Abstract
Autophagic elimination of defective mitochondria suppresses oxidative stress and preserves mitochondrial function. Here, the essential autophagy gene Atg7 was deleted in a mouse model of BRAFV600E-induced lung cancer in the presence or absence of the tumor suppressor TRP53. Atg7 deletion initially induced oxidative stress and accelerated tumor cell proliferation in a manner indistinguishable from Nrf2 ablation. Compound deletion of Atg7 and Nrf2 had no additive effect suggesting that both genes modulate tumorigenesis by regulating oxidative stress, revealing a potential mechanism of autophagy-mediated tumor suppression. At later stages of tumorigenesis, Atg7 deficiency resulted in an accumulation of defective mitochondria, proliferative defects, reduced tumor burden, conversion of adenomas and adenocarcinomas to oncocytomas, and increased mouse lifespan. Autophagy-defective tumor-derived cell lines were impaired in their ability to respire, survive starvation and were glutamine-dependent, suggesting that autophagy-supplied substrates from protein degradation sustains BRAFV600E-tumor growth and metabolism.
Keywords: autophagy, BRAF, lung cancer, mitochondria, metabolism, NRF2
Introduction
Autophagy is a catabolic process activated by metabolic stress whereby cellular proteins and organelles are engulfed and targeted to lysosomes for degradation. This process sustains metabolism by recycling intracellular components for use in macromolecular synthesis and prevents the accumulation of damaged proteins and organelles that can be toxic. The role of autophagy in cancer is complex and still poorly understood (1). Mice with allelic loss of the essential autophagy gene Beclin1 are tumor prone and mosaic or liver-specific deletion of Atg5 or Atg7 produces benign hepatomas, suggesting a role for autophagy in tumor suppression (2–4). However, autophagy localizes to metabolically stressed, hypoxic regions of solid tumors where it enables tumor cell survival (5–7). Indeed, KRASG12D-driven tumors upregulate autophagy which is required for maintenance of functional mitochondria, survival during starvation, and tumor growth (8, 9). In the absence of autophagy, defective mitochondria accumulate, triggering a reduction in mitochondrial respiration that is ultimately incompatible with proliferation. This compromises stress tolerance, prompting us to label these tumors “autophagy-addicted” (8–10). Autophagy addiction was recently confirmed in genetically engineered mouse models of KRASG12D–driven non-small-cell lung cancer (NSCLC) where autophagy deletion converted adenomas and adenocarcinomas to oncocytomas, a benign tumor type associated with an accumulation of defective mitochondria (11–14). Thus, the emerging picture is that the role of autophagy in cancer is context-dependent. In some settings autophagy may limit tumorigenesis by preserving protein and organelle quality control (preventing oxidative stress and tissue damage) while in other contexts autophagy promotes metabolism, adaptation to stress and tumor growth. A comprehensive understanding of the role of the pathway in different tissue types as well as the spatial and temporal variations in autophagy-dependence in tumors driven by specific oncogenic events will be required before modulation of autophagy can become an effective therapeutic strategy for cancer (15–17).
Since KRASG12D driven tumors are exquisitely sensitive to autophagy inhibition, we turned our attention to its downstream effector BRAF. Mutations in BRAF are found in many human tumors including melanoma (65%) lung (3%) papillary thyroid (40%) ovarian (4%) and colorectal cancers (18%) (18–20) with the most common alteration being a valine to glutamic acid substitution at residue 600 (BRAFV600E) leading to constitutive activation of the kinase and RAS independence. To examine the role of autophagy in BRAFV600E–driven lung cancers, mice with Cre-activatible BRAFV600E with and without conditional alleles of the essential autophagy gene Atg7 were generated, lung tumorigenesis was induced via intra-nasal administration of adenoviral Cre recombinase, and the resulting tumors were followed over time. Additionally, conditional alleles of the tumor suppressor Trp53 were bred into the Braf and Atg7 compound mutant mice to assess the consequences of Trp53 loss on the need for autophagy to sustain metabolism and tumorigenesis in the mouse lung.
We report here that autophagy is required for growth of BRAFV600E–driven lung tumors and identify the metabolic alterations underlying autophagy addiction. Moreover, we provide direct evidence that the tumor suppressive function of autophagy can be explained by reactive oxygen species (ROS) mitigation early in tumorigenesis. Most importantly, the dominant tumor promoting function of autophagy can be explained by preservation of mitochondrial function and the supply of metabolic substrates that confer stress tolerance likely necessary for sustained tumorigenesis. Thus, autophagy prevents metabolic dysfunction and ensures the health of BRAFV600E-driven lung tumors.
Results
Deletion of Atg7 blocks autophagy in BRAFV600E–driven lung cancer
To examine the role of autophagy in tumor establishment and progression, mice homozygous for a Cre-activatible BrafCA allele (21) were mated to Atg7flox/flox mice (22), generating animals in which intranasal administration of adenoviral Cre recombinase simultaneously enabled expression of the oncogenic BRAFV600E variant while deleting the floxed allele of Atg7 in the mouse lung. The BRAFV600E model was chosen both for its rapid progression (hyperplasia in 2–4 weeks with discrete adenoma formation by 6–8 weeks) and for the capacity to give rise to a spectrum of malignancies when mated to loss- and gain-of-function tumor models. The BRAFV600E mouse lung tumor model yields benign adenomas unless crossed to mice with deletions of known tumor suppressors INK4A/ARF or TRP53, at which point the lesions progress to adenocarcinomas (21). Thus, the BRAFV600E mouse lung tumor model enables direct testing of the hypothesis that autophagy functions as a regulator of lung tumorigenesis with the potential to reveal both promotion and inhibition of tumor growth by autophagy.
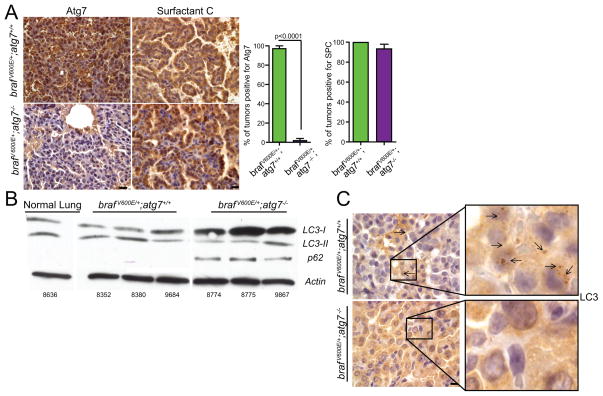
In this genetically engineered mouse model of lung cancer, all tumors are initiated by the constitutively active BRAFV600E protein (21). Atg7 deletion and loss of ATG7 protein expression was confirmed by polymerase chain reaction (PCR) (data not shown) and at early (5 and 7 weeks) and late stages (20 and 30 weeks) of tumorigenesis in the BRAFV600E-derived tumors by immunohistochemistry (IHC) (Fig. 1A and Fig. S1). ATG7 deletion was detected in tumor and not normal lung tissues as adenoviral-Cre infects only a small fraction of lung cells, and only those with activated BRAFV600E form tumors and expand the population of Atg7 wild type or deleted cells. Thus, assessment of autophagy function in normal lung is not addressable in this setting. Atg7 deletion alone by adenoviral-Cre did not cause tumors (Fig. S2 and (13). All tumors were positive for surfactant protein c, a marker of type II alveolar epithelial cells, confirming tumor initiation in the lung (Fig. 1A). Tumor lysates from BrafV600E/+; Atg7−/− mice failed to convert LC3-I to LC3-II, and accumulated the autophagy substrate p62 (Fig. 1B). Similarly, LC3 puncta were visible in the tumor tissue of BrafV600E/+; Atg7+/+ mice while only diffuse cytoplasmic staining was detected in tumor tissues from the BrafV600E/+; Atg7−/− mice (Fig. 1C). Taken together, these data demonstrate that Atg7 deletion is sufficient to block autophagy in BRAFV600E-driven lung tumors.
