Abstract
Background
Etiolated seedlings initiate grana stacking and chlorophyll biosynthesis in parallel with the first exposure to light, during which phytochromes play an important role. Functional phytochromes are biosynthesized separately for two components. One phytochrome is biosynthesized for apoprotein and the other is biosynthesized for the chromophore that includes heme oxygenase (HO).
Methodology/Principal Finding
We isolated a ho1 homolog by map-based cloning of a maize elongated mesocotyl2 (elm2) mutant. cDNA sequencing of the ho1 homolog in elm2 revealed a 31 bp deletion. De-etiolation responses to red and far-red light were disrupted in elm2 seedlings, with a pronounced elongation of the mesocotyl. The endogenous HO activity in the elm2 mutant decreased remarkably. Transgenic complementation further confirmed the dysfunction in the maize ho1 gene. Moreover, non-appressed thylakoids were specifically stacked at the seedling stage in the elm2 mutant.
Conclusion
The 31 bp deletion in the ho1 gene resulted in a decrease in endogenous HO activity and disrupted the de-etiolation responses to red and far-red light. The specific stacking of non-appressed thylakoids suggested that the chlorophyll biosynthesis regulated by HO1 is achieved by coordinating the heme level with the regulation of grana stacking.
Introduction
Chlorophyll (Chl) plays a central role in the harvesting of light energy for photosynthesis. The regulation of Chl metabolism includes all levels of control to balance the metabolic flow in response to external and endogenous variation during plant development [1-5]. In the dark, germinating seedlings utilize all of the nutrients contained in the seeds to establish conditions suited for harvesting light signals. This results in the dramatic elongation of the hypocotyl. Within the cotyledons, proplastids differentiate into etioplasts. A large supply of the Chl precursor protochlorophyllide (Pchlide) is also built up in the prolamellar bodies [1,6-9].
During the first exposure to light, etiolated seedlings initiate grana stacking and reduce the accumulated Pchlide to Chl; these changes occur in parallel and collectively lead to rapid greening [6,10]. This process is called photomorphogenesis [11,12], during which phytochromes play an important role and are solely responsible for the perception of red and far-red light [13-15]. In Arabidopsis, the phytochrome family consists of five genes, PHYA-E [16]. Most monocots typically have the following three phytochromes: PhyA, PhyB, and PhyC [15,17]. In maize, an ancestral genomic duplication has enlarged the total family size to at least six genes [18-22].
Holophytochrome biosynthesis requires the convergence of two separate pathways, one for synthesis of the apoprotein and another for the synthesis of the chromophore phytochromobilin (PΦB) [21]. Heme oxygenase (HO; EC1.14.99.3) belongs to the PΦB synthetic branch and is responsible for oxidizing heme to biliverdin (BV) [12,23-28]. BV is then reduced to PΦB by PΦB synthase and is subsequently isomerized [29]. Although phytochrome apoproteins are encoded by a multigene family, it is likely that all plant apophytochromes bind the same chromophore [30,31]. Therefore, the genetic disruption of chromophore PΦB synthesis could inactivate the entire phytochrome system. Such disruption usually leads to a yellowish phenotype. There are a number of known mutants in which the synthesis of chromophore PΦB is disrupted, such as the Arabidopsis mutants hy1 and hy2 [25,26,32,33], the tobacco (Nicotiana plumbaginifolia) mutants pew1 and pew2 [34], the pea (Pisum sativum) mutants pcd1 and pcd2 [24,35], the tomato (Lycopersicon esculentum) mutants au and yg-2 [23,36], the rice mutant se-5 [27,37], and the maize mutant elm1 [38,39]. All of the above mutants are defective in either HO or PΦB synthesis.
There are four putative HO genes in Arabidopsis; these genes are known as HY1 (AtHO1), AtHO2, AtHO3 and AtHO4. Genetic analysis demonstrated that HY1 is responsible for the majority of BV synthesis; hy1 mutant plants have long hypocotyls and decreased accumulation of chlorophyll [25,26]. Further research indicated that family members other than the AtHO1 gene also play a role in synthesizing BV during photomorphogensis, but the effects of these genes are subtler [12,28]. Inactivation of HO1 in the tomato yg-2 mutant enhances the heme level; this post-translationally inhibits the first enzyme committed to tetrapyrrole biosynthesis, Glu-tRNA reductase (designated HEMA) and decreases Chl biosynthesis [40,41]. HO is a mutifunctional enzyme that is involved in many biological processes. Beside its role in oxidizing heme, this protein also participates in programmed cell death (PCD) [42], adventitious root formation [43-45], and protection from oxidative damage [46-51].
In our current research, we isolated a ho1 homolog by map-based cloning in the maize elongated mesocotyl2 (elm2) mutant. elm2 seedlings displayed a disruption in de-etiolation responses under red and far-red light conditions, with a pronounced elongation of the mesocotyl. The endogenous HO activity in elm2 decreased remarkably. The chloroplast ultrastructure showed that the elm2 mutant was delayed in the stacking of the grana but not in the stacking of the non-appressed thylakoids in the seedling stage. In conclusion, we believe that ELM2 encodes a HO1 homolog in maize. The specific stacking of non-appressed thylakoids in the elm2 mutant suggests that chlorophyll biosynthesis is regulated by HO1 via the coordination of the heme level with the regulation of grana stacking.
Materials and Methods
Plant materials and the mapping population
The maize elm2 mutant is a spontaneous yellow-green leaf mutant isolated from an open-pollinated population. This mutant was self-pollinated for more than 8 generations before mapping based cloning was undertaken. The F2 mapping population was generated by crossing the elm2 mutant with the normal green Zheng58 maize inbred line. In total, 8,350 recessive individuals from the F2 generation with a yellow-green phenotype were selected for mapping.
Marker development, gene annotation and cDNA sequencing
The well-developed SSR markers were obtained from the Maize Genetics and Genomics Database (http://www.maizegdb.org/). To narrow down the region of the targeted locus, BAC sequences on chromosome 9 were used for developing new SSR, InDel and CAPS markers (Table S1). SSR sequences were analyzed using SSRHunter 1.3 software. InDel and CAPS markers were developed by comparing elm2 and Zheng58 sequences after PCR amplification and sequencing.
Gene annotation within the located region was obtained from the Maizesequence Database (http://www.maizesequence.org). cDNA sequences of the HO1 homolog from the wild-type line Zheng58 and the mutant elm2 were amplified with the forward primer 5’-GTCGCTTCCCCGGCACCGTAC-3’ and the reverse primer 5’-CTCACCAGTAATAAAGTTTTAACAG-3’. The resulting amplicons were sequenced and compared. All of the above primers were designed using Primer 5.0 software.
