Abstract
We identified novel gene fusions in patients with lung cancer harboring the kinase domain of the NTRK1 gene that encodes the TRKA receptor. Both the MPRIP-NTRK1 and CD74-NTRK1 fusions lead to constitutive TRKA kinase activity and are oncogenic. Treatment of cells expressing NTRK1 fusions with inhibitors of TRKA kinase activity inhibited autophosphorylation of TRKA and cell growth. Three of 91 lung cancer patients (3.3%), without known oncogenic alterations, assayed by NGS or FISH demonstrated evidence of NTRK1 gene fusions.
Orally active kinase inhibitors crizotinib and erlotinib or gefitinib are superior to standard chemotherapy in lung cancer patients with ALK fusions or EGFR mutations, respectively.1,2 Additional oncogenes such as ROS1, RET, and FGFR1-3 fusions have been identified in lung cancer and demonstrate great potential for therapeutic intervention.3-9 These oncogenes also occur in several other common malignancies expanding the potential relevance of this therapeutic approach.9-12
We performed a targeted next generation sequencing (NGS) assay on tumor samples from 36 patients with lung adenocarcinoma whose tumors did not contain known genetic alterations using standard clinical assays (Supplementary Table 1).10
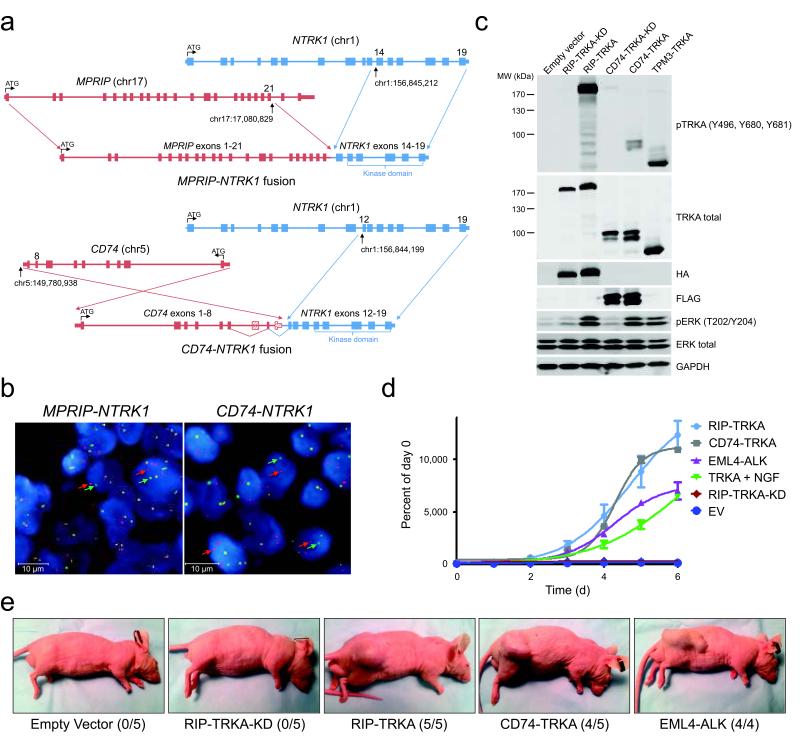
We detected evidence of an in-frame gene fusion event, in 2 of 36 patients, involving the kinase domain of the NTRK1 gene, which encodes the TRKA receptor tyrosine kinase (Fig. 1a, Supplementary Fig. 1). In the first case, the 5′ end of the myosin phosphatase Rho interacting protein (MPRIP) gene is joined with the 3′ end of NTRK1. MPRIP is involved in actin cytoskeleton regulation and has been implicated in a gene fusion in small cell lung cancer, putatively causing early termination of TP53.13 A second case harbored a CD74-NTRK1 gene fusion. Confirmation of the exon junctions and mRNA expression was achieved by RT-PCR and cloning of the entire cDNA (Supplementary Fig. 2-4). We detected expression of the fusion protein, RIP-TRKA (encoded by MPRIP-NTRK1) in both a malignant pleural effusion sample and early passage cells (CUTO-3) growing in culture (Supplementary Fig. 4). CUTO-3 cells demonstrated autophosphorylation of this novel protein at critical TRKA tyrosine residues.14 MPRIP harbors three coiled-coil domains, a common feature of 5′ fusion gene partners whose function is likely to mediate dimerization and consequently activation of the TRKA kinase domain (Supplementary Fig. 5).15,16 CD74, which encodes the MHC class II invariant chain, is a known activating fusion partner of ROS1 and the CD74-TRKA protein is predicted to be localized in the plasma membrane (Supplementary Fig. 5).3,17-19
Figure 1. Discovery and validation of oncogenic NTRK1 gene fusions in lung cancer samples.