Figure 1. Atg7 deletion blocks autophagy in a mouse model of brafV600E–driven lung tumorigenesis.
A. Representative IHC for ATG7 and Surfactant Protein C at 7 (BrafV600E/+; Atg7 +/+ and BrafV600E/+; Atg7−/−) post Cre. Quantification of these data is shown. IHC for ATG7 expression at late time points (20, 30 weeks) is provided in Figure S1. Scale bar= 10μm
B. Western blot analysis of LC3-I to LC3-II conversion, and assessment of p62 accumulation in BrafV600E/+; Atg7 +/+ and BrafV600E/+; Atg7−/− tumor lysates 10 weeks post Cre. Numbers refer to individual mice. A blank lane was intentionally included between the normal lung and tumor lysates. An additional Atg7 null tumor lysate was spliced out for aesthetic reasons.
C. Representative IHC staining of LC3 puncta in BrafV600E/+; Atg7 +/+ and BrafV600E/+; Atg7 −/− tumors 8 weeks post infection. Scale bar= 10μm. Arrows denote puncta. Note the absence of puncta and accumulation of diffuse staining in the Atg7 null tumors.
Autophagy ablation causes robust early tumor establishment in the lung
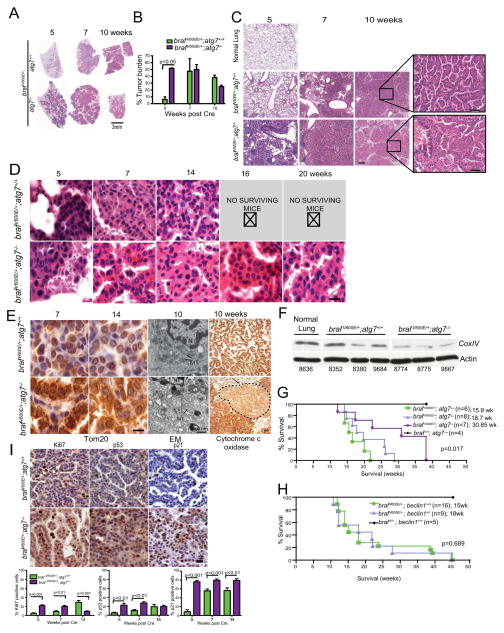
Next, we conducted time course analyses to assess tumor burden over time in mice bearing BrafV600E/+ and either Atg7+/+ or Atg7−/− lung tumors. A marked difference in tumor burden was revealed upon examination of the lung histology. At 5 weeks post Cre, Atg7 deficient BrafV600E/+ tumors increased tumor burden by nearly 40% in comparison to Atg7+/+ tumors (Fig. 2A–C). This increased early tumorigenesis in the BrafV600E/+; Atg7−/− mice was also observed in micro-CT analysis of the mice at 3 and 5 weeks post Cre (Fig. S3). Thus, Atg7 deficiency promotes early BRAFV600E–driven tumor growth.
Figure 2. Atg7 deficiency has distinct consequences for tumor establishment and maintenance in BrafV600E–driven lung tumors and is associated with the emergence of oncocytomas and failure of mitochondrial function.
A. Representative H&E staining of lung lobes at the indicated times post Cre. Scale bar= 3mm. Scanned H&E slides of the full time course are provided in Figure S2.
B. Quantification of tumor burden (two mice/group, all lobes analyzed) from H&E slides across the indicated time points via Matlab. Error bars indicate SEM. Scanned H&E slides of the full time course are provided in Figure S2.
C. Representative low power images of H&E slides 5, 7, 14 weeks post Cre. Scale bar= 100 μm. Higher magnification images are provided in the inset. Scale bar= 50 μm.
D. Representative high magnification images of H&E staining of tumor for the indicated time course. Note the increasing large cytoplasm in the Braf V600E/+; Atg7−/− tumors indicative of oncocytoma. Scale bar=10 μm
E. Braf V600E/+; Atg7−/− tumors have an accumulation of defective mitochondria. Representative images of Tom20 IHC, electron micrographs, and the enzymatic cytochrome c oxidase assay at the indicated time post Cre. For Tom20, Scale bar= 10μm. For EM, N=nucleus MT= mitochondria. Scale bar=500 nm. Arrows point to swollen mitochondria in Braf V600E/+; Atg7−/− tumors. For cytochrome c oxidase, arrows point to tumor regions, indicated by dashed line, with defective mitochondria in the Braf V600E/+; Atg7−/− tumors. Scale bar =100 μm.
F. Western blot of Cox-IV in tumor lysates of Braf V600E/+; Atg7+/+ and Braf V600E/+; Atg7−/− mice 10 weeks post Cre. Numbers refer to individual mice. A blank lane was intentionally included between the normal lung and tumor lysates. An additional Atg7 null tumor lysate was spliced out for aesthetic reasons.
G. Kaplan Meier analysis of overall survival of Braf V600E/+; Atg7+/+ and Braf V600E/+; Atg7−/− mice post Cre. Numbers of animals per group and median survival are indicated.
H. Kaplan Meier analysis of overall survival of Braf V600E/+; Beclin1+/+ and Braf V600E/+; Beclin1+/− mice post Cre. Numbers of animals per group and median survival are indicated.
I. Representative IHC of Ki67, p53 and p21 from Braf V600E/+; Atg7+/+ and Braf V600E/+; Atg7−/− mice 14 weeks post Cre. Scale bar=10 μm. Quantification of IHC at 5, 7, 14 weeks post Cre is shown below. Images for entire time course are provided in Figure S4.
Histologically, BrafV600E/+; Atg7+/+ tumors displayed mild hyperplasia at 5 weeks, progressed to discrete adenomas at 7 weeks, grew steadily into papillary adenomas at 14 weeks post Cre, and mice died shortly thereafter (Fig. 2D). Allelic loss of Atg7 in BrafV600E/+; Atg7+/− tumors modestly accelerated growth, illustrated by the presence of small focal adenomas 5 weeks post Cre (data not shown). Importantly, the BrafV600E/+; Atg7−/− tumors exhibited profound adenoma formation as early as 5 weeks post Cre, but by 10 weeks post Cre the tumors displayed signs of blunted growth (Figs. 2A–C and S2).
Autophagy deficiency alters BRAFV600E tumor fate from adenomas to oncocytomas
Examination of the histology revealed that BrafV600E/+; Atg7+/+ tumors were adenomas, as expected, whereas BrafV600E/+; Atg7−/− tumors were composed of tumor cells with an enlarged, granular cytoplasm characteristic of oncocytomas (Fig. 2D). Oncocytomas are a rare but predominantly benign tumor type characterized by an accumulation of defective mitochondria (12). Indeed, Atg7 deficiency similarly altered progression of KRASG12D-driven non-small-cell lung cancers to oncocytomas (13).
Consistent with Atg7 deficiency altering tumor fate from adenomas to oncocytomas, the autophagy-deficient tumors accumulated significantly more mitochondria than their Atg7-expressing counterparts as measured by IHC for Tom20 (Fig. 2E). Electron microscopy (EM) revealed that the abundant mitochondria in the BrafV600E/+; Atg7−/− tumors were swollen and misshapen in contrast to the normal mitochondria observed in the BrafV600E/+; Atg7+/+ tumors. Moreover, mitochondria in the BrafV600E/+; Atg7−/− tumors were poorly functional, measured by markedly decreased enzymatic activity of cytochrome c oxidase, an essential component of the electron transport chain, compared to their Atg7-expressing counterparts (Fig. 2E). Reduced expression of Cox IV was observed in tumor lysates from Atg7-deficient tumors compared to those expressing Atg7, indicating that autophagy ablation results in a functional defect in the mitochondria of these tumors (Fig. 2E, F). Thus, autophagy deficiency is sufficient to alter tumor fate from adenomas to oncocytomas in BRAFV600E lung tumors similarly to those with KRASG12D mutations.