Transmission electron microscopy analysis
Leaf samples from the wild-type Zheng58 inbred line and the elm2 mutant were harvested from 4-week-old seedlings and 2-month-old adults grown under natural conditions. Only the top fully expanded leaves (except the flag leaf) were taken as samples. Fresh leaves were quickly sliced into strips with a width of 2 mm. The strips were then fixed in a 3% glutaraldehyde solution containing 0.1 M potassium phosphate (pH 7.2) and further fixed in 1% OsO4. The samples were then dehydrated in an ethanol series and embedded in Spurr resin prior to thin sectioning [52,53]. After staining with lead citrate, the samples were examined using a transmission electron microscope (JEM-1230; JEOL).
Mesocotyl measurement and RT-PCR analysis
Seeds were grown in nutrient soil in artificial-climate chambers under continuous light or in constant darkness at 28°C for 7 d. The light intensities were set to 100 μmol m-2 s-1 for white light, 30 μmol m-2 s-1 for red light, 2.5 μmol m-2 s-1 for far-red light, and 10 μmol m-2 s-1 for blue light [38]. On day 7, the parts above the mesocotyl were sampled for RNA isolation and cDNA synthesis before RT-PCR analysis [52,53]. Meanwhile, the mesocotyl length was measured to nearest millimeter.
Total RNA was isolated from 100 mg (fresh weight) of maize seedling tissue from above the mesocotyl using an RNA Isolation Kit (Tiangen Biotech). High-quality first-strand cDNA was generated from 5 μg total RNA using Invitrogen's cDNA Synthesis Kit. Specific primers were designed using Primer 5.0 and were used for RT-PCR analysis (Table S2). PCR was performed using 2 µl of a 5-fold dilution of the cDNA, 1 µl each of 10 pmol forward and reverse primer solutions, and 1 unit of Taq polymerase (TransGen Biotech) in a 20 µl reaction volume. The relative abundance of β-actin was used as an internal standard.
Chloroplast isolation and heme oxygenase assay
The seeds were grown in nutrient soil in artificial-climate chambers under continuous 100 μmol m-2 s-1 white light at 28°C for 10 d. Approximately 0.3 g of the top well-expanded leaves were used for chloroplast isolation according to the methods of Muramoto et al. [26], Balestrasse et al. [54], and Xuan et al. [43] with minor modifications.
An HO assay was performed as previously described with minor modifications [26,43,55]. The reaction was initiated by adding NADPH. One unit of activity was calculated as the quantity of enzyme needed to produce 1 nM BV per 30 min at 37°C.
Transgenic complementation
For transgenic identification, the Elm2 and elm2 genes were transformed into the Arabidopsis ho1 mutant hy1-100 using a pCAMBIA3301 plasmid; this was performed because Agrobacterium-mediated transformation is difficult in maize. This plasmid contains kanamycin and phosphinothricin resistance for bacteria selection and plant selection, respectively. The transformed gene is driven by a CaMV 35S promoter. The full-length cDNA fragments encoding Elm2 and elm2 were obtained by PCR amplification using the forward primer 5’-AGATATCGTCCGTCATCGGTGCCGTCG-3’ and the reverse primer 5’-CGGATCCCCCCCTCACCAGTAATAAAG-3’. The genes and the vector were digested with EcoRV and BamHI before sub-cloning. The Agrobacterium strain GV3101 harbored the constructs and was used to transform the Arabidopsis hy1-100 mutant. Homozygous transgenic lines of the T3 generation were used for the comparison of the hypocotyl length.
Determination of pigments
Sampling for the Chl and carotenoid (Car) determination was performed in parallel with the transmission electron microscopy analysis. Total Chl and Cars were extracted with 80% acetone and detected with DU 800 UV/Vis Spectrophotometers (Beckman Coulter). Total Chl and total Cars were calculated according to the absorbance of the extract at 663 nm, 646 nm and 470 nm using equations established by Lichtenthaler and Wellburn [56]: Ca=12.21A663-2.81A646, Cb=20.13A646-5.03A663, and Cx+=(1000A470-3.27Ca-104Cb)/229).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: AtHO1 (AT2G26670), AtHO2 (AT2G26550), AtHO3 (AT1G69720), AtHO4 (AT1G58300), SbHO1 (XP_002438642), OsHO1 (NP_001058011), B73-Chr9 (KC404968), B73-Chr6 (AFW76417), Zheng58-Chr9 (KC404967), Zheng58-Chr6 (KC731572), elm2-Chr9 (KC404965), elm2-Chr6 (KC404966), and EU962994.
Results
Genetic characterization of the elm2 mutant
The elm2 mutant was a spontaneous mutant with yellow-green leaves; this mutant was isolated from an open-pollinated population (Figure 1). The mutant line was self-pollinated for more than 8 generations before the following research was undertaken. In addition to yellow-green leaves, the mutant also presented with an elongated mesocotyl under continuous red or far-red light conditions (Figure 2C, D, 3). A similar phenotype in maize was first reported by Sawers et al. [38]; accordingly, we designated the current mutant as elm2.
Figure 1. Phenotypes of the wild-type line Zheng58, the mutant elm2, and the F2 plants.
A, 10 d elm2 mutant seedlings (left), Zheng58 wild-type seedlings (middle), and F2 seedlings (right). B, 4-week-old Zheng58 seedlings (left) and elm2 seedlings (right). C, 2-month-old Zheng58 adult plants (left) and elm2 adult plants (right) at the tasseling stage. Scale bar=5 cm.
Figure 2. De-etiolation responses in Zheng58 wild-type plants and elm2 mutants.

Representative seedlings were photographed after 7 d of growth in continuous darkness (A), white light (B), red light (C), far-red light (D) or blue light (E) conditions. Left, Zheng58 wild-type seedlings; Right, elm2 mutants. Scale bar=1 cm.
Figure 3. Statistical measurement of mesocotyl length in Zheng58 wild-type plants and elm2 mutant plants.
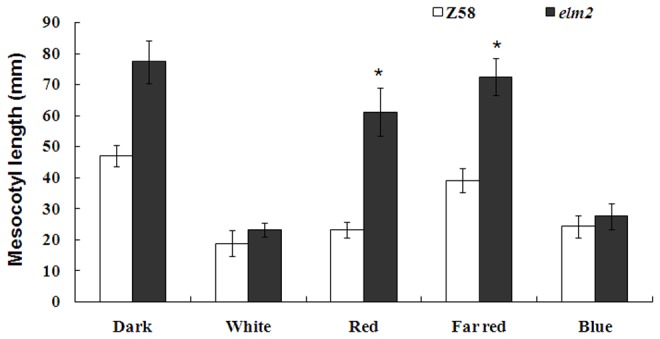
Zheng58 wild-type seedlings and mutant elm2 seedlings were grown for 7 d in continuous darkness, white light, red light, far-red light, or blue light conditions. The sample size was 16-18 seedlings per treatment/genotype. Asterisk indicates significant difference as compared with Zheng58 wild-type at P<0.01 (Student’s t test).
