, to appreciate quantitatively the extent in which a “pathological glycome” could be distinguished from normal conditions and how such quantitative measurements could potentially be used in clinics. Common physiological fluids, such as blood serum or plasma, can nominally be analyzed, but other biological samples (cell lines, cysts, tumor biopsies, etc.) are also applicable.
Figure 1.
Workflow for high-sensitivity glycoprotein analysis used in our laboratory for complex biological samples.
In-depth investigations of mammalian glycomes and glycoproteomes in complex biological samples are complicated by the very extensive concentration range in which various (glyco)proteins occur [18]. Whereas “total” (or “global”) glycomic profiling of small aliquots of blood serum from cancer patients can yield diagnostically or prognostically useful information [19], it will be necessary to cover a multitude of glycoproteins spanning the range of nearly 10 orders of magnitude. This task is difficult to accomplish without developing effective preconcentration strategies.
A more complete understanding of the roles of glycosylation in the immune system has also been important. While it is established that both the innate and adaptive immunity involve glycoproteins, the use of highly sensitive glycomic and glycoproteomic techniques will be needed to address the intricacies of the immune system dealing with inflammation and cancer [20]. Glycomic methodologies can be utilized in the structural characterization of neutrophils [21], eosinophils, basophils, and mast cells [22]. With reference to bodily fluids, the abundant IgGs and their different chains have been the obvious target of current investigations ; as the microisolation techniques further evolve [3,23], less abundant, albeit biologically important, minor immunoglobulins can also be measured. Glycan isomerism in sialylation and fucosylation could potentially be involved in the intricate functions in immunity. Recent reviews specifically endorse glycomic and glycoproteomic techniques [3,24].
Preconcentration of Glycoproteins from Complex Mixtures
Structural characterization of trace glycoproteins in complex biological materials continues to rely on the methodologies that can selectively isolate the target molecules or groups of glycosylated species from non-glycosylated molecules which otherwise mask their presence during analytical measurements. A typical example of such interferences is encountered during a coincidental chromatographic elution of glycopeptides with non-glycosylated peptides, in which competitive ionization results in a signal suppression during MS interrogations. During analyses of serum or plasma, it is common to deplete the major proteins by one of the commercially available immunoaffinity chromatographic columns prior to glycomic or glycoproteomic measurements. Immunoaffinity steps can also facilitate identification and quantification of a priori known minor glycoprotein target molecules through a design of specialized capture materials [25]. A recent example includes isolation of haptoglobin [26], followed by MS measurements demonstrating increased glycan fucosylation in pancreatic cancer, as well as similar measurements in hepatocellular cancer [27]. However, the immunoaffinity approach is somewhat limited by the availability of specific antibodies and their price.
For high-sensitivity measurements, the availability of miniaturized reactors constructed from optimal solid supports with minimum irreversible and nonspecific interactions is essential to sample recovery. A miniaturized serial trapping of the major and minor immunoglobulins has been demonstrated [23] prior to their glycomic profiling from small (a few µL) volumes of blood serum. While glycosylation of major proteins encountered in biological fluids could be informative in different health-related situations, measurements of tissue-derived glycoproteins (putative disease biomarkers) will undoubtedly require a new set of detection approaches [28].
Whereas different lectins have long been utilized in biochemical and histological practice, their analytical (quantitative) explorations are relatively recent. The degree of selectivity toward different glycan structures (differently linked sialic acids, high-mannose structures, fucosylation, etc.) can be utilized in complex fractionation and glycoprotein preconcentration schemes, as well as in the fabrication of lectin arrays (L. Mahal, this issue). Microscale affinity techniques have been essential in numerous investigations to characterize the subglycoproteomes from a wide array of biological matrices, including human urine, saliva, ovarian tissue [29], pancreatic cysts [30] and blood serum [31]. In the quantitative uses of lectin preconcentration, the immobilized lectins should ideally be bound to pressure-resistant support materials, such as silica particles [32] or monolithic polymers [33], which can then be used in conjunction with LC/MS-MS as a part of an integrated on-line system capable of comparing quantitatively hundreds of different glycoproteins [31,34] through bottom-up proteomic analysis. A recent review [35] estimates that there are 160 easily obtainable lectins, some of which could be selectively chosen for different enrichment schemes. The currently available lectins feature both overlapping and fairly unique interactions with glycoproteins [33].