(a) Schematic of genomic rearrangement from tumor samples harboring MPRIP-NTRK1 and CD74-NTRK1 using the FoundationOne Next Generation Sequencing Assay including chromosomal breakpoints for each gene rearrangement. (b) Break-apart FISH analysis of MPRIP-, CD74-, and unknown-NTRK1 tumor samples showing clear separation of green (5′) and red (3′) signals corresponding to the NTRK1 gene. (c) TRKA (NTRK1) fusions are autophosphorylated and activate key downstream signaling pathways. Representative immunoblot analyses (n = 3) of cell lysates from 293T cells expressing RIP-TRKA and CD74-TRKA, but not their kinase dead (KD) variants display phosphorylation of critical tyrosine residues and activation of pERK. TPM3-TRKA was expressed in 293T cells as a positive control. (d) NTRK1 fusions support cellular proliferation. MTS assay of Ba/F3 demonstrates that cells expressing RIP-TRKA, CD74-TRKA, EML4-ALK, or full length TRKA supplemented with NGF proliferate in the absence of IL-3, whereas Ba/F3 cells expressing EV or the kinase dead variant of RIP-TRKA do not proliferate (n = 3). Values represent the mean ± SEM. (e) MPRIP- or CD74-NTRK1 gene fusions induce tumorigenesis. NIH3T3 cells expressing RIP-TRKA, RIP-TRKA kinase dead (KD), CD74-TRKA, and EML4-ALK or empty vector were injected into the flanks of nude mice and observed for tumor growth. Representative pictures taken at day 12 following injection are shown. The numbers of tumors induced in the injected animals are shown in parentheses.
We developed a fluorescence in situ hybridization (FISH) assay to detect chromosomal rearrangements within the NTRK1 gene (Supplementary Fig. 6a). Hybridization of these probes showed clear separation of the 5′ and 3′ probes in the tumor samples containing the MPRIP- or CD74-NTRK1 gene fusions, but not in a control sample (Fig. 1b and Supplementary Fig. 6b). Fusions between NTRK1 and TPM3, TFG, or TPR have previously been identified in colorectal and thyroid cancers.11,20 Although TPM3 (1q22-23) lies in close proximity to NTRK1 (1q21-22), FISH could detect a separation in signals in the KM12 colorectal cell line that harbors a TPM3-NTRK1 fusion (Supplementary Fig. 6c and 7).21 Using this FISH assay, 56 additional lung adenocarcinoma samples without detectable oncogenic alterations were screened for NTRK1 rearrangements and one additional positive case was identified (Supplementary Table 2, Fig. 6d). Quantitative PCR demonstrated high NTRK1 kinase domain expression only in the tumors with the known NTRK1 rearrangements or in the KM12 cell line (Supplementary Fig. 8). Analysis of transcriptome data from The Cancer Genome Atlas of 230 lung adenocarcinomas failed to detect evidence of NTRK1 fusions (data not shown). The recent transcriptome study of 87 lung adenocarcinoma tumor samples also did not identify oncogenic fusions involving NTRK1 (J.S.Seo, personal communication).22
To formally prove that these novel fusion proteins are oncogenic, MPRIP- and CD74-NTRK1 cDNA constructs were expressed in 293T cells, NIH3T3 fibroblasts and Ba/F3 cells. We observed expression of the appropriate-sized chimeric proteins and TRKA autophosphorylation, as in the CUTO-3 cells (Fig. 1c, Supplementary Fig. 4, 9).14 Introduction of a NTRK1 kinase dead mutation did not result in TRKA autophosphorylation or to increased ERK1/2 and AKT phosphorylation (Fig. 1c, 2a and Supplementary Fig. 14). MPRIP- and CD74-NTRK1, but not their kinase-dead counterparts, induced IL-3 independent proliferation of Ba/F3 cells (Fig. 1d). Similarly, MPRIP- and CD74-NTRK1 supported anchorage-independent growth of NIH3T3 cells, formed tumors in nude mice, and CD74-NTRK1 induced a refractory appearance of NIH3T3 cells (Fig. 1e, Supplementary Fig. 10 and 11). Knockdown of TPM3-NTRK1 in KM12 cells reduced proliferation, further supporting the role of NTRK1 fusions as oncogenes (Fig. 2a, Supplementary Fig. 12).
