Abstract
Mitochondrial transcription factor A (TFAM), the first well-characterized transcription factor from vertebrate mitochondria, is closely related to mitochondrial DNA (mtDNA) maintenance and repair. Recent evidence has shown that the ratio of mtDNA to nuclearDNA (nDNA) is increased in both human cells and murine tissues after ionizing radiation (IR). However, the underlying mechanism has not as yet been clearly identified. In the present study, we demonstrated that in human lung adenocarcinoma A549 cells, expression of TFAM was upregulated, together with the increase of the relative mtDNA copy number and cytochrome c oxidase (COX) activity after α-particle irradiation. Furthermore, short hairpin RNA (shRNA)-mediated TFAM knockdown inhibited the enhancement of the relative mtDNA copy number and COX activity caused by α-particles. Taken together, our data suggested that TFAM plays a crucial role in regulating mtDNA amplification and mitochondrial biogenesis under IR conditions.
Keywords: mitochondrial transcription factor A, ionizing radiation, mitochondrial DNA, mitochondrial biogenesis
INTRODUCTION
Ionizing radiation (IR) can generate excessive reactive oxygen/nitrogen species (ROS/RNS) leading to DNA damage and genomic instability [1]. Most of this IR-induced ROS/RNS is largely produced in the mitochondria [2, 3]. Due to the close proximity to the electron transport chain and the low efficiency of repair systems, mitochondrial DNA (mtDNA) is more sensitive to oxidative stress than nuclear DNA (nDNA), and the mtDNA damage is more extensive and persists longer than the nDNA damage [4, 5]. MtDNA damage and the subsequent mitochondrial genomic instability are closely related to retinal degeneration, neurodegeneration, aging, and carcinogenesis [6–9]. Therefore, it is of special importance to maintain mtDNA and mitochondrial functions, especially under oxidative stress conditions. Recent evidence has shown that the ratio of mtDNA/nDNA is increased in both human cells [10] and murine tissues [11–13] after IR. However, the mechanisms underlying the mtDNA amplification induction after irradiation are as yet poorly understood.
Mitochondrial transcription factor A (TFAM), a 25-kDa nDNA-encoded protein and the major protein component of mitochondrial DNA (mtDNA) nucleiods [14], is a member of the high mobility group (HMG) superfamily of DNA-binding proteins [15]. Previous studies have shown that the TFAM protein directly regulates mtDNA copy number [16, 17]. The TFAM protein is translocated into the mitochondria and binds to mtDNA without sequence specificity, playing an important role in the maintenance of mtDNA [16]. TFAM also activates mtDNA transcription initiation in the presence of mitochondrial RNA polymerase and transcription factor B (TFBM1 or TFBM2) [18]. Recently, Thomas et al. reported that mitochondrial biogenesis was stimulated in mice treated with recombinant human TFAM [19]. Nishiyama et al. also showed an increase in the mtDNA copy number in mitochondrial disease mouse models by over-expression of TFAM [20]. Recent reports have suggested that TFAM is involved in mitochondrial biogenesis [21, 22]. Additionally, TFAM recognizes the structural alterations of DNA, cisplatin-damaged DNA and oxidatively damaged DNA [23], suggesting that TFAM is one of the mtDNA-repairing proteins [24, 25]. Given the role of TFAM in mtDNA repair and maintenance, we hypothesized that under IR conditions, TFAM plays a crucial role in the regulation of mtDNA copy number and mitochondrial biogenesis.
In this study, we investigated relative mtDNA copy number and cytochrome c oxidase (COX) activity to assess mitochondrial biogenesis after α-particle irradiation. Our data showed that mitochondrial biogenesis is induced after α-particle irradiation in human lung adenocarcinoma A549 cells, and demonstrated that TFAM regulates this IR-induced mitochondrial biogenesis.
MATERIALS AND METHODS
Cell culture, kits and reagents
A549 cells were maintained in Dulbecco's Modified Eagle Medium (Gibco, Life Technologies, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS) (Hyclone, Thermo Fisher Scientific, Beijing, China) at 37°C in a humidified 5% CO2 incubator. Succinate dehydrogenase (SDH) activity and COX activity were detected with assay kits from GENMED (Shanghai, China) according to the manufacturer's instructions. Protein was quantified with BCA reagent from Sangon (Shanghai, China). Total DNA was extracted using an AxyPrep gDNA extraction kit purchased from Axygen Biosciences (Hangzhou, China). LY294002 was purchased from Sigma-Aldrich (St Louis, MO, USA).
