Abstract
Alterations in DNA repair promote tumor development, but the impact on tumor progression is poorly understood. Here, discovery of a biochemical circuit linking hormone signaling to DNA repair and therapeutic resistance is reported. Findings demonstrate that androgen receptor (AR) activity is induced by DNA damage, and promotes expression and activation of a gene expression program governing DNA repair. Subsequent investigation revealed that activated AR promotes resolution of double-strand breaks and resistance to DNA damage both in vitro and in vivo. Mechanistically, DNAPKcs was identified as a key target of AR after damage, controlling AR-mediated DNA repair and cell survival after genotoxic insult. Finally, DNAPKcs was shown to potentiate AR function, consistent with a dual role in both DNA repair and transcriptional regulation. Combined, these studies identify the AR-DNAPKcs circuit as a major effector of DNA repair and therapeutic resistance, and establish a new node for therapeutic intervention in advanced disease.
Keywords: DNA damage, DNAPK, androgen receptor, prostatic adenocarcinoma, double strand break
INTRODUCTION
DNA damage response (DDR) pathways are intricate networks that sense DNA damage and activate repair mechanisms to maintain genomic integrity. Distinct DDR pathways exist to resolve different classes of DNA lesions, including mismatch repair, nucleotide excision repair, base excision repair, and the Fanconi anemia pathway (FA, which resolves collapsed replication forks and associated interstrand crosslinks) [1]. Finally, DNA double strand breaks, such as those induced by ionizing radiation (IR), are repaired through one of two major pathways: non-homologous end joining (NHEJ) or homologous recombination (HR) [2]. Predictably, dysregulation of such highly controlled repair processes can result in catastrophic cellular outcomes. Failure to efficiently repair DNA lesions reduces cellular viability via either apoptosis or senescence [3]; alternatively, incomplete or inaccurate DNA repair promotes sustained pro-tumorigenic DNA alterations (i.e. mutations, chromosomal losses/gains, and/or translocations) [4].
Consonantly, DDR alterations are linked to tumor development and progression. Primarily, DDR and subsequent checkpoint activation serve as crucial mechanisms that maintain genomic stability and protect against gross genomic rearrangements that can lead to malignant conversion of normal cells [5, 6]. However, deregulation of cell cycle checkpoints in the presence of incomplete or inaccurate DDR results in inappropriate cell proliferation and survival; as such, mutations in DDR-associated genes are tightly associated with tumorigenesis [7]. Failure to efficiently repair damage promotes errors in DNA replication and chromosome segregation, thus facilitating genomic instability in both hereditary and sporadic cancers [8]. Direct ligation of double strand DNA breaks through NHEJ also promotes deleterious rearrangements at the break site and chromosomal translocations associated with malignancy [9]. Distinct from effects on tumor development, it is increasingly appreciated that heightened DDR can drive cancer progression and promote therapeutic resistance in established tumors, as DDR checkpoints are often disabled [5, 6]. Notably, cancer cells that have acquired radioresistance demonstrate markedly enhanced DNA repair capacity [10]. Additionally, increased levels of key NHEJ components, including DNAPKcs (DNA-dependent protein kinase catalytic subunit), Ku70, and Ku80, correlate with recurrence after radiation therapy (RT) and progression to advanced disease [11, 12]. Recent findings across multiple tumor types showed that DNA repair genes are upregulated in primary tumors predicted to metastasize, indicating that heightened DNA repair capacity is associated with increased risk of distant metastasis [13]. On balance, emerging evidence strongly supports the contention that escalated DDR function contributes to disease progression and therapeutic resistance. As such, concentrated effort has been put forth to determine how these alterations may lead to therapeutic opportunities, and to delineate the molecular mechanisms controlling DNA damage repair in human malignancies.
Unexpectedly, clinical observations suggest that steroid hormones might modulate the response to DNA damage in hormone-responsive cancers. In breast cancer (BCa), chemotherapeutics that induce genotoxic stress are utilized to treat all known subtypes, including those that remain dependent on estrogen receptor (ER) alpha [14]. Clinically, suppression of ER signaling by tamoxifen enhances the response to radiation [15]; although the underlying mechanisms have not been completely defined, ER can interact with a subset of known DNA damage response factors [16] and modulate expression of several genes associated with DDR [17]. Prostatic adenocarcinoma (PCa), another example of a hormone-responsive cancer, is classified according to disease stage and is dependent on androgen receptor (AR) activity at all stages for growth, survival, and progression [18]. Upon binding of ligand (androgens), AR is activated, releases from inhibitory heat-shock proteins, homodimerizes, and translocates to the nucleus where it binds to androgen response elements (AREs) on DNA and induces a gene expression program that is requisite for PCa maintenance [19]. AR-directed therapeutics play a central role for all stages of PCa. In the context of disseminated PCa, while the vast majority of patients respond initially to androgen deprivation therapy (ADT), the response is unfortunately transient, and recurrent, castration resistant PCa (CRPC) tumors arise due to inappropriate AR reactivation within 24–36 months [20]. Importantly, while AR frequently is activated in CRPC under castrate conditions, the receptor still responds to androgen, including those produced intratumorally [21]. Treatment of CRPC also includes DNA-damaging agents such as radium-223 chloride, which targets to bone metastases and emits alpha radiation; however, as monotherapy, this agent affords only a modest survival advantage [22], clearly defining the need for improved and synergistic approaches to effectively treat advanced disease.
Among men diagnosed with localized PCa, one of the primary treatment modalities is RT; however, similar to metastatic disease, it is clear that those patients with high-risk or locally advanced disease have suboptimal outcomes when treated with radiation alone [23]. In the context of locally aggressive disease, the combination of ADT and radiation is the standard of care, based on numerous phase III randomized trials, encompassing thousands of patients, which demonstrate large survival benefits with this combination therapy compared to either treatment alone [24–26]. While these trials indicate that AR activity may impact tumor sensitivity to radiation, the mechanism underlying this cooperativity has not been defined. Molecularly, AR activation has been associated with creation of DNA double strand breaks at sites of transcriptional regulation, incurred as a way to release torsional strain and facilitate gene expression [27]. Moreover, DNAPKcs and Ku70/80 have been preliminarily proposed using in vitro models to serve as AR coactivators [28]. Thus, despite clinical evidence to suggest crosstalk between AR and DDR pathways, the mechanism and consequence of this communication is not understood.
Given these observations and the urgent need to identify durable mechanisms to manage advanced disease, the present study aimed to investigate the influence of activated AR on the DNA damage response and resultant biological outcomes. Findings herein reveal that activated AR promotes expression and activity of key factors involved in DDR. Critical observations demonstrate that RT induces AR activity, and that AR promotes resistance to DNA damage, using both ADT-sensitive and CRPC models of disease. Elucidation of the underlying mechanism revealed that AR activity promotes DNA double strand break resolution, independent of effects on cell cycling, by regulating expression and activity of key players in DDR. AR is recruited to regulatory regions of the genes indentified, directly implicating AR as a transcriptional regulator of components of the DDR. Further mechanistic investigation identified AR-activated DNAPKcs expression and function as a critical component of AR-mediated DNA repair and cellular survival after genomic insult. Moreover, accumulated DNAPKcs feeds back to AR and supports transcriptional transactivation potential of the receptor. The collective findings suggest a model wherein androgen induced DNAPKcs expression and activity enhances AR activity, creating a positive circuit through which androgens promote DNA repair and tumor cell resistance to DNA damage-inducing therapeutics. As will be demonstrated, these studies identify activated AR as a key regulator of the DNA damage response, and characterize a novel mechanism of regulation that reveals new opportunities for precision therapies in AR-dependent tumors.
RESULTS
Suppression of AR activity enhances the response to DNA damage
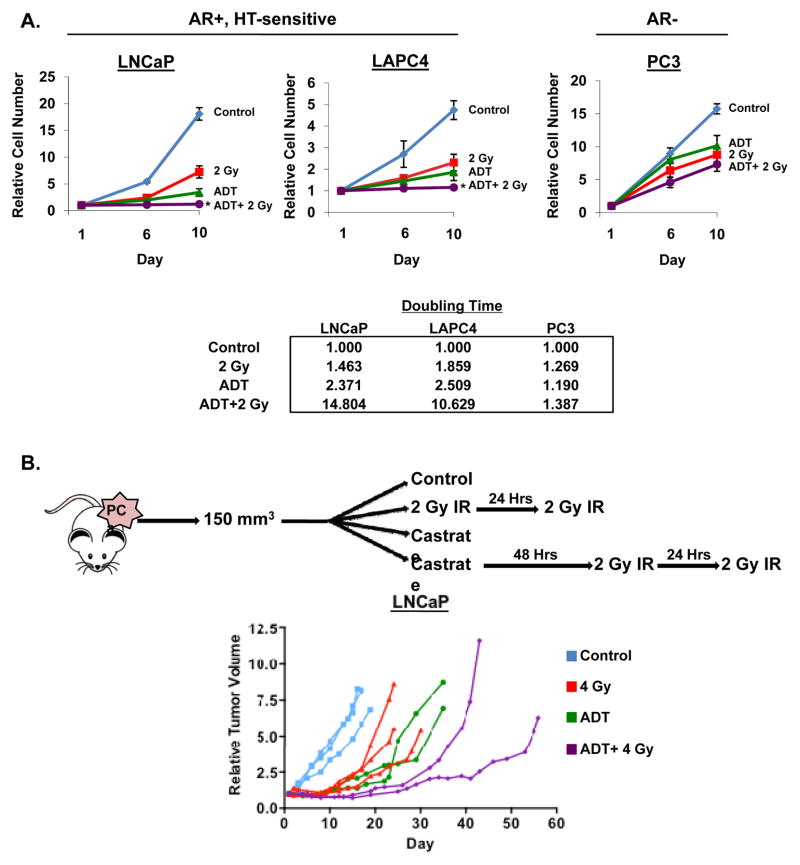
To explore the impact of androgens on DDR, PCa cells were exposed to IR in combination with androgen replete (control) or androgen deprived conditions mimicking androgen deprivation therapy (ADT). In ADT-sensitive, AR-positive models of human disease (LNCaP and LAPC4), control cells exhibited expected growth kinetics in the presence of androgen, but exposure to IR treatment alone reduced cell survival and doubling by ~50%, consistent with the known role of IR in suppressing PCa growth (Figure 1A, top). In parallel, the ADT-sensitive cancer models showed a strong anti-proliferative response to ADT alone, exemplified by a 2.3 fold increase in cell doubling time (Figure 1A, bottom). Despite the robust response to ADT alone, combining ADT and IR further modestly suppressed cell growth and survival at both 6 and 10 days post treatment compared to untreated control, ADT, or IR alone (Figure 1A, top). Consonantly, combination treatment further increased cell doubling time, demonstrating a potential cooperative effect between ADT and IR (Figure 1A, bottom). Alteration of treatment schedule did not significantly alter cell number (Supplemental Figure 1A, C), similar to what was observed clinically [29]. By contrast, combination therapy resulted in no significant alteration in cell number (as compared to IR alone) in AR-negative PCa cells (PC3) (Figure 1A, top and bottom). Combined, these studies suggest that androgen deprivation in ADT-sensitive cells and resultant suppression of AR activity modestly enhances the response to radiation in vitro. To interrogate this concept in vivo, xenografts of ADT-sensitive disease were randomized into one of four treatment arms (control, IR, castrate, or castrate and IR), treated, and tumor volume monitored (Figure 1B, top). While all three treatment arms resulted in delayed tumor growth compared to control, the combination of ADT and IR resulted in more effective tumor suppression, as compared to either modality alone (Figure 1B, bottom). Together, these data demonstrate that androgen ablation can cooperate with IR to modestly decrease tumor cell growth and survival in ADT-sensitive, AR-positive PCa.