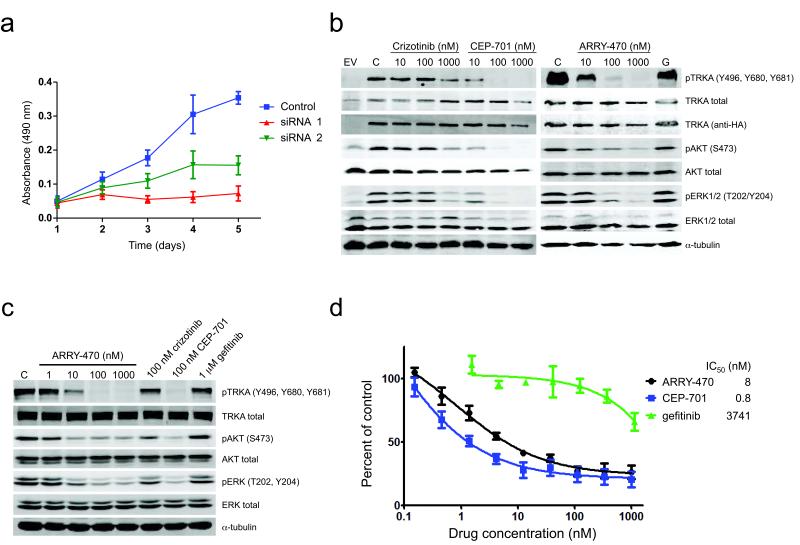
Figure 2. Drug treatment inhibits activation of TRKA, downstream signaling, and proliferation in cells expressing NTRK1 fusions.
(a) RNAi knockdown of NTRK1 inhibits cell proliferation in a cell line harboring TPM3-NTRK1. KM12 cells were analyzed by MTS proliferation assay 1-5 days after siRNA transfection (left, n = 3). (b) Ba/F3 cells expressing MPRIP-NTRK1 (RIP-TRKA) or empty vector (EV) were lysed after 5h of treatment with the indicated doses of drugs (G = gefitinib 1000nM) or DMSO control (C). (c) CUTO-3 lung cancer cells harboring the MPRIP-NTRK1 gene fusion were treated with the indicated doses and drugs and subjected to immunoblot analysis. (d) Treatment of Ba/F3 cells expressing the MPRIP-TRKA fusion with TRKA inhibitors inhibits cell proliferation as measured by MTS assay (a-c, n = 5). Values represent the mean ± SEM. Ba/F3 cells expressing the MPRIP-TRKA fusion demonstrate inhibition of proliferation by the pan-TRK inhibitor, ARRY-470, and the multi-kinase inhibitor, CEP-701, but not the EGFR inhibitor, gefitinib.
Given the prior success of treating ALK and ROS1 fusion positive cancer patients with kinase inhibitors, we asked whether NTRK1 fusions might provide a similar target in patients with lung cancer or other malignancies. ARRY-470 is a selective kinase inhibitor with nanomolar activity against TRKA/B/C but no other significant kinase inhibition below 1000nM (Supplementary Fig. 13 and Supplementary Table 3). CEP-701 and crizotinib also have activity against TRKA in addition to other kinases.23,24 Treatment of cells expressing MPRIP- or CD74-NTRK1 with ARRY-470, CEP-701 and, to a lesser extent, crizotinib, inhibited autophosphorylation of RIP-TRKA and CD74-TRKA (Fig. 2b and Supplementary Fig. 9, 14a). Activation of the MAPK and AKT pathways was also inhibited in Ba/F3 cells (Fig. 2b and Supplementary Fig. 14). Phosphorylation of endogenously expressed RIP-TRKA in CUTO-3 and TPM3-TRKA in KM12 cells was similarly inhibited by all three drugs (Fig. 2c and Supplementary Fig. 15a).