All crosses of elm2 to the wild-type inbred lines Zheng58, Mo17 and B73 yielded only wild-type F1 plants. The F2 segregation ratio of crosses with Zheng58 and Mo17 tended to be 3:1 (green: yellow-green plants), while the ratio of crosses with B73 tended to be 15:1 (as tested by Chi-square, χ2<χ2 (0.05, 1)=3.84; P>0.05; Table 1). This result indicated that the yellow-green phenotype most likely results from a recessive mutation at a single locus and that another locus exists in B73 that can complement this mutation.
Table 1. Genetic segregation of F2 generations.
| Cross | Green leaf | Yellow Leaf | Theoretical Ratio | χ2 |
|---|---|---|---|---|
| elm2×Mo17 | 229 | 66 | 3: 1 | 0.9503 |
| elm2×Zheng58 | 220 | 73 | 3: 1 | 0.0011 |
| elm2×B73 | 691 | 48 | 15: 1 | 0.0398 |
The F2 segregation ratio was tested by Chi-square: χ2<χ2 (0.05, 1)=3.84, P>0.05.
Determination of pigments and chloroplast development in the elm2 mutant
The elm2 mutation resulted in a reduction in total Chl as well as in Car content (Table 2). The total Chl content in elm2 was only 16.4% (0.35/2.14) of that found in the wild-type line Zheng58 at the seedling stage; the Chl content in elm2 increased to 47.2% (1.49/3.16) of the wild type at the tasseling stage (Table 2). The Car content was relatively stable at the two different stages; Car content in elm2 was maintained at approximately 60% (0.21/0.34; 0.24/0.40) of the level observed in the wild-type line Zheng58 (Table 2). In addition, the elm2 mutant showed a substantial increase in the Chl a/b ratio. This ratio was 8.99 at the seedling stage and declined to 4.44 at the tasseling stage (Table 2). These results suggest that the elm2 mutation mainly affected Chl content rather than Car accumulation, and this delayed de-etiolation during photomorphgenesis.
Table 2. Pigment measurements.
| Total Chl (mg g-1 FW) | Chl a:b Ratio | Car (mg g-1 FW) | Growth Stage | |
|---|---|---|---|---|
| elm2 | 0.35±0.02 | 8.99±0.26* | 0.21±0.01 | Seedling |
| Zheng58 | 2.14±0.23 | 3.70±0.06 | 0.34±0.02 | Seedling |
| Mo17 | 2.10±0.19 | 3.69±0.10 | 0.34±0.03 | Seedling |
| elm2 | 1.49±0.09 | 4.44±0.17* | 0.24±0.01 | Tasseling |
| Zheng58 | 3.16±0.04 | 3.43±0.14 | 0.40±0.03 | Tasseling |
| Mo17 | 3.19±0.22 | 3.21±0.11 | 0.39±0.01 | Tasseling |
Chl and Car were measured in 80% acetone extracts from the top fully expanded leaves (except the flag leaf) from 4-week-old seedlings or tasseling adults grown in field. Mean and SE values were calculated from five independent determinations. Asterisk indicates significant difference as compared with Zheng58 wild-type at P<0.01 (Student’s t test).
Detection of the chloroplast ultrastructure revealed that thylakoid stacking was abnormal in elm2 compared to the wild-type line Zheng58. The elm2 mutant specifically stacked non-appressed thylakoids at the seedling stage (Figure 4B). The wild-type line Zheng58 had more and larger granal stacks at the seedling stage, which became much denser at the tasseling stage (Figure 4A, C). Only rare granal stacks appeared in the elm2 mutant at the tasseling stage (Figure 4D). This indicated that the elm2 mutation apparently delayed thylakoid stacking, especially grana stacking, during photomorphogenesis.
Figure 4. Electron microscope images of chloroplast structures.
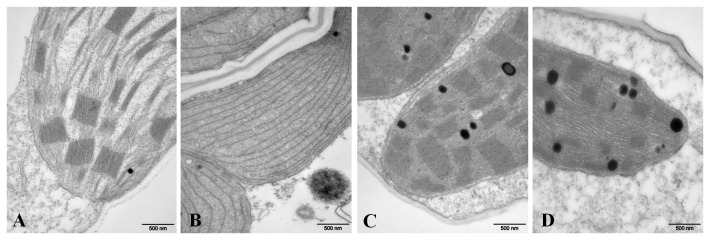
A and C, Zheng58 wild-type plants at the seedling and tasseling stages, respectively; B and D, elm2 mutant plants at the seedling and tasseling stages, respectively. Scale bar=500 nm.
To map the elm2 locus, an F2 mapping population was generated from a cross between elm2 and the wild-type line Zheng58. The elm2 locus was first mapped to the 9.03 Bin of chromosome 9 between the SSR markers P2 and P4 in 570 F2 recessive individuals with yellow-green leaves (Table S1; Figure 5A). To narrow down the mapping region of the elm2 gene, a larger F2 mapping population consisting of 8,350 recessive individuals segregated from more than 33,000 plants were used for fine mapping. Seventy and sixty-seven recombinant individuals were identified from the 8,350 recessive individuals by the use of the markers P2 and P4, respectively. Through further genotyping of the recombinant plants using the markers P5-P12 (Table S1), we localized the elm2 locus to the interval between markers P6 and P9 and co-segregating with P10-P12 (Table S1; Figure 5B). The candidate region of 920 kb on chromosome 9 was near the centromere; therefore, further mapping would be difficult.
Figure 5. Map-based cloning of elm2.
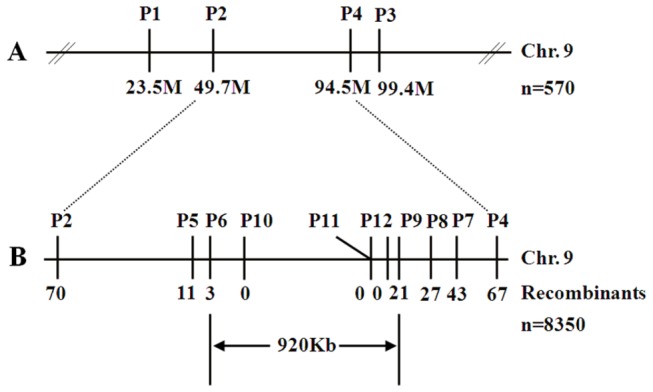
The F2 population was generated by crossing elm2 to the wild-type line Zheng58. 8,350 recessive individuals with a yellow-green phenotype were selected for mapping. The candidate region was finally located to a 920 kb region on chromosome 9, which was near the centromere.