Glycopeptide Enrichment
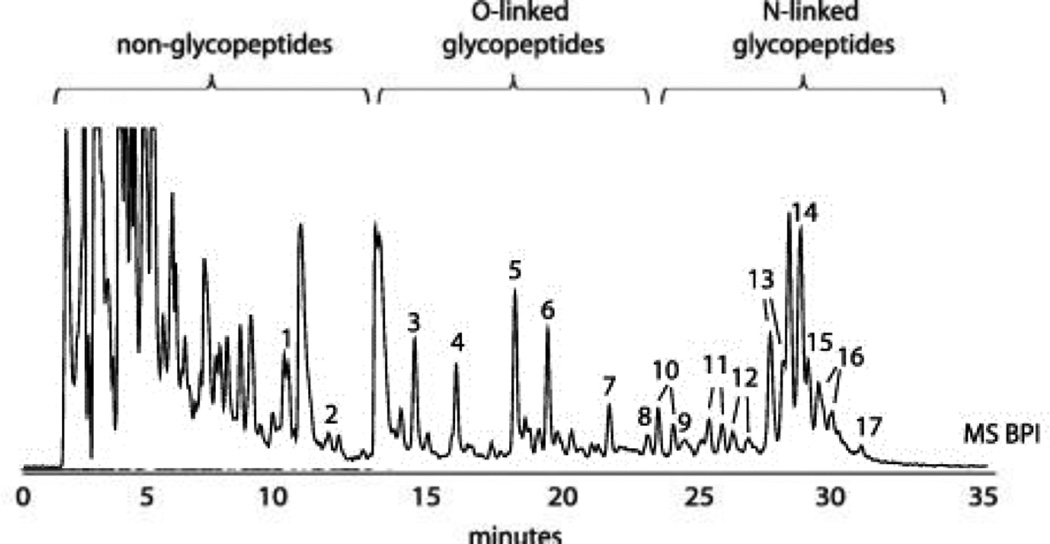
Specific enrichments or fractionations of peptidase digests are generally advisable before high-sensitivity MS or LC/MS-MS measurements of glycopeptides in order to reduce interferences from other peptides. Enrichments can be broad in their specificity, as is exemplified by the use of hydrophilic interaction chromatography (HILIC) [36,37], demonstrated in Figure 2, which depticts the fractionation and enrichment of both O- and N-glycopeptides in a single chromatographic experiment. Glycopeptides exhibiting certain saccharide motifs may also be enriched through lectin chromatography [38], although these procedures appear more effective for intact glycoprotein mixtures. Sialylated glycopeptides are quite selectively enriched with TiO2 particles using very acidic buffer conditions [39]. An innovative approach to enrich cell-surface glycoproteins is to incorporate unnatural azide-modified carbohydrates into the glycan structures through metabolic labeling [40]. The azide (or alkyne) moiety is subsequently reacted via a “click” mechanism to a phosphine-FLAG peptide, and the so-labeled glycoconjugates are, in turn, isolated using an anti-FLAG antibody [41].
Figure 2.
An example of the power of HILIC enrichment for tryptic glycopeptides (derived from bovine fetuin). Adapted from Ref. 37.
Non-specific enrichment procedures, in which the carbohydrate becomes covalently attached to appropriately functionalized solid supports, have become increasingly common in the practice of proteomic measurements. Since the initial report on the “hydrazide capture” [42], a modified procedure was reported [43] where samples are tryptically digested prior to periodate oxidation of carbohydrates’ vicinal diols to aldehydes and formation of hydrazones with the surface of functionalized beads. Subsequent variations in the protocols for the oxidation/hydrazide capture [44] aim at further optimization for quantification purposes. Boronic acid-functionalized materials provide another option for glycopeptide enrichment through utilization of the vicinal diol chemistry [45], although they seem less popular, presumably due to the weak binding constants.
Mass Spectrometry Advances
MS capabilities, particularly the tandem MS modes, have further evolved for the benefits of both glycomics and glycoproteomics. While collision-induced dissociation (CID) is still extensively used, the tandem MS innovations emphasizing electron-transfer dissociation (ETD) hold significant potential for the characterization of glycopeptides. This radical-initiated process is generally advantageous for post-translationally modified peptides, as the modification is largely unaffected by the fragmentation process, allowing ETD to assess the site of attachment and yield information on the multiple glycans associated with each site [46,47]. Another recent development in tandem MS is the higher-energy collisional dissociation (HCD) method that is performed in the octopole of Orbitrap instruments [48]. One of the most intense product ions is the so-called Y1-fragment (peptide + GlcNAc ion), which can be re-isolated, and in a subsequent HCD experiment, allowed to dissociate, yielding the amino-acid sequence [49]. Moreover, this approach can be used to selectively “trigger” ETD procedures if the oxonium ions (carbohydrate-specific fragments) are detected [50].
Tandem MS of glycans also continues to mature toward the negative-ion mode techniques, permitting extensive cross-ring fragmentations and thus yielding linkage-specific and branching information. While first reported some time ago [51], the general approach continues to be refined and combined with nano-scale LC [52] using porous graphitic carbon (PGC), a medium known for its ability to resolve isomeric glycans, as the stationary phase, with fluoride ions as a mobile-phase additive.
Ion-mobility spectrometry (IMS) coupled to MS is becoming more widely adapted within the glycoscience community due to the recent availability of commercial instrumentation. IMS holds potential as a gas-phase separative technique to resolve some isomeric structures, based on their unique collisional cross-sections. Moreover, in a recent report [53], the potential of IMS/MS has eloquently been demonstrated by the application to crude enzymatic digests (containing both peptides and glycans without desalting) to minimize sample preparation and handling steps. Peptides and carbohydrates could be separately detected, as they occupied different regions of the IMS drift space, with good sensitivity and reproducibility.