Figure 1.
Androgen ablation cooperates with IR in HT-sensitive PCa. (A) Top: Cells were cultured in hormone proficient (FBS, full serum control) media for 24 hours then treated with 2 Gy IR, steroid deprived conditions (ADT, androgen deprivation therapy), or a concurrent combination of ADT and 2 Gy IR. Cell number was determined on days 6 and 10 post-treatment and set relative to day 1 (control, untreated). Bottom: Relative cell doubling time was calculated on day 10 for each treatment using the formula Td= (t2−t1)*(ln(2)/ln(q2/q1)), t=time and q=quantity. (B) Top: Schematic representing xenograft treatment cohorts. Bottom: LNCaP cells were injected into the flanks of nude mice and randomized into one of the four treatment arms as shown. Tumor volume was determined periodically and relative volume reported. *<0.05 p value compared to all other treatment conditions.
AR suppression sensitizes CRPC to DNA damage
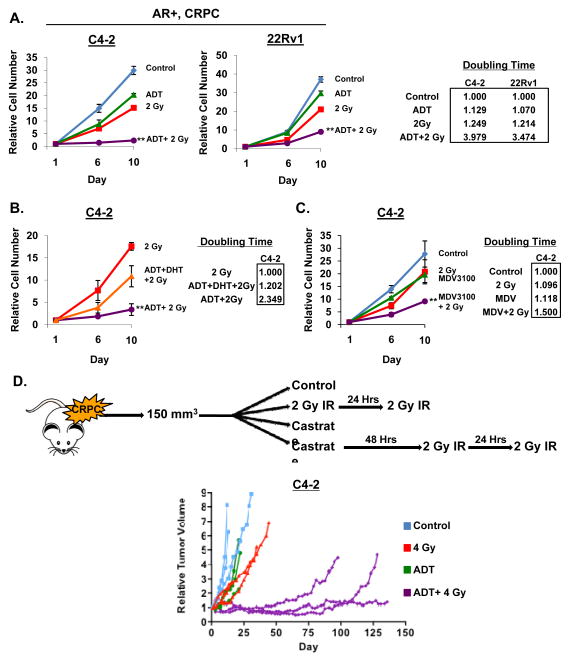
While the observation that suppression of AR activity can enhance the response to radiation demonstrates in vitro and in vivo mimicry of clinical outcomes, it is well established that ADT induces a potent G1 arrest in hormone therapy-sensitive cells [30], which could therefore impact efficacy of the radiation-induced DNA damage response. Consequently, the subsequent studies were conducted using ADT-refractory prostate cancer cells (CRPC, castrate-resistant prostate cancer cells), which fail to undergo G1 arrest upon androgen ablation and/or AR suppression, and represent advanced disease. Initial in vitro growth analyses were performed in CRPC models that attained ADT-resistance through variant mechanisms. Notably, whereas C4-2 cells express full-length AR, 22Rv1 cells express full length and short forms of AR (derived from alternative splicing) that lack the ligand (androgen) binding domain and are constitutively active [31]. Predictably, ADT alone had little influence on CRPC cells, whereas IR resulted in an approximate 50% reduction in overall cell survival relative to control (Figure 2A, left). Strikingly, combining ADT and IR resulted in dramatically enhanced growth suppression (Figure 2A, left) and increased cell doubling time by more than 3-fold in both model systems as compared to ADT alone (Figure 2A, right). Alteration in treatment schedule resulted in only minimal changes in cell number (Supplemental Figure 1B, C). These findings indicate that the impact of androgens and AR on the response to radiation occurs independently of AR-dependent cell cycle control, and is highly significant in models of late stage, castration-resistant disease.
Figure 2.
AR suppression sensitizes CRPC to DNA damage. (A) Left: Cells were cultured in hormone proficient media for 24 hours then treated with 2 Gy IR, ADT, or a concurrent combination of ADT and 2 Gy IR. Cell number was determined on days 6 and 10 post-treatment and set relative to day 1 control. Right: Relative cell doubling time was calculated on day 10 for each treatment. (B) Cells were treated as per panel A, but treatments were 2 Gy IR, concurrent ADT and 2 Gy IR, or concurrent ADT supplemented with 0.1nM DHT and 2 Gy IR. (C) Cells were treated as per panel A, but treatments were 2 Gy IR, 1uM MDV3100, or concurrent 1uM MDV3100 and 2 Gy IR. (D) Top: Schematic representing xenograft treatment cohorts. Bottom: C4-2 cells were injected into the flanks of nude mice and randomized into one of the four treatment arms as shown. Tumor volume was determined periodically and relative volume reported. **<0.01 p value compared to all other treatment conditions.
To further assess the role of AR activity on the response to DNA damage, dihydrotestosterone (DHT) re-supplementation studies were performed. As shown in Figure 2B, cells subjected to steroid deprivation and then supplemented with DHT prior to radiation demonstrated extensive rescue of cell growth and survival, as shown by a nearly 2 fold reduction in doubling time compared to combined ADT and IR, and only a slight increase compared to IR alone (Figure 2B). These provocative findings strongly suggest that androgens and AR activity promote resistance to genotoxic insult. To further assess this posit, additional studies were performed using the next-generation AR antagonist MDV3100 (enzalutamide), which not only competes with DHT for binding to the receptor, but promotes cytoplasmic sequestration of AR [32]. In these studies, CRPC cells treated with MDV3100 responded similarly to ADT alone, exhibiting only a marginal decrease in cell number compared to control (Figure 2C). Consistent with results obtained from ADT treatment, combining MDV3100 and IR resulted in substantially decreased cell number and subsequent increase in cell doubling time compared to control or either treatment alone (Figure 2C). Further, xenografts of CRPC models were randomized into one of four treatment arms (control, IR, castrate, or castrate and IR) as depicted in Figure 2D, treated, and tumor volume monitored. Consistent with in vitro findings, castration alone had minimal effect on tumor volume while combination of castration and IR resulted in profound growth suppression (Figure 2D, bottom). Collectively, these results indicate that active AR promotes resistance to radiation, using both in vitro and in vivo models of disease progression.
DNA damage induces AR activity
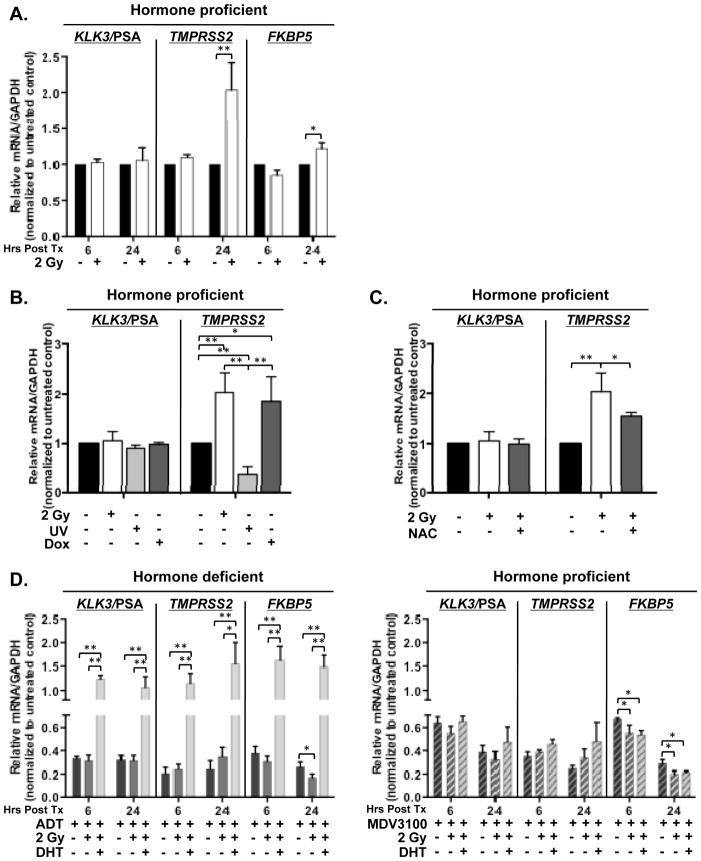
To begin to explore the mechanism by which AR promotes radiation resistance, AR activity was assessed in CRPC cells after IR treatment by analyzing expression of clinically relevant AR target genes (KLK3/PSA, TMPRSS2, and FKBP5) whose expression is known to be under stringent AR regulation. As shown in Figure 3A, both TMPRSS2 and FKBP5 transcript showed marked induction after radiation, whereas KLK3/PSA levels were generally unchanged (Figure 3A). Further, treatment with increasing dosage of IR resulted in elevated expression of AR target genes (Supplemental Figure 2), further suggesting that AR activity is selectively enhanced in response to genomic insult. To address specificity of this response, the impact of doxorubicin (known to result in double strand DNA breaks), was assessed. As shown, marked induction of AR activity occurred, similar to that observed in response to IR (Figure 3B). Interestingly, treatment with UV, which primarily results in single strand DNA breaks, failed to alter AR target gene expression (Figure 3B). Finally, as DNA damage is known to induce reactive oxygen species (ROS), the impact of a ROS scavenger, N-acetylcysteine, was assessed. Pre-treatment of cells with N-acetylcysteine prior to IR reversed the observed induction of AR target genes, thus implicating ROS and free radicals generated by DNA damage as being involved in the response (Figure 3C). Combined, these observations unexpectedly demonstrate that AR activity is selectively induced in response to DNA double strand breaks in a dose dependent manner, and that this event involves ROS generation. Specificity of this event was further assessed in parallel studies, and it was observed that androgen deprivation nullified the DNA-damage-induced upregulation of AR target gene expression (Figure 3D, left). Consistent with effects on cell growth and survival, DHT re-supplementation rescued gene expression after IR (Figure 3D, left). By contrast, cells cultured in hormone proficient conditions and treated with MDV3100 showed gene expression profiles similar to those cultured under conditions of androgen deprivation, with no detectable increase in AR target gene expression observed after IR treatment (Figure 3D, right). DHT supplementation was unable to influence AR target gene expression in cells treated with MDV3100, likely because the low level of DHT added (0.1 nM) was unable to outcompete MDV3100 (1 μM) for binding to AR [32]. Combined, these results demonstrate that AR activity is induced by DNA damage in models of CRPC.