Inhibition of proliferation of Ba/F3 cells expressing NTRK1 gene fusions, was greatest with CEP-701 and ARRY-470 (Fig. 2d and Supplementary Fig. 14b). Crizotinib was a less potent inhibitor, although in a similar range seen for inhibition of EML4-ALK or SDC4-ROS1 (Supplementary Fig. 16).3 The less potent effects of crizotinib on cell proliferation are consistent with decreased inhibition of pTRKA and downstream pERK1/2 (Fig. 2d and Supplementary Fig. 14). All three drugs also inhibited colony formation of NIH3T3 cells expressing NTRK1 fusions in soft agar (Supplementary Fig. 10). KM12 cells were similarly sensitive to ARRY-470 and CEP-701, but less so to crizotinib (Supplementary Fig. 15b). ARRY-470 did not inhibit proliferation of Ba/F3 cells expressing other oncogene targets (EGFR, ALK, or ROS1) or of lung and colorectal cell lines that do not harbor an NTRK1 fusion (Supplementary Fig. 17). All three drugs induced cell-cycle arrest in G1 and apoptosis of KM12 cells (Supplementary Fig. 18).
The index patient (MPRIP-NTRK1) had no standard therapies and no clinical trials of potentially effective TRKA inhibitors available; therefore the patient consented to treatment with crizotinib (250 mg twice daily) outside of a clinical trial. She experienced a minor radiographic response with a decrease in her serum levels of CA125, but experienced disease progression after ~3 months (Supplementary Fig. 19). This modest clinical activity of crizotinib is consistent with the in vitro activity and could be due to non-TRKA kinase effects.
We have identified novel, recurrent oncogenic NTRK1 fusions in a subset of patients (3/91; 3.3%) with lung adenocarcinoma that did not contain other common oncogenic alterations. Our study further highlights the utility of targeted NGS to discover novel drug sensitive genetic alterations in lung cancer. Based on our data, clinical studies of selective TRKA inhibitors in NTRK1 rearranged NSCLC are warranted.
Online Methods
Patients
Colorado Multiple Institution Review Board (IRB) or the Dana-Farber Cancer Institute IRB approval was obtained for all patients in this study. FoundationOne testing and FISH analyses were performed in CLIA-certified laboratories. The index patient who underwent treatment with crizotinib consented to treatment outside of a clinical trial.
Next Generation DNA Sequencing
DNA was extracted from 40 μm of FFPE or frozen tissue using the Maxwell 16 FFPE Plus LEV DNA Purification kit (Promega) and quantified using the PicoGreen fluorescence assay (Invitrogen). Library Construction was performed as previously described using 50-200ng of DNA sheared by sonication to ~100-400bp prior to end-repair, dA addition and ligation of indexed, Illumina sequencing adaptors.25 Enrichment of target sequences (3,320 exons of 182 cancer-related genes and 37 introns from 14 genes recurrently rearranged in cancer representing ~1.1 Mb of the human genome) was achieved by solution-based hybrid capture with a custom Agilent SureSelect biotinylated RNA baitset.25 The libraries were sequenced on an Illumina HiSeq 2000 platform using 49×49 paired-end reads. Sequence data from genomic DNA was mapped to the reference human genome (hg19) using the Burrows-Wheeler Aligner and were processed using the publicly available SAMtools, Picard, and Genome Analysis Toolkit.26,27 Genomic rearrangements were detected by clustering chimeric reads mapped to targeted introns.
RNA extraction from FFPE and Frozen tissues
RNA was isolated from FFPE or frozen tumor samples as described previously.3 Briefly, FFPE samples were processed using the RecoverAll™ Total Nucleic Acid Isolation Kit (Ambion) following deparaffinization in xylene and washed with 100% ethanol prior to Protease K digest. Extraction of RNA from frozen tissue was accomplished using TriReagent (Ambion). Alternatively, tumors from NSCLC patients obtained at surgery were snap-frozen in liquid nitrogen, embedded in OCT and sectioned. RNA was prepared using Trizol (Invitrogen) followed by RNeasy MinElute cleanup kit (Qiagen).