Alpha particle irradiation
Cells were seeded into each specially designed circular dish (inside diameter: 3 mm) with a 3.0-μm thick Mylar film bottom to which the cells were attached. Culture medium was refreshed every other day and the cells were irradiated by α-particles when the confluence reached 60–70%. Alpha particles from the 241Am irradiation source traversed the 65-mm-long vacuum chamber, the exit window, the 3.5-μm-thick Mylar film mounted in the exit window, the 1-mm-thick air layer, the 3.0-μm-thick Mylar bottom, and finally reached the cells which were attached to the bottom. The dose rate was 1.0 cGy/s measured using a silicon surface barrier detector and a CR39 particle track detector [26]. The source activity was 7.4 MBq and the active area 50 mm in diameter. The energy of the source was measured to be an average of 3.5 MeV at the cell layer.
Quantification of the relative mtDNA copy number
The relative mtDNA copy number was quantified by SYBR Green quantitative PCR on a StepOneTM Real-Time PCR System (Applied Biosystems, Life Technologies, Foster City, CA, USA) using a relative standard curve method [27]. MtDNA content was determined by amplification of 12S rDNA coded by mtDNA with primers 5′TAACCCAAGTCAATAGAAGCC and 5'CTAGAGGGATATGAAGCACC. nDNA content was determined by amplification of the β-actin coding sequence with primers 5'GAGCGGGAAATCGTGCGTGAC and 5'GGAAGGAAGGCTGGAAGAGTG. The mtDNA/nDNA ratio was used to estimate the relative mtDNA copy number.
Western blotting
Total proteins were separated by 10–12% SDS-PAGE and transferred to a PVDF membrane. After blocking in PBST with 1% skimmed milk, the PVDF membrane was incubated with primary antibody overnight at 4°C. Then the membrane was washed with PBST and incubated with the corresponding HRP-linked secondary antibody for 2 h at room temperature. Protein bands were visualized using enhanced ECL substrates purchased from CWBIO (Beijing, China). Primary antibodies used in this work were as follows: TFAM (1:2000, Gene Tex, Irvine, CA, USA), Akt (1:10000, Epitomics, Burlingame, CA, USA), phosphorylated Akt (Ser473) (1:500, Cell Signaling Technology, Beverly, MA, USA), and β-Actin (1:2000, Abmart, Shanghai, China).
Generation of TFAM-knockdown A549 cells
TFAM-short hairpin RNA (shRNA) and Scrambled-shRNA were purchased from OriGene Technologies (Rockville, MD, USA). A549 cells were transfected with TFAM-shRNA or Scrambled-shRNA using Mega Tran 1.0 Transfection Reagent (OriGene Technologies, Rockville, MD, USA) according to the manufacturer's recommendations. DMEM with 1 µg/ml puromycin was used as the selective medium. Transfectants were isolated and maintained in DMEM with 0.2 µg/ml puromycin for further experiments. Knockdown of TFAM was confirmed by western blotting.
Statistical analysis
All experimental results reported here represent at least three independent replications. Data are presented as mean values ± standard deviation (SD). Significant levels were assessed using Student's t-test. A P-value of < 0.05 between groups was considered to be statistically significant.
