Abstract
Single-cell imaging mass spectrometry (IMS) is a powerful technique used to map the distributions of endogenous biomolecules with sub-cellular resolution. Currently, secondary ion mass spectrometry is the predominant technique for single-cell IMS, thanks to its sub-micron lateral resolution and surface sensitivity. However, recent methodological and technological developments aimed at improving the spatial resolution of matrix assisted laser desorption ionization (MALDI) have made this technique a potential platform of single-cell IMS. MALDI opens the field of single-cell IMS to new possibilities, including single cell proteomic imaging and atmospheric pressure analyses; however, sensitivity is a challenge. In this report, we estimate the availability of proteins and lipids in a single cell and discuss strategies employed to improve sensitivity at the single-cell level.
Keywords: SIMS, MALDI, nanoSIMS, Single-cell, Imaging Mass Spectrometry
Introduction
There is a fundamental flaw associated with analytical measurements of cell ensembles; the chemistry of an individual cell is often lost in the average chemistry of the cell population.[1,2] The same principle can be applied to the sub-cellular environment, as chemical heterogeneities within a single cell may be lost with single-cell analyses. Fortunately, single-cell imaging mass spectrometry (IMS) can elucidate intercellular chemical heterogeneity. The techniques ability to elucidate fundamental cellular processes by connecting chemical distributions with cell morphology makes it a valuable tool for molecular biology, cancerology, and pharmacology.
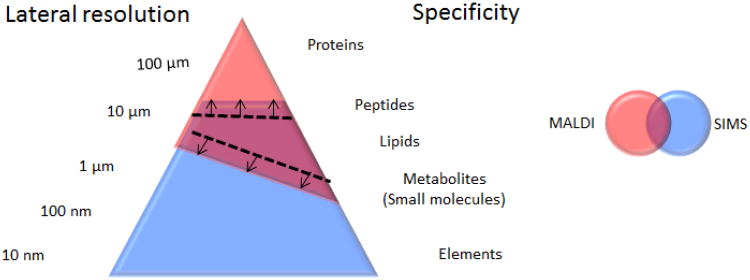
IMS is a popular and rapidly growing bio-analytical method that encompasses a number of ionization techniques, including desorption electrospray ionization (DESI) matrix-assisted laser desorption ionization (MALDI) and secondary ion mass spectrometry (SIMS).[3-7] SIMS is a label-free, matrix-free technique that utilizes a focused primary ion beam to desorb and ionize analyte molecules. The high surface sensitivity and sub-micron lateral resolution of the technique make it ideal for single- cell imaging. [8,9] However, recent methodological and technological developments in the field of MALDI have pushed its spatial resolution from the 20-100 μm range to the 1-10 μm range (Figure 1). With the average mammalian cell having a diameter of approximately 20 μm, this advancement has allowed MALDI to cross the barrier from single-cell ‘profiling’ to single-cell ‘imaging’. In this report, we examine the progress and the potential advantages of the SIMS and MALDI platforms in single-cell IMS.
Fig. 1.
The overlap between MALDI and SIMS methodologies is growing, in part by the various strategies independently developed to improve the lateral resolution of MALDI and extend the dynamic range of SIMS.
Sensitivity challenge
Sensitivity is a two-fold challenge in single-cell IMS. First, there is a sensitivity limitation related to the nature of the sample itself. A single cell contains a diverse population of biomolecules with varying abundances. A typical mammalian cell is about 1 to 20 pL in volume and the abundance of a particular biomolecule can range from a few to a few million copies per cell.
In order to elucidate the demand single-cell analysis places on mass spectrometry detection, we have estimated the abundance of proteins and lipids in a single mammalian cell (Table 1). Based on a comprehensive proteomic map of the mammalian cell line, U2OS, reported by Beck et al., the most abundant cellular proteins were present within a range of 4.5 × 103 to 2.5 × 106 copy numbers per cell.[10] Similarly, lipid abundances were extracted from data collected the murine cell line RAW 264.7 by LipidMAPS.[11] When directly compared, these data reveal that the most abundance lipid species is approximately 10 times more abundant than the most abundance protein species. Therefore, even with an attomole detection limit, a reasonable limit of detection (LOD) for most commercial mass spectrometers, only the most abundant proteins and lipids are detectable.