RT-PCR and Sequencing of MPRIP- and CD74-NTRK1
RT-PCR of MPRIP-NTRK1 was carried out using the SuperScript® III First-Strand Synthesis System (SSIII RT) from Invitrogen with a NTRK1 primer located in exon 15 (‘NTRK1 Y490R1’) for reverse transcription by PCR using the same reverse primer, ‘NTRK1 Y490R1’, and a primer to MPRIP located in its 3rd coil-coiled domain (‘MPRIP CC3F1’). PCR products were resolved on an agarose gel and the fragments were excised and treated with ExoSapIT (Affymetrix) prior to sequencing by the University of Colorado Cancer Center DNA Sequencing and Analysis Core using the BigDye Terminator Cycle Sequencing Ready Reaction kit version 1.1 (Applied Biosystems) using the same forward and reverse primer in the RT-PCR reaction. For CD74-NTRK1, reverse transcription was carried out using the QuanTitec Reverse Transcription kit (Qiagen). PCR of the resulting cDNA was performed using the primers ‘CD74 Exon 3 FOR’ and ‘NTRK1 Exon 15 REV’. Primers used for RT-PCR and sequencing are available in Supplementary Table 4. The reference sequences used for exon alignment are NCBI Reference Sequences: NM_002529.3 (NTRK1), NM_015134.3 (MPRIP), and NM_001025159.2 (CD74).
Cloning full length MPRIP-, CD74-, and TPM3-NTRK1
cDNA was generated from the patient using the SSIII RT kit describe above along with a primer located at the end of NTRK1 (NTRK1stopR2). This cDNA was used to amplify two separate overlapping fragments that were used to generate full length MPRIP-NTRK1 by overlap extension PCR using the two fragments alone for 10 cycles and then adding the MPRIPStart and NTRK1stopR1 primers for an additional 30 cycles of PCR amplification. The resulting 4kb PCR product was gel isolated and confirmed by Sanger sequencing. A 3′ hemagglutinin (HA) tag was added to MPRIP-NTRK1 using PCR amplification with primers harboring the HA encoding sequence.. The amplified product was subsequently cloned into the pCDH-CMV-MSC1-EF1-Puro lentiviral plasmid (System Biosciences). Full length TPM3-NTRK1 was amplified from KM12 cDNA using TPM3Start RI and NTRKStopNotI primers and cloned into the lentiviral plasmid as described above. The NCBI Reference Sequence used for TPM3 is NM_153649.3. For the CD74-NTRK1 construct, cDNA was transcribed with Quantiscript Reverse Transcriptase (Qiagen). Full length CD74-NTRK1 was amplified using the primers ‘CD74 FOR’ and ‘NTRK1 REV’ using AccuPrime™ Taq DNA Polymerase (Invitrogen) and cloned into the pDNR-Dual vector (BD Biosciences) and recombined into JP1520 retroviral vector as previously described.28 The full-length cDNA of each gene was confirmed by sequencing. Primers used for cloning are available in Supplementary Table 4.
Quantitative PCR of NTRK1
Relative Quantification Polymerase Chain Reaction (RQ-PCR) assay of the NTRK1 tyrosine-kinase domain (Hs01021011_m1; Applied Biosystems) was used to evaluate its level of mRNA expression. The relative quantification method (ΔΔCT) in the StepOnePlus Real-time PCR system (Applied Biosystems) was used with GUSB (Applied Biosystems) as an endogenous control. All samples were evaluated in triplicate.
RNA Sequencing
Paired-end RNA sequencing was performed as previously described.29 RNA FASTQ files were aligned and splice junctions mapped using TopHat30 and analyzed for fusion reads using the Broad Institute Cancer Genome Analysis Tools Suite (www.broadinstitute.org/cancer/cga) and (www.broadinstitute.org/cancer/software/genepattern/modules/RNA-seq/).
Cell lines and reagents
NIH3T3, HEK-293T, and Ba/F3 were previously described.31 The lung cancer cell lines A549, H3122, H1650, H1299, and HCC78 were previously described.31-34 The colorectal cancer cell lines, KM12, HCT116, HCT15, HT29, and SW837 were previously described.35 Ba/F3 cells expressing the mutant EGFR allele, E746_A750del were previously described.28 The lymphoblastoid cell line, GM09948 (Coriell Cell Repository), was used for genomic mapping in FISH studies.