RESULTS
TFAM gene expression was upregulated after α-particle irradiation
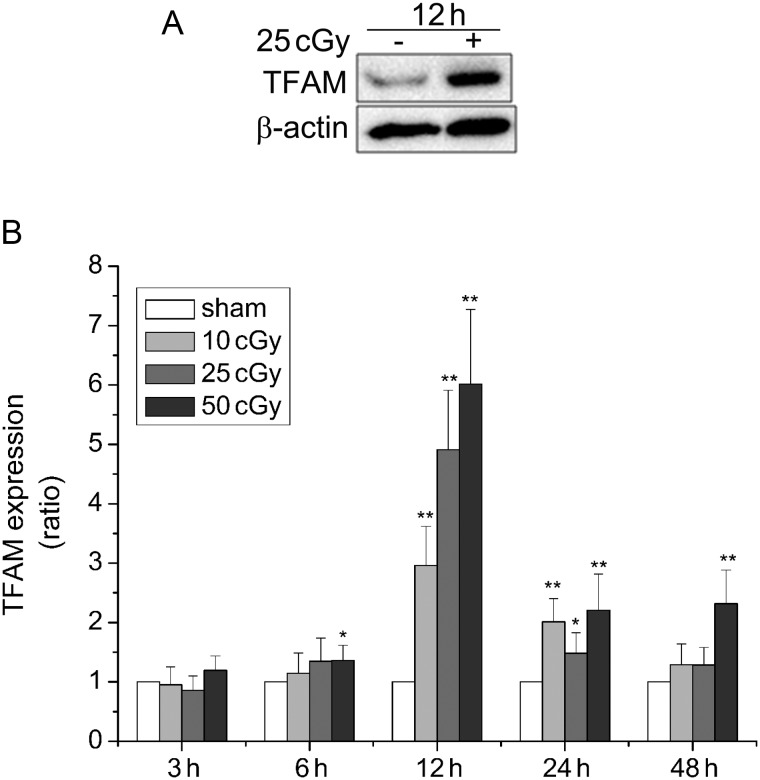
The expression of TFAM in A549 cells after α-particle irradiation was determined by western blotting. Increased expression of TFAM was observed at 12 h after irradiated by 25 cGy α-particles (Fig 1A). Quantitative data of TFAM expression was shown in Fig. 1B. TFAM gene expression reached the maximal level at 12 h after irradiation with each dose (10 cGy, 2.96-fold; 25 cGy, 4.91-fold; 50 cGy, 6.01-fold), and then declined with the extension of post-irradiation time. At 48 h, TFAM expression level in A549 cells irradiated with 10 cGy and 25 cGy returned to a level approximate to that observed in sham-irradiated cells. In addition, the effect of different radiation doses (10 cGy, 25 cGy, 50 cGy) on TFAM expression was also indicated in Fig. 1B; the expression level of TFAM was significantly upregulated with increase in the dose of α-particles, especially at 12 h after irradiation.
Fig. 1.
Upregulation of TFAM expression in A549 cells after exposure to α–particles. (A) Western blotting analysis of TFAM protein level in A549 cells at 12 h after 25cGy α–particle irradiation. (B) Quantitative analysis of TFAM expression level detected by western blotting (normalized to β-actin). A549 cells were irradiated by 10, 25, 50 cGy α–particles respectively and incubated for 3, 6, 12, 24 and 48 h after irradiation. Sham-irradiated cells were used as controls and values were standardized to those of controls. (*P < 0.05, **P < 0.01.)
Mitochondrial biogenesis was stimulated after α-particle irradiation
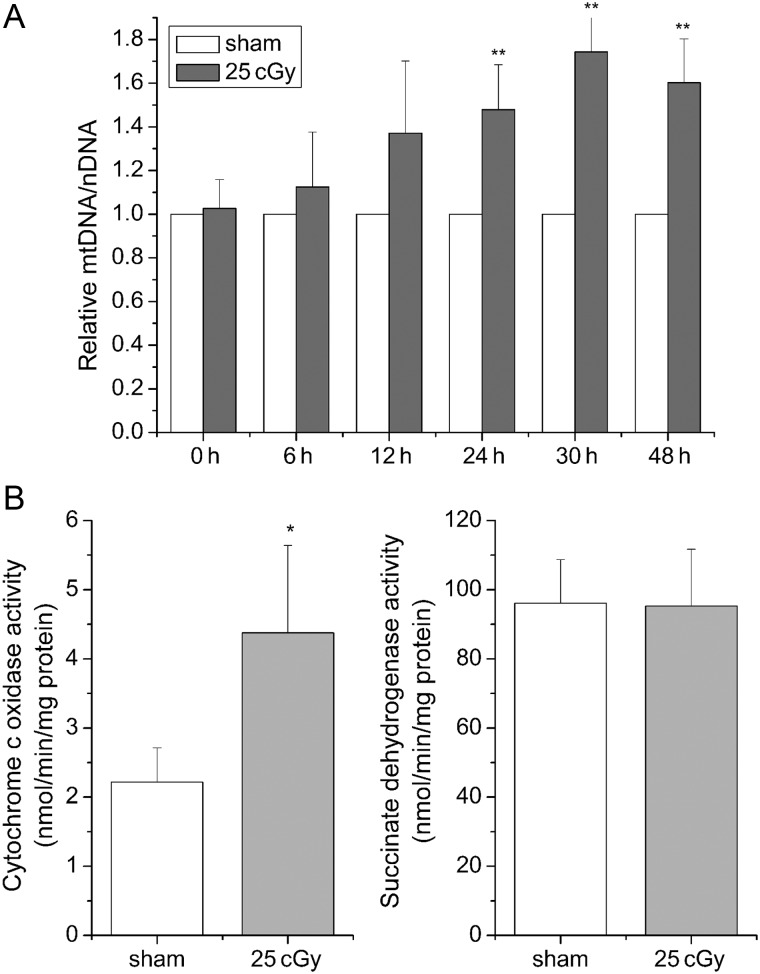
To examine whether mitochondrial biogenesis was stimulated by α-particle irradiation, the relative mtDNA copy number was analyzed in A549 cells. As shown in Fig. 2A, the relative mtDNA copy number was increased by 0.48-, 0.74- and 0.6-fold at 24, 30 and 48 h post-irradiation compared to the sham-irradiated control, indicating mitochondrial biogenesis was stimulated by α-particles. To further verify this result, COX activity and SDH activity were measured, since the COX complex was jointly encoded by mtDNA and nDNA, while the succinate dehydrogenase complex was encoded entirely by nDNA [28]. At 24 h after irradiation, the activity of COX was increased 0.98-fold in A549 cells compared to sham-irradiated cells, whereas the activity of SDH was only slightly affected by 25 cGy α-particles (Fig. 2B). This further confirmed the induction of mitochondrial biogenesis in α-particle-irradiated A549 cells.