According to the Maizesequence Database (www.maizesequence.org), there are 14 predicted genes within this 920 kb region (Table S2). One of the 14 genes encoded a protein with high homology to HO1 (Table S2, No 6; Figure S1) [57], which has been extensively analyzed and shown to regulate Chl biosynthesis in Arabidopsis [26], pea [24], tomato [23], and rice [27]. Another HO1-like gene was found to reside next to this HO1 homolog (Table S2, No 7). The remaining 12 candidate genes were not reported to participate in Chl metabolism, or their functions were unknown (Table S2). RT-PCR analysis revealed that no bright bands of the HO1-like sequence were detected after 35 cycles amplification in elm2 and the wild-type line Zheng58 (Figure S2). Genomic sequencing of this HO1-like gene showed many inserts and deletions compared to its counterpart in B73 (Figure S3), indicating that this gene most likely does not function during de-etiolation. We then focused on sequencing the HO1 homolog (Table S2, No 6) in elm2; we obtained the complete transcriptional sequence of HO1 by RT-PCR. The predicted protein consisted of 290 amino acids, while the counterpart in B73 and Zheng58 contained 285 amino acids. An homologous alignment of cDNA with Zheng58 and B73 counterparts showed a 31 bp deletion in elm2 that caused a translational frame-shift (Figure 6, S4).
Figure 6. Sequence alignment of HO1 homologs from elm2, B73 and Zheng58.
The start (ATG) or stop (TGA/TAA) codons are enclosed with boxes. The dotted lines indicate the 31 bp deletion present in elm2. Clustal X software was used for the sequence alignment.
A BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) search with the full-length amino acid sequence of Elm2 (KC404967) found another HO1 loci in the B73 genome, on chromosome 6. We also sequenced the paralog on chromosome 6 in elm2 and Zheng58. The CDS in elm2 (KC404966) was only 711 bp, which was caused by a premature termination mutation. Amino acid sequence alignment showed that the paralog in elm2 on chromosome 6 (KC404966) terminated prematurely. In addition, there are at least 3 amino acid variations in the paralog of Zheng58 on chromosome 6 (KC731572) that are conserved in cereal crops (Figure S4).
Disruption of de-etiolation responses in elm2 seedlings
It has been reported that HO is responsible for converting heme to BV, which is the first committed step in the synthesis of PΦB [12,23-29]. The linear tetrapyrrole chromophore PΦB covalently binds a phytochrome apoprotein and generates the photoactive holophytochrome [30]. According to these mapping and cDNA sequencing results, the elm2 mutant should show disrupted responses during de-etiolation under monochromatic light if the ho1 mutation caused the elm2 phenotype. The de-etiolation responses of elm2 seedlings indicated that it was developmentally insensitive to red and far-red light, showing a pronounced elongation of the mesocotyl (equivalent to the hypocotyl in Arabidopsis; Figure 2C, D, 3). While under white and blue light conditions, elm2 showed a moderately elongated mesocotyl corresponding to that observed in the wild-type line Zheng58 (Figure 2B, E, 3), indicating that the blue light receptors do function. Therefore, the morphology of elm2 suggests a disruption in functional phytochromes. Considering the mapping and cDNA sequencing results, we suggest that the elm2 mutant was defective in HO1 activity.
Assay of the endogenous HO activity in elm2
As the above results pointed to a ho1 mutation responsible for the elm2 phenotype, we directly assayed whether the endogenous level of HO activity had changed. The results demonstrate that the activity in elm2 was lower than that in the wild-type lines Zheng58 or Mo17. However, the enzyme activity in elm2 was not inactivated completely and still retained 68% (6.35/9.31) of the activity observed in Zheng58 (Table 3). We assayed the enzyme activity in the F2 plants and found a linkage between enzyme activity and leaf color; namely, the enzyme activity in the yellow plants of the F2 population (elm2 cross to Zheng58) was low and the activity in the green plants was high (Table 3). Therefore, we suggest that the ho1 mutation is responsible for the elm2 phenotype. However, it is worth noting that the activity assay essentially reflected the ability of the HO to oxidize heme to the BV end product. Other HO members in addition to HO1 could participate in this process in elm2, although their effects may be subtler [12,28]. We suspected that the HO1 mutation could affect other physiological reactions or that the regulation of Chl biosynthesis was combinational, just as many reports demonstrate that HO is a multifunctional enzyme [12,23-28,42-51].
Table 3. HO activity assay.
| Plant | Zheng58 | Mo17 | elm2 | F2/green | F2/yellow |
|---|---|---|---|---|---|
| Enzyme Activity (U mg protein-1) | 9.31±0.70 | 8.57±0.32 | 6.35±0.59* | 10.70±0.52 | 6.82±0.68* |
One unit of activity was calculated as the quantity of enzyme needed to produce 1 nM BV per 30 min 37°C. Mean and SE values were calculated from four independent determinations. Asterisk indicates significant difference as compared with Zheng58 wild-type or F2 individuals with green phenotype at P<0.01 (Student’s t test).
Transgenic complementation
The HO1 homolog from the wild-type line Zheng58 or the ho1 allele from elm2, both driven by the CaMV 35S promoter, were transformed into the corresponding Arabidopsis hy1-100 mutant. The goal was to determine if the ELM2 or elm2 genes could complement the defective responses of the mutant during de-etiolation. To quantify the complementation, we measured the hypocotyl (equivalent to the mesocotyl in maize) length in the transgenic lines under continuous light or constant darkness at 22 °C for 7 d. The light intensity was equivalent to that used when the maize mesocotyls were measured. The results revealed that there were no statistically differences between the Arabidopsis wild-type line Columbia and the transgenic line over-expressing the Zheng58 HO1 homolog; this indicated that ELM2 could completely rescue the de-etiolation response (Figure S5, S6). Further measurements demonstrated that there were statistically significant differences between the transgenic lines expressing ELM2 or elm2 under continuous white, red or far-red light conditions (Figure S5, S6). Taken together, it appears that the Zheng58 HO1 homolog could rescue the hy1-100 phenotype completely. The ho1 homolog from elm2 can partially complement this phenotype, although the gene was over transcribed by the CaMV 35S promoter. We therefore suggest that the ho1 homolog in elm2 was functionally abnormal and that its mutation caused the defective responses during de-etiolation in elm2.