Atg7 deficiency extends lifespan of mice with BRAFV600E–driven lung tumors
Aggressive tumors, such as those driven by oncogenic H or KRAS are dependent on autophagy for survival (8, 9, 13, 15). In keeping with these findings and consistent with the conversion of Atg7-deficient tumors to a more benign fate, we observed that complete blockade of autophagy was sufficient to extend and nearly double the survival of mice with BRAFV600E driven lung tumors (p=0.017) (Fig. 2G). Mice with BrafV600E/+; Atg7+/+ tumors were the first to succumb, exhibiting a median survival of 16 weeks post Cre. In comparison, BrafV600E/+; Atg7−/− tumor-bearing mice exhibited a median survival of 31 weeks. Tumor-specific allelic loss of Atg7 or whole body allelic loss of the essential autophagy gene Beclin1, which significantly impairs but does not ablate autophagy (23) did not alter survival of mice bearing BRAFV600E lung tumors (Fig. 2G, H), suggesting that complete and not partial ablation of autophagy is necessary to suppress tumor growth and increase mouse survival (23). Of note, the median survival of the Beclin1+/− lung tumor bearing mice (15.1 weeks) was nearly identical to that of mice bearing Atg7+/− tumors (15.8 weeks). Importantly, the accumulation of the autophagy substrate p62 was accelerated in Braf V600E/+; Atg7−/− tumors in comparison to Braf V600E/+; Beclin1+/− tumors, consistent with a more profound autophagy defect induced by Atg7 deficiency (data not shown).
Increased survival of mice bearing Atg7−/− compared to Atg7+/+; BrafV600E/+ tumors was accompanied by reduced tumor cell proliferation assessed by nuclear Ki67staining, and increased p53, p21 and γ-H2AX levels at 14 weeks post Cre administration (Figs. 2I, S4, S5). Comparable numbers of cells in wild type and Atg7-deficient tumors were positive for senescence-associated β-galactosidase (Fig. S6A), eliminating senescence as the mechanism for growth arrest and reduced tumor burden with Atg7 deficiency at 14 weeks. Similarly, no significant changes in active caspase-3 were detected 14 weeks post Cre administration (Fig. S6B).
p53 induction in Atg7-deficient BrafV600E lung tumors limits tumor growth
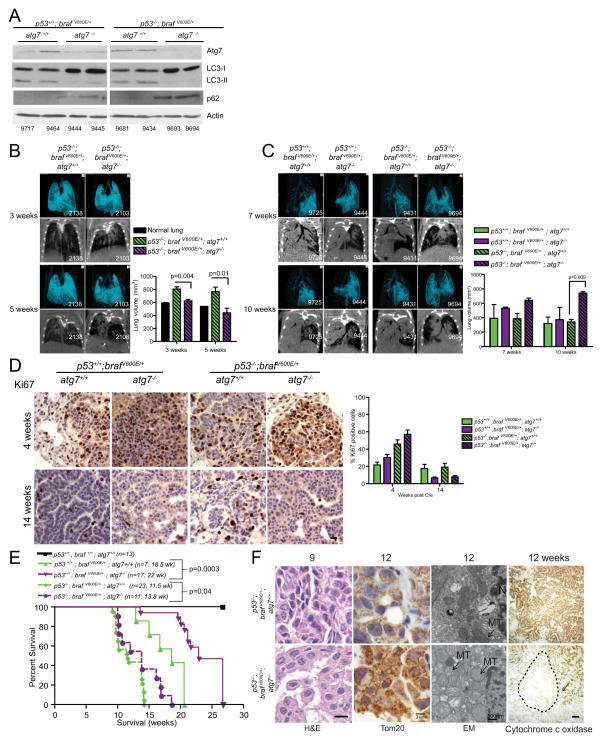
The induction of p53 and p21 in Atg7-deficient tumors led us to question whether the mechanism by which autophagy deficiency suppressed tumor proliferation was through induction of a p53 response. To test this hypothesis, we deleted Trp53 in Braf lung tumors with and without Atg7 and followed tumorigenesis in the resulting mice. Deletion of Atg7 in Trp53-deficient, BRAFV600E lung tumors caused a loss of ATG7 protein expression (residual ATG7 protein is to due the stromal component of the tumor that is not deleted for Atg7), the accumulation of LC3, and the autophagy substrate p62 indicative of autophagy inactivation (Fig. 3A). Ablation of Trp53 did not alter the increased early tumorigenesis of the Atg7-deficient tumors, shown as decreased normal/healthy lung volume in the quantitation of the micro-CT analysis at 3 and 5 weeks (Fig. 3B), or the eventual reduction in tumor burden, shown as increased normal/healthy lung volume at 10 weeks, compared to Atg7 wild type tumors (Fig. 3C). As observed in mice bearing Trp53-intact BRAFV600E-activated tumors (Fig. 2I), this phenotype was associated with a proliferative defect shown via reduction in nuclear Ki67 staining (Fig. 3D). Kaplan-Meier analysis demonstrated that autophagy deficiency extended the lifespan of tumor-bearing mice independent of p53 status. However, the kinetics are different; mice with Trp53-deficient tumors succumb to their tumors more rapidly than those with intact Trp53 (Fig. 3E). Thus, loss of ATG7 causes premature p53 induction in BRAFV600E lung tumors that suppresses proliferation and tumor growth. Atg7-deficiency suppresses growth of Trp53-deficient tumors in a statistically significant manner (p=0.04), revealing a second, Trp53 independent, growth suppressive mechanism in response to tumor-specific loss of Atg7. However, we note that Trp53 has a major role in suppressing tumor growth and promoting overall survival of mice bearing Atg7 deleted tumors.
Figure 3. Loss of p53 does not ablate the increased early tumorigenesis or life span extension of mice carrying Braf V600E/+; Atg7−/− tumors.
A. Western blot analysis of ATG7 expression, LC3-I to LC3-II conversion, and accumulation of p62 in tumor lysates from Trp53+/+; BrafV600E/+; Atg7+/+, Trp53+/+; BrafV600E/+; Atg7−/− and Trp53−/−; BrafV600E/+; Atg7+/+, Trp53−/−; BrafV600E/+; Atg7−/− 14 weeks post Cre. Numbers refer to individual mice.
B. Representative micro-CT images and 3D reconstructions of Trp53−/−; BrafV600E/+; Atg7+/+ and Trp53−/−; BrafV600E/+; Atg7−/− tumors at 3 and 5 weeks post Cre demonstrating increased tumor in the Trp53−/−; BrafV600E/+; Atg7−/− mice at early time points. Numbers refer to individual mice. Quantification of normal lung volume is shown at right.
C. Representative micro-CT images and 3D reconstructions of Trp53+/+; BrafV600E/+; Atg7+/+, Trp53+/+; BrafV600E/+; Atg7−/− and Trp53−/−; BrafV600E/+; Atg7+/+, Trp53−/−; BrafV600E/+; Atg7−/− tumors at 7 and 10 weeks post Cre demonstrating reduced tumor in the Trp53−/−; BrafV600E/+; Atg7−/− mice at late time points. Numbers refer to individual mice. Quantification of normal lung volume is shown at right.
D. Representative images of Ki67 staining of Trp53+/+; BrafV600E/+; Atg7+/+, Trp53+/+; BrafV600E/+; Atg7−/− and Trp53−/−; BrafV600E/+; Atg7+/+, Trp53−/−; BrafV600E/+; Atg7−/− tumors at 4 and 14 weeks post Cre. Scale bar= 10 μm Quantification is shown at right.