Figure 3.
DNA damage induces AR activity. (A) C4-2 cells were cultured in hormone proficient media for 24 hours then treated with 2 Gy IR. (B, C) C4-2 cells were cultured in hormone proficient media for 24 hours then treated with 2 Gy IR, 100 J/m2 UV, 1uM doxorubicin, or 1 hour pre-treatment of 1uM N-acetylcysteine followed by 2 Gy IR. (D) Left: C4-2 cells were cultured in hormone deficient or hormone deficient supplemented with 0.1nM DHT media for 24 hours then treated as indicated. Right: C4-2 cells were cultured in hormone proficient media for 24 hours then treated with 1uM MDV3100, concurrent 1uM MDV3100 and 2 Gy IR, or concurrent 1uM MDV3100, 2 Gy IR, and 0.1nM DHT. Relative expression of indicated transcript levels were analyzed and normalized to GAPDH mRNA then set relative to untreated control at indicated time points post treatment. *<0.05; **<0.01 p value.
AR promotes DNA double strand break resolution
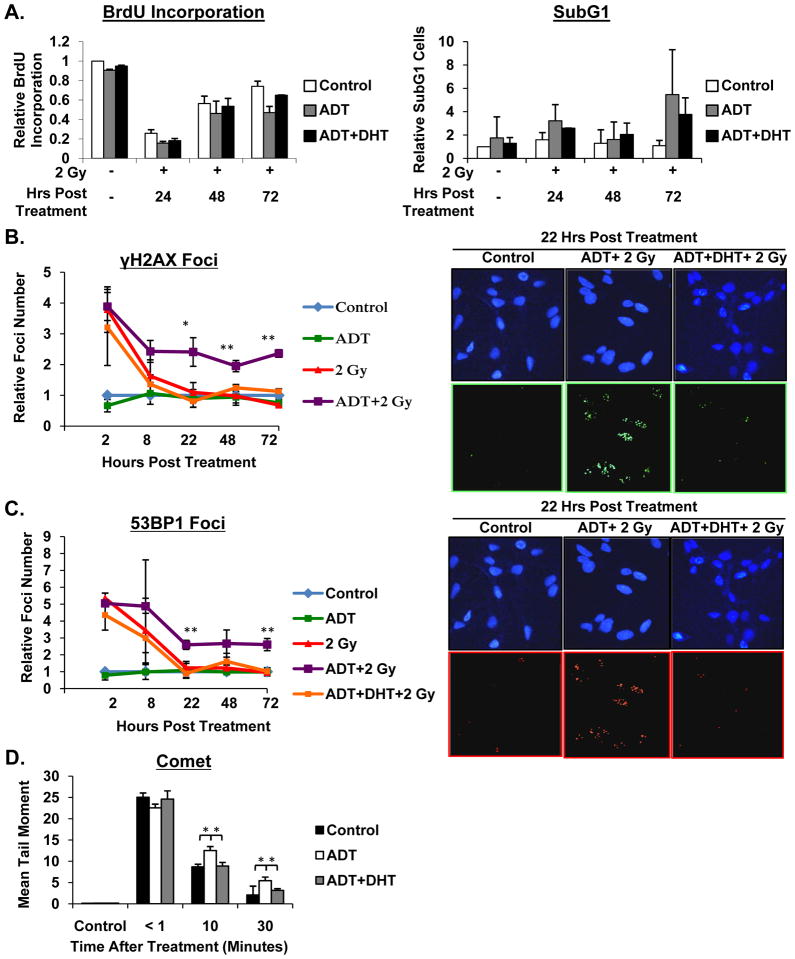
It is well established that cell cycle checkpoints are induced in response to radiation that allow for evaluation of damage extent, and subsequent DNA repair or cell death ensues [33]. Given the robust observations that AR activity promotes cell survival after DNA damage, it was critical to assess the impact of AR on radiation-induced cell cycle alteration and cell death. First, to assess checkpoint control, BrdU incorporation indices and cell cycle profiles were assessed after radiation exposure, both in the presence and absence of androgen, and after androgen deprivation with DHT re-supplementation. Reduction in BrdU incorporation and cell cycle arrest were observed as an initial response to radiation in all three conditions (Figure 4A, left, and Supplemental Figure 3A, B). Similarly, re-entry into cell cycle, as observed by monitoring BrdU kinetics after radiation, was relatively equivalent amongst the three conditions. Next, to assess the impact on cell death, subG1 cell populations were quantified for each condition after radiation (Figure 4A, right). No significant differences in subG1 cells were observed, suggesting that AR activation status does not significantly influence rates of cell death in CRPC cells. This is distinct from what has been previously observed in a model of hormone-therapy sensitive disease [34], revealing distinctions in CRPC after radiation. Thus, in advanced disease the function of AR in promoting cell survival after DNA damage appears to occur independently of alterations in cell cycle control or cell death. As such, the impact of AR on DNA repair was determined.
Figure 4.
AR promotes DNA double strand break resolution. (A) C4-2 cells were cultured in hormone proficient media for 24 hours then treated as indicated. At indicated time points cells were pre-treated with BrdU for 2 hours then fixed. Cells were incubated with BrdU antibody and propidium iodide and processed for FACS analysis. Data is graphed as normalized to untreated control. (B, C) C4-2 cells were cultured in hormone proficient media for 24 hours then treated as indicated. Cells were fixed at the indicated time points, stained with α-γH2AX and α-53BP1 antibodies, and imaged by confocal microscopy. Foci were counted and plotted as average foci per cell set relative to control at each time point. (D) C4-2 cells were cultured in hormone proficient, hormone deficient, or hormone deficient supplemented with 0.1nM DHT media for 24 hours then treated with 15 Gy IR and harvested at indicated timepoints. Single-cell gel electrophoresis was performed and tail moments assessed. *<0.05; **<0.01 p value.
A proximal consequence of radiation is DNA damage, the most lethal of which is DNA double strand breaks [35]. Inability to repair DNA double strand breaks is known to be detrimental to cell growth and survival [3, 36]. To determine the impact of AR on this process, the proficiency of CRPC cells to repair DNA double strand breaks in the presence or absence of androgen, or after steroid deprivation and DHT resupplementation, was assessed using γH2AX and 53BP1 foci as markers of double strand breaks [37]. As expected, steroid deprivation alone did not alter γH2AX and 53BP1 foci formation, as compared to untreated controls (Figure 4B, C). By contrast, radiation exposure resulted in rapid induction of γH2AX (~11.5 foci/cell at 2 hours post IR, consistent with levels previously reported for similar doses and time courses [37]) and 53BP1 foci (indicative of DNA double strand breaks) compared to control (Figure 4B, C). Cells treated with IR alone (in hormone proficient conditions) showed double strand break resolution at 22 hrs post-treatment (Figure 4B, C), consistent with what has previously been reported for similar doses of IR [38]. Remarkably, cells deprived of hormones just prior to radiation and held in androgen-free conditions after radiation showed a markedly diminished capacity to repair double stand breaks 22 hrs post-treatment. In fact, elevated levels of both γH2AX and 53BP1 foci persisted up to 72 hours (Figure 4B, C), indicating that cells subjected to androgen deprivation were severely compromised in the ability to repair radiation-induced double strand breaks. The specificity of this event was further demonstrated in that cells subjected to steroid deprivation but resupplemented with physiologically relevant levels of DHT showed restoration of double strand break repair at 22 hrs post treatment (Figure 4B, C). To further interrogate the hypothesis that activated AR promotes DNA break repair, analysis of DNA fragmentation by neutral Comet assay revealed significantly elevated DNA fragmentation after high dose IR (15 Gy) in cells deprived of hormone, an effect that was reversed by resupplementation with DHT (Figure 4D). Combined, these data confirm that AR activation results in repair of DNA damage. These unexpected findings reveal for the first time that AR promotes DNA double strand break repair, independent of the capacity of AR to regulate cell cycle progression and cell survival.