Discussion
The elm2 mutant was a spontaneous mutant with yellow-green leaves (Figure 1). Genetic characterization of elm2 indicated that its mutation resulted from a recessive mutation at a single locus compared with wild-type line Zheng58. Another locus exists in B73 that can complement elm2 mutation (Table 1). Pigment measurements demonstrated that the Chl level decreased significantly in elm2, especially at the seedling stage. Car content was also affected, but was maintained a relatively high level at the two different growth stages in comparison with Zheng58 (Table 2). Therefore, the elm2 mutation mainly interfered with Chl biosynthesis. Through map-based cloning, the location of the elm2 allele was found to be within a 920 Kb region on chromosome 9 that contains 14 predicted genes (Figure 5). Among these 14 candidate genes, the HO1 homolog was most likely related to the elm2 phenotype; this was determined according to previous reports [23-25,27]. cDNA sequencing of this homolog showed a 31 bp deletion that resulted in a frame-shift mutation. The predicted CDS encodes 290 amino acids and is 15 base pairs longer than the CDSs found in Zheng58 and B73 (Figure 6).
It was reported that HO belongs to the biosynthesis pathway of the phytochrome chromophore; its mutation would cause a phytochrome deficiency and show a disruption in the de-etiolation response [12,25]. Furthermore, HO1 is responsible for the majority of BV synthesis during photomorphogensis. The other gene family members play a role in synthesizing BV, but their effects are subtler [12,25,26,28]. When grown under continuous red or far-red light conditions, elm2 seedlings showed elongated mesocotyls as expected (Figure 2). This result indicated a phytochrome deficiency in elm2. Because phytochrome apoproteins are encoded by a multigene family with at least six copies in maize [18-22] that all bind the same chromophore [30,31], the chromophore synthetic pathway is more likely to cause a functional phytochrome deficiency. Considering this with the cDNA sequencing results (Figure 6, S4), we suggest that the elm2 mutant was defective in HO1 activity.
An assay of the endogenous HO activity revealed that the HO level in elm2 was significantly decreased compared with the wild-type lines Zheng58 and Mo17 (Table 3). Upon further assaying of the F2 plants, we found there was a linkage between enzyme activity and leaf color; the enzyme activity in the yellow plants of F2 population was low, while the activity in the green plants was high (Table 3). The linkage indicated that the elm2 phenotype was most likely caused by a defect in HO activity. Genetic transformation showed that over-expressing the HO1 homolog from Zheng58 could completely rescue the defective responses during de-etiolation of the corresponding Arabidopsis hy1-100 mutant (Figure S5, S6). Arabidopsis plants over-expressing the mutant ho1 homolog from elm2 showed statistically significant differences compared to plants over-expressing HO1 homolog from Zheng58 under continuous white, red or far-red light conditions (Figure S5, S6). Taken together, we suggest that elm2 encoded a mutant ho1 homolog.
A BLAST search with the full-length amino acid sequence of Elm2 (KC404967) found two HO1 loci in B73 genome, with one loci on chromosome 9 (within our located region) and another on chromosome 6. The F2 segregation ratio of crosses to B73 and Zheng58 indicated that the two paralogs in B73 and the one paralog in Zheng58 could function normally (Table 1). The two paralogs in elm2 are both defective, which is consistent with the subsequent cDNA sequencing results. The complete cDNA of ho1 in elm2 on chromosome 9 (KC404965) showed a 31 bp deletion that resulted in a translational frame-shift (Figure 6). The elm2 CDS on chromosome 6 (KC404966) was only 711 bp; this truncated sequence was caused by a premature termination mutation (Figure S4). Although the CDS length of the paralog in Zheng58 on chromosome 6 (KC731572) was as expected, there are at least 3 amino acid variations in this paralog that are conserved in cereal crops (Figure S4). Because maize is often cross-pollinated, long-term self-pollination for breeding may result in genetic changes [58]. Deleterious mutations are accumulated until the persistence of the phenotype is threatened [59]; this phenomenon is usually referred to as inbreeding depression [60]. This phenomenon is commonly observed in the conservation of homozygous inbred lines by self-pollination [61]. The genetic variation in the HO1 gene in maize presents a good example of in-depth inbreeding depression. In addition, the HO1 gene belongs to a small family [12]; therefore, the genetic divergency can potentially be used as a selection index in future breeding programs to reduce the time and effort required in large-scale field tests [58].
Examination of the chloroplast ultrastructure revealed that non-appressed thylakoids were specifically stacked in elm2 at seedling stage (Figure 4B). Even at the tasseling stage, only rare granal stacks appeared in elm2 (Figure 4D). This phenotype was different than that observed in Chl biosynthetic mutants, such as the Arabidopsis porB and porC single mutants and the porB porC double mutant [7] and the rice ygl1 mutant (encoding Chl synthase) [53]. Chloroplasts in these mutants contained primarily unstacked thylakoids, with occasional distributed double membrane stacks. While the granal stacks in the rice 824ys mutant (encoding divinyl reductase) [62] appeared relatively normal, they appeared was less dense than in the wild type. Previous studies have shown that Chl biosynthesis appears to be feedback-inhibited by HEMA activity; this is performed by controlling the heme level, as HEMA is the first enzyme committed to tetrapyrrole biosynthesis [40,41]. The abnormal grana stacking in elm2 indicated that the regulation of Chl biosynthesis by HO1 may be a combinational result, which is achieved by coordinating the inhibition of HEMA activity with the regulation of grana stacking. The ho1 mutants will otherwise show grana stacking characteristics similar to the Chl biosynthetic mutants. The specific regulation of grana stacking most likely was achieved by regulating the expression of Lhcb via phytochrome signaling [63].
In conclusion, we isolated a ho1 homolog by map-based cloning from the maize elm2 mutant and characterized its function comprehensively. The 31 bp deletion in the ho1 gene resulted in a decrease in endogenous HO activity and disrupted the de-etiolation responses to red and far-red signal; this resulted in a yellowish phenotype. The divergence of the HO1 gene in different inbred lines is a good example of in-depth inbreeding depression, and this gene has potential as a selection index in future breeding programs. Furthermore, the specific stacking of non-appressed thylakoids demonstrated that HO1 may also regulate grana stacking, which contributed to the yellowish phenotype observed in the ho1 mutants.
Supporting Information
Phylogenetic analysis of ELM2 and the previously reported HO enzymes. The neighbor-joining method designed in the MEGA 4.0 software program was used to construct the phylogenetic tree. The branch length indicates the extent of divergence according to the scale at the bottom.