E. Kaplan Meier analysis of overall survival of Trp53+/+ and Trp53−/−; Braf V600E/+; Atg7+/+ and Braf V600E/+; Atg7−/− mice post Cre. Numbers of mice per group and median survival are indicated on graph. Note: this study was ended at 22 weeks once the p value was highly significant (p=0.0003).
F. Representative images of histology (H&E), Tom20 IHC, electron micrographs, and the enzymatic cytochrome c oxidase assay at the indicated time post Cre. For H&E, Scale bar=10 μm For Tom20, Scale bar= 2 μm. For EM, N=nucleus MT= mitochondria. Scale bar=500 nm. Arrows point to swollen mitochondria in Trp53−/−; Braf V600E/+; Atg7−/− tumors. For cytochrome c oxidase, arrows point to tumor regions, indicated by dashed lines, with defective mitochondria in the Trp53−/−; Braf V600E/+; Atg7−/− tumors. Scale bar =100 μm
Consistent with the emergence of oncocytomas upon Atg7 deletion in the Trp53 −/−; BrafV600E/+ lung tumors, these tumors were characterized by the accumulation of defective mitochondria (indicated by increased staining with the mitochondrial marker Tom20), ultrastructural mitochondrial abnormalities, and loss of enzymatic activity for cytochrome c oxidase (Fig. 3F). The fact that Trp53 deficiency did not influence the conversion of BRAFV600E-driven adenocarcinomas to oncocytomas induced by loss of Atg7 suggests that autophagy is critical to prevent the accumulation of defective mitochondria and maintain adenocarcinoma fate. Importantly, despite the early growth advantage provided by Atg7 deficiency, tumor growth was ultimately impaired, indicating that the requirement for autophagy to promote tumorigenesis is dominant. Therefore autophagy inhibition may be an important therapeutic tool to alter lung tumor progression to a more benign fate.
Atg7 deficiency induces Nrf2 accumulation in BrafV600E lung tumors
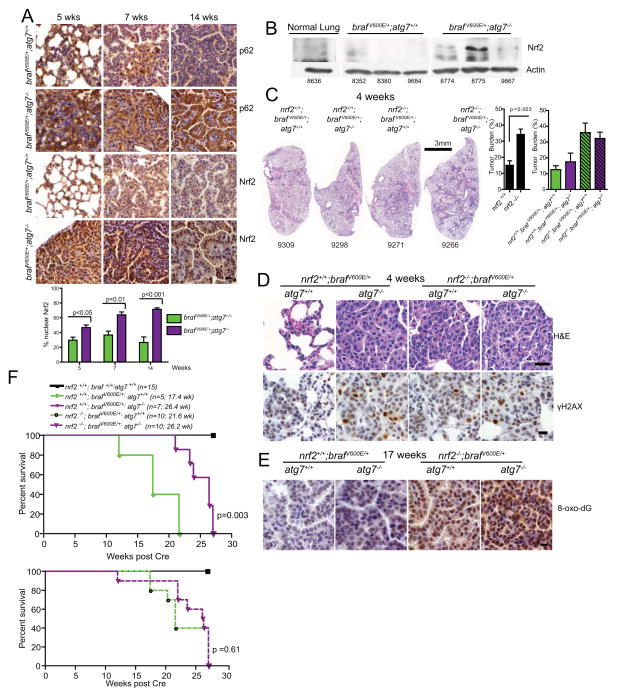
Having established opposing tumor growth suppressing and promoting roles for autophagy in BRAFV600E-driven lung tumors, we turned our attention to identifying the underlying mechanisms. One explanation for the early enhancement of tumor growth upon loss of Atg7 was that increased ROS generated by abnormal mitochondria was responsible. To test this hypothesis, we first examined the levels of the master regulator of antioxidant defense, NRF2, in BrafV600E/+; Atg7+/+ and BrafV600E/+; Atg7−/− tumors. NRF2 is normally ubiquitinated and degraded via the proteasome through its interaction with KEAP1 (24). Autophagy deficiency results in the accumulation of the autophagy substrate p62, which competes with NRF2 for binding sites on KEAP1. Thus, under autophagy deficient conditions, NRF2 accumulates and translocates to the nucleus where it activates the cellular antioxidant defense program (25–29). Indeed, induction of NRF2 was observed in Atg7-deficient tumors both by IHC examining levels of nuclear NRF2 and by Western blotting of tumor lysates (Fig. 4A, B). Induction of nuclear NRF2 correlated with p62 accumulation in Atg7-deficient tumors as expected (Fig. 4A). However, it was unclear whether this induction was caused by Atg7 deletion as a consequence of the accumulation of p62, or rather resulted from increased ROS production due to failure of mitochondrial quality control that also induces NRF2 (26). To resolve this question, we examined the functional consequences of Nrf2 loss in BRAFV600E-driven lung tumorigenesis in the presence or absence of Atg7.
Figure 4. Loss of Nrf2 phenocopies Atg7 deletion and is associated with increased lifespan.
A. Representative IHC staining for p62 and NRF2 in lung tumors of BrafV600E/+; Atg7+/+ and BrafV600E/+; Atg7 −/− mice at the indicated time post Cre. Scale bar=10 μm. Quantification is shown below. Error bars are SEM.
B. Western blot analysis of NRF2 accumulation in lysates of tumors harvested from BrafV600E/+; Atg7 +/+ and BrafV600E/+; Atg7−/− mice 10 weeks post Cre. Numbers refer to individual animals. A blank lane was intentionally included between the normal lung and tumor lysates. An additional Atg7 null tumor lysate was spliced out for aesthetic reasons.
C. H&E staining of single lung lobes showing dramatic increase in size and number of tumors in Nrf2−/− mice independent of autophagy status 4 weeks post Cre. Scale bar= 3 mm. Quantification of tumor burden shown at right. The Nrf2+/+ and Nrf2−/− graph is the aggregate of autophagy competent and deficient braf tumors. These data are plotted individually in the next graph. The difference in tumor burden between the Nrf2 −/−;Braf V600E/+; Atg7+/+ and Nrf2 −/−;Braf V600E/+; Atg7−/− mice is not statistically significant (p=0.63). The difference in tumor burden between the Nrf2 +/+;Braf V600E/+; Atg7+/+ and Nrf2 +/+;Braf V600E/+; Atg7−/− mice is not statistically significant in this experiment (p=0.49). Please refer to similar experiments in Figs. 2A–C, S3, and 3B.
D. Representative images of H&E and IHC analysis of γ-H2AX 4 weeks post Cre. Scale bar=10μm
E. Representative images of IHC for 8-oxo-dG 17 weeks post Cre. Scale bar= 10μm
F. Kaplan Meier analysis of overall survival of the Nrf2; Braf; Atg7 compound mutant mice. The number of mice per group and median survival are indicated. Note that the increased survival between the Nrf2−/−;Braf V600E/+; Atg7+/+ and Nrf2 +/+;Braf V600E/+; Atg7+/+ mice just misses statistical significance (p= 0.06).
Nrf2 deficiency enhances early and impedes late BRAFV600E lung tumor growth
Mice with Cre-activatible BrafV600Ewith and without conditional alleles of Atg7 were mated to Nrf2 knockout mice, tumorigenesis was induced in the lung via intra-nasal administration of adenoviral-Cre recombinase, and the resulting tumors were followed over time. Surprisingly, Nrf2 ablation caused a profound yet comparable increase in early tumor growth independent of autophagy status, indicating that the early tumor growth is driven by loss of anti-oxidant defense (Fig. 4C). Compound deficiency in both Nrf2 and Atg7 produced no additional increase in early tumor growth compared to loss of either gene alone (Fig. 4C). Stimulation of early tumorigenesis induced by Nrf2 deficiency was associated with activation of the DNA damage response (γ-H2AX positivity) consistent with elevated ROS production and oxidative stress (Fig. 4D). Compound loss of both Nrf2 and Atg7 failed to produce significant increases in the levels of phosphorylated γ-H2AX compared to loss of either alone, suggesting that they function in the same pathway (Fig. 4D). These findings are consistent with increased positivity for the ROS-specific DNA adduct 8-oxo-2′-deoxiguanosine (8-oxo-dG) in tumor sections upon loss of either or both Atg7 and Nrf2 but not in Atg7 and Nrf2 wild type tumors at 17 weeks post Cre administration (Figs. 4D, E).