AR activity regulates genes required for DNA damage repair
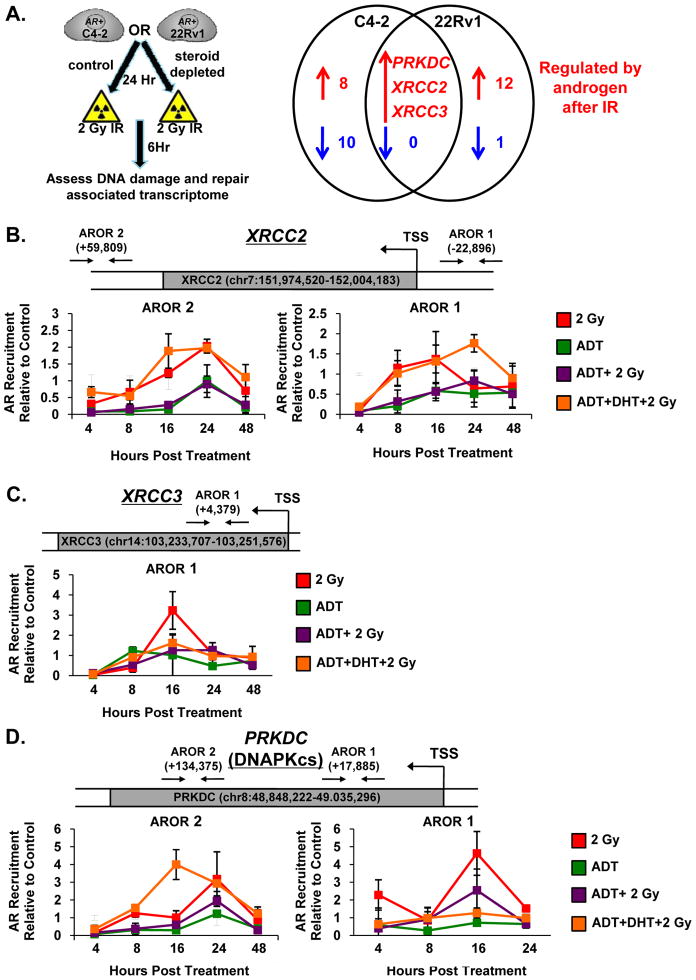
Given the well-defined role of AR as a transcription factor [39] combined with the new observations herein that: i) DNA damage induces AR activity; ii) active AR promotes cell survival after DNA damage; and iii) AR facilitates double strand DNA break repair, the impact of AR on expression of genes encoding the DNA repair machinery was determined. A rigorous screen was developed, wherein a custom qPCR array was used to assess changes in the DNA damage and repair-associated transcriptome after radiation, in the presence and absence of hormone (Figure 5A, left). The screen was implemented so as to identify genes significantly up- or down-regulated by both radiation and androgens (see Supplemental Figure 4A), and genes with identical patterns in both the CRPC models tested were prioritized for further analyses. Using this strategy, 3 lead candidates were identified: PRKDC (encoding DNAPKcs), XRCC2, and XRCC3 (Figure 5A, right). These results were notable, as DNAPKcs binds broken DNA ends through the Ku 70/80 heterodimer and is critical for proper recruitment of repair factors during NHEJ [1, 40], while XRCC2 and XRCC3 are RAD51 paralogs that promote strand transfer at sites of DNA damage during HR [41]. Since each was induced by androgen in the presence of DNA damage, previously published ChIP-seq data [39], in addition to our own ChIP-seq analyses (McNair et al., in preparation), was utilized to identify AR binding within 50 Kb of the transcriptional start site for each gene, as would be expected based on current knowledge of AR function [42]. Using these combined datasets, AR occupied regions (ARORs) were identified in the putative regulatory regions of all three genes, and validation studies were initiated using this information. For each potential AROR, chromatin immunoprecipitation (ChIP) analyses were performed at multiple time points post treatment in either hormone proficient or hormone deficient conditions. For XRCC2, two ARORs were identified, both of which contained an ARE half site [43]. As shown, AR was recruited in a time-dependent manner (post IR) both in hormone proficient conditions and in cells which had been deprived of steroids and re- supplemented with DHT (Figure 5B, red and orange lines). Recruitment to these sites was diminished in hormone deficient conditions after IR (Figure 5B, purple line), indicating that increasing AR activity promotes binding after IR. For XRCC3, only one AROR was identified (contained an ARE half site), and while the recruitment of AR was not as striking as that exhibited by XRCC2, elevated AR recruitment at 16 hrs post-IR was observed only in hormone proficient conditions (Figure 5C, red line). Finally, similar to XRCC2, two ARORs were identified within proximity of the PRKDC (encoding the protein product DNAPKcs) locus; however, neither contained an ARE half site. This is consistent with previous studies, which reveal that AREs are present at only approximately 40% of known AR binding sites [44]. Again, AR recruitment to these sites was observed in a time dependent fashion after IR treatment in both hormone proficient and hormone deficient supplemented with DHT conditions (Figure 5D, red and orange lines). AR recruitment was reduced in hormone deficient conditions after IR (Figure 5D, purple line), suggesting activated AR acts as a critical regulator of gene expression in response to IR. No substantial recruitment of AR relative to control was detected at a region of the KLK3/PSA locus known to be devoid of AR binding sites [45] (Supplemental Figure 4B), suggesting that elevated levels of AR recruitment to the DNA damage loci represents specific recruitment of AR in response to DNA damage. Collectively, the DNA damage gene expression profile combined with the AR chromatin binding data support the novel concept that AR activity regulates genes required for DNA damage repair.
Figure 5.
AR activity regulates genes required for DNA damage repair. (A) Left: Schematic showing experimental design. Right: Array results depicting genes regulated by androgen after 2 Gy IR in C4-2 and 22Rv1 cells. (B–D) C4-2 cells were cultured in hormone proficient, hormone deficient, or hormone deficient supplemented with 0.1nM DHT media for 24 hours then treated as indicated. Samples were harvested for ChIP-qPCR analysis and percent (input) occupancy of AR set relative to control at each time point is reported. Arrows and numbers on diagrams depict location of identified AROR in relation to transcriptional start site (TSS).
AR induces DNAPKcs expression and activity
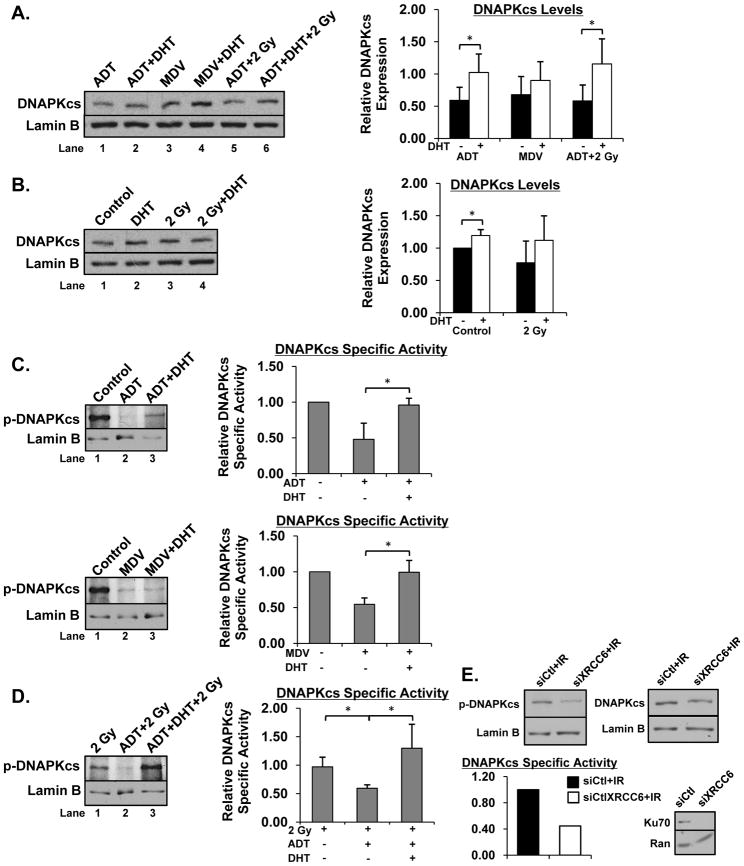
Of the genes identified, PRKDC is the most critical for efficient DNA repair, exhibiting a well-defined role in the early stages of the NHEJ double strand break repair pathway [1, 40]. As cells were unable to efficiently repair double strand breaks when AR activity was suppressed, the impact of AR status on DNAPKcs expression was investigated. As shown, compared to ADT or combination ADT and IR, supplementation with DHT in either treatment, respectively, resulted in induction of overall DNAPKcs levels (Figure 6A, compare lanes 1,2 and 5,6, quantification on right). Treatment with MDV3100 also modestly diminished DNAPKcs levels compared to combination MDV3100 and DHT treatment (Figure 6A, compare lanes 3, 4, quantification on right). Further, CRPC cells treated exclusively with DHT showed increased expression of DNAPKcs compared to untreated control (Figure 6B, compare lanes 1, 2, quantification on right). Cells treated with IR showed a similar trend regarding DNAPKcs levels compared to cells treated with IR and DHT despite not achieving statistical significance (Figure 6B, compare lanes 3, 4, quantification on right). These data indicate that activated AR promotes DNAPKcs expression in both the presence and absence of IR.
Figure 6.
AR induces DNAPKcs expression and activity. (A, B) C4-2 cells were cultured in hormone proficient, hormone deficient, or hormone deficient supplemented with 0.1nM DHT media for 24 hours then treated. Cells were harvested and expression of DNAPKcs was analyzed and quantified. Representative image of at least 3 independent experiments is shown. (C–D) Same as (A, B), but expression of phospho (Ser2056)-DNAPKcs was analyzed and quantified. DNAPKcs specific activity was determined by dividing phospho-DNAPKcs by total DNAPKcs. (E) C4-2 cells were transfected with a pool of siRNAs directed against the XRCC6 transcript or control siRNA for 96 hours then treated with 2 Gy IR. Cells were harvested and expression of DNAPKcs, phospho-DNAPKcs, and Ku70 analyzed and quantified. *<0.05 p value.