(TIF)
RT-PCR analysis of the 14 candidate genes. Z58-D and elm2-D are seedlings (above the mesocotyl) from the Zheng58 wild-type and the elm2 mutant, respectively, under constant darkness for 7 d; Z58-W and elm2-W are seedlings (above the mesocotyl) from the Zheng58 wild type and the elm2 mutant, respectively, under continuous white light for 7 d. The 14 candidate genes are labeled G1-G14. β-actin was amplified as a control.
(TIF)
Genomic sequence comparisons of the HO1-like gene on chromosome 9 in elm2 and B73. The predicted exons are underlined. The start codon and termination codon are indicated by black arrows. The predicted CDS in elm2 terminated prematurely; the length in elm2 is 552 bp whereas its counterpart in B73 is 729 bp.
(TIF)
Amino acid sequence comparison of HO1 paralogs in maize, sorgum, and rice. The black triangles denote the amino acid mutations in Zheng58-Chr6. The white triangle marks the premature termination in elm2-Chr6. The black arrow indicates the frame shift in elm2-Chr9.
(TIF)
De-etiolation responses in transgenic lines. The seedlings are, from left to right, the Columbia wild-type, the hy1-100 mutant, the transgenic lines hy1-100/ZmHO1-ox (with HO1 from Zheng58), and hy1-100/Zmho1-ox (with ho1 from elm2). A, constant darkness; B, white light; C, red light; D, far-red light; E, blue light. The sample size is 15-18 seedlings per treatment/genotype. Scale bar=1 mm.
(TIF)
Statistical measurement of hypocotyl length in transgenic lines. hy1-100/ZmHO1-ox and hy1-100/Zmho1-ox are transgenic lines with the HO1 sequence from Zheng58 and ho1 from elm2, respectively. The sample size is 15-18 seedlings per treatment/genotype. Bars denoted by the different letters were different significantly at P<0.01 according to Tukey's multiple range test.
(TIF)
The PCR-based molecular markers designed for fine mapping.
(PDF)
Gene annotation within the identified region.
(PDF)
Acknowledgments
We thank Dr. Yufeng Yang (Henan Academy of Agricultural Sciences, China), and Dr. Panfeng Zhao (Beijing DBN Biotechnology Center, China) for their helpful discussions.
Funding Statement
This work was supported by the National Science Foundation of China (31025018) and the Ministry of Science and Technology (2012AA10A305). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Von Wettstein D, Gough S, Kannangara CG (1995) Chlorophyll Biosynthesis. Plant Cell 7: 1039-1057. doi: 10.2307/3870056. PubMed: 12242396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beale SI (1999) Enzymes of chlorophyll biosynthesis. Photosynth Res 60: 43-73. doi: 10.1023/A:1006297731456. [DOI] [Google Scholar]
- 3. Hörtensteiner S (2006) Chlorophyll degradation during senescence. Annu Rev Plant Biol 57: 55-77. doi: 10.1146/annurev.arplant.57.032905.105212. PubMed: 16669755. [DOI] [PubMed] [Google Scholar]
- 4. Tanaka R, Tanaka A (2007) Tetrapyrrole biosynthesis in higher plants. Annu Rev Plant Biol 58: 321-346. doi: 10.1146/annurev.arplant.57.032905.105448. PubMed: 17227226. [DOI] [PubMed] [Google Scholar]
- 5. Masuda T, Fujita Y (2008) Regulation and evolution of chlorophyll metabolism. Photochem Photobiol Sci 7: 1131-1149. doi: 10.1039/b807210h. PubMed: 18846277. [DOI] [PubMed] [Google Scholar]
- 6. Reinbothe S, Reinbothe C (1996) Regulation of chlorophyll biosynthesis in angiosperms. Plant Physiol 111: 1-7. PubMed: 12226272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Frick G, Su Q, Apel K, Armstrong GA (2003) An Arabidopsis porB porC double mutant lacking light-dependent NADPH: protochlorophyllide oxidoreductases B and C is highly chlorophyll-deficient and developmentally arrested. Plant J 35: 141-153. doi: 10.1046/j.1365-313X.2003.01798.x. PubMed: 12848821. [DOI] [PubMed] [Google Scholar]
- 8. Masuda T, Fusada N, Oosawa N, Takamatsu Ki Yamamoto YY et al. (2003) Functional analysis of isoforms of NADPH: protochlorophyllide oxidoreductase (POR), PORB and PORC, in Arabidopsis thaliana. Plant Cell Physiol 44: 963-974. doi: 10.1093/pcp/pcg128. PubMed: 14581621. [DOI] [PubMed] [Google Scholar]
- 9. Reinbothe C, Bakkouri ME, Buhr F, Muraki N, Nomata J et al. (2010) Chlorophyll biosynthesis: spotlight on protochlorophyllide reduction. Tr Plant Sci 15: 614-624. doi: 10.1016/j.tplants.2010.07.002. PubMed: 20801074. [DOI] [PubMed] [Google Scholar]
- 10. Green BR, Durnford DG (1996) The chlorophyll-carotenoid proteins of oxygenic photosynthesis. Annu Rev Plant Physiol Plant Mol Biol 47: 685-714. doi: 10.1146/annurev.arplant.47.1.685. PubMed: 15012305. [DOI] [PubMed] [Google Scholar]
- 11. von Arnim AG, Deng XW (1994) Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79: 1035-1045. doi: 10.1016/0092-8674(94)90034-5. PubMed: 8001131. [DOI] [PubMed] [Google Scholar]
- 12. Davis SJ, Bhoo SH, Durski AM, Walker JM, Vierstra RD (2001) The heme-oxygenase family required for phytochrome chromophore biosynthesis is necessary for proper photomorphogenesis in higher plants. Plant Physiol 126: 656-669. doi: 10.1104/pp.126.2.656. PubMed: 11402195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Smith H (2000) Phytochromes and light signal perception by plants--an emerging synthesis. Nature 407: 585-591. doi: 10.1038/35036500. PubMed: 11034200. [DOI] [PubMed] [Google Scholar]
- 14. Quail PH (2002) Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol 3: 85-93. doi: 10.1038/nrg731. PubMed: 11836510. [DOI] [PubMed] [Google Scholar]
- 15. Takano M, Inagaki N, Xie X, Kiyota S, Baba-Kasai A et al. (2009) Phytochromes are the sole photoreceptors for perceiving red/far-red light in rice. Proc Natl Acad Sci U S A 106: 14705-14710. doi: 10.1073/pnas.0907378106. PubMed: 19706555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Clack T, Mathews S, Sharrock RA (1994) The phytochrome apoprotein family inArabidopsis is encoded by five genes: the sequences and expression ofPHYD andPHYE. Plant Mol Biol 25: 413-427. doi: 10.1007/BF00043870. PubMed: 8049367. [DOI] [PubMed] [Google Scholar]