Fig. 2.
Increases of relative mtDNA copy number and COX activity in A549 cells after exposure to 25 cGy α–particles. (A) DNA samples were prepared at 0, 6, 12, 24, 30 and 48 h, respectively after irradiation. Relative mtDNA copy number was calculated using mtDNA/nDNA. (B) COX and SDH activity at 24 h after irradiation. (*P < 0.05, **P < 0.01.)
TFAM knockdown diminished IR-induced mitochondrial biogenesis
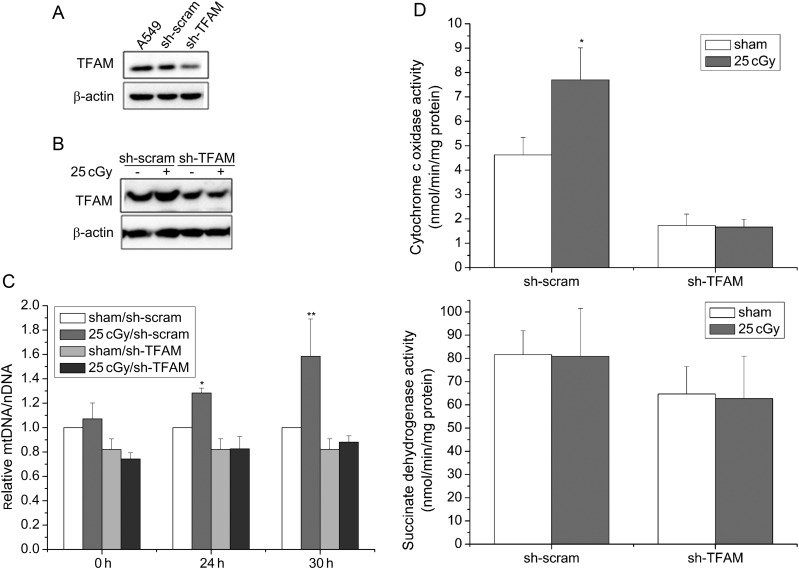
Since both TFAM expression and mitochondrial biogenesis were significantly promoted in A549 cells by 25 cGy α-particles, we then tested whether the mitochondrial biogenesis was regulated by TFAM after irradiation. TFAM knockdown A549 cells (sh-TFAM A549 cells) and its control cells (sh-scram A549 cells) were established by transfecting wild type A549 cells with TFAM-shRNA plasmids and scrambled-shRNA plasmids, respectively. TFAM knockdown efficiency was detected by western blotting and shown in Fig. 3A. After 25 cGy α-particles, TFAM level was increased in sh-scram A549 cells, which was similar to the observation made in wild type A549 cells. However, in sh-TFAM A549 cells, no obvious upregulation of TFAM was observed after irradiation (Fig 3B). Further, changes in the relative mtDNA copy number, along with the COX and SDH activity, were investigated in both sh-TFAM and sh-scram A549 cells after irradiation. After 25cGy α-particles, the relative mtDNA copy number in sh-scram A549 cells was significantly increased, by 0.28- and 0.58-fold compared with sham-irradiated sh-scram cells at 24 h and 30 h, respectively (Fig 3C). However, with respect to sh-TFAM A549 cells, 25 cGy α-particles exerted no effect on mtDNA copy number (Fig 3C). Results of COX and SDH activity were shown in Fig. 3D. After irradiation with 25 cGy α-particles, no evident change was observed in SDH activity in either sh-TFAM or sh-scram A549 cells. However, 24 h after irradiation, a 