Table 1. The estimated number and concentration of various biomolecules in a single mammalian cell. The concentration was estimated based on a homogeneous distribution of the biomolecules in a 4.2-picoliter cell volume (20-μm sphere).
| ‘Omics | Species | Units per cell | Concentration |
|---|---|---|---|
| Lipidomic* | PC(36:1) | 200 attomole | 50 μM |
| Cholesterol | 4.3 femtomole | 1.0 mM | |
|
|
|||
| Proteomic** | High abundance proteins | 33 attomole | 1.65 μM |
| Low abundance proteins | 830 yotomole | 198 pM | |
Information taken from LipidMAPS. The data have been pooled from the control cases at various time points from the KDO2-Lipid A experimental dataset, available at LipidMAPS.org.[11] To estimate the phosphocholine contribution, data have been pooled from 35 glycerophoscholine and 16 sphingomylin species. The lipidomic information is reported in pmol per microgram of DNA, we assume 6 pg of DNA per diploid cell.
Information obtained from proteomiccommons.org. [10]
The second major challenge is the inherent limitation associated with the imaging modality itself. Sensitivity declines with increasing lateral resolution as reducing the analysis area or pixel diameter to improve the lateral resolution reduces the amount of material available to measure. McDonnell and coworkers calculated the biomolecule concentration would have to be in the millimolar (mM) range in order to be detected, assuming an attomole limit of detection.[12] Again, this restricts detection to the most abundant biomolecules or localized regions of high concentration.
Improving the sensitivity of ToF-SIMS and its impact on single cell-IMS
There are two regimes in SIMS, static and dynamic SIMS. These two regimes have independently developed methods to improve the sensitivity of SIMS.
Static SIMS
Static SIMS instruments typically employ a pulsed ion source and a ToF mass analyzer. The development of cluster ion sources has significantly improved the sensitivity of ToF-SIMS. The advantage of cluster sources is two-fold; cluster ion beams improve the molecular ion yields of organic biomolecules and reduce the damage accumulation cross-section, allowing molecular information to be retained during depth profiling.[13*, 14**, 15] The energy from cluster projectiles is deposited closer to the surface with minimal damage to the sub-surface compared to traditional atomic ion sources. The reduced altered layer improves the retention of molecular information during sputtering and allows molecular depth profiling and 3D mass spectrometry imaging to be carried out. This is a unique characteristic of SIMS-based IMS.
The sample preparation step provides a valuable opportunity to improve sensitivity by pre-treating the sample before analysis. A variety of matrices, including organic acids, ionic liquids and deposited metals, have been used to improve bimolecular ion yields. In addition, washing procedures designed to remove salts (i.e. ammonium formate) have improved biomolecule signals by mitigating saltrelated ion suppression effects. Unfortunately, this step is also susceptible to artifacts, as perturbing the sample can introduce variances into the analysis.[16*]. The need to introduce and maintain the cells in a vacuum environment is an unfortunate drawback of SIMS analyses. The development of flash freezing methods combined with analyses of cells in a frozen hydrated state, not only preserves the integrity of the cell, but also improves secondary ion yields compared to freeze dried cells.[17]
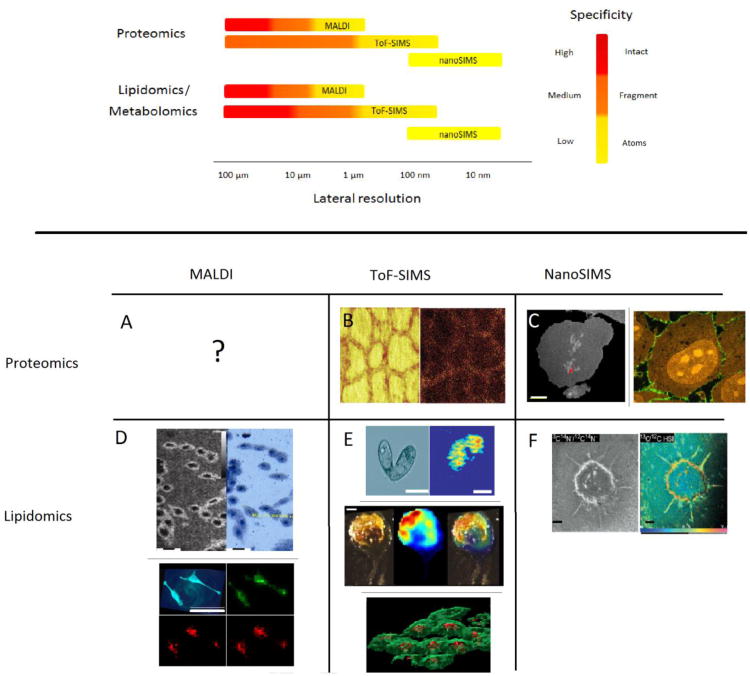
Sensitivity and image contrast have been improved by employing data processing procedures to reduce the dimensionality of the dataset. One of the most common techniques is principal component analysis (PCA). In this technique, mass spectral peaks are compiled based on commonalities in the variance. This method has been successful in differentiating cell types in a heterogeneous culture.[18] Similarly, the chemical signature of vitamin E obtained from an in situ tandem MS spectrum has been used to extract and compile vitamin E-related fragment peaks from a ToF-SIMS image of an Aplysia neuron (Figure 2E).[19**]
Fig. 2.