Deficiency in Nrf2 phenocopies Atg7 loss in survival analyses
Despite the marked promotion of early tumorigenesis, loss of Nrf2 extended survival of mice in a similar manner to what was observed with Atg7 deletion previously: median survival of Nrf2 intact mice was 17.4 weeks compared to 21.6 weeks for the Nrf2 deleted mice (p=0.003) (Fig. 4F and 2G). Critically, there was no further increase in survival when both Atg7 and Nrf2 were deleted (p=0.61) indicating that Nrf2 ablation phenocopies Atg7 loss and that the two genes are epistatic to one another. Further strengthening this argument is the comparable survival of Nrf2+/+; BrafV600E/+; Atg7−/− (26.4 weeks) and Nrf2−/−; BrafV600E/+; Atg7 +/+ (21.6 weeks) mice (p= 0.55). Thus, loss of either Nrf2 or Atg7 or loss of both genes reduces tumor growth and increases survival as previously reported for Nrf2 deletion in BRAFV600E-induced lung tumors (30). Taken together, this suggests that although oxidative stress through loss of antioxidant defense or defective autophagy, or both, stimulates early tumor growth this early promotion of tumor cell proliferation is not sustained and eventually results in a tumor growth defect.
Loss of Atg7 impairs mitochondrial metabolism and survival during starvation
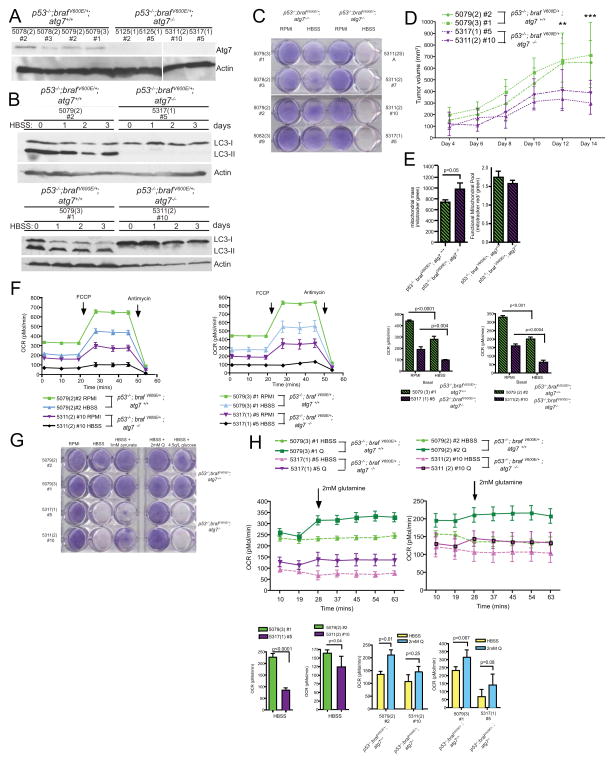
We hypothesized that the autophagy-deficient tumors eventually fail to thrive because they evolve into oncocytomas that are metabolically compromised. To test this hypothesis, we generated Atg7-wild type and -deficient Trp53−/−; BrafV600E/+ tumor-derived cell lines (TDCLs) and directed our efforts toward the identification of the metabolic defects responsible for the blunted tumorigenesis observed in the autophagy-deficient setting. Western blot analysis confirmed deficiency in ATG7 in the Atg7−/− TDCLs but not in those derived from Atg7+/+ tumors (Fig. 5A). During starvation in HBSS, the autophagy block in the Atg7−/− TDCLs manifested as a persistent accumulation of LC3-I that was not processed to LC3-II (Fig. 5B), confirming that Atg7−/− TDCLs were autophagy deficient (Fig. 5B).
Figure 5. Loss of Atg7 impairs mitochondrial metabolism and survival during starvation.
A. Western blot analysis of ATG7 expression in TDCLs isolated 9 weeks post Cre from Trp53−/−; BrafV600E/+; Atg7 +/+ or Trp53−/−; BrafV600E/+; Atg7 −/− mice. Numbers refer to individual clones. An additional Atg7 null tumor lysate was spliced out for aesthetic reasons.
B. Western blot analysis of conversion of LC3-I to LC3-II during a 3 day HBSS starvation time course. Numbers refer to individual clones. Two pairs of ATG7 wild type and ATG7 null derived TDCLs are shown.
C. Clonogenic assay of autophagy-competent or -deficient TDCLs following 3 days of HBSS starvation and 4 day recovery in normal growth media.
D. Allograft growth of Trp53−/−; BrafV600E/+; Atg7 −/− and Trp53−/−; BrafV600E/+; Atg7 +/+ TDCLs. n=8 tumors from each genotype were monitored.
E. FACS analysis of Total Mitochrondrial Mass and Relative Mitochondrial Membrane Potential (MMP) of TDCLs.
F. Seahorse measurement of basal and reserve oxygen consumption rates (OCR) of autophagy-competent and -deficient TDCLs under normal growth conditions or following 4hr HBSS starvation.
G. Clonogenic survival of TDCLs incubated with RPMI, HBSS, or HBSS supplemented with exogenous glutamine (2mM), sodium pyruvate (1mM) or glucose (4.5g/L) during 3 day HBSS starvation and 4 day recovery in normal growth media.
H. Seahorse measurement of OCR in the TDCLs incubated with HBSS before and after the addition of 2mM glutamine (Q). Injection timing is indicated by arrow.
Deficiency in Atg7 dramatically impaired both starvation survival and tumor growth in nude mice compared to the Atg7 wild type counterparts (Fig. 5C, D) recapitulating the findings from the genetically engineered mice (Fig. 2G). Consistent with the mitochondrial defects observed with Atg7 deficiency in tumors in the genetically engineered mice, the autophagy-deficient TDCLs possessed greater numbers of mitochondria yet had a smaller fraction of functional mitochondria than their wild type counterparts (Fig. 5E). Next, we examined the oxygen consumption rates (OCR) of autophagy-competent and -deficient TDCLs maintained in normal growth media (RPMI) or following short-term starvation with HBSS (4 hours) as a first approximation of their metabolic fitness. Basal and starvation-induced OCR levels of the autophagy-deficient TDCLs were significantly lower than the rates measured with the autophagy-competent TDCLs (Fig. 5F). Importantly, the mitochondrial reserve capacity (OCR post-injection of the uncoupler Trifluorocarbonylcyanide Phenylhydrazon FCCP) of the autophagy-deficient TDCLs was nearly absent following starvation, lending further support to the hypothesis that autophagy supplies metabolites which maintain mitochondrial function (Fig. 5F).
We next identified the metabolic component supplied by autophagy that enabled survival during starvation. Addition of exogenous glutamine or to a lesser extent sodium pyruvate, but not glucose, or the ROS scavenger N-acetyl-cysteine (NAC), was sufficient to rescue survival of the Atg7-deficient TDCLs in response to starvation (HBSS) in a clonogenic assay (Figs. 5G and S7). This suggests that autophagy deficiency renders TDCLs glutamine-dependent. The inability of NAC to protect Atg7-deficient TDCLs from starvation-induced death suggests that loss of viability does not result from excess ROS production but rather resulted from metabolic insufficiency (Fig. S7). OCR analysis revealed that the autophagy-deficient TDCLs were capable of utilizing glutamine to turn the tricarboxylic acid (TCA) cycle, resulting in an increased OCR compared to glutamine starved conditions, although they were not as efficient at doing so as their autophagy competent counterparts (Fig. 5H). Taken together this data suggests that the failure of the late stage Atg7-deficient tumors to thrive is due both to an impairment of overall mitochondrial function and substrate limitation (particularly glutamine) of the remaining functional mitochondria, which together lead to metabolic crisis that is incompatible with continued tumor growth.