0.67-fold increase in COX activity was measured in sh-scram A549 cells, which was consistent with the observation made in wild type A549 cells, although no increase in COX activity was observed in sh-TFAM A549 cells. The above results suggest that TFAM is a key regulator of mitochondrial biogenesis in A549 cells after IR stimulation. Moreover, as shown in Fig. 3C and D, knockdown of TFAM negatively influences the basal levels of relative mtDNA copy number and COX activity. This result further confirms our conclusion.
Fig. 3.
Knockdown of TFAM in A549 cells attenuated the increases of mtDNA copy number and COX activity caused by 25 cGy α-particles. (A) TFAM expression in wild type, sh-scram, sh-TFAM A549 cells was determined by western blotting. (B) TFAM expression in sh-scram, sh-TFAM A549 cells at 12 h after irradiation. (C) Relative mtDNA copy number in sh-scram, sh-TFAM A549 cells at 0, 24 and 30 h after irradiation. (D) COX and SDH activity in sh-scram, sh-TFAM A549 cells at 24 h after irradiation. (*P < 0.05, **P < 0.01.)
Phosphatidylinositol 3,4,5-triphosphate kinase/protein kinase B was involved in TFAM upregulation after α-particle irradiation
Phosphatidylinositol 3,4,5-triphosphate kinase/protein kinase B (PI3K/Akt) has been reported to be activated after IR [29]. To determine the relationship between PI3K/Akt activation and TFAM upregulation after irradiation, LY294002, a specific chemical inhibitor of PI3K/Akt, was used, and Akt phosphorylation was detected by western blotting. As shown in Fig. 4 (left two lanes), the phosphorylation of Akt (Ser473) was stimulated by 25 cGy α-particles, accompanied by the upregulation of TFAM. However, the use of 100 µM LY294002 suppressed the activation of Akt and resulted in attenuation of TFAM (Fig 4, right two lanes). This indicated that the PI3K/Akt signaling pathway was involved in the increase of TFAM expression in A549 cells after irradiation.
Fig. 4.
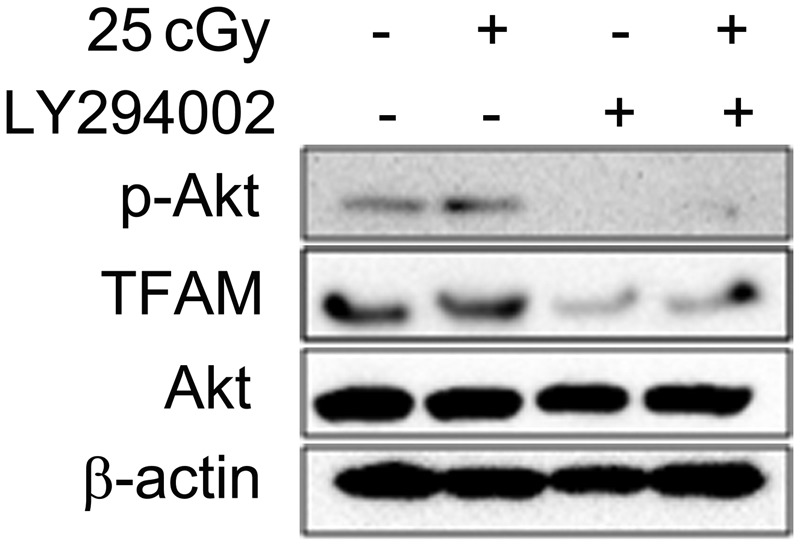
Influence of PI3K/Akt inhibitor LY294002 on IR-induced TFAM upregulation. 100 µM LY294002 was added into culture medium 1 h before irradiation. The expression of phosphorylated Akt (Ser473), Akt, TFAM and β-actin at 12 h after irradiation was detected by western blotting.