Top: In general chemical specificity is lost at high lateral resolution. MALDI has the unique ability to detect intact proteins and peptides; however, sensitivity limits this ability with sub-cellular spatial resolution. ToF-SIMS has a higher spatial resolution, but the bombardment physics preclude the desorption of intact proteins. Information on all three ‘omic categories can be provided with NanoSIMS, despite the reduced specificity associated with the technique. Bottom: MS images of single cells obtained with MALDI (A and D), ToF-SIMS (B and E) and nanoSIMS (C and F) methodologies. A: Single cell MS images mapping the distribution of intact proteins in a single cell may be in the near future. B: ToF-SIMS image of the protein fragment of thyroglobulin in thyroid cells after trypsin digest.[44] C: One enriched pixel shown in red (arrow) on an MS image of 12C14N indicates the position of 15N-enriched TDG/Provectin in a single COS-7 cell after 4 h incubation; scale bar is 5 μm.[32] High resolution dynamic SIMS images of 15N-enriched Shiga toxin B-subunit (15N-STxB) protein vector in a single HeLa cell after incubation (Red: 12C14N, green:12C15N, 100 nm working beam diameter).[31] D: Top, MALDI MS image of intact lipid (m/z 782) from HEK-293 cells (spatial resolution ∼2 μm) and corresponding optical image.[37*] Bottom, AP-MALDI MS image of intact lipids (m/z 782, red)), exogenous fluorescent tag (m/z 445, green) and optical image of HeLa cells (spatial resolution ∼7 μm).[38*] E: ToF-SIMS image of phosphocholine headgroup (m/z 184) in tetrahymenia (top 2),[21**] and intact vitamin E (m/z 430.3) in Aplysia california neuron (middle 3),[19**] and 3D image of phosphocholine headgroup (m/z 184, green) and adenine (m/z 136, red) in HeLa cells.[17] F: High resolution dynamic SIMS images of adiopcytes reveas the uptake of enriched free fatty acid 13C-oleate after incubation.[30]
Sensitivity is gained by reducing specificity and this strategy has been applied to lipids and proteins in SIMS-based cellular imaging. For example, instead of monitoring a single intact lipid species (100 attomoles per cell), a common moiety, such as phosphocholine (m/z 184) is monitored (7 femtomoles per cell). In this case, two-orders of magnitude are gained by reducing the specificity of the targeted analyte. Ostrowski et al. and Kurczy et al. used the gain in sensitivity to image the distribution of lipid domains during the mating of tetrahymena cells (Figure 2E).[20,21**]
Laser post ionization is a strategy, still under development, with immense potential to improve the sensitivity of SIMS analyses.[22] The technique employs a femtosecond laser system to ionize the large population of neutral species (106) sputtered by the primary ion beam. A breakthrough in post laser ionization would greatly enhance SIMS-based single-cell IMS.