Discussion
The role of autophagy in cancer is complex; it can either suppress or promote tumorigenesis depending on cellular context (1, 15). Whether these differential effects of autophagy are due to the artificiality of the models examined or occur in human cancer, remain to be determined. Factors with the potential to influence the role of autophagy in cancer include tissue type, nature of the driver mutations, the metabolic demands imposed by growth and proliferation, level of inherent metabolic stress as well as the particular metabolic pathways engaged. It is clear that physiologic, genetic, and in vivo approaches are needed to resolve these issues.
To begin to address the context-dependent role of autophagy in cancer, we ablated Atg7 in the Cre-activatible mouse model of BRAFV600E-driven lung tumorigenesis that normally results in benign tumors (21). We demonstrated that defective autophagy through loss of Atg7 and the concomitant loss of mitochondrial quality control that increases oxidative stress, promotes early tumorigenesis, revealing a potential mechanism by which autophagy can limit tumor growth. Persistent mitochondrial dysfunction, however, eventually overshadows this initial tumor growth advantage, resulting in metabolic impairment, reduction in tumor burden, the emergence of oncocytomas, and an increase in the overall mouse survival.
Several important points emerge from these studies. First, Atg7 deficiency in the context of BRAFV600E-driven lung tumorigenesis promotes early tumor growth. This initial stimulation of tumor cell proliferation was associated with increased oxidative stress suggesting that it may be a ROS-mediated event. Similarly, loss of the master regulator of antioxidant defense, Nrf2, initially promoted proliferation of BRAFV600E-driven lung tumors, and loss of both Atg7 and Nrf2 provided no additional growth advantage. This suggests that oxidative stress from loss of either autophagy or antioxidant defense may stimulate early BRAFV600E lung tumor growth and that Atg7 and Nrf2 are genetically epistatic.
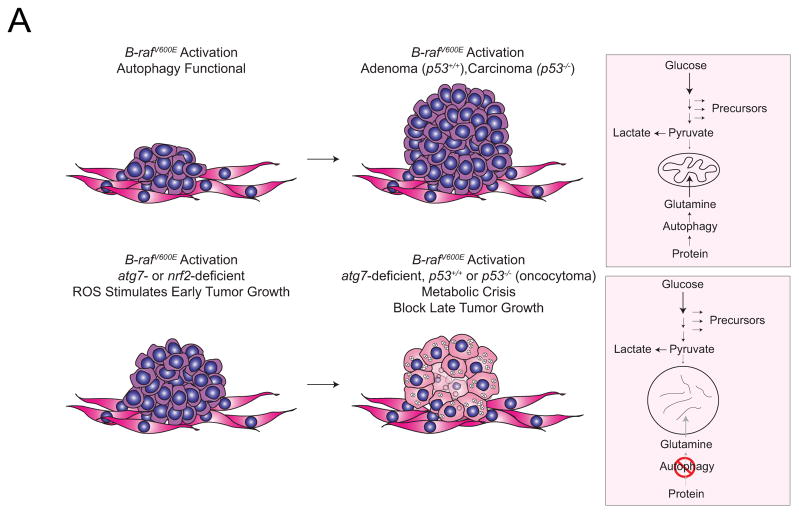
It is well established that ROS induces genomic damage and mutations that can promote cancer, and that autophagy deficiency increases ROS levels, mutation rates, and genome instability (6, 7). However, these consequences of ROS production are unlikely to be playing a role here since the growth promotion induced by Atg7 or Nrf2 loss is temporary. ROS activates oncogenic signaling pathways including HIF, Wnt/β-catenin, Notch, and ERK and it is more likely that these signaling events are responsible for the transient growth stimulation observed in the BRAFV600E lung tumors deficient for Atg7 or Nrf2. It remains to be seen whether the eventual diminution of tumor burden observed with Atg7 or Nrf2 loss corresponds to the progressive deterioration of mitochondrial function and loss of ROS production. We propose that autophagy deficiency results in the accumulation of malfunctioning mitochondria giving rise to oxidative stress (as is the case with loss of Nrf2-mediated antioxidant defense) that initially stimulates the growth of BRAFV600E-driven lung tumors by activating growth promoting signaling pathways. The potential signaling events activated in this setting remain to be determined, although no difference was observed in the levels of phospho-ERK in Atg7 wild type and deficient BrafV600E lung tumors (data not shown). Alternatively, insufficient levels of autophagy-supplied substrates, such as glutamine from protein degradation, may impair mitochondrial metabolism, ROS production and metabolic fitness (Fig. 6).
Figure 6. Role of Atg7 in the growth of BrafV600E-driven lung tumors.
See text for details.
Second, autophagy is required for sustaining the growth of BRAFV600E-driven lung tumors and for dictating their fate. Specifically, autophagy ablation shifts tumor progression from adenomas (Trp53 intact tumors) and adenocarcinomas (Trp53 null tumors) to oncocytomas. This data suggests that altering tumor fate to benign oncocytomas by inhibiting autophagy could be a novel therapeutic approach to BRAFV600E-driven lung cancer and may extend to other tumors driven by MAP kinase signaling. Importantly, deficiency in either of the essential autophagy genes Atg7 or Atg5 in RAS-transformed cancer cell lines impairs tumor growth and causes the accumulation of defective mitochondria (8, 9) as seen here for BRAFV600E-induced tumors. Thus, the oncocytoma phenotype and defective tumor growth are a consequence of defective autophagy and not related to any autophagy-independent function of Atg7. Although Atg7 deficiency also alters the fate of KRASG12D-driven adenomas and carcinomas to oncocytomas (13), in BRAFV600E lung tumors this occurs more rapidly and confers a greater survival advantage, suggesting that MAP kinase signaling may be a key driver for autophagy-dependence. It will be of great interest to test the role of autophagy in other BRAFV600E driven malignancies. It will also be very important to determine whether acute autophagy ablation is sufficient to convert established tumors to oncocytomas.
Production of oncocytic tumors by autophagy ablation was associated with reduced proliferation indicating that cell cycle inhibition was the main cause of reduced tumor burden. Enhanced p53 induction caused by Atg7-deficiency contributed to but was not essential for the proliferative arrest in response to Atg7 deficiency, revealing both p53-dependent and p53-independent mechanisms by which autophagy promotes tumor cell proliferation as is the case with KRASG12D-driven lung tumors (13). Autophagy deficiency (either ablation or allelic loss) activates the DNA damage response (6, 23). Recently, allelic loss of Beclin1 was shown to activate a p53-mediated block to mammary tumorigenesis (31). Tumors apparently activate autophagy to suppress the p53 response as part of a tumor-promotion mechanism. These findings are consistent with autophagy being of central importance for maintenance of mitochondrial metabolism in RAS-driven tumors (8–10, 13) and extend the generalizability of the phenomenon to include BRAFV600E-driven lung tumors and possibly those driven by other oncogenic events (32).