DISCUSSION
It has been widely accepted that IR has detrimental effects on nuclear DNA, which may lead to cell death and tumorigenesis. However, extra-nuclear parts that cannot avoid being targeted by IR have not drawn enough attention until recent years. Using a microbeam facility, Wu et al. have previously proved that cytoplasm-targeted irradiation results in genotoxic effects [2], indicating the necessity of investigating the effects of IR on extra-nuclear organelles. Mitochondria, where the majority of IR-induced ROS/RNS come from, and which directly suffer the oxidative damage in turn, are inevitable targets of IR among the extra-nuclear parts. Our results show that the relative mtDNA copy number was increased in human lung adenocarcinoma A549 cells after α-particle irradiation (Fig. 2A). This observation is consistent with the previous studies in murine tissues and malignantly transformed human small airway epithelial cells (SAECs) [10–13]. Zhang et al. reported a decrease in COX activity in SAEC cells after α-particle irradiation [10], whereas a significant increase in COX activity in A549 cells was evident in our results (Fig. 2B). This variance could be owing to the difference in physiological requirements between these two cell lines. That is, in human SAECs, IR-induced ROS might alter mitochondrial capacity and promote subsequent neoplastic transformation [10], while in human lung adenocarcinoma A549 cells, because of the low efficiency of repair systems, enhanced mitochondrial capacity may be needed for urgent energy demand.
It has been proposed that TFAM, the first well-characterized transcription factor from vertebrate mitochondria, is closely associated with mitochondrial biogenesis [21, 22]. However, under IR conditions such as in the radiotherapy of cancer, the relationship between TFAM and mitochondrial biogenesis remains incompletely defined. In the present study, we proved that TFAM is a key regulator of the increase of mtDNA copy number and COX activity in A549 cells after α-particle irradiation (Fig. 3), suggesting involvement of TFAM in IR-induced mitochondrial biogenesis. It has been demonstrated that TFAM plays an important role in recognizing and binding oxidatively damaged DNA [23], and participates in the repair of damaged mtDNA [7, 30]. In addition, mtDNA is a critical cellular target of IR-induced mitochondrial ROS [4]. Point mutations [31], deletions [32] and supercoiling formation changes [5] caused by IR have recently been identified in mtDNA. Thus, one possible explanation for our results is that, after IR, mtDNA damage triggered TFAM-involved mtDNA repair. The total mitochondrial content of irradiated cells may have been increased, through TFAM-mediated mitochondrial biogenesis, to guarantee the overall energy supply, which might be compensatory machinery for damaged mitochondria. Further investigations are needed in order to identify the role of TFAM in the coordination of IR-induced mitochondrial biogenesis and damaged mtDNA repair. It is worth mentioning that, although both our results and Zhang's research show that mtDNA copy number is increased after irradiation, the expression patterns of TFAM differ a lot. Zhang and his colleagues observed an increase of Pol-γ expression and a decrease of TFAM expression in murine intestinal tissues at 4 days, 21 days and 30 days after 5 Gy γ-ray whole-body irradiation [13]. The authors suggested that increased Pol-γ was responsible for mtDNA synthesis, and that TFAM was downregulated to prevent the expression of proteins encoded by damaged mitochondrial genomes in normal murine intestinal tissues after irradiation [13]. However, as a type of cancer cells, A549 cells might require a sustained energy supply to overcome the damage caused by radiation and thus survive. This might lead to upregulation of TFAM for both the replication of mitochondrial genomes and the expression of mtDNA-encoded proteins within a relatively short time after irradiation. At the same time, the different responses of single cells and tissues to cell irradiation and whole-body irradiation, and the different corresponding effects on the regulation of TFAM and mitochondrial functions should not be neglected. Since the available information is limited, it is of great importance to perform further investigations. Our results have also shown that TFAM is activated by PI3K/Akt following α-particle irradiation (Fig 4). Piantadosi et al. demonstrated that PI3K/Akt regulates TFAM by directly phosphorylating nuclear respiratory factor-1 (NRF-1), which facilitates NRF-1 nuclear translocation and TFAM activation [27]. However, further investigations are needed to clarify the details of how PI3K/Akt stimulates TFAM after IR.