- 17. Mathews S, Sharrock RA (1996) The phytochrome gene family in grasses (Poaceae): a phylogeny and evidence that grasses have a subset of the loci found in dicot angiosperms. Mol Biol Evol 13: 1141-1150. doi: 10.1093/oxfordjournals.molbev.a025677. PubMed: 8865668. [DOI] [PubMed] [Google Scholar]
- 18. Christensen AH, Quail PH (1989) Structure and expression of a maize phytochrome-encoding gene. Gene 85: 381-390. doi: 10.1016/0378-1119(89)90431-9. PubMed: 2628175. [DOI] [PubMed] [Google Scholar]
- 19. Childs KL, Miller FR, Cordonnier-Pratt MM, Pratt LH, Morgan PW et al. (1997) The sorghum photoperiod sensitivity gene, Ma3, encodes a phytochrome B. Plant Physiol 113: 611-619. doi: 10.1104/pp.113.2.611. PubMed: 9046599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Basu D, Dehesh K, Schneider-Poetsch HJ, Harrington SE, McCouch SR et al. (2000) Rice PHYC gene: structure, expression, map position and evolution. Plant Mol Biol 44: 27-42. doi: 10.1023/A:1006488119301. PubMed: 11094977. [DOI] [PubMed] [Google Scholar]
- 21. Sheehan MJ, Farmer PR, Brutnell TP (2004) Structure and expression of maize phytochrome family homeologs. Genetics 167: 1395-1405. doi: 10.1534/genetics.103.026096. PubMed: 15280251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schnable PS, Ware D, Fulton RS, Stein JC, Wei F et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112-1115. doi: 10.1126/science.1178534. PubMed: 19965430. [DOI] [PubMed] [Google Scholar]
- 23. Terry MJ, Kendrick RE (1996) The aurea and yellow-green-2 mutants of tomato are deficient in phytochrome chromophore synthesis. J Biol Chem 271: 21681-21686. doi: 10.1074/jbc.271.35.21681. PubMed: 8702958. [DOI] [PubMed] [Google Scholar]
- 24. Weller JL, Terry MJ, Rameau C, Reid JB, Kendrick RE (1996) The Phytochrome-Deficient pcd1 Mutant of Pea Is Unable to Convert Heme to Biliverdin IX [alpha]. Plant Cell 8: 55-67. doi: 10.1105/tpc.8.1.55. PubMed: 12239355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Davis SJ, Kurepa J, Vierstra RD (1999) The Arabidopsis thaliana HY1 locus, required for phytochrome-chromophore biosynthesis, encodes a protein related to heme oxygenases. Proc Natl Acad Sci U S A 96: 6541-6546. doi: 10.1073/pnas.96.11.6541. PubMed: 10339624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Muramoto T, Kohchi T, Yokota A, Hwang I, Goodman HM (1999) The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. Plant Cell 11: 335-347. doi: 10.2307/3870864. PubMed: 10072395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Izawa T, Oikawa T, Tokutomi S, Okuno K, Shimamoto K (2000) Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant). Plant J 22: 391-399. doi: 10.1046/j.1365-313X.2000.00753.x. PubMed: 10849355. [DOI] [PubMed] [Google Scholar]
- 28. Emborg TJ, Walker JM, Noh B, Vierstra RD (2006) Multiple heme oxygenase family members contribute to the biosynthesis of the phytochrome chromophore in Arabidopsis. Plant Physiol 140: 856-868. doi: 10.1104/pp.105.074211. PubMed: 16428602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Terry MJ, McDowell MT, Lagarias JC (1995) (3Z)- and (3E)-phytochromobilin are intermediates in the biosynthesis of the phytochrome chromophore. J Biol Chem 270: 11111-11118. doi: 10.1074/jbc.270.19.11111. PubMed: 7744741. [DOI] [PubMed] [Google Scholar]
- 30. Terry M (1997) Phytochrome chromophore-deficient mutants. Plant Cell Environ 20: 740-745. doi: 10.1046/j.1365-3040.1997.d01-102.x. [DOI] [Google Scholar]
- 31. Terry MJ, Wahleithner JA, Lagarias JC (1993) Biosynthesis of the plant photoreceptor phytochrome. Arch Biochem Biophys 306: 1-15. doi: 10.1006/abbi.1993.1473. PubMed: 8215388. [DOI] [PubMed] [Google Scholar]
- 32. Koornneef M, Rolff E, Spruit C (1980) Genetic Control of Light-inhibited Hypocotyl Elongation in Arabidopsis thaliana (L.) Heynh. Z Pflanzenphysiol 100: 147-160. [Google Scholar]
- 33. Kohchi T, Mukougawa K, Frankenberg N, Masuda M, Yokota A et al. (2001) The Arabidopsis HY2 gene encodes phytochromobilin synthase, a ferredoxin-dependent biliverdin reductase. Plant Cell 13: 425-436. doi: 10.2307/3871286. PubMed: 11226195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kraepiel Y, Jullien M, Cordonnier-Pratt MM, Pratt L (1994) Identification of two loci involved in phytochrome expression in Nicotiana plumbaginifolia and lethality of the corresponding double mutant. Mol Gen Genet 242: 559-565. doi: 10.1007/BF00285279. PubMed: 8121413. [DOI] [PubMed] [Google Scholar]
- 35. Weller JL, Terry MJ, Reid JB, Kendrick RE (1997) The phytochrome-deficient pcd2 mutant of pea is unable to convert biliverdin IXα to 3 (Z)-phytochromobilin. Plant J 11: 1177-1186. doi: 10.1046/j.1365-313X.1997.11061177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Koornneef M, Cone J, Dekens R, O'Herne-Robers E, Spruit C et al. (1985) Photomorphogenic responses of long hypocotyl mutants of tomato. J Plant Physiol 120: 153-165. doi: 10.1016/S0176-1617(85)80019-5. [DOI] [Google Scholar]
- 37. Yokoo M, Okuno K (1993) Genetic analysis of early mutations induced in the rice cultivar Norin 8. Japan J Breed 43: 1-11. doi: 10.1270/jsbbs1951.43.1. [DOI] [Google Scholar]