Dynamic SIMS
In dynamic SIMS, sensitivity is gained by surpassing the static limit (1013 primary ions per cm2) and integrating the signal as a function of depth. By turning a pixel into a voxel, it is possible to improve the sensitivity of the technique by five orders of magnitude.[14**]
New hybrid SIMS instruments, the J105 and the C60-QSTAR, combine the detection range and multiplexing abilities of a ToF mass analyzer with the molecular sensitivity and throughput of a continuous cluster ion source.[23,24] In both instruments the ionization process is decoupled from the mass analyzer, which makes the design compatible with a continuous ion beam and allows spectral information to be collected during the sputtering process. With this technology, three-dimensional renderings reveal the distribution of lipids and nucleotides in a single cell (Figure 2E).
Sub-cellular imaging is routinely performed in the dynamic SIMS community.[25] The Cameca nanoSIMS instrument has several design features aimed towards optimizing sensitivity. For instance, the instrument employs a reactive primary ion beams (i.e. O2), a continuous ion source for high throughput and low duty cycle losses, a co-axial extraction which reduces the working distance between the ion optics and the sample, and a high transmission magnetic sector mass analyzer.[26,27] The trade-off between mass resolution and transmission is an unfortunate caveat of the design, therefore sensitivity may be improved, but at the expense of mass resolution (e.g. transmission is 60 % at M/ΔM=5000 and 10 % at M/ΔM=15,000).[26]
Despite the highly destructive ion beam, which limits the detection to atomic or diatomic species, nanoSIMS has been successfully utilized in a wide range of applications, including proteomics, lipidomics, metabolomics and genomics all at the single cell level. These endeavors are possible due to the use of a multi-isotope imaging mass spectrometer (MIMS).[28] In this method, the metabolic turnover of protein vectors, lipids, and nucleotides with stable heavy isotopic labels can be measured. Recently, Lechene et al. used MIMS and the nanometer spatial resolution of nanoSIMS to rebut the immortalized DNA strand theory and provide evidence in support of random DNA segregation during cell division.[29**] In another noteworthy application, tracking the journey of isotopically enriched lipids (free fatty acids in adipocytes), proteins (thymine DNA glycosylase (TDG) and retinoid × receptor α (RXRα) in COS-7 cells) and peptide vectors (STxB in HeLa cells) has provided insight into cellular metabolism, transport, and catabolism (Figure 2C and 2F).[30-31,32**] The sensitivity of the system allowed Delaune et al. to track individual proteins within a cell.[32**]
Advances in MALDI Imaging
MALDI utilizes laser radiation to ablate and ionize analyte molecules cocrystallized in a matrix.[33,34] Images are obtained by moving the sample plate between laser shots. The desorption and ionization mechanisms of MALDI allow the detection of large molecules, such as proteins and peptides, making it a potentially powerful tool for single-cell proteomics.
In the early days of MALDI IMS, the spatial resolution of the technique was limited by the laser beam diameter and the size of the matrix crystal. Various developments targeting these shortcomings have been highlighted in a recent review.[35*] Briefly, the development of solvent-free matrix application methods, such as the sublimation technique developed in the Murphy laboratory, has significantly reduced the crystal size of matrix.[36] Unfortunately, without solvent to extract the analytes into the matrix, the gain in lateral resolution is often accompanied by a reduction in sensitivity.
Both the sensitivity and spatial resolution of MALDI IMS have been improved by positioning the laser behind the sample and irradiating from the backside of the sample.[37*] The geometry reduces the working distance between the sample and laser optics allowing sub-micrometer laser beam focus, while the axial ablation path improves sensitivity. In a proof-of-concept experiment, Zavalin et al. imaged the distribution of intact lipids (m/z 782) in cultured cells (HEK-293 and RKO) at sub-micrometer lateral resolution and obtaining sub-cellular profiles of intact peptides in human pancreatic islet cells (Figure 2D).[37*]
One of the greatest advantages of MALDI is its ability to be interfaced with a sample outside of a vacuum environment (i.e. atmospheric pressure MALDI). Schober et al. utilized atmospheric pressure MALDI to image a number of metabolites and lipids of individual HeLa cells with 7- μm resolution (Figure 2D).[38*] Sensitivity loses associated with atmosphere pressure interface have been mitigated by the development of the pulsed dynamic focusing (MassTech, Inc.), this technique improves ion transmission into the mass spectrometer by removing the electric field between the sample plate and the ion inlet.[39]
Boutaghou et al. recently introduced the matrix 9,10- diphenylanthracene for MALDI.[40*] The new matrix utilizes secondary electron-transfer reactions to improve the ionization of non-polar, aprotic compounds. This compound not only improves the sensitivity, it also expands the analytical capabilities of MALDI by increasing the breadth of detectable molecules.