In addition to examining DNAPKcs expression, phosphorylation of DNAPKcs on Ser2056 (indicative of activated DNAPKcs) was interrogated in all treatment conditions, as DNAPKcs activity is known to be induced in response to DNA damage [46]. CRPC cells treated with ADT or MDV3100 showed decreased phospho-DNAPKcs levels and decreased DNAPKcs specific activity (phospho-DNAPKcs/ total DNAPKcs) compared to control, which was rescued by supplementation with DHT (Figure 6C compare lanes 1,2 and 2,3). Finally and most intriguingly, the combination of ADT and IR elicited decreased phospho-DNAPKcs and DNAPKcs specific activity compared to IR alone, a result that was reversed by supplementation with DHT (Figure 6D compare lanes compare lanes 1,2 and 2,3). To further address the mechanism(s) by which AR controls DNAPKcs activity, Ku70 was depleted by siRNA, as Ku70 has been shown to recruit DNAPKcs and contribute to DNAPKcs activation [47]. Moreover, the gene encoding Ku70 (XRCC6) was identified as a potential AR target gene in the qPCR array (Supplemental Figure 4A). As shown, Ku70 depletion resulted in decreased phospho-DNAPKcs in response to IR treatment (Figure 6E), revealing one mechanism by which AR likely controls DNAPKcs activation. Overall, these data reveal that an activated AR signaling axis induces both DNAPKcs expression (through binding to and regulating the PRKDC locus) and activity (requiring Ku70, both in the presence and absence of DNA damage.
DNAPKcs is required for DNA repair in the presence of androgen
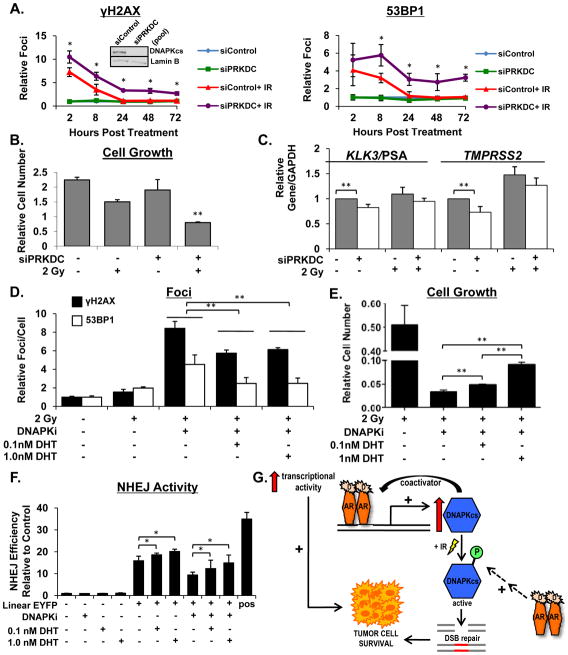
To determine if altered DNAPKcs expression is sufficient to account for the inefficient double strand break repair and decreased cell number observed after combination ADT and IR in CRPC cells, DNAPKcs was depleted by RNAi and double strand break resolution assessed by quantification of γH2AX and 53BP1 foci formation (Figure 7A). DNAPKcs knockdown alone did not induce DNA double strand breaks (Figure 7A, compare green line to blue line). Consistent with Figure 4, control cells treated with IR exhibited increased levels of both γH2AX and 53BP1 foci immediately following IR treatment, and the lesions were largely resolved within 24 hrs (Figure 7A, red line). However, cells with diminished DNAPKcs expression exhibited both an increase in foci immediately following IR and a significantly compromised ability to repair double strand breaks, noted by elevated levels of foci that persisted for up to 72 hrs post-IR (Figure 7A, compare purple, red lines). These results are consistent with the concept that AR-mediated double strand DNA break repair is dependent on AR-induced DNAPKcs expression and activity. DNAPKcs is also required for AR-mediated cell survival after DNA damage, as combined DNAPKcs knockdown and IR exhibited decreased cell number compared to siControl, siControl and IR, or DNAPKcs knockdown alone (Figure 7B). Importantly, DNAPKcs knockdown persisted throughout the course of the experiment (Supplemental Figure 5), and these results mimic growth suppression seen in CRPC cells after combination ADT and IR treatment (Figure 2A), indicating that loss of DNAPKcs expression at least partially accounts for the observed decreased in cell number. Additionally, DNAPKcs knockdown caused significant decreases in expression of AR target genes (Figure 7C), consistent with previous reports identifying DNAPKcs as a transcriptional coactivator of AR [28]. Consistent with what has previously been reported [48], DNAPKcs depletion resulted in decreased expression of ATM and concurrent decrease in phospho-ATM levels (Supplemental Figure 6A).
Figure 7.
DNAPK is required for DNA repair in the presence of androgen. (A) C4-2 cells were transfected with a pool of siRNAs directed against the PRKDC transcript or control siRNA for 96 hours then treated with 2 Gy IR. Cells were fixed at the indicated timepoints, stained with α-γH2AX and α-53BP1 antibodies, and analyzed by confocal microscopy. Foci were counted and plotted as average foci per cell set relative to control at each time point. (B) Same as (A), but cells were counted 5 days post treatment and cell number set relative to control. (C) Same as (A), but relative expression of transcript levels were analyzed and normalized to GAPDH mRNA at 24 hours post treatment (D) C4-2 cells were cultured in hormone proficient media for 24 hours then treated with combinations of DHT (1 hour pre-treatment), 2 Gy IR, and 1uM DNAPKcs inhibitor. Cells were fixed at the indicated timepoints, stained with α-γH2AX and α-53BP1 antibodies, and analyzed by confocal microscopy. Foci were counted and plotted as average foci per cell set relative to control at each time point. (E) Same as (D), but cells were counted 5 days post treatment and cell number set relative to control. (F) C4-2 cells transfected with linearized pEYFP plasmid were treated as indicated and activity assessed. (G) Model describing the feedback circuit of AR and DNAPKcs expression and activity controlling DNA repair and tumor cell survival *<0.05; **<0.01 p value.
If AR-driven DNAPKcs expression and activity is critical for AR-mediated repair and survival post IR, heightened AR activity may be able to overcome DNAPKcs inhibition. To further address this hypothesis, a highly specific DNAPKcs inhibitor (Nu7441) [49] was assessed for the impact on AR-dependent DNA repair and cellular outcomes. First, cells cultured in hormone proficient conditions were exposed to DHT for 1 hr followed by treatment with the DNAPKcs inhibitor and IR. At 24 hrs post-treatment, cells treated with the DNAPKcs inhibitor and IR exhibited elevated levels of γH2AX and 53BP1 foci compared to cells treated with IR alone (Figure 7D). Remarkably, cells pre-treated with DHT showed a significant reduction in foci levels (though rescue of repair capacity was only partial), indicating that an active AR signaling axis resulted in enhanced DNA double strand break repair (Figure 7D). Accordingly, cells pre-treated with DHT exhibited modestly increased cell number 5 days post treatment compared to cells that were treated with DNAPKcs inhibitor and IR alone (Figure 7E). DNAPKcs inhibition also resulted in increased levels of phospho-ATM without significantly altering overall ATM levels (Supplemental Figure 6B), consistent with what has been reported regarding redundancy between DNAPKcs and ATM in activating the damage response [50]. To further assess the requirement of DNAPKcs as a modulator of AR-mediated outcomes, clonogenic assays were performed (Supplemental Figures 7 A-E). Similar to in vitro cell doubling assays in Figure 1, the impact of androgen deprivation on the response to IR was modest in the hormone-therapy sensitive models (Panel A), consistent with previous reports [34], and no effect of variant hormone status was observed in AR-negative cells (Panel B). By contrast, a robust effect was observed in CRPC cells, which was rescued by DHT supplementation (Panel C, D). Strikingly, suppression of DNAPKcs activity eliminated the radioprotective impact of androgens in these models, though pre-treatment with DHT again partially restored the protective effect (Panel E). Additional analysis demonstrated that pre-treatment with DHT resulted in enhanced repair within 2 hours post IR exposure (Supplemental Figure 8A, B), demonstrating an immediate impact on repair kinetics. Analysis of HR activity, monitored by Rad51 foci, revealed a significant increase in HR activity at 2, 5, and 8 hours after IR treatment (Supplemental Figure 8C), demonstrating that AR activation results in a rapid increase in HR mediated repair when DNAPKcs is inhibited. Further, utilization of a plasmid based NHEJ activity assay demonstrated that activation of AR resulted in a significant increase in NHEJ activity, but only a modest increase in NHEJ when DNAPKcs is inhibited (Figure 7F). In sum, these results identify DNAPKcs as an important effector of AR function whose activity is critical for efficient AR-mediated DNA repair. Combined, the data presented suggest an overall model wherein AR signaling directly regulates DNAPKcs expression and indirectly influences DNAPKcs activity, resulting in a positive feedback circuit controlling efficient DNA double strand break repair after DNA damage and tumor cell survival (Figure 7G).
DISCUSSION
Understanding the mechanisms by which altered DDR promotes tumor progression and aggressive phenotypes is crucial for development of effective therapeutic strategies for advanced cancers. The present study identifies for the first time a critical link between hormone action and DNA repair. Key findings show that (i) DNA damage enhances AR activity; (ii) androgens promote double strand DNA break repair and resistance to genotoxic insult both in vitro and in vivo; (iii) DNAPKcs expression and activity is induced by androgens; (iv) AR-mediated DNA repair requires DNAPKcs activity; and (v) DNAPKcs enhances AR activity, thereby creating a positive feedback circuit through which androgens promote DNA repair. Together, these findings uncover a powerful and unexpected mechanism through which AR promotes tumor cell survival and therapeutic resistance, and identify a new node for therapeutic intervention.