Glycan Profiling
Scientists who emphasize the integrated roles of the glycomes in different biological phenomena are beginning to appreciate new methodologies that enable entire profiles of glycolipids, oligosaccharides released from glycoprotein mixtures, or other glycoconjugates to be displayed quantitatively. These “glycan mapping” efforts can be increasingly correlated with screening the genes coding for corresponding glycosyltranferases [54], or even in vivo imaging of glycosylation through metabolic labeling [55] in model biological systems. How complex can the individual glycomes be for different species in their entire extent? From the nearly endless combinations of monosaccharide units and isomerism to form hypothetical glycans [56], functional arguments seem to restrict the number to less than 10,000 structures [57], This is still a respectable task for analytical profiling measurements, but knowing precisely the structural details of even trace carbohydrates will likely advance the knowledge of the binding protein interactions and the means to control them pharmaceutically. Consequently, the effective ways of glycan profiling should be: (a) as inclusive as possible; (b) structurally informative; (c) highly sensitive and quantitative; and (d) facilitating detection and measurements in a wide dynamic concentration range.
MS in its different ionization modes and mass-analyzing capabilities, provides a nearly ideal and the most sensitive means to cover the expected range of structures for N- and O-linked oligosaccharides [58]. However, MS is not always informative concerning the frequent isomerism of glycans and its biological significance, unless particular isomeric species are selectively derivatized [59]. Fortunately, capillary liquid-chromatographic and electrophoretic methods can often resolve structural isomers, and when used in combination with MS, they lead to effective analytical platforms for glycan profiling.
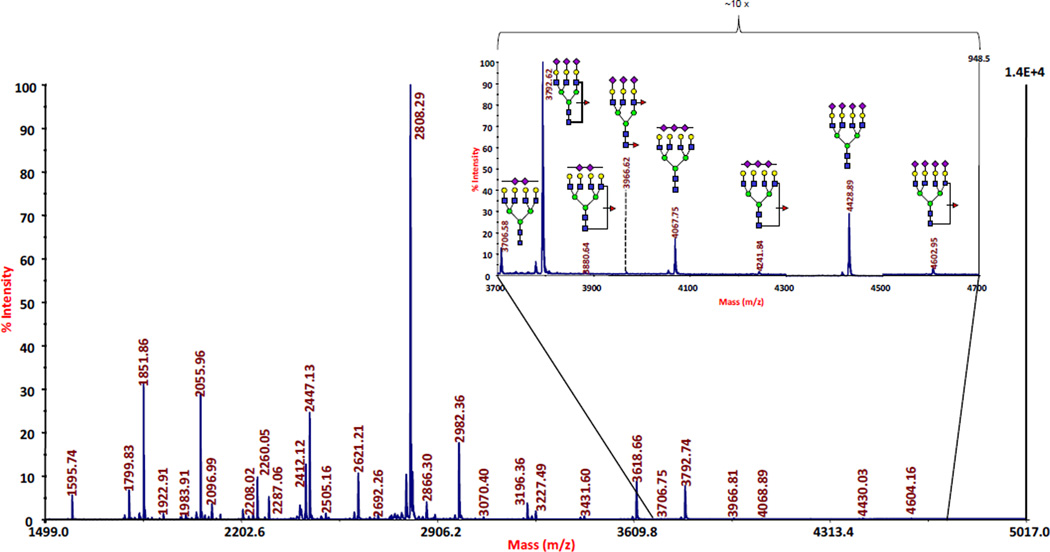
To begin with, glycans must be liberated quantitatively from the glycoproteins of interest. A quantitative and reproducible release of oligosaccharides has acquired great importance with today’s emphasis on high-sensitivity measurements. The traditionally-used chemical release procedures relying on hydrazinolysis or Carlson-type β-elimination have now been replaced by enzymatic N-deglycosylations (through the use of N-glycanases, largely the commercially available peptide N-glycosidases F and A), and microscale β-elimination procedures for threonine/serine-linked oligosaccharides. Largely due to the availability of N-glycanases, N-glycans are generally easier to analyze than O-glycans. Reducing digestion times without sacrificing digestion efficiency has been pursued in the recent studies involving ultra-high pressure cycling [60] or microwave radiation [61], and the use of immobilized reactors [62]. In the area of O-glycans, alternative chemical cleavage procedures were actively sought, including the use of dimethylamine coupled with microwave radiation [63] and the O-glycan recovery utilizing Pronase for a complete protein digestion, followed by a solid-phase permethylation and its coincident β-elimination from serine and threonine sites [64]. Profiles of released glycans can be quantitatively displayed through MALDI-MS, as exemplified by Figure 3 for blood-serum originated N-glycans, or ESI-MS, with the latter typically combined with liquid chromatography (LC). Both approaches are finding their utilization across different biochemical areas. The distinct advantage of MALDI-based techniques is their sensitivity and relative tolerance to sample impurities. ESI has the disadvantage of producing a complex set of differently-charged ions for the same parent mass. With regards to MS-fragile glycans (due to sialylation, or fucosylation, and the occasional presence of sulfate or phosphate groups), it is often recommended to derivatize glycans prior to their MS analysis. The time-honored approach of permethylation has now been widely utilized [3] to (a) stabilize the analytes; (b) include both the neutral and acidic glycans in a single profile; and (c) enhance sensitivity and create more discernible tandem MS fragmentation patterns. Permethylation is also desirable in making the glycans of interest sufficiently hydrophobic for reversed-phase LC separations in combination with MS [65]. Isotopic labeling through permethylation [66,67] further enhances the use of this derivatization for comparative profiling measurements.