Nanostructured initiated mass spectrometry (NIMS) is a highly sensitivity matrix-free method with the potential to impact single-cell IMS. In this technique, functionalized surfaces are used to absorb the laser, eliminating the need for matrices. Recently, nanofabricated nanopost arrays, surfaces with highly ordered columns of silicon geometrically arranged to reduce the ionization threshold, have been used to demonstrate ultra-trace analyses (e.g., LOD 800 zeptamole of verapamil).[41*] Similarly, ion-NIMS is a technique that combines functionalized surfaces with a focused ion beam.[42*] The technique has the potential to achieve 150-nm spatial resolution and a femtomole detection limit for peptides. Thus far, sub-cellular resolution has not been demonstrated with this technique; however, the sensitivity and lateral resolution have the potential to detect low abundant lipids and proteins within a cell.
The future: Single cell 'omics: Single-cell MS imaging of proteins, lipids and metabolites
Proteins are a challenging target for single-cell IMS. Thus far, ToF-SIMS has been successful at imaging lipids and protein fragments (e.g. thyroglobulin in thyroid cells) at the single-cell level (Figure 2B).[43, 44] Unfortunately, the heterogeneous collision dynamics of SIMS bombardment prevents the ejection of high mass intact proteins.[45] With MIMS, dynamic SIMS has been able to provide information on all three ‘omics categories, but the destructive nature of the technique also prevents the desorption of intact proteins. MALDI is a potential platform for single-cell protein imaging. Thus far, proteomic ‘profiles’ have been successfully extracted from large Aplysia neurons.[46,47] Recent developments in MALDI IMS, provide an opportunity for detection of lipids and metabolites with sub-cellular lateral resolution. However, further development is need to obtain the combined lateral resolution and sensitivity required to achieve single-cell IMS of intact proteins.
Highlights.
SIMS is the predominant imaging mass spectrometry (IMS) technique for single-cell MS imaging
SIMS at sub-cellular spatial resolution has led to new biological observations.
Sensitivity is the ‘new challenge’ limiting MALDI spatial resolution.
MALDI is a potential platform for proteomics-based single-cell IMS
Acknowledgments
The Swedish Research Council, the European Research Council (Advanced grant), the Knut and Alice Wallenberg Foundation (Wallenberg Fellow), and the USA National Institutes of Health are gratefully acknowledged for support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
(*) special interest and (**) outstanding interest
- 1.Amantonico A, Urban PL, Fagerer SR, Balabin RM, Zenobi R. Single-cell MALDI-MS as an analytical tool for studying intrapopulation metabolic heterogeneity of unicellular organisms. Analytical chemistry. 2010;82:7394–7400. doi: 10.1021/ac1015326. [DOI] [PubMed] [Google Scholar]
- 2.Trouillon R, Passarelli MK, Wang J, Kurczy ME, Ewing AG. Chemical Analysis of Single Cells. Analytical chemistry. 2012;85:522–542. doi: 10.1021/ac303290s. [DOI] [PubMed] [Google Scholar]
- 3.McDonnell LA, Heeren RMA. Imaging mass spectrometry. Mass Spectrometry Reviews. 2007;26:606–643. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- 4.Amstalden van Hove ER, Smith DF, Heeren R. A concise review of mass spectrometry imaging. Journal of chromatography A. 2010;1217:3946–3954. doi: 10.1016/j.chroma.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 5.Ifa DR, Wiseman JM, Song QY, Cooks RG. Development of capabilities for imaging mass spectrometry under ambient conditions with desorption electrospray ionization (DESI) International Journal of Mass Spectrometry. 2007;259:8–15. [Google Scholar]
- 6.Cornett DS, Reyzer ML, Chaurand P, Caprioli RM. MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nature Methods. 2007;4:828–833. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- 7.Vickerman JC, Gilmore IS. Surface analysis: the principal techniques. Vol. 2. Wiley Online Library; 2009. [Google Scholar]