CONCLUSION
In conclusion, we have shown that both mitochondrial biogenesis and TFAM expression are promoted in A549 cells after α-particle irradiation. Furthermore, our data indicate that TFAM is a key regulator of IR-induced mitochondrial biogenesis. These findings suggest that under IR conditions, TFAM plays a crucial role in the signaling pathways regulating mtDNA amplification and mitochondrial biogenesis.
FUNDING
This work was supported by grants from the National Natural Science Foundation of China, grant numbers 10935009 and 31200644.
REFERENCES
- 1.Azzam EI, Jay-Gerin JP, Pain D. Ionizing radiation-induced metabolic oxidative stress and prolonged cell injury. Cancer Lett. 2012;327:48–60. doi: 10.1016/j.canlet.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu LJ, Randers-Pehrson G, Xu A, et al. Targeted cytoplasmic irradiation with alpha particles induces mutations in mammalian cells. Proc Natl Acad Sci U S A. 1999;96:4959–64. doi: 10.1073/pnas.96.9.4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamamori T, Yasui H, Yamazumi M, et al. Ionizing radiation induces mitochondrial reactive oxygen species production accompanied by upregulation of mitochondrial electron transport chain function and mitochondrial content under control of the cell cycle checkpoint. Free Radic Biol Med. 2012;53:260–70. doi: 10.1016/j.freeradbiomed.2012.04.033. [DOI] [PubMed] [Google Scholar]
- 4.Yakes FM, VanHouten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci U S A. 1997;94:514–9. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou X, Li N, Wang Y, et al. Effects of X-irradiation on mitochondrial DNA damage and its supercoiling formation change. Mitochondrion. 2011;11:886–92. doi: 10.1016/j.mito.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Jarrett SG, Lin HJ, Godley BF, et al. Mitochondrial DNA damage and its potential role in retinal degeneration. Prog Retin Eye Res. 2008;27:596–607. doi: 10.1016/j.preteyeres.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Jeppesen DK, Bohr VA, Stevnsner T. DNA repair deficiency in neurodegeneration. Prog Neurobiol. 2011;94:166–200. doi: 10.1016/j.pneurobio.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen JH, Hales CN, Ozanne SE. DNA damage, cellular senescence and organismal ageing: causal or correlative? Nucl Acids Res. 2007;35:7417–28. doi: 10.1093/nar/gkm681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandra D, Singh KK. Genetic insights into OXPHOS defect and its role in cancer. Biochim Biophys Acta. 2011;1807:620–5. doi: 10.1016/j.bbabio.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang S, Wen G, Huang SX, et al. Mitochondrial alteration in malignantly transformed human small airway epithelial cells induced by alpha-particles. Int J Cancer. 2013;132:19–28. doi: 10.1002/ijc.27656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malakhova L, Bezlepkin VG, Antipova V, et al. The increase in mitochondrial DNA copy number in the tissues of gamma-irradiated mice. Cell Mol Biol Lett. 2005;10:721–32. [PubMed] [Google Scholar]
- 12.Zhang H, Maguire D, Swarts S, et al. Replication of murine mitochondrial DNA following irradiation. In: Liss P, Hansell P, Bruley DF, et al., editors. Oxygen Transport to Tissue XXX. New York: Springer Science+Business Media, LLC; 2009. pp. 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H, Maguire DJ, Zhang M, et al. Elevated mitochondrial DNA copy number and POL-gamma expression but decreased expression of TFAM in murine intestine following therapeutic dose irradiation. In: LaManna JC, Puchowicz MA, Xu K, et al., editors. Oxygen Transport to Tissue XXXII. New York: Springer Science+Business Media, LLC; 2011. pp. 201–206. [DOI] [PubMed] [Google Scholar]