Figure 3.
A MALDI-MS profile of permethylated glycans derived from a blood serum sample originated from an ovarian cancer patient. The series of glycans marked in the high-mass region are diagnostically important. Adapted from Ref. 19.
Glycomic profiling can also be performed, and often significantly enhanced, through the use of separation techniques such as HPLC, capillary LC, or capillary or chip-based electrophoresis (CE). Fluorescence derivatization at the microscale is usually important for the detection of profile constituents. This is perhaps best demonstrated by successful applications of HPLC/fluorescence detection in search for disease biomarkers [68,69], and the increasing use of hydrophilic interaction chromatography (HILIC) featuring smaller particles [36,37], as well as the demonstrations of HILIC in resolution of glycan isomers [37].
The rising popularity of a unique chromatographic material, porous graphitized carbon (PGC), can largely be attributed its effectiveness in separating isomeric glycans [70]. PGC small columns, combined with MS, have been utilized in the highly sensitive analyses of very small biological samples [71].
Microfabricated devices (microchips) have recently become popular in various glycan separations. Not only can the separatory channels within some microchips be tightly packed with chromatographic particles, but it is also possible to include various trapping columns, microreactors, and switching valves into an integrated analytical unit; this reduces the number of manual operations and sample transfers and, moreover, allows the microchip to be connected as a specialized “MS inlet”. The LC-based microchips, employing different packing materials have been demonstrated with the ESI-MS analysis of permethylated glycans extracted from blood serum [65]. Using graphite chips, both serum [72] and breast milk samples [73] were extensively characterized by the Lebrilla group. Another separation technique, CE, has also been applicable to very efficient separations of glycan mixtures (including isomeric structures) which had been labeled with a suitable fluorophore for detection. Here, too, the originally-used capillary formats quite easily translate to the microchip designs [74,75], achieving greater separation efficiencies and shorter run times. Unfortunately, CE is not easily combined with MS for the needed positive identification of separated glycans. The general lack of authentic glycan standards has also been a serious hindrance to positive identification of CE-separated glycan mixtures. Advances in coupling CE with MS have recently been reviewed [76,77].
Conclusions
In structural terms, the current and future explorations of glycomes and glycoproteomes are strongly dependent on the further evolution of MS instrumentation. The emphasis on high measurement sensitivity and information content is justified by the needs to characterize trace-level constituents of complex biological mixtures, as exemplified by the search for disease biomarkers. Selective sample preconcentration, glycan derivatization at the microscale, stable-isotopic labeling, and small-scale separations, such as capillary HILIC and CE, will continue to be used to resolve isomeric structures and simplify the task of MS measurements. There is also a significant trend to integrate the entire glycomic and glycoproteomic analytical platforms into a microchip format. The lack of authentic glycans still hinders the field of analytical glycobiology; it will hopefully be overcome by the current efforts in carbohydrate synthesis.
Highlights.
In-depth structural details of biologically diverse glycomes and glycoproteomes remain important to study.
Mass spectrometry has become the most important tool in structural glycobiology.
Quantitative glycomic profiling has been greatly assisted through the use of capillary and microfabricated separation devices.
Acknowledgments
We acknowledge financial support from the National Institute of General Medical Sciences through grant No. R01-GM024349.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Szymanski CM, Wren BW. Protein glycosylation in bacterial mucosal pathogens. Nat Rev Microbiol. 2005;3:225–237. doi: 10.1038/nrmicro1100. [DOI] [PubMed] [Google Scholar]
- 2.Comstock LE, Kasper DL. Bacterial glycans: key mediators of diverse host immune responses. Cell. 2006;126:847–850. doi: 10.1016/j.cell.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 3. Alley WR, Jr, Mann BF, Novotny MV. High-sensitivity analytical approaches for the structural characterization of glycoproteins. Chem Rev. 2013 doi: 10.1021/cr3003714. In press. A comprehensive review with 654 references.