- 8.Colliver TL, Brummel CL, Pacholski ML, Swanek FD, Ewing AG, Winograd N. Atomic and molecular imaging at the single-cell level with TOF-SIMS. Analytical Chemistry. 1997;69:2225–2231. doi: 10.1021/ac9701748. [DOI] [PubMed] [Google Scholar]
- 9.Fletcher JS. Cellular imaging with secondary ion mass spectrometry. Analyst. 2009;134:2204–2215. doi: 10.1039/b913575h. [DOI] [PubMed] [Google Scholar]
- 10.Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, Aebersold R. The quantitative proteome of a human cell line. Molecular systems biology. 2011;7 doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dennis EA, Deems RA, Harkewicz R, Quehenberger O, Brown HA, Milne SB, Myers DS, Glass CK, Hardiman G, Reichart D, et al. A Mouse Macrophage Lipidome. Journal of Biological Chemistry. 2010;285:39976–39985. doi: 10.1074/jbc.M110.182915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDonnell LA, Corthal GL, Willems SM, van Remoortere A, van Zeijl RJM, Deelder AM. Peptide and protein imaging mass spectrometry in chancer research. Journal of Proteomics. 2010;73:1941–1944. doi: 10.1016/j.jprot.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 13*.Winograd N. The magic of cluster SIMS. Analytical Chemistry. 2005;77:142A–149A. [Google Scholar]
- 14**.Winograd N. Molecular Depth Profiling. Surface and Interface Analysis. 2013;45:3–8. doi: 10.1002/sia.4913. The article describes protocols and the theoretical framework associated with molecular depth profiling, the foundation for 3D MS imaging. Also discussed are the advantages of cluster ion beams, the grand challenges, and the future direction of molecule depth profiling in the field of SIMS analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuo J, Okubo C, Seki T, Aoki T, Toyoda N, Yamada I. A new secondary ion mass spectrometry (SIMS) system with high-intensity cluster ion source. Nuclear Instruments and Methods in Physics Research Section B: Beam Interactions with Materials and Atoms. 2004;219:463–467. [Google Scholar]
- 16*.Goodwin RJ. Sample preparation for mass spectrometry imaging: Small mistakes can lead to big consequences. Journal of proteomics. 2012 doi: 10.1016/j.jprot.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 17.Fletcher JS, Rabbani S, Henderson A, Lockyer NP, Vickerman JC. Three-dimensional mass spectral imaging of HeLa-M cells - sample preparation, data interpretation and visualisation. Rapid Communications in Mass Spectrometry. 2011;25:925–932. doi: 10.1002/rcm.4944. [DOI] [PubMed] [Google Scholar]
- 18.Barnes CA, Brison J, Robinson M, Graham DJ, Castner DG, Ratner BD. Identifying Individual Cell Types in Heterogeneous Cultures Using Secondary Ion Mass Spectrometry Imaging with C60 Etching and Multivariate Analysis. Analytical chemistry. 2012;84:893–900. doi: 10.1021/ac201179t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19**.Passarelli MK, Ewing AG, Winograd N. Single Cell Lipidomics: Characterizing and Imaging Lipids on the Surface of Individual Aplysia Californica Neurons with Cluster SIMS. Analytical chemistry. 2013 doi: 10.1021/ac303038j. The C60-QSTAR instrument was used to map the distibution of intact lipid species on a single neuron. Lipid assignment were obtained using in situ tandem MS. The chemical signiture of Vitamin E obtained from the tandem MS analysis was used to extract and compile fragment peaks from the ToF-MS image dataset. This stratagy improved senitivity and image contract and provides a clearer rendition of Vitamin E distribution in the neuron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ostrowski SG, Van Bell CT, Winograd N, Ewing AG. Mass spectrometric imaging of highly curved membranes during Tetrahymena mating. Science. 