The concept that DNA damage activates AR and promotes AR binding to genes whose products promote DNA repair was unexpected. With few exceptions, there is little understanding of the cause and consequence of DNA damage-induced transcription factor activation. The p53 tumor suppressor is phosphorylated and activated by multiple DDR factors in response to DNA damage [51], and alters the cellular response to genotoxic insult. Although the effects of AR activity on the response to DNA damage were consistent in both p53-positive (e.g. LNCaP) and p53-deficient models (e.g. LAPC4), p53 has been proposed to influence AR activity [52], and may therefore contribute to altered AR activity after genotoxic insult. The E2F1 transcription factor is also induced in response to DNA damage [53], and can promote double-strand DNA break repair [53]; strikingly, E2F1 positively regulates AR, and is frequently upregulated in CRPC [54], providing an additional means by which AR activity may be induced in response to DNA damage. Alternatively, the histone acetyltransferases CBP and p300, known AR coactivators, promote expression of the HR genes BRCA1 and RAD51 [55], indicating that AR likely utilizes a distinct cohort of cofactors to alter gene expression after DNA damage. Finally, post-translational modifications of nucleosomes are known to influence DDR [56], and it is also tempting to speculate that AR may be recruited to genes that promote DDR via altered histone modification. Combined, these collective studies strongly suggest that DNA damage-induced transcription factor activation plays a major role in DDR and the cellular response to genotoxic damage.
DNA damage-activated AR was found herein to promote expression of numerous genes associated with DNA repair (Figure 5, Supplemental Figure 2). FEN1 (Flap endonuclease 1), which is implicated in base excision repair and oxidative stress response [57], is associated with aggressive PCa [58]. Since IR induces oxidative damage in addition to DNA double strand breaks, increased FEN1 expression after DNA damage may contribute to RT resistance via modulation of the oxidative stress response, a concept supported by the finding that ROS and free radicals generated by damage play a role in the response of AR to damage (Figure 3C). More proximal to DNA double strand break repair, XRCC6 (protein product Ku70) is induced by AR after DNA damage. Ku70 binds the free ends of DNA double strand breaks as a heterodimer with Ku80 and recruits DNAPKcs to facilitate NHEJ [1, 40]. Recent reports show that Ku70 expression is decreased after castration and correlates with PSA in clinical specimens [59], providing additional evidence that AR regulates Ku70 in human disease. In addition to FEN1 and XRCC6, genes whose products are central to DNA double strand break repair were identified herein as regulated by AR after DNA damage. XRCC2 and XRCC3, each RAD51 paralogs required for RAD51 recruitment and accumulation during HR, are both induced in the presence of androgens after DNA damage. Additional analysis revealed AR recruitment to regulatory regions of both genes in a time dependent fashion post IR only when androgens were present. While AR regulated expression of XRCC2 has not been previously reported, XRCC3 was recently identified as an AR target gene in CRPC tissue [60]. Further, XRCC3 was identified as part of a 16 gene signature that predicts CRPC [60], indicating that AR mediated expression of XRCC3 (and possibly XRCC2, as both genes have similar functions) drives resistance to therapy and development of late stage disease. Time dependent recruitment of AR to XRCC2 and XRCC3 regulatory regions observed herein in response to IR reveals a novel signaling network where AR induces XRCC2 and XRCC3 as a direct response to DNA damage. While ongoing investigation will delineate the contribution of these AR regulated genes to AR-mediated DNA repair and therapeutic resistance, amongst the AR targets identified herein, PRKDC (protein product DNAPKcs) plays the most prominent role in the DDR.
DNAPKcs initiates NHEJ after recruitment to DNA double strand breaks, and the present study clearly identifies AR-mediated DNAPKcs expression and activity as essential for AR-mediated DNA repair and therapeutic resistance. Strikingly little is understood with regard to DNAPKcs regulation-- basal levels of genes involved in NHEJ (including PRKDC) are expressed throughout all stages of cell cycle [61]. In BCa, ER was recently demonstrated to alter DNAPKcs expression, in that activated ER binds sites within regulatory regions of PRKDC; however, whether this event occurs after DNA damage remains uncertain. [17]. By contrast, the data presented herein demonstrate for the first time that activated AR resulted in increased DNAPKcs expression, and that AR was recruited to regulatory regions of PRKDC in a time dependent manner after DNA damage. These findings reveal the crucial mechanistic insight that AR is a transcriptional regulator of DNAPKcs expression. Moreover, the critical role of DNAPKcs in DNA double strand break repair indentifies AR driven DNAPKcs expression as an important mediator of therapeutic resistance. While AR directly binds and promotes expression of the PRKDC locus, active AR also stimulates DNAPKcs activity, as determined by increased phospho-DNAPKcs and DNAPKcs specific activity. Further, inhibition of DNAPKcs activity resulted in inability to repair DNA damage and decreased cell growth. However, both effects are partially reversed by increased AR mediated induction of HR, as demonstrated by increased Rad51 foci, and to a minor extent, NHEJ activity. The ability of AR to induce HR is not entirely surprising, given the finding that AR induces XRCC2 and XRCC3, two genes involved in HR mediated repair, in response to IR (Figure 5B, C). Additionally, as HR is known to require a template for repair, AR-mediated cell cycle progression stimulated by DHT provides an environment that is required for HR, ultimately further implicating AR in coordination of DDR pathways. Combined, these findings reveal that though AR mediated DNA repair is complex, DNAPKcs activity is essential for efficient AR-mediated repair of damaged DNA and resultant cell survival. DNAPKcs is typically autophophorylated in response to IR and recruited to broken DNA ends by the Ku70/Ku80 heterodimer, where additional phosphorylation events occur [62]. Interaction between Ku70 and DNAPKcs suggests a possible mechanism of AR mediated DNAPKcs regulation; in addition to recruiting DNAPKcs to broken DNA ends, Ku70 can facilitate DNAPKcs phosphorylation [47], and this is supported by the finding that Ku70 depletion results in decreased phospho-DNAPKcs in response to IR treatment in CRPC cells (Figure 6E). Further, the observations that DNAPKcs inhibition and DNAPKcs knockdown both resulted in inefficient DNA double strand break repair despite exerting opposite effects on phospho-ATM levels strongly suggest that AR-mediated DNA repair is dependent on DNAPKcs but not directly on ATM. On balance, the data herein intriguingly demonstrate that AR controls both DNAPKcs expression and activity, and that efficient AR-mediated DNA repair requires DNAPKcs activity.
While the main function of DNAPKcs is as a mediator of DDR, it is becoming well appreciated that DNAPKcs impinges on other cellular processes [63]. Specifically, DNAPKcs can function as a transcriptional coactivator for numerous hormone receptors, including ER and PR (progesterone receptor) [16, 64]. DNAPKcs directly phosphorylates ER, facilitating protein stability and transcriptional activation [16]. While little is known with regard to AR, DNAPKcs can induce AR transcriptional activity in a luciferase reporter system [28], suggesting that DNAPKcs may serve as an AR coactivator. Consonantly, DNAPKcs downregulation resulted in significant decrease in expression of numerous AR target genes, indicating that DNAPKcs is necessary for robust AR activity. Combined, these data demonstrate that AR driven DNAPKcs expression and activity is required for AR-mediated DNA repair and that DNAPKcs enhances AR transcriptional activation, resulting in a positive feedback circuit that drives tumor survival and therapeutic resistance.
Given the role of DDR in cancer progression, and recent revelations that advanced stage prostate cancers show significant chromosomal alterations [65], considerable efforts have been put forth to develop therapeutic strategies targeting double strand break repair, focusing on precision approaches based on pathways driving different tumor types [66]. The discovery of a positive feedback circuit involving AR-mediated expression and activity of DNAPKcs and DNAPKcs activation of AR reveals that targeting AR in combination with DNAPKcs inhibition and a DNA damaging agent may be an effective therapeutic strategy in CRPC as well as other AR driven cancers including liver [67] and molecular apocrine BCa [68]; this postulate is currently under investigation. Targeting the AR-DDR crosstalk is dependent on ability to decrease AR activity and disrupt DDR by targeting factors critical for damage repair, though diminishing AR activity in CRPC has proven challenging. However, next generation AR antagonists such as enzalutamide and ARN-509 have demonstrated significantly enhanced efficacy in late stage disease [32, 69]. Additionally, intratumoral androgen synthesis is targeted using the CYP17A inhibitor abiraterone acetate, resulting in decreased AR activity in CRPC [70]. Related to the present findings, recently developed DNAPKcs inhibitors are now in phase I trial for multiple, advanced, solid tumors [66]. In the context of AR-dependent cancers, it is hypothesized that DNAPKcs inhibitors would both diminish DDR capacity and reduce the transcriptional activation of AR-dependent cell survival. Thus, targeting the dual roles of DNAPKcs is likely to suppress multiple pathways that promote advanced tumor phenotypes.
In summary, the present study identifies a positive feedback circuit linking hormone action to the DNA damage response, and demonstrates the significant impact of this process on tumor progression and therapeutic response. These provocative findings provide the foundation for development of novel nodes of therapeutic intervention for advanced disease.
MATERIALS AND METHODS
Cell culture and treatment
See Supplemental Materials and Methods. Cell lines used were not cultured longer than 6 months after receipt from the original source of American Type Culture Collection.
Cell growth assays
Assays were preformed as described previously [71]. Media was changed on day 6 for all 10 day cell growth studies.
Xenograft analysis
Cells were resuspended in 100uL of saline with 50% Matrigel (BD Biosciences) and injected subcutaneously into the flank of 6 week old athymic nude male mice (NCI Frederick). Treatment was initiated when tumors reached ~150 mm3 as described in Supplemental Materials and Methods. Tumor volume was measured periodically and at time of sacrifice with digital calipers. Mice were sacrificed once their tumors reached an approximate size of 1000 mm3. Mouse weight was assessed at time of sacrifice, and no mice lost more than 5% of their initial body weight.
Gene expression analysis
Cells were seeded at equal densities in hormone proficient or hormone deficient conditions. RNA was isolated using TRIzol and cDNA generated using SuperScript III (Invitrogen). Quantitative PCR was performed using an ABI StepOne machine and PowerSybr in accordance with the manufacturer’s specifications using primers described in Supplemental Table 1.
Flow cytometry
Cells were seeded in hormone proficient media and labeled with BrdU (1:1000) 2 hours prior to harvest. Cells were fixed in ice-cold 100% ethanol and stained with FITC-conjugated anti-BrdU antibody (BD Biosciences, #556028). Ethanol fixed cell pellets were gently resuspended in 1 mL PBS containing 0.02 mg/mL propidium iodide and 0.04 mg/mL RNase A, incubated for 15 min at room temperature in the dark, and processed using a Beckman FACS Calibur (at least 10,000 events per sample). Analysis was performed using FlowJo (v8.8) for BrdU incorporation and cell cycle profile.