- 4.Novotny MV, Alley WR, Jr, Mann BF. Analytical glycobiology at high sensitivity: Current approaches and directions. Glycoconj J. 2013;30:89–117. doi: 10.1007/s10719-012-9444-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hicks MG, Kettner C, editors. Glyco-bioinformatics: Cracking the sugar code by navigating the glycospace. Proceedings of the International Beilstein Symposium; Beistein-Institut, Frankfurt, Germany. 2012 [Google Scholar]
- 6.Young NM, Brisson JR, Kelly J, Watson DC, Tessier L, Lanthier PH, Jarrell HC, Cadotte N, St Michael F, Aberg E, et al. Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J Biol Chem. 2002;277:42530–42539. doi: 10.1074/jbc.M206114200. [DOI] [PubMed] [Google Scholar]
- 7.Balonova L, Mann BF, Cerveny L, Alley WR, Jr, Chovancova E, Forslund AL, Salomonsson EN, Forsberg A, Damborsky J, Novotny MV, et al. Characterization of protein glycosylation in Francisella tularensis subsp. holarctica: identification of a novel glycosylated lipoprotein required for virulence. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.015016. M111 015016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang CC, Lam SN, Acharya P, Tang M, Xiang SH, Hussan SS, Stanfield RL, Robinson J, Sodroski J, Wilson IA, et al. Structures of the CCR5 N-terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science. 2007;317:1930–1934. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson IBH, Paschinger K, Rendic D. Glycosylation of model and "lower" organisms. In: Gabius HJ, editor. The sugar code: Fundamentals of glycosciences. Hoboken, NJ, USA: Wiley-Blackwell; 2009. pp. 139–154. [Google Scholar]
- 10.Aoki K, Porterfield M, Lee SS, Dong B, Nguyen K, McGlamry KH, Tiemeyer M. The diversity of O-linked glycans expressed during drosophila melanogaster development reflects stage- and tissue-specific requirements for cell signaling. J Biol Chem. 2008;283:30385–30400. doi: 10.1074/jbc.M804925200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Die I, Cummings RD. Glycomics in unraveling glycan-driven immune responses by parasitic helminths. In: Cummings RD, Pierce JM, editors. Handbook of Glycomics. Waltham, Massachusetts, USA: Academic Press; 2009. pp. 367–396. [Google Scholar]
- 12.Vancova M, Sterba J, Dupejova J, Simonova Z, Nebesarova J, Novotny MV, Grubhoffer L. Uptake and incorporation of sialic acid by the tick Ixodes ricinus. J Insect Physiol. 2012;58:1277–1287. doi: 10.1016/j.jinsphys.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 13.Flintegaard TV, Thygesen P, Rahbek-Nielsen H, Levery SB, Kristensen C, Clausen H, Bolt G. N-glycosylation increases the circulatory half-life of human growth hormone. Endocrinology. 2010;151:5326–5336. doi: 10.1210/en.2010-0574. [DOI] [PubMed] [Google Scholar]
- 14.Shinkawa T, Nakamura K, Yamane N, Shoji-Hosaka E, Kanda Y, Sakurada M, Uchida K, Anazawa H, Satoh M, Yamasaki M, et al. The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J Biol Chem. 2003;278:3466–3473. doi: 10.1074/jbc.M210665200. [DOI] [PubMed] [Google Scholar]
- 15.Maeda E, Kita S, Kinoshita M, Urakami K, Hayakawa T, Kakehi K. Analysis of nonhuman N-glycans as the minor constituents in recombinant monoclonal antibody pharmaceuticals. Anal Chem. 2012;84:2373–2379. doi: 10.1021/ac300234a. [DOI] [PubMed] [Google Scholar]
- 16.Huhn C, Selman MH, Ruhaak LR, Deelder AM, Wuhrer M. IgG glycosylation analysis. Proteomics. 2009;9:882–913. doi: 10.1002/pmic.200800715. [DOI] [PubMed] [Google Scholar]
- 17.Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of glycobiology. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2009. [PubMed] [Google Scholar]
- 18.Surinova S, Schiess R, Huttenhain R, Cerciello F, Wollscheid B, Aebersold R. On the development of plasma protein biomarkers. J Proteome Res. 2011;10:5–16. doi: 10.1021/pr1008515. [DOI] [PubMed] [Google Scholar]
- 19.Alley WR, Jr, Vasseur JA, Goetz JA, Svoboda M, Mann BF, Matei DE, Menning N, Hussein A, Mechref Y, Novotny MV. N-linked glycan structures and their expressions change in the blood sera of ovarian cancer patients. J Proteome Res. 2012;11:2282–2300. doi: 10.1021/pr201070k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dube DH, Bertozzi CR. Glycans in cancer and inflammation--potential for therapeutics and diagnostics. Nat Rev Drug Discov. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 21.Babu P, North SJ, Jang-Lee J, Chalabi S, Mackerness K, Stowell SR, Cummings RD, Rankin S, Dell A, Haslam SM. Structural characterisation of neutrophil glycans by ultra sensitive mass spectrometric glycomics methodology. Glycoconj J. 2009;26:975–986. doi: 10.1007/s10719-008-9146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.North SJ, von Gunten S, Antonopoulos A, Trollope A, MacGlashan DW, Jr, Jang-Lee J, Dell A, Metcalfe DD, Kirshenbaum AS, Bochner BS, et al. Glycomic analysis of human mast cells, eosinophils and basophils. Glycobiology. 2012;22:12–22. doi: 10.1093/glycob/cwr089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Svoboda M, Mann BF, Goetz JA, Novotny MV. Examination of glycan profiles from IgG-depleted human immunoglobulins facilitated by microscale affinity chromatography. Anal Chem. 2012;84:3269–3277. doi: 10.1021/ac203336u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolarich D, Lepenies B, Seeberger PH. Glycomics, glycoproteomics and the immune system. Curr Opin Chem Biol. 2012;16:214–220. doi: 10.1016/j.cbpa.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 25.Hage DS. Handbook of affinity chromatography. 2nd edn. Boca Raton: Taylor & Francis; 2006. [Google Scholar]
- 26.Lin Z, Simeone DM, Anderson MA, Brand RE, Xie X, Shedden KA, Ruffin MT, Lubman DM. Mass spectrometric assay for analysis of haptoglobin fucosylation in pancreatic cancer. J Proteome Res. 2011;10:2602–2611. doi: 10.1021/pr200102h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pompach P, Brnakova Z, Sanda M, Wu J, Edwards N, Goldman R. Site specific glycoforms of haptoglobin in liver cirrhosis and hepatocellular carcinoma. Mol Cell Proteomics. 2013 doi: 10.1074/mcp.M112.023259. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Liu AY, Loriaux P, Wollscheid B, Zhou Y, Watts JD, Aebersold R. Mass spectrometric detection of tissue proteins in plasma. Mol Cell Proteomics. 2007;6:64–71. doi: 10.1074/mcp.M600160-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Abbott KL, Lim JM, Wells L, Benigno BB, McDonald JF, Pierce M. Identification of candidate biomarkers with cancer-specific glycosylation in the tissue and serum of endometrioid ovarian cancer patients by glycoproteomic analysis. Proteomics. 2010;10:470–481. doi: 10.1002/pmic.200900537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mann BF, Goetz JA, House MG, Schmidt CM, Novotny MV. Glycomic and proteomic profiling of pancreatic cyst fluids identifies hyperfucosylated lactosamines on the N-linked glycans of overexpressed glycoproteins. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.015792. M111 015792. Structurally unusual hyperfucosylated glycans have been found in pancreatic cysts with high malignant potential through glycomic profiling of several proteins that appear associated with this aberrant glycosylation.