All cancer cell lines were maintained in RPMI media with 10% calf serum. NIH3T3 and Ba/F3 cells transduced with full length NTRK1 were supplemented with 100 ng/ml and 200 ng/ml β-NGF (R&D Systems), respectively. Crizotinib and gefitinib were purchased from Selleck Chemicals, CEP-701 from Sigma Aldrich or Santa Cruz Biotechnology, K252a from Tocris, and ARRY-470 was supplied by Array BioPharma. Total AKT, AKT pSer473, total ERK, ERK pThr202/Tyr204, total STAT3, STAT3 pY705, PARP, and TRK pY490 and pY674/675 (corresponding to Y496, Y680, and Y681 in TRKA, respectively) antibodies were purchased from Cell-Signaling Technologies. Total TRKA (C-14), GAPDH, and α-tubulin were purchased from Santa Cruz Biotechnologies Inc.
Lentivirus or retrovirus production and cell transduction
MPRIP-NTRK1 or the kinase dead variant was introduced into cells via lentivirus as previously described.31 NIH3T3 cells transduced with lentivirus were cultured in DMEM medium with 5% calf serum and 0.75 μg/ml puromycin. Ba/F3 cells transduced with lentivirus were cultured as above with 2 μg/ml puromycin, and with or without 1 ng/ml IL-3 (R&D Systems). Alternatively, CD74-NTRK1 was introduced into cells using retrovirus as previously described.28 Polyclonal cell lines were established by puromycin selection. Cell proliferation and growth were performed as previously described.28,36
Mouse Xenograft Studies
NIH3T3 cells (106) harboring the indicated expression vectors were resuspended in Matrigel (BD Biosciences) and injected subcutaneously into athymic nude mice (kind gift of James DeGregori). Mice were monitored three times weekly for tumor formation and sacrificed when tumors reached approximately 2 cm × 2 cm. Approval for the use of animals in this study was granted by the University of Colorado Institutional Animal Care and Use Committee (IACUC).
Immunoblotting
Immunoblotting was performed as previously described.31 Briefly, cells were lysed in RIPA buffer with Halt protease and phosphatase inhibitor cocktail (Thermo-Scientific) and diluted in loading buffer (LI-COR Biosciences). Membranes were scanned and analyzed using the Odyssey Imaging System and software (LI-COR). Alternatively, immunoblotting was performed according to the antibody manufacturer’s recommendations using chemiluminescent detection (Perkin Elmer). All western blot images are representative of at least 3 independent experiments.
Proliferation assays
All assays were performed as previously described by seeding 1000 cells/well, drug treatments were performed 24 hours after seeding, and Cell Titer 96 MTS (Promega) was added 72 hours later or as described previously.28,31,36 IL-3 was removed from Ba/F3 cells 48 hours prior to seeding.
Soft agar assays
Anchorage-independent growth was measured by seeding 100,000 cells per well of soft agar in 6 well plates as previously described.31 Media was changed every 4 days for 2 weeks. Quantification was performed with MetaMorph Offline Version 7.5.0.0 (Molecular Devices).
Fluorescence In-Situ Hybridization
Formalin-fixed, paraffin-embedded (FFPE) tissue sections were submitted to a dual-color FISH assay using the laboratory developed NTRK1 break-apart probe (3′ NTRK1 [SpectrumRed] and 5′ NTRK1 [SpectrumGreen]) or the fusion MPRIP [SpectrumGreen]-NTRK1 [SpectrumRed] probe. The pre-hybridization treatment was performed using the reagents from the Vysis Paraffin Kit IV (Abbott Molecular). Hybridization and analysis was performed as previously described.3,31 Samples were deemed positive for NTRK1 rearrangement if ≥15% of tumor cells demonstrated an isolated 3′ signal or a separation of 5′ and 3′ signals that was greater than one signal diameter.
siRNA Transfection
KM12 cells were transfected with 30nM NTRK1 Silencer Select siRNAs (Life Technologies) using siPORT NeoFX transfection reagent (Life Technologies) at 4μL/mL.
Flow Cytometry
Cell cycle analysis was performed as previously described.3 Apoptosis was measured using the Vybrant apoptosis YO-PRO/PI kit (Invitrogen). Briefly, KM12 cells were seeded 24 hours prior to treatment at 500,000 cells/well prior to trypsinization and staining.