2004;305:71–73. doi: 10.1126/science.1099791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21**.Kurczy ME, Piehowski PD, Van Bell CT, Heien ML, Winograd N, Ewing AG. Mass spectrometry imaging of mating Tetrahymena show that changes in cell morphology regulate lipid domain formation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2751–2756. doi: 10.1073/pnas.0908101107. Researchers use ToF-SIMS to elucidate the pore formation process that occurs during the mating of tetrahymenia. A decrease in lamellar lipids is detected in the fussion pores linking mating cells. The time-based study that shows that changes in the lipid chemsitry follow structural changes in the membrane, indicating that local structure dictates membrane chemistry and not visa versa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willingham D, Kucher A, Winograd N. Molecular depth profiling and imaging using cluster ion beams with femtosecond laser postionization. Applied Surface Science. 2008;255:831–833. [Google Scholar]
- 23.Carado A, Passarelli M, Kozole J, Wingate J, Winograd N, Loboda A. C60 secondary ion mass spectrometry with a hybrid-quadrupole orthogonal time-of-flight mass spectrometer. Analytical chemistry. 2008;80:7921–7929. doi: 10.1021/ac801712s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fletcher JS, Rabbani S, Henderson A, Blenkinsopp P, Thompson SP, Lockyer NP, Vickerman JC. A New Dynamic in Mass Spectral Imaging of Single Biological Cells. Analytical Chemistry. 2008;80:9058–9064. doi: 10.1021/ac8015278. [DOI] [PubMed] [Google Scholar]
- 25.Lechene C, Hillion F, McMahon G, Benson D, Kleinfeld A, Kampf JP, Distel D, Luyten Y, Bonventre J, Hentschel D, et al. High-resolution quantitative imaging of mammalian and bacterial cells using stable isotope mass spectrometry. Journal of Biology. 2006;5:20. doi: 10.1186/jbiol42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guerquin-Kern JL, Wu TD, Quintana C, Croisy A. Progress in analytical imaging of the cell by dynamic secondary ion mass spectrometry (SIMS microscopy) Biochimica Et Biophysica Acta-General Subjects. 2005;1724:228–238. doi: 10.1016/j.bbagen.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 27.Boxer SG, Kraft ML, Weber PK. Advances in Imaging Secondary Ion Mass Spectrometry for Biological Samples. Annual Review of Biophysics. 2009;38:53–74. doi: 10.1146/annurev.biophys.050708.133634. [DOI] [PubMed] [Google Scholar]
- 28.Peteranderl R, Lechene C. Measure of carbon and nitrogen stable isotope ratios in cultured cells. Journal of the American Society for Mass Spectrometry. 2004;15:478–485. doi: 10.1016/j.jasms.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 29**.Steinhauser ML, Bailey AP, Senyo SE, Guillermier C, Perlstein TS, Gould AP, Lee RT, Lechene CP. Multi-isotope imaging mass spectrometry quantifies stem cell division and metabolism. Nature. 2012;481:516–519. doi: 10.1038/nature10734. In this article, the high resolution imaging cabilities of dynamic SIMS combined with the quantitative nature of multi-isotope imaging mass spectrometry (MIMS) was used to elucidate cellular processes. The technique provides convincing evidence that DNA segregation during cell division is random, measured the rate of dietary lipid turnover, and time-based studies reveal the gestation period of lymphocytes in human bone marrow. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleinfeld AM, Kampf JP, Lechene C. Transport of C-13-oleate in adipocytes measured using multi imaging mass Spectrometry. Journal of the American Society for Mass Spectrometry. 2004;15:1572–1580. doi: 10.1016/j.jasms.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 31.Römer W, Wu TD, Duchambon P, Amessou M, Carrez D, Johannes L, Guerquin-Kern JL. Subcellular localisation of a 15 N-labelled peptide vector using NanoSIMS imaging. Applied surface science. 2006;252:6925–6930. [Google Scholar]