- 31.Madera M, Mechref Y, Klouckova I, Novotny MV. Semiautomated high-sensitivity profiling of human blood serum glycoproteins through lectin preconcentration and multidimensional chromatography/tandem mass spectrometry. J Proteome Res. 2006;5:2348–2363. doi: 10.1021/pr060169x. [DOI] [PubMed] [Google Scholar]
- 32.Mann BF, Mann AK, Skrabalak SE, Novotny MV. Sub 2-mm macroporous silica particles derivatized for enhanced lectin affinity enrichment of glycoproteins. Anal Chem. 2013;85:1905–1912. doi: 10.1021/ac303274w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bedair M, El Rassi Z. Affinity chromatography with monolithic capillary columns. II. Polymethacrylate monoliths with immobilized lectins for the separation of glycoconjugates by nano-liquid affinity chromatography. J Chromatogr A. 2005;1079:236–245. doi: 10.1016/j.chroma.2005.02.084. [DOI] [PubMed] [Google Scholar]
- 34.Madera M, Mechref Y, Novotny MV. Combining lectin microcolumns with high-resolution separation techniques for enrichment of glycoproteins and glycopeptides. Anal Chem. 2005;77:4081–4090. doi: 10.1021/ac050222l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fanayan S, Hincapie M, Hancock WS. Using lectins to harvest the plasma/serum glycoproteome. Electrophoresis. 2012;33:1746–1754. doi: 10.1002/elps.201100567. This article discusses the use of different lectins, each with somewhat unique carbohydrate selectivies, to counter the dynamic range challenge associated with blood serum/plasma for enriching glycoproteins present in these fluids.
- 36.Ahn J, Bones J, Yu YQ, Rudd PM, Gilar M. Separation of 2-aminobenzamide labeled glycans using hydrophilic interaction chromatography columns packed with 1.7 micron sorbent. J Chromatogr B Analyt Technol Biomed Life Sci. 2010;878:403–408. doi: 10.1016/j.jchromb.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 37. Gilar M, Yu YQ, Ahn J, Xie H, Han H, Ying W, Qian X. Characterization of glycoprotein digests with hydrophilic interaction chromatography and mass spectrometry. Anal Biochem. 2011;417:80–88. doi: 10.1016/j.ab.2011.05.028. The power of new chromatographic technologies is nicely demonstrated through the applications of glycoproteomic interest.