Immunohistochemistry
Immunohistochemical studies for TTF-1 and thyroglobulin were performed using standard procedures to exclude the possibility of a thyroid carcinoma, which can also express TTF-1, (Supplementary Fig. 20). Antibody against TTF-1 (Cell Marque, Cat#CMC-573) was applied at 1:100 dilution and thyroglobulin (Signet, Cat#228-13) was applied at 1:25 dilution and incubated at 37°C for 32 min. Detection for TTF-1 was performed using Ventana multiview (UltraView) and detection for thyroglobulin using Ventana Avidin-Biotin (iView).
Supplementary Material
Acknowledgements
This work was supported by a Colorado Bioscience Discovery Evaluation Grant Program, Colorado Clinical and Translational Sciences Institute CO-Pilot, and the Boettcher Foundation’s Webb-Waring Biomedical Research Program grants (R.C.D), The Dana Farber/Harvard Cancer Center Lung Cancer SPORE P50 CA090578 (P.A.J.), the Cammarata Family Foundation Research Fund (M.C. and P.A.J.), the Nirenberg Fellowship at the Dana-Farber Cancer Institute (M.C. and P.A.J.) and the University of Colorado Lung Cancer SPORE grant P50CA058187 (M.V.G.) and the NCI CCSG P30CA46934 (M.V.G.).
Footnotes
Competing financial interests
D.L., P.J.S., and V.A.M. are employees of Foundation Medicine, Inc., J.H. and S.W.A. are employees of Array BioPharma, Inc.. R.C.D. has received research funding and R.C.D and P.A.J. have received consulting fees from Pfizer.
Contributions
R.C.D., P.A.J., and V.A.M. conceived of the research idea. R.C.D and P.A.J. designed experiments, took responsibility for the oversight of the project and wrote the manuscript. A.V. and D.E. designed and performed immunoblot, generated Ba/F3 and NIH-3T3 cells, performed flow cytometry and MTS assays and contributed to writing of the manuscript. D.L., and P.J.S. designed and performed NGS assays. M.V.G. designed FISH probes and interpreted FISH experiments. S.K. and S.M. performed and analyzed FISH experiments. A.T.L. and M.C. performed cloning of RT-PCR fusion constructs. D.L.A. analyzed tumor samples and performed IHC. K.D.D. performed in vitro analyses and contributed to interpretation of the data. A.B.P. performed in vivo experiments and contributed to interpretation of data. P.S.H., L.A.G., and G.K. performed bioinformatics analyses. J.H. and S.W.A designed and profiled ARRY-470. E.M.B. and M.B. collected clinical data. J.K., H.S., and S.P. provided subject specimens. All authors contributed to revision of the manuscript.
References
- 1.Mok TS, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 2.Shaw AT, et al. Crizotinib versus Chemotherapy in Advanced ALK-Positive Lung Cancer. N Engl J Med. 2013 doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 3.Davies KD, et al. Identifying and targeting ROS1 gene fusions in non-small cell lung cancer. Clin Cancer Res. 2012;18:4570–4579. doi: 10.1158/1078-0432.CCR-12-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeuchi K, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18:378–381. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 5.Ju YS, et al. A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res. 2012;22:436–445. doi: 10.1101/gr.133645.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohno T, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 2012;18:375–377. doi: 10.1038/nm.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drilon A, et al. Response to Cabozantinib in Patients with RET Fusion-Positive Lung Adenocarcinomas. Cancer Discov. 2013 doi: 10.1158/2159-8290.CD-13-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Majewski IJ, et al. Identification of recurrent FGFR3 fusion genes in lung cancer through kinome-centred RNA sequencing. J Pathol. 2013;230:270–276. doi: 10.1002/path.4209. [DOI] [PubMed] [Google Scholar]
- 9.Wu YM, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3:636–647. doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lipson D, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18:382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alberti L, Carniti C, Miranda C, Roccato E, Pierotti MA. RET and NTRK1 proto-oncogenes in human diseases. J Cell Physiol. 2003;195:168–186. doi: 10.1002/jcp.10252. [DOI] [PubMed] [Google Scholar]