- 32**.Delaune A, Cabin-Flaman A, Legent G, Gibouin D, Smet-Nocca C, Lefebvre F, Benecke A, Vasse M, Ripoll C. 50nm-Scale Localization of Single Unmodified, Isotopically Enriched, Proteins in Cells. PloS one. 2013;8:e56559. doi: 10.1371/journal.pone.0056559. The article describes a strategy which allows for single molecule detection within a cell. High resolution dynamic SIMS images capture the journey of enriched proteins, TDG and RXRa, from the nucleus to the cytoplasm. The technique was also used to test the efficiency of the Provectin-Imgenex protein delivery reagent system (Cliniscience, Montrouge, France) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cornett DS, Reyzer ML, Chaurand P, Caprioli RM. MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nature Methods. 2007;4:828–833. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- 34.Walch A, Rauser S, Deininger SO, Hofler H. MALDI imaging mass spectrometry for direct tissue analysis: a new frontier for molecular histology. Histochemistry and Cell Biology. 2008;130:421–434. doi: 10.1007/s00418-008-0469-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35*.Boggio KJ, Obasuyi E, Sugino K, Nelson SB, Agar NY, Agar JN. Recent advances in single-cell MALDI mass spectrometry imaging and potential clinical impact. Expert Review of Proteomics. 2011;8:591–604. doi: 10.1586/epr.11.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hankin JA, Barkley RM, Murphy RC. Sublimation as a method of matrix application for mass spectrometric imaging. Journal of the American Society for Mass Spectrometry. 2007;18:1646–1652. doi: 10.1016/j.jasms.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37*.Zavalin A, Todd EM, Rawhouser PD, Yang J, Norris JL, Caprioli RM. Direct imaging of single cells and tissue at sub-cellular spatial resolution using transmission geometry MALDI MS. Journal of Mass Spectrometry. 2012;47:1473–1481. doi: 10.1002/jms.3108. [DOI] [PubMed] [Google Scholar]
- 38.Schober Y, Guenther S, Spengler B, Römpp A. Single cell matrix-assisted laser desorption/ionization mass spectrometry imaging. Analytical chemistry. 2012;84:6293–6297. doi: 10.1021/ac301337h. [DOI] [PubMed] [Google Scholar]
- 39.Tan PV, Laiko VV, Doroshenko VM. Atmospheric pressure MALDI with pulsed dynamic focusing for high-efficiency transmission of ions into a mass spectrometer. Analytical chemistry. 2004;76:2462–2469. doi: 10.1021/ac0353177. [DOI] [PubMed] [Google Scholar]
- 40*.Nazim Boutaghou M, Cole RB. 9, 10-Diphenylanthracene as a matrix for MALDI-MS electron transfer secondary reactions. Journal of Mass Spectrometry. 2012;47:995–1003. doi: 10.1002/jms.3027. [DOI] [PubMed] [Google Scholar]
- 41.Walker BN, Stolee JA, Vertes A. Nanophotonic Ionization for Ultratrace and Single-Cell Analysis by Mass Spectrometry. Analytical chemistry. 2012;84:7756–7762. doi: 10.1021/ac301238k. [DOI] [PubMed] [Google Scholar]
- 42.Northen TR, Yanes O, Northen MT, Marrinucci D, Uritboonthai W, Apon J, Golledge SL, Nordström A, Siuzdak G. Clathrate nanostructures for mass spectrometry. Nature. 2007;449:1033–1036. doi: 10.1038/nature06195. [DOI] [PubMed] [Google Scholar]
- 43.Passarelli MK, Winograd N. Lipid imaging with time-of-flight secondary ion mass spectrometry (ToF-SIMS) Biochim Biophys Acta. 2011 doi: 10.1016/j.bbalip.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nygren H, Malmberg P. High-resolution imaging and proteomics of peptide fragments by TOF-SIMS. Proteomics. 2010;10:1694–1698. doi: 10.1002/pmic.200900782. [DOI] [PubMed] [Google Scholar]
- 45.Garrison BJ, Delcorte A, Zhigilei L, Itina TE, Krantzman KD, Yingling YG, McQuaw CM, Smiley EJ, Winograd N. Big molecule ejection - SIMS vs. MALDI. Applied Surface Science. 2003;203:69–71. [Google Scholar]
- 46.Rubakhin SS, Churchill JD, Greenough WT, Sweedler JV. Profiling signaling peptides in single mammalian cells using mass spectrometry. Analytical chemistry. 2006;78:7267–7272. doi: 10.1021/ac0607010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li L, Garden RW, Sweedler JV. Single-cell MALDI: a new tool for direct peptide profiling. Trends in biotechnology. 2000;18:151–160. doi: 10.1016/s0167-7799(00)01427-x. [DOI] [PubMed] [Google Scholar]