- 38.Kim JY, Kim SK, Kang D, Moon MH. Dual lectin-based size sorting strategy to enrich targeted N-glycopeptides by asymmetrical flow field-flow fractionation: profiling lung cancer biomarkers. Anal Chem. 2012;84:5343–5350. doi: 10.1021/ac300772w. [DOI] [PubMed] [Google Scholar]
- 39.Palmisano G, Lendal SE, Engholm-Keller K, Leth-Larsen R, Parker BL, Larsen MR. Selective enrichment of sialic acid-containing glycopeptides using titanium dioxide chromatography with analysis by HILIC and mass spectrometry. Nat Protoc. 2010;5:1974–1982. doi: 10.1038/nprot.2010.167. [DOI] [PubMed] [Google Scholar]
- 40.Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA, Bertozzi CR. Copper-free click chemistry for dynamic in vivo imaging. Proc Nat Acad Sci USA. 2007;104:16793–16797. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hubbard SC, Boyce M, McVaugh CT, Peehl DM, Bertozzi CR. Cell surface glycoproteomic analysis of prostate cancer-derived PC-3 cells. Bioorg Med Chem Lett. 2011;21:4945–4950. doi: 10.1016/j.bmcl.2011.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 43.Wollscheid B, Bausch-Fluck D, Henderson C, O'Brien R, Bibel M, Schiess R, Aebersold R, Watts JD. Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat Biotechnol. 2009;27:378–386. doi: 10.1038/nbt.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parker BL, Palmisano G, Edwards AV, White MY, Engholm-Keller K, Lee A, Scott NE, Kolarich D, Hambly BD, Packer NH, et al. Quantitative N-linked glycoproteomics of myocardial ischemia and reperfusion injury reveals early remodeling in the extracellular environment. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.006833. M110 006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin S, Cheng Y, Reid S, Li M, Wang B. Carbohydrate recognition by boronolectins, small molecules, and lectins. Med Res Rev. 2010;30:171–257. doi: 10.1002/med.20155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alley WR, Jr, Mechref Y, Novotny MV. Characterization of glycopeptides by combining collision-induced dissociation and electron-transfer dissociation mass spectrometry data. Rapid Commun Mass Spectrom. 2009;23:161–170. doi: 10.1002/rcm.3850. [DOI] [PubMed] [Google Scholar]
- 47.Wu SL, Huhmer AF, Hao Z, Karger BL. On-line LC-MS approach combining collision-induced dissociation (CID), electron-transfer dissociation (ETD), and CID of an isolated charge-reduced species for the trace-level characterization of proteins with post-translational modifications. J Proteome Res. 2007;6:4230–4244. doi: 10.1021/pr070313u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olsen JV, Macek B, Lange O, Makarov A, Horning S, Mann M. Higher-energy C-trap dissociation for peptide modification analysis. Nat Methods. 2007;4:709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- 49.Segu ZM, Mechref Y. Characterizing protein glycosylation sites through higher-energy C-trap dissociation. Rapid Commun Mass Spectrom. 2010;24:1217–1225. doi: 10.1002/rcm.4485. [DOI] [PubMed] [Google Scholar]
- 50.Steentoft C, Vakhrushev SY, Vester-Christensen MB, Schjoldager KT, Kong Y, Bennett EP, Mandel U, Wandall H, Levery SB, Clausen H. Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat Methods. 2011;8:977–982. doi: 10.1038/nmeth.1731. [DOI] [PubMed] [Google Scholar]
- 51.Harvey DJ. Fragmentation of negative ions from carbohydrates: part 1. Use of nitrate and other anionic adducts for the production of negative ion electrospray spectra from N-linked carbohydrates. J Am Soc Mass Spectrom. 2005;16:622–630. doi: 10.1016/j.jasms.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 52.Ni W, Bones J, Karger BL. In-Depth Characterization of N-Linked Oligosaccharides Using Fluoride-Mediated Negative Ion Microfluidic Chip LC-MS. Anal Chem. 2013;85:3127–3135. doi: 10.1021/ac3031898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harvey DJ, Sobott F, Crispin M, Wrobel A, Bonomelli C, Vasiljevic S, Scanlan CN, Scarff CA, Thalassinos K, Scrivens JH. Ion mobility mass spectrometry for extracting spectra of N-glycans directly from incubation mixtures following glycan release: application to glycans from engineered glycoforms of intact, folded HIV gp120. J Am Soc Mass Spectrom. 2011;22:568–581. doi: 10.1007/s13361-010-0053-0. [DOI] [PubMed] [Google Scholar]
- 54.Nairn AV, Aoki K, dela Rosa M, Porterfield M, Lim JM, Kulik M, Pierce JM, Wells L, Dalton S, Tiemeyer M, et al. Regulation of glycan structures in murine embryonic stem cells: Combined transcript profiling of glycan-related genes and glycan structural analysis. J Biol Chem. 2012;287:37835–37856. doi: 10.1074/jbc.M112.405233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR. In vivo imaging of membrane-associated glycans in developing zebrafish. Science. 2008;320:664–667. doi: 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laine RA. A calculation of all possible oligosaccharide isomers both branched and linear yields 1.05 × 1012 structures for a reducing hexasaccharide: The isomer barrier to development of single-method saccharide sequencing or synthesis systems. Glycobiology. 1994;4:759–767. doi: 10.1093/glycob/4.6.759. [DOI] [PubMed] [Google Scholar]
- 57.Cummings RD. The repertoire of glycan determinants in the human glycome. Mol Biosyst. 2009;5:1087–1104. doi: 10.1039/b907931a. [DOI] [PubMed] [Google Scholar]
- 58. Zaia J. Mass spectrometry and glycomics. OMICS. 2010;14:401–418. doi: 10.1089/omi.2009.0146. A review discussing comprehensively recent developments in mass spectrometry of glycoconjugates.