- 12.Davies KD, Doebele RC. Molecular Pathways: ROS1 Fusion Proteins in Cancer. Clin Cancer Res. 2013 doi: 10.1158/1078-0432.CCR-12-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peifer M, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stephens RM, et al. Trk receptors use redundant signal transduction pathways involving SHC and PLC-gamma 1 to mediate NGF responses. Neuron. 1994;12:691–705. doi: 10.1016/0896-6273(94)90223-2. [DOI] [PubMed] [Google Scholar]
- 15.Surks HK, Richards CT, Mendelsohn ME. Myosin phosphatase-Rho interacting protein. A new member of the myosin phosphatase complex that directly binds RhoA. J Biol Chem. 2003;278:51484–51493. doi: 10.1074/jbc.M305622200. [DOI] [PubMed] [Google Scholar]
- 16.Soda M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 17.Busch R, Doebele RC, Patil NS, Pashine A, Mellins ED. Accessory molecules for MHC class II peptide loading. Curr Opin Immunol. 2000;12:99–106. doi: 10.1016/s0952-7915(99)00057-6. [DOI] [PubMed] [Google Scholar]
- 18.Rikova K, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 19.Awad MM, et al. Acquired Resistance to Crizotinib from a Mutation in CD74-ROS1. N Engl J Med. 2013 doi: 10.1056/NEJMc1309091. [DOI] [PubMed] [Google Scholar]
- 20.Martin-Zanca D, Hughes SH, Barbacid M. A human oncogene formed by the fusion of truncated tropomyosin and protein tyrosine kinase sequences. Nature. 1986;319:743–748. doi: 10.1038/319743a0. [DOI] [PubMed] [Google Scholar]
- 21.Bouhana KS, et al. Identification of pan-Trk inhibitors for the treatment of Trk-driven cancers; Proceedings of the 103rd Annual Meeting of the American Association for Cancer Research; Chicago, IL. 2012 Mar 31-Apr 4; Philadelphia (PA): AACR; Cancer Res 2012;72(8 Suppl):Abstract nr 1798. [Google Scholar]
- 22.Seo JS, et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res. 2012 doi: 10.1101/gr.145144.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.George DJ, et al. Sustained in vivo regression of Dunning H rat prostate cancers treated with combinations of androgen ablation and Trk tyrosine kinase inhibitors, CEP-751 (KT-6587) or CEP-701 (KT-5555) Cancer Res. 1999;59:2395–2401. [PubMed] [Google Scholar]
- 24.Cui JJ, et al. Structure based drug design of crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal-epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK) J Med Chem. 2011;54:6342–6363. doi: 10.1021/jm2007613. [DOI] [PubMed] [Google Scholar]
- 25.Gnirke A, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou W, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berger MF, et al. Integrative analysis of the melanoma transcriptome. Genome Res. 2010;20:413–427. doi: 10.1101/gr.103697.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doebele RC, et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res. 2012;18:1472–1482. doi: 10.1158/1078-0432.CCR-11-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Helfrich BA, et al. Antitumor activity of the epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor gefitinib (ZD1839, Iressa) in non-small cell lung cancer cell lines correlates with gene copy number and EGFR mutations but not EGFR protein levels. Clin Cancer Res. 2006;12:7117–7125. doi: 10.1158/1078-0432.CCR-06-0760. [DOI] [PubMed] [Google Scholar]
- 33.Davies KD, et al. Identifying and Targeting ROS1 Gene Fusions in Non-Small Cell Lung Cancer. Clin Cancer Res. 2012 doi: 10.1158/1078-0432.CCR-12-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marek L, et al. Fibroblast growth factor (FGF) and FGF receptor-mediated autocrine signaling in non-small-cell lung cancer cells. Mol Pharmacol. 2009;75:196–207. doi: 10.1124/mol.108.049544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pitts TM, et al. Association of the epithelial-to-mesenchymal transition phenotype with responsiveness to the p21-activated kinase inhibitor, PF-3758309, in colon cancer models. Front Pharmacol. 2013;4:35. doi: 10.3389/fphar.2013.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sasaki T, et al. A novel ALK secondary mutation and EGFR signaling cause resistance to ALK kinase inhibitors. Cancer Res. 2011;71:6051–6060. doi: 10.1158/0008-5472.CAN-11-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.