Third, we identify a potential mechanism to explain the eventual growth defect in Atg7-deficient BRAFV600E-driven lung tumors. Autophagy was originally identified in yeast as a pathway required for survival to nitrogen starvation (33). Mammals also require autophagy to survive starvation (34) as is the case with many cancer cells (1). Autophagy degrades intracellular components to recycle them in metabolic and biosynthetic pathways in times of deprivation (16). Indeed, autophagy deficient BRAFV600E TDCLs were impaired in tumorigenesis, and failed to survive starvation but were dramatically rescued by glutamine supplementation. Thus, Atg7 deficiency causes dependency on exogenously supplied glutamine likely due to the absence of internally supplied glutamine recycled from autophagy-mediated protein degradation. Thus, autophagy addiction may just be a means to provide the tumor cells with glutamine. It will be important to determine whether the Atg7-deficient tumor cells are metabolically exhausted due to substrate limitation, or due to dysfunctional mitochondria, or both. Moreover, it will be important to determine whether the metabolic frailty of Atg7-deficient TDCLs in vitro is the direct cause of defective tumor growth in vivo.
Deletion of Atg7 in Trp53-deficient and KRASG12D activated lung tumors resulted in significant inflammation causing pneumonia, and an accumulation of defective mitochondria and neutral lipids secondary to impaired fatty acid oxidation (13). We did not observe inflammation or lipid accumulation with deletion of Atg7 in the Trp53-deficient, BRAFV600E-activated lung tumors or in TDCLs in vitro; nor did we see the appearance of lipidic cysts in tumor allografts (Fig. S8 and data not shown) suggesting a fundamental difference in metabolism and mitochondrial utilization distinguishing KRASG12D and BRAFV600E lung tumors. Indeed, in melanoma, BRAFV600E activates pyruvate dehydrogenase promoting pyruvate utilization in the TCA cycle, which drives oncogene-induced senescence (35). It is also well established in melanoma models that BRAFV600E suppresses mitochondrial biogenesis via regulation of the transcription factor MITF and peroxisome proliferator-activated receptor gamma, coactivator 1 alpha (PGC1α (36). Mitochondrial quality control may be critically important for BrafV600E malignancies given the context of an already reduced pool of functioning mitochondria undergoing enhanced respiration due to increased pyruvate utilization. Taken together, this work provides valuable insights into where and how autophagy should be modulated in BRAFV600E-driven lung tumors and lends further support to the clinical utility of autophagy inhibition for cancer therapy.
Materials and Methods
Animal husbandry and infection
All experiments were conducted in compliance with IACUC guidelines. Braf CA/CA mice were bred to Atg7flox/flox (provided by Dr. M. Komatsu) or Beclin1+/− (provided by Dr. Z. Yue) to generate BrafCA/+; Atg7 +/+, BrafCA/+; Atg7 −/− and BrafCA/+; Beclin1 +/+ or BrafCA/+; Beclin1 +/− mice. Compound Braf; Atg7 mice were bred to Trp53 lox/flox (Jackson Laboratory) or Nrf2−/− (YW Kan) generating triple mutant mice. For adenoviral Cre infection, mice 6–8 weeks of age were anesthetized with isoflourane, and high titer replication defective adenoviral-Cre (4× 107 pfu/mouse) (University of Iowa Gene Vector Core) was administered intra-nasally as a calcium precipitate. Mice were monitored until fully recovered from the anesthetic before returning to their housing. For allograft studies of the TDCLs, 107 cells were injected bilaterally into the flank of 6 week old nude mice (Taconic). Tumor size was measured every 2 days. Mice were sacrificed at 14d post injection. Tumors were fixed for histology as described below.
Micro-CT analysis
Respiratory gated, low resolution CT images of maximally ventilated mice were obtained using a Siemens Inveon PET/CT and the INVEON Acquisition Workplace software. Using INVEON Research Workplace software, reconstructed data were processed through a gaussian filter and tissue determined to be pulmonary was segmented and its volume calculated. 3D reconstructions of normal lung tissue and transverse sections were generated using Osirix software.
Antibodies
The following antibodies were used for Western blotting or IHC analysis: ATG7 (Sigma, Cat#: A2856), Surfactant C protein (Seven Hills Bioreagents, Cat#: WARAB-SPC), LC-3 (Nano Tools, Cat#: LC3-5F10), p62 (antisera raised against MBP full length p62 (8) or purchased from Enzo Life Sciences PW9860-0100), Ki67 (Abcam., Cat#: ab-15580), p53 (Leica, Product Code: NCL-p53-CM5P), p21 (BD Biosciences, Cat#: 56431), active caspase-3 (Cell Signaling, Cat#: 9661), γ–H2AX (Cell Signaling, Cat#: 2577), Nrf2 (Epitomics, Cat#: 2178-1), Tom20 (Santa Cruz Biotechnology, Cat# sc-11415, Cox-IV (Molecular Probes), 8-oxo-dG (clone 2E2, Trevigen), phospho-Erk (Cell Signaling Cat #4376), NRF2 (Epitomics Cat #2178) Beta actin (Calbiochem Cat #CP01).
Histology
For paraffin sections, lungs were dissected into individual lobes, washed 2x in PBS, then fixed for 4h at RT in 10% buffered formalin solution (Formaldefresh, Fisher Scientific) prior to transfer to 70% EtOH and sectioning. For frozen sections, lung tissue was fixed in 10% formalin, transferred to 15% sucrose for 2 hours before final transfer to 30% sucrose and processing.
Cytochrome C Oxidase Staining
A modified Nadi reaction was conducted as follows. Unfixed frozen sections were stained in Incubating Solution (75mg sucrose in 7.5ml water, 0.2M phosphate buffer pH 7.6, 5mg 3′ diaminobenzidine tetrachloride, 10 mg cytochrome c, 20ug catalase) for 1hr at RT. Catalase (C-10), Cytochrome C (C-2506), and DAB (D-5637) were purchased from Sigma. Slides were washed three times in deionized water, dehydrated in ascending alcohols (50%, 70%, 80%, 95% twice, then 100% twice) prior to mounting and imaging.
Senesence-Associated βGalactosidase Assessment
The β-galactosidase staining was performed per manufacturer’s recommendations (Cell Signaling Cat#9860). Briefly, sections were fixed for 10mins in 1X fixative, washed twice with PBS before incubation with β-galactosidase staining buffer overnight at 37C in the absence of CO2. Samples were briefly counterstained with Eosin to facilitate visualization of tissue.
TDCLs
TDCLs were generated from p53−/−; Braf V600E/+ tumors with or without atg7 at 9 weeks post Cre. TDCLs were cultured in RPMI 1640 medium containing 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, 1 mM sodium pyruvate, and 1 mM sodium bicarbonate at 38.5°C with 8.5% CO2. Atg7 status in these lines was confirmed by PCR and western blot. TDCLs were further characterized for LC3 and p62 accumulation following starvation prior to usage. No additional authentication was performed.
Bodipy staining
0.6 ×106 cells were plated on coverslips in 6 well plates, starved for 24h in HBSS. After starvation, cells were rinsed twice with PBS prior to incubation with Bodipy (1mg/ml) for 15 min and counterstained with DAPI. Slides were rinsed in water and mounted with Prolong Anti-fade Gold and imaged.
Western Blotting
Tumor pieces were snap frozen, then ground in liquid nitrogen with mortar and pestle. Material was lysed in RIPA buffer, and assessed for protein concentration with the Bio-Rad BCA reagent and analyzed via SDS-PAGE gel electrophoresis. For TDCLs, whole cell lysates were prepared as described previously (5).
Mitochondrial Mass and Potential Measurements
Live TDCLs were co-labeled with 25nM Mitotracker-Red CMXRos (Molecular Probes/Invitrogen) to assess Mitochondrial Membrane Potential (MMP) and 25nM Mitotracker –Green FM (Molecular Probes/Invitrogen) to measure mitochondrial mass for 30 mins under normal growth conditions. Post labeling, samples were washed twice in growth medium, trypsinized and analyzed by flow cytometry (BD Influx Cell Sorter, BD Biosciences). The mean mitochondrial mass fluorescence is shown. Relative MMP was calculated by dividing the mean MMP fluorescence (red) with the mean of mitochondrial mass fluorescence (green).