- 59.Alley WR, Jr, Novotny MV. Glycomic analysis of sialic acid linkages in glycans derived from blood serum glycoproteins. J Proteome Res. 2010;9:3062–3072. doi: 10.1021/pr901210r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Szabo Z, Guttman A, Karger BL. Rapid release of N-linked glycans from glycoproteins by pressure-cycling technology. Anal Chem. 2010;82:2588–2593. doi: 10.1021/ac100098e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tzeng YK, Chang CC, Huang CN, Wu CC, Han CC, Chang HC. Facile MALDI-MS analysis of neutral glycans in NaOH-doped matrixes: microwave-assisted deglycosylation and one-step purification with diamond nanoparticles. Anal Chem. 2008;80:6809–6814. doi: 10.1021/ac801137g. [DOI] [PubMed] [Google Scholar]
- 62.Jmeian Y, Hammad LA, Mechref Y. Fast and efficient online release of N-glycans from glycoproteins facilitating liquid chromatography-tandem mass spectrometry glycomic profiling. Anal Chem. 2012;84:8790–8796. doi: 10.1021/ac301855v. [DOI] [PubMed] [Google Scholar]
- 63.Maniatis S, Zhou H, Reinhold V. Rapid de-O-glycosylation concomitant with peptide labeling using microwave radiation and an alkyl amine base. Anal Chem. 2010;82:2421–2425. doi: 10.1021/ac902734w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goetz JA, Novotny MV, Mechref Y. Enzymatic/chemical release of O-glycans allowing MS analysis at high sensitivity. Anal Chem. 2009;81:9546–9552. doi: 10.1021/ac901363h. [DOI] [PubMed] [Google Scholar]
- 65.Alley WR, Jr, Madera M, Mechref Y, Novotny MV. Chip-based reversed-phase liquid chromatography-mass spectrometry of permethylated N-linked glycans: a potential methodology for cancer-biomarker discovery. Anal Chem. 2010;82:5095–5106. doi: 10.1021/ac100131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kang P, Mechref Y, Kyselova Z, Goetz JA, Novotny MV. Comparative glycomic mapping through quantitative permethylation and stable-isotope labeling. Anal Chem. 2007;79:6064–6073. doi: 10.1021/ac062098r. [DOI] [PubMed] [Google Scholar]
- 67.Alvarez-Manilla G, Warren NL, Abney T, Atwood J, 3rd, Azadi P, York WS, Pierce M, Orlando R. Tools for glycomics: relative quantitation of glycans by isotopic permethylation using 13CH3I. Glycobiology. 2007;17:677–687. doi: 10.1093/glycob/cwm033. [DOI] [PubMed] [Google Scholar]
- 68.Arnold JN, Saldova R, Galligan MC, Murphy TB, Mimura-Kimura Y, Telford JE, Godwin AK, Rudd PM. Novel glycan biomarkers for the detection of lung cancer. J Proteome Res. 2011;10:1755–1764. doi: 10.1021/pr101034t. [DOI] [PubMed] [Google Scholar]
- 69. Marino K, Bones J, Kattla JJ, Rudd PM. A systematic approach to protein glycosylation analysis: a path through the maze. Nat Chem Biol. 2010;6:713–723. doi: 10.1038/nchembio.437. This paper briefly reviews different research strategies and complementary analytical methodologies toward a complete characterization of glycosylated proteins.
- 70.Davies MJ, Smith KD, Carruthers RA, Chai W, Lawson AM, Hounsell EF. Use of a porous graphitized carbon column for the high-performance liquid chromatography of oligosaccharides, alditols, and glycopeptides with subsequent mass spectrometry analysis. J Chromatogr. 1993;646:317–326. doi: 10.1016/0021-9673(93)83344-r. [DOI] [PubMed] [Google Scholar]
- 71.Wilson NL, Schulz BL, Karlsson NG, Packer NH. Sequential analysis of N- and O-linked glycosylation of 2D-PAGE separated glycoproteins. J Proteome Res. 2002;1:521–529. doi: 10.1021/pr025538d. [DOI] [PubMed] [Google Scholar]
- 72. Aldredge D, An HJ, Tang N, Waddell K, Lebrilla CB. Annotation of a serum N-glycan library for rapid identification of structures. J Proteome Res. 2012;11:1958–1968. doi: 10.1021/pr2011439. This paper discusses the development of a library with over 300 hundred entries for the rapid identification of N-linked glycans derived from blood-serum glycoproteins.
- 73.Wu S, Tao N, German JB, Grimm R, Lebrilla CB. Development of an annotated library of neutral human milk oligosaccharides. J Proteome Res. 2010;9:4138–4151. doi: 10.1021/pr100362f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhuang Z, Mitra I, Hussein A, Novotny MV, Mechref Y, Jacobson SC. Microchip electrophoresis of N-glycans on serpentine separation channels with asymmetrically tapered turns. Electrophoresis. 2011;32:246–253. doi: 10.1002/elps.201000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhuang Z, Starkey JA, Mechref Y, Novotny MV, Jacobson SC. Electrophoretic analysis of N-glycans on microfluidic devices. Anal Chem. 2007;79:7170–7175. doi: 10.1021/ac071261v. [DOI] [PubMed] [Google Scholar]
- 76.Mechref Y, Novotny MV. Glycomic analysis by capillary electrophoresis-mass spectrometry. Mass Spectrom Rev. 2009;28:207–222. doi: 10.1002/mas.20196. [DOI] [PubMed] [Google Scholar]
- 77. Cortes DF, Kabulski JL, Lazar AC, Lazar IM. Recent advances in the MS analysis of glycoproteins: Capillary and microfluidic workflows. Electrophoresis. 2011;32:14–29. doi: 10.1002/elps.201000394. This reference reviews recent progress in microfluidic instrumentation, emphasizing integrated systems with improved sample handling capabilities.