- 14.Kukat C, Wurm CA, Spahr H, et al. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc Natl Acad Sci U S A. 2011;108:13534–9. doi: 10.1073/pnas.1109263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gangelhoff TA, Mungalachetty PS, Nix JC, et al. Structural analysis and DNA binding of the HMG domains of the human mitochondrial transcription factor A. Nucl Acids Res. 2009;37:3153–64. doi: 10.1093/nar/gkp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanki T, Ohgaki K, Gaspari M, et al. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol Cell Biol. 2004;24:9823–34. doi: 10.1128/MCB.24.22.9823-9834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ekstrand MI, Falkenberg M, Rantanen A, et al. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13:935–44. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 18.Malarkey CS, Bestwick M, Kuhlwilm JE, et al. Transcriptional activation by mitochondrial transcription factor A involves preferential distortion of promoter DNA. Nucl Acids Res. 2011;40:614–24. doi: 10.1093/nar/gkr787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas RR, Khan SM, Portell FR, et al. Recombinant human mitochondrial transcription factor A stimulates mitochondrial biogenesis and ATP synthesis, improves motor function after MPTP, reduces oxidative stress and increases survival after endotoxin. Mitochondrion. 2011;11:108–18. doi: 10.1016/j.mito.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishiyama S, Shitara H, Nakada K, et al. Over-expression of Tfam improves the mitochondrial disease phenotypes in a mouse model system. Biochem Biophys Res Commun. 2010;401:26–31. doi: 10.1016/j.bbrc.2010.08.143. [DOI] [PubMed] [Google Scholar]
- 21.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–38. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 22.Ljubicic V, Joseph AM, Saleem A, et al. Transcriptional and post-transcriptional regulation of mitochondrial biogenesis in skeletal muscle: effects of exercise and aging. Biochim Biophys Acta. 2010;1800:223–34. doi: 10.1016/j.bbagen.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 23.Yoshida Y, Izumi H, Ise T, et al. Human mitochondrial transcription factor A binds preferentially to oxidatively damaged DNA. Biochem Biophys Res Commun. 2002;295:945–51. doi: 10.1016/s0006-291x(02)00757-x. [DOI] [PubMed] [Google Scholar]
- 24.Larsson NG, Wang JM, Wilhelmsson H, et al. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–6. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 25.Ueta E, Sasabe E, Yang Z, et al. Enhancement of apoptotic damage of squamous cell carcinoma cells by inhibition of the mitochondrial DNA repairing system. Cancer Sci. 2008;99:2230–7. doi: 10.1111/j.1349-7006.2008.00918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu BR, Wu JF, Han W, et al. Development of a dose-adjustable α-particle irradiation facility for radiobiological studies. Nucl Sci Techn. 2005;16:102–7. [Google Scholar]
- 27.Piantadosi CA, Suliman HB. Mitochondrial transcription factor A induction by redox activation of nuclear respiratory factor 1. J Biol Chem. 2005;281:324–33. doi: 10.1074/jbc.M508805200. [DOI] [PubMed] [Google Scholar]
- 28.Huang SX, Partridge MA, Ghandhi SA, et al. Mitochondria-derived reactive intermediate species mediate asbestos-induced genotoxicity and oxidative stress-responsive signaling pathways. Environ Health Perspect. 2012;120:840–7. doi: 10.1289/ehp.1104287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Viniegra JG, Martinez N, Modirassari P, et al. Full activation of PKB/Akt in response to insulin or ionizing radiation is mediated through ATM. J Biol Chem. 2005;280:4029–36. doi: 10.1074/jbc.M410344200. [DOI] [PubMed] [Google Scholar]
- 30.Canugovi C, Maynard S, Bayne ACV, et al. The mitochondrial transcription factor A functions in mitochondrial base excision repair. DNA Repair (Amst.) 2010;9:1080–9. doi: 10.1016/j.dnarep.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy JEJ, Nugent S, Seymour C, et al. Mitochondrial DNA point mutations and a novel deletion induced by direct low-LET radiation and by medium from irradiated cells. Mutat Res. 2005;585:127–36. doi: 10.1016/j.mrgentox.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 32.Zhao XT, Feng JB, Li YW, et al. Identification of two novel mitochondrial DNA deletions induced by ionizing radiation. Biomed Environ Sci. 2012;25:533–41. doi: 10.3967/0895-3988.2012.05.006. [DOI] [PubMed] [Google Scholar]