Immunofluorescence analysis
Cells were seeded onto coverslips and fixed in 3.7% formaldehyde at indicated timepoints. Cells were permeabilized in 0.3% TritonX-100 at 37°C for 30 min then stained with primary antibody (1:500) for 1 hour at 37°C followed by secondary antibody (1:1000) for 1 hour at room temperature utilizing antibodies described in Supplemental Materials and Methods. All immunofluoresence was counterstained with DAPI to visualize nuclei, and coverslips were mounted on slides using Gelvatol. Foci were imaged on a Zeiss Confocal Laser Scanning microscope, counted for at least 50 cells per treatment condition, and set relative to untreated control.
Comet assay
Single-cell gel electrophoresis was performed according to the manufacturer’s instructions (Trevigen, Gaithersburg, MD). See Supplemental Materials and Methods for experimental details.
PCR array analysis
Cells were seeded in hormone proficient or hormone deficient conditions and treated as indicated. RNA was isolated using TRIzol and cDNA generated using the RT2 First Strand Kit (SABiosciences) according to manufacturer’s specifications. cDNA was analyzed by real-time PCR using either the DNA Repair (#PAHS-042Z) or DNA Damage Signaling Pathway (#PAHS-029Z) RT2 Profiler PCR array (SABiosciences) and results quantified according to manufacturer’s specifications.
ChIP analysis
Cells were seeded in hormone proficient or hormone deficient conditions and treated as specified. Cells were fixed with 1% formaldehyde at indicated timepoints and ChIP analyses performed as previously described [54]. Genomic DNA was purified and quantitative PCR performed for indicated loci using primers described in Supplemental Table 1. Data were analyzed as percentage of input of total samples calculated as previously described [54].
Immunoblotting
Cells were seeded in hormone proficient or hormone deficient conditions and treated as specified. Cell lysates (30–40 μg) were generated as previously described [72], resolved by SDS-page gel electrophoresis, transferred to PVDF membranes, and analyzed using antibodies described in Supplemental Materials and Methods. Quantification was performed using a Bio-Rad ChemiDoc MP Imaging System.
RNA interference
Cells were seeded at a density of 1×105 in hormone proficient conditions (complete media) for 24 hours. Cells were then transfected (6–8 hours) in serum-free conditions with either control, PRKDC, or XRCC6 siRNA pools (Thermo Scientific, D-001810-10-20, L-005030-00-0020, or L-005084-00-0005 respectively) according to the manufacturer’s specifications. Cells were maintained in complete media for an additional 96 hours then treated as specified and harvested at indicated time points. For growth analysis, a day 1 count was taken 96 hours post transfection for both siPRKDC and siControl cells to which day 5 cell counts were compared. Immunoblot analysis to confirm DNAPKcs or Ku70 knockdown was performed on cells harvested at time of treatment (96 hours post transfection).
NHEJ activity assay
Linearized pEYFP vector was transfected into C4-2 cell lines. The cells were treated with either DHT (0.1 nM or 1nM) or DNAPK inhibitor (NU7441 1μM) at the time of transfection. 24 hours after transfection, genomic DNA was extracted with Puregene Cell and Tissue kit (Cat. No.158388). Quantitative PCR was performed using an ABI StepOne machine and PowerSybr in accordance with the manufacturer’s specifications using primers described in Supplemental Table 1. The relative efficiency of NHEJ was calculated by the comparative CT method according to the CT values of the internal control as described previously [73].
Clonogenic survival assays
Supplementary Material
STATEMENT OF SIGNIFICANCE.
The present study identifies for the first time a positive feedback circuit linking hormone action to the DNA damage response, and demonstrates the significant impact of this process on tumor progression and therapeutic response. These provocative findings provide the foundation for development of novel nodes of therapeutic intervention for advanced disease.
Acknowledgments
The authors thank members of the K. Knudsen laboratory for input and commentary. The authors specifically acknowledge M. Augello and C. McNair for insightful discussions.
Footnotes
Financial support: This work was supported by NIH grants (R01 CA159945 and R01 CA099996 to K.E. Knudsen), a Prostate Cancer Foundation Mazzone Challenge Award (to K.E. Knudsen and F.Y. Feng), a Physician Research Training Award from the DOD (PC094231 to F.Y. Feng), a Prostate Cancer Foundation Young Investigator Award (to M.J. Schiewer), and Predoctoral Fellowships from the DOD (PC073287 to J.F. Goodwin and PC094195 to M.J. Schiewer), and the Kimmel Cancer Center was supported by the NIH/NCI Cancer Center Core grant P30-CA-56036.
Conflict of interest: The authors disclose no potential conflicts of interest.
References
- 1.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 3.Gewirtz DA, Holt SE, Elmore LW. Accelerated senescence: an emerging role in tumor cell response to chemotherapy and radiation. Biochem Pharmacol. 2008;76:947–57. doi: 10.1016/j.bcp.2008.06.024. [DOI] [PubMed] [Google Scholar]
- 4.Huang L, Snyder AR, Morgan WF. Radiation-induced genomic instability and its implications for radiation carcinogenesis. Oncogene. 2003;22:5848–54. doi: 10.1038/sj.onc.1206697. [DOI] [PubMed] [Google Scholar]
- 5.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–70. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 6.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–13. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 7.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–8. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 8.Shen Z. Genomic instability and cancer: an introduction. J Mol Cell Biol. 2011;3:1–3. doi: 10.1093/jmcb/mjq057. [DOI] [PubMed] [Google Scholar]
- 9.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynam-Lennon N, Reynolds JV, Pidgeon GP, Lysaght J, Marignol L, Maher SG. Alterations in DNA repair efficiency are involved in the radioresistance of esophageal adenocarcinoma. Radiat Res. 2010;174:703–11. doi: 10.1667/RR2295.1. [DOI] [PubMed] [Google Scholar]
- 11.Beskow C, Skikuniene J, Holgersson A, Nilsson B, Lewensohn R, Kanter L, et al. Radioresistant cervical cancer shows upregulation of the NHEJ proteins DNA-PKcs, Ku70 and Ku86. Br J Cancer. 2009;101:816–21. doi: 10.1038/sj.bjc.6605201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouchaert P, Guerif S, Debiais C, Irani J, Fromont G. DNA-PKcs expression predicts response to radiotherapy in prostate cancer. Int J Radiat Oncol Biol Phys. 2012;84:1179–85. doi: 10.1016/j.ijrobp.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 13.Sarasin A, Kauffmann A. Overexpression of DNA repair genes is associated with metastasis: a new hypothesis. Mutat Res. 2008;659:49–55. doi: 10.1016/j.mrrev.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 14.Higgins MJ, Baselga J. Targeted therapies for breast cancer. J Clin Invest. 2011;121:3797–803. doi: 10.1172/JCI57152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allred DC, Anderson SJ, Paik S, Wickerham DL, Nagtegaal ID, Swain SM, et al. Adjuvant tamoxifen reduces subsequent breast cancer in women with estrogen receptor-positive ductal carcinoma in situ: a study based on NSABP protocol B-24. J Clin Oncol. 2012;30:1268–73. doi: 10.1200/JCO.2010.34.0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Medunjanin S, Weinert S, Schmeisser A, Mayer D, Braun-Dullaeus RC. Interaction of the double-strand break repair kinase DNA-PK and estrogen receptor-alpha. Mol Biol Cell. 2010;21:1620–8. doi: 10.1091/mbc.E09-08-0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Medunjanin S, Weinert S, Poitz D, Schmeisser A, Strasser RH, Braun-Dullaeus RC. Transcriptional activation of DNA-dependent protein kinase catalytic subunit gene expression by oestrogen receptor-alpha. EMBO Rep. 2010;11:208–13. doi: 10.1038/embor.2009.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, Sawyers CL, Scher HI. Targeting the androgen receptor pathway in prostate cancer. Curr Opin Pharmacol. 2008;8:440–8. doi: 10.1016/j.coph.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shang Y, Myers M, Brown M. Formation of the androgen receptor transcription complex. Mol Cell. 2002;9:601–10. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- 20.Knudsen KE, Scher HI. Starving the addiction: new opportunities for durable suppression of AR signaling in prostate cancer. Clin Cancer Res. 2009;15:4792–8. doi: 10.1158/1078-0432.CCR-08-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montgomery RB, Mostaghel EA, Vessella R, Hess DL, Kalhorn TF, Higano CS, et al. Maintenance of intratumoral androgens in metastatic prostate cancer: a mechanism for castration-resistant tumor growth. Cancer Res. 2008;68:4447–54. doi: 10.1158/0008-5472.CAN-08-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker CHD, O’Sullivan JM, et al. Overall survival benefit and safety profile of radium-223 chloride, a first-in-class alpha-pharmaceutical: results from a phase III randomized trial (ALSYMPCA) in patients with castration-resistant prostate cancer (CRPC) with bone metastases. J Clin Oncol. 2012;30(Suppl 5):Abstr 8. [Google Scholar]
- 23.Klein EA, Ciezki J, Kupelian PA, Mahadevan A. Outcomes for intermediate risk prostate cancer: are there advantages for surgery, external radiation, or brachytherapy? Urol Oncol. 2009;27:67–71. doi: 10.1016/j.urolonc.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Bolla M, Gonzalez D, Warde P, Dubois JB, Mirimanoff RO, Storme G, et al. Improved survival in patients with locally advanced prostate cancer treated with radiotherapy and goserelin. N Engl J Med. 1997;337:295–300. doi: 10.1056/NEJM199707313370502. [DOI] [PubMed] [Google Scholar]
- 25.Widmark A, Klepp O, Solberg A, Damber JE, Angelsen A, Fransson P, et al. Endocrine treatment, with or without radiotherapy, in locally advanced prostate cancer (SPCG-7/SFUO-3): an open randomised phase III trial. Lancet. 2009;373:301–8. doi: 10.1016/S0140-6736(08)61815-2. [DOI] [PubMed] [Google Scholar]