Oxygen consumption rate measurement
OCR rates were measured with the Seahorse Biosciences extracellular flux analyzer (XF24) as described previously (8). 0.05 × 106 cells were seeded per well in normal growth media (RPMI with 10% FBS) and allowed to attach 24–30h at 38.5°C and 8.5% CO2 prior to study. Basal measurements were collected following 4h incubation with either RPMI or HBSS. SRC (maximal respiratory capacity-basal respiration rate) and total reserve capacity were measured by injecting the mitochondrial uncoupler FCCP (1.5μM) and Complex III inhibitor Antimycin (20 μM) from the XF24 ports as indicated. For glutamine utilization experiments, cells were plated as above, incubated with HBSS for 30 minutes, transferred to the Seahorse for analysis where l-glutamine (2mM) was injected via the XF24 port. Measurements were collected pre/post glutamine addition as indicated.
Electron microscopy
Tumors were fixed in 2.5% gluteraldehyde / 4% paraformaldehyde/8 mM calcium chloride in 0.1M cacodylate buffer pH 7.4. Samples were stored at 4C overnight prior to delivery for further processing. They were post-fixed in buffered 1% osmium tetroxide and subsequently dehydrated in a graded series of acetone prior to embedding in epon resin. 90nm thin sections were cut on a Leica EM UC6 ultra-microtome. Sectioned grids were stained with saturated solution of uranyl acetate and lead citrate. Images were captured with an AMT XR41 digital camera at 80 Kv on a JEOL 1200EX transmission electron microscope as described previously (5).
Clonogenic Survival Assays
0.07× 106 TDCL were seeded in 12 well plates in normal growth media and allowed to attach overnight. The cells were washed twice with PBS prior to incubation with either normal growth media, HBSS, or HBSS supplemented with 1mM sodium pyruvate, 2mM L-glutamine, 4.5g/L glucose, or 4mM NAC as indicated for 3 days. Following starvation, cells were allowed to recover in normal growth media for 4 days before methanol fixation and Giemsa staining. For Figure S7, cells were starved for 3 days and allowed to recover in normal media for 3 days prior to fixation and staining.
Statistics
Statistical analyses were carried out with GraphPad Prism version 5.0. Statistical significance was calculated by 2-way ANOVA with Bonferroni post-test. Significance in the Kaplan-Meier analyses was calculated using the log-rank test. For quantification of tumor burden from IHC slides, specimens were digitized with a Trestle MedScan whole slide scanner. An automated image processing protocol was built on Matlab 2011b. Analysis was performed on whole slide map of each specimen to produce tumor area and total tissue area measurements. Tumor and whole tissue masks were created for each slide. The segmentation masks were used for generation of ratios of tumor burden. For quantitation of IHC a minimum of 200 cells were scored from multiple images for each genotype.
Supplementary Material
Statement of Significance.
The essential autophagy gene Atg7 functions to promote BRAFV600E-driven lung tumorigenesis by preserving mitochondrial glutamine metabolism. This suggests that inhibiting autophagy is a novel approach to treat BRAFV600E-driven cancers.
Acknowledgments
This work was supported by grants from the NIH (R37 CA53370, R01 CA130893, RO1 CA163591, RCI CA147961 to EW and CA131261 to MM) and a postdoctoral fellowship from NJCCR to AMS (09-2406-CCR-E0)
We thank M. Komatsu for the conditional Atg7 mice, Z. Xue for the Beclin1+/− mice, and Y. W. Kan for the Nrf2 knockout mice. We also thank R. Patel for the electron microscopy, N. Campbell for micro-CT, W. Chen for histology quantification, A. Roberts for FACS analysis, CINJ Tissue Analytic Services staff, P. Chin for technical assistance, and members of the White lab for helpful discussions.
Abbreviations
- IHC
Immunohistochemistry
- ROS
Reactive Oxygen Species
- NSCLC
Non Small Cell Lung Cancer
- TDCLs
tumor derived cell lines
- NAC
N-acetyl-cysteine
- Q
glutamine
- OCR
Oxygen consumption rate
- FCCP
Trifluorocarbonylcyanide Phenylhydrazone
- TCA cycle
The Citric Acid Cycle
- EM
Electron Microscopy
- PCR
Polymerase Chain Reaction
Footnotes
The authors disclose no potential conflicts of interest
References
- 1.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–10. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, et al. Autophagy-deficient mice develop multiple liver tumors. Genes & development. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes & development. 2007;21:1621–35. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes & development. 2007;21:1367–81. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes & development. 2011;25:460–70. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes & development. 2011;25:717–29. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, et al. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Molecular biology of the cell. 2011;22:165–78. doi: 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasparre G, Porcelli AM, Lenaz G, Romeo G. Relevance of mitochondrial genetics and metabolism in cancer development. Cold Spring Harb Perspect Biol. 2013:5. doi: 10.1101/cshperspect.a011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasparre G, Romeo G, Rugolo M, Porcelli AM. Learning from oncocytic tumors: Why choose inefficient mitochondria? Biochim Biophys Acta. 2011;1807:633–42. doi: 10.1016/j.bbabio.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 13.Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes & development. 2013;27:1447–61. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iommarini L, Calvaruso MA, Kurelac I, Gasparre G, Porcelli AM. Complex I impairment in mitochondrial diseases and cancer: parallel roads leading to different outcomes. Int J Biochem Cell Biol. 2013;45:47–63. doi: 10.1016/j.biocel.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 15.Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, et al. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17:654–66. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–8. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White E, Karp C, Strohecker AM, Guo Y, Mathew R. Role of autophagy in suppression of inflammation and cancer. Curr Opin Cell Biol. 2010;22:212–7. doi: 10.1016/j.ceb.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 19.Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, et al. High frequency of BRAF mutations in nevi. Nature genetics. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 20.Puxeddu E, Moretti S, Elisei R, Romei C, Pascucci R, Martinelli M, et al. BRAF(V599E) mutation is the leading genetic event in adult sporadic papillary thyroid carcinomas. The Journal of clinical endocrinology and metabolism. 2004;89:2414–20. doi: 10.1210/jc.2003-031425. [DOI] [PubMed] [Google Scholar]
- 21.Dankort D, Filenova E, Collado M, Serrano M, Jones K, McMahon M. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes & development. 2007;21:379–84. doi: 10.1101/gad.1516407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–75. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–9. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, et al. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol. 2011;193:275–84. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nature cell biology. 2010;12:213–23. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 27.Kwon J, Han E, Bui CB, Shin W, Lee J, Lee S, et al. Assurance of mitochondrial integrity and mammalian longevity by the p62-Keap1-Nrf2-Nqo1 cascade. EMBO Rep. 2012;13:150–6. doi: 10.1038/embor.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, et al. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30:3275–85. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li QK, Singh A, Biswal S, Askin F, Gabrielson E. KEAP1 gene mutations and NRF2 activation are common in pulmonary papillary adenocarcinoma. J Hum Genet. 2011;56:230–4. doi: 10.1038/jhg.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–9. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huo Y, Cai H, Teplova I, Bowman-Colin C, Chen G, Price S, et al. Autophagy opposes p53-mediated tumor barrier to facilitate tumorigenesis in a model of PALB2-associated hereditary breast cancer. Cancer discovery. 2013 doi: 10.1158/2159-8290.CD-13-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei H, Wei S, Gan B, Peng X, Zou W, Guan JL. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes & development. 2011;25:1510–27. doi: 10.1101/gad.2051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS letters. 1993;333:169–74. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 34.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 35.Kaplon J, Zheng L, Meissl K, Chaneton B, Selivanov VA, Mackay G, et al. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature. 2013;498:109–12. doi: 10.1038/nature12154. [DOI] [PubMed] [Google Scholar]
- 36.Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer cell. 2013;23:302–15. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.