- 26.Warde P, Mason M, Ding K, Kirkbride P, Brundage M, Cowan R, et al. Combined androgen deprivation therapy and radiation therapy for locally advanced prostate cancer: a randomised, phase 3 trial. Lancet. 2011;378:2104–11. doi: 10.1016/S0140-6736(11)61095-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 28.Mayeur GL, Kung WJ, Martinez A, Izumiya C, Chen DJ, Kung HJ. Ku is a novel transcriptional recycling coactivator of the androgen receptor in prostate cancer cells. J Biol Chem. 2005;280:10827–33. doi: 10.1074/jbc.M413336200. [DOI] [PubMed] [Google Scholar]
- 29.Weller MA, Tendulkar RD, Reddy CA, Stephans KL, Kupelian P. Adjuvant versus neoadjuvant androgen deprivation with radiation therapy for prostate cancer: Does sequencing matter?. Poster session presented at: 2013 Genitourinary Cancers Symposium; 2013 Feb 14–16; Orlando, FL, USA. [DOI] [PubMed] [Google Scholar]
- 30.Knudsen KE, Arden KC, Cavenee WK. Multiple G1 regulatory elements control the androgen-dependent proliferation of prostatic carcinoma cells. J Biol Chem. 1998;273:20213–22. doi: 10.1074/jbc.273.32.20213. [DOI] [PubMed] [Google Scholar]
- 31.Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68:5469–77. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324:787–90. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–8. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pollack A, Salem N, Ashoori F, Hachem P, Sangha M, von Eschenbach AC, et al. Lack of prostate cancer radiosensitization by androgen deprivation. Int J Radiat Oncol Biol Phys. 2001;51:1002–7. doi: 10.1016/s0360-3016(01)01750-3. [DOI] [PubMed] [Google Scholar]
- 35.Ward JF. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Prog Nucleic Acid Res Mol Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- 36.Al-Ejeh F, Kumar R, Wiegmans A, Lakhani SR, Brown MP, Khanna KK. Harnessing the complexity of DNA-damage response pathways to improve cancer treatment outcomes. Oncogene. 2010;29:6085–98. doi: 10.1038/onc.2010.407. [DOI] [PubMed] [Google Scholar]
- 37.Sak A, Stuschke M. Use of gammaH2AX and other biomarkers of double-strand breaks during radiotherapy. Semin Radiat Oncol. 2010;20:223–31. doi: 10.1016/j.semradonc.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 38.Dean JL, McClendon AK, Knudsen ES. Modification of the DNA damage response by therapeutic CDK4/6 inhibition. J Biol Chem. 2012;287:29075–87. doi: 10.1074/jbc.M112.365494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–56. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417:639–50. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thacker J. The RAD51 gene family, genetic instability and cancer. Cancer Lett. 2005;219:125–35. doi: 10.1016/j.canlet.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 42.Ni M, Chen Y, Lim E, Wimberly H, Bailey ST, Imai Y, et al. Targeting androgen receptor in estrogen receptor-negative breast cancer. Cancer Cell. 2011;20:119–31. doi: 10.1016/j.ccr.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Claessens F, Denayer S, Van Tilborgh N, Kerkhofs S, Helsen C, Haelens A. Diverse roles of androgen receptor (AR) domains in AR-mediated signaling. Nucl Recept Signal. 2008;6:e008. doi: 10.1621/nrs.06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu J, Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443–54. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell. 2005;19:631–42. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 46.Chen BP, Chan DW, Kobayashi J, Burma S, Asaithamby A, Morotomi-Yano K, et al. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J Biol Chem. 2005;280:14709–15. doi: 10.1074/jbc.M408827200. [DOI] [PubMed] [Google Scholar]
- 47.Chan DW, Chen BP, Prithivirajsingh S, Kurimasa A, Story MD, Qin J, et al. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 2002;16:2333–8. doi: 10.1101/gad.1015202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng Y, Woods RG, Beamish H, Ye R, Lees-Miller SP, Lavin MF, et al. Deficiency in the catalytic subunit of DNA-dependent protein kinase causes down-regulation of ATM. Cancer Res. 2005;65:1670–7. doi: 10.1158/0008-5472.CAN-04-3451. [DOI] [PubMed] [Google Scholar]
- 49.Zhao Y, Thomas HD, Batey MA, Cowell IG, Richardson CJ, Griffin RJ, et al. Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Res. 2006;66:5354–62. doi: 10.1158/0008-5472.CAN-05-4275. [DOI] [PubMed] [Google Scholar]
- 50.Stiff T, O’Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004;64:2390–6. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- 51.Velez-Cruz R, Johnson DG. E2F1 and p53 Transcription Factors as Accessory Factors for Nucleotide Excision Repair. Int J Mol Sci. 2012;13:13554–68. doi: 10.3390/ijms131013554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guseva NV, Rokhlin OW, Glover RA, Cohen MB. P53 and the proteasome regulate androgen receptor activity. Cancer Biol Ther. 2012;13:553–8. doi: 10.4161/cbt.19605. [DOI] [PubMed] [Google Scholar]
- 53.Biswas AK, Johnson DG. Transcriptional and nontranscriptional functions of E2F1 in response to DNA damage. Cancer Res. 2012;72:13–7. doi: 10.1158/0008-5472.CAN-11-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharma A, Yeow WS, Ertel A, Coleman I, Clegg N, Thangavel C, et al. The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression. J Clin Invest. 2010;120:4478–92. doi: 10.1172/JCI44239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ogiwara H, Kohno T. CBP and p300 histone acetyltransferases contribute to homologous recombination by transcriptionally activating the BRCA1 and RAD51 genes. PLoS One. 2012;7:e52810. doi: 10.1371/journal.pone.0052810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar R, Horikoshi N, Singh M, Gupta A, Misra HS, Albuquerque K, et al. Chromatin modifications and the DNA damage response to ionizing radiation. Front Oncol. 2012;2:214. doi: 10.3389/fonc.2012.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Asagoshi K, Tano K, Chastain PD, 2nd, Adachi N, Sonoda E, Kikuchi K, et al. FEN1 functions in long patch base excision repair under conditions of oxidative stress in vertebrate cells. Mol Cancer Res. 2010;8:204–15. doi: 10.1158/1541-7786.MCR-09-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Urbanucci A, Sahu B, Seppala J, Larjo A, Latonen LM, Waltering KK, et al. Overexpression of androgen receptor enhances the binding of the receptor to the chromatin in prostate cancer. Oncogene. 2012;31:2153–63. doi: 10.1038/onc.2011.401. [DOI] [PubMed] [Google Scholar]
- 59.Al-Ubaidi FL, Schultz N, Egevad L, Granfors T, Loseva O, Helleday T. Castration therapy results in decreased Ku70 levels in prostate cancer. Clin Cancer Res. 2013 doi: 10.1158/1078-0432.CCR-12-2795. [DOI] [PubMed] [Google Scholar]
- 60.Sharma NL, Massie CE, Ramos-Montoya A, Zecchini V, Scott HE, Lamb AD, et al. The androgen receptor induces a distinct transcriptional program in castration-resistant prostate cancer in man. Cancer Cell. 2013;23:35–47. doi: 10.1016/j.ccr.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 61.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–47. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 62.Collis SJ, DeWeese TL, Jeggo PA, Parker AR. The life and death of DNA-PK. Oncogene. 2005;24:949–61. doi: 10.1038/sj.onc.1208332. [DOI] [PubMed] [Google Scholar]
- 63.Kong X, Shen Y, Jiang N, Fei X, Mi J. Emerging roles of DNA-PK besides DNA repair. Cell Signal. 2011;23:1273–80. doi: 10.1016/j.cellsig.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 64.Weigel NL, Carter TH, Schrader WT, O’Malley BW. Chicken progesterone receptor is phosphorylated by a DNA-dependent protein kinase during in vitro transcription assays. Mol Endocrinol. 1992;6:8–14. doi: 10.1210/mend.6.1.1738374. [DOI] [PubMed] [Google Scholar]
- 65.Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–77. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Basu B, Yap TA, Molife LR, de Bono JS. Targeting the DNA damage response in oncology: past, present and future perspectives. Curr Opin Oncol. 2012;24:316–24. doi: 10.1097/CCO.0b013e32835280c6. [DOI] [PubMed] [Google Scholar]
- 67.Kalra M, Mayes J, Assefa S, Kaul AK, Kaul R. Role of sex steroid receptors in pathobiology of hepatocellular carcinoma. World J Gastroenterol. 2008;14:5945–61. doi: 10.3748/wjg.14.5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hickey TE, Robinson JL, Carroll JS, Tilley WD. Minireview: The androgen receptor in breast tissues: growth inhibitor, tumor suppressor, oncogene? Mol Endocrinol. 2012;26:1252–67. doi: 10.1210/me.2012-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Clegg NJ, Wongvipat J, Joseph JD, Tran C, Ouk S, Dilhas A, et al. ARN-509: a novel antiandrogen for prostate cancer treatment. Cancer Res. 2012;72:1494–503. doi: 10.1158/0008-5472.CAN-11-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stein MN, Goodin S, Dipaola RS. Abiraterone in prostate cancer: a new angle to an old problem. Clin Cancer Res. 2012;18:1848–54. doi: 10.1158/1078-0432.CCR-11-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schiewer MJ, Goodwin JF, Han S, Brenner JC, Augello MA, Dean JL, et al. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov. 2012;2:1134–49. doi: 10.1158/2159-8290.CD-12-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sharma A, Comstock CE, Knudsen ES, Cao KH, Hess-Wilson JK, Morey LM, et al. Retinoblastoma tumor suppressor status is a critical determinant of therapeutic response in prostate cancer cells. Cancer Res. 2007;67:6192–203. doi: 10.1158/0008-5472.CAN-06-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deans AJ, Khanna KK, McNees CJ, Mercurio C, Heierhorst J, McArthur GA. Cyclin-dependent kinase 2 functions in normal DNA repair and is a therapeutic target in BRCA1-deficient cancers. Cancer Res. 2006;66:8219–26. doi: 10.1158/0008-5472.CAN-05-3945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.