- 38. Sawers RJ, Linley PJ, Farmer PR, Hanley NP, Costich DE et al. (2002) elongated mesocotyl1, a phytochrome-deficient mutant of maize. Plant Physiol 130: 155-163. doi: 10.1104/pp.006411. PubMed: 12226496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sawers RJ, Linley PJ, Gutierrez-Marcos JF, Delli-Bovi T, Farmer PR et al. (2004) The Elm1 (ZmHy2) gene of maize encodes a phytochromobilin synthase. Plant Physiol 136: 2771-2781. doi: 10.1104/pp.104.046417. PubMed: 15347785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Terry MJ, Kendrick RE (1999) Feedback inhibition of chlorophyll synthesis in the phytochrome chromophore-deficient aurea and yellow-green-2 mutants of tomato. Plant Physiol 119: 143-152. doi: 10.1104/pp.119.1.143. PubMed: 9880355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cornah JE, Terry MJ, Smith AG (2003) Green or red: what stops the traffic in the tetrapyrrole pathway? Tr Plant Sci 8: 224-230. doi: 10.1016/S1360-1385(03)00064-5. PubMed: 12758040. [DOI] [PubMed] [Google Scholar]
- 42. Wu M, Huang J, Xu S, Ling T, Xie Y et al. (2011) Haem oxygenase delays programmed cell death in wheat aleurone layers by modulation of hydrogen peroxide metabolism. J Exp Bot 62: 235-248. doi: 10.1093/jxb/erq261. PubMed: 20797999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xuan W, Zhu F-Y, Xu S, Huang B-K, Ling T-F et al. (2008) The heme oxygenase/carbon monoxide system is involved in the auxin-induced cucumber adventitious rooting process. Plant Physiol 148: 881-893. doi: 10.1104/pp.108.125567. PubMed: 18689445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cao Z, Geng B, Xu S, Xuan W, Nie L et al. (2011) BnHO1, a haem oxygenase-1 gene from Brassica napus, is required for salinity and osmotic stress-induced lateral root formation. J Exp Bot 62: 4675-4689. doi: 10.1093/jxb/err190. PubMed: 21673093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Han B, Xu S, Xie YJ, Huang JJ, Wang LJ et al. (2012) ZmHO-1, a maize haem oxygenase-1 gene, plays a role in determining lateral root development. Plant Sci 184: 63-74. doi: 10.1016/j.plantsci.2011.12.012. PubMed: 22284711. [DOI] [PubMed] [Google Scholar]
- 46. Noriega GO, Balestrasse KB, Batlle A, Tomaro ML (2004) Heme oxygenase exerts a protective role against oxidative stress in soybean leaves. Biochem Biophys Res Commun 323: 1003-1008. doi: 10.1016/j.bbrc.2004.08.199. PubMed: 15381099. [DOI] [PubMed] [Google Scholar]
- 47. Yannarelli GG, Noriega GO, Batlle A, Tomaro ML (2006) Heme oxygenase up-regulation in ultraviolet-B irradiated soybean plants involves reactive oxygen species. Planta 224: 1154-1162. doi: 10.1007/s00425-006-0297-x. PubMed: 16703357. [DOI] [PubMed] [Google Scholar]
- 48. Xie Y, Ling T, Han Y, Liu K, Zheng Q et al. (2008) Carbon monoxide enhances salt tolerance by nitric oxide-mediated maintenance of ion homeostasis and up-regulation of antioxidant defence in wheat seedling roots. Plant Cell Environ 31: 1864-1881. doi: 10.1111/j.1365-3040.2008.01888.x. PubMed: 18811735. [DOI] [PubMed] [Google Scholar]
- 49. Xie Y, Xu D, Cui W, Shen W (2012) Mutation of Arabidopsis HY1 causes UV-C hypersensitivity by impairing carotenoid and flavonoid biosynthesis and the down-regulation of antioxidant defence. J Exp Bot 63: 3869-3883. doi: 10.1093/jxb/ers078. PubMed: 22419743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xie YJ, Xu S, Han B, Wu MZ, Yuan XX et al. (2011) Evidence of Arabidopsis salt acclimation induced by up-regulation of HY1 and the regulatory role of RbohD-derived reactive oxygen species synthesis. Plant J 66: 280-292. doi: 10.1111/j.1365-313X.2011.04488.x. PubMed: 21205037. [DOI] [PubMed] [Google Scholar]
- 51. Santa-Cruz DM, Pacienza NA, Polizio AH, Balestrasse KB, Tomaro ML et al. (2010) Nitric oxide synthase-like dependent NO production enhances heme oxygenase up-regulation in ultraviolet-B-irradiated soybean plants. Phytochemistry 71: 1700-1707. doi: 10.1016/j.phytochem.2010.07.009. PubMed: 20708206. [DOI] [PubMed] [Google Scholar]
- 52. Jiang H, Li M, Liang N, Yan H, Wei Y et al. (2007) Molecular cloning and function analysis of the stay green gene in rice. Plant J 52: 197-209. doi: 10.1111/j.1365-313X.2007.03221.x. PubMed: 17714430. [DOI] [PubMed] [Google Scholar]
- 53. Wu Z, Zhang X, He B, Diao L, Sheng S et al. (2007) A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol 145: 29-40. doi: 10.1104/pp.107.100321. PubMed: 17535821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Balestrasse KB, Yannarelli GG, Noriega GO, Batlle A, Tomaro ML (2008) Heme oxygenase and catalase gene expression in nodules and roots of soybean plants subjected to cadmium stress. Biometals 21: 433-441. PubMed: 18228149. [DOI] [PubMed] [Google Scholar]
- 55. Muramoto T, Tsurui N, Terry MJ, Yokota A, Kohchi T (2002) Expression and biochemical properties of a ferredoxin-dependent heme oxygenase required for phytochrome chromophore synthesis. Plant Physiol 130: 1958-1966. doi: 10.1104/pp.008128. PubMed: 12481078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lichtenthaler HK, Wellburn AR (1983) Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem Soc T 11: 591-592. [Google Scholar]
- 57. Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596-1599. doi: 10.1093/molbev/msm092. PubMed: 17488738. [DOI] [PubMed] [Google Scholar]
- 58. Jiao Y, Zhao H, Ren L, Song W, Zeng B et al. (2012) Genome-wide genetic changes during modern breeding of maize. Nat Genet 44: 812-815. doi: 10.1038/ng.2312. PubMed: 22660547. [DOI] [PubMed] [Google Scholar]
- 59. Byers D, Waller D (1999) Do plant populations purge their genetic load? Effects of population size and mating history on inbreeding depression. Annu Rev Ecol Syst: 479-513. [Google Scholar]
- 60. Charlesworth D, Willis JH (2009) The genetics of inbreeding depression. Nat Rev Genet 10: 783-796. doi: 10.1038/nrg2664. PubMed: 19834483. [DOI] [PubMed] [Google Scholar]
- 61. Angeloni F, Ouborg N, Leimu R (2011) Meta-analysis on the association of population size and life history with inbreeding depression in plants. Biol Conserv 144: 35-43. doi: 10.1016/j.biocon.2010.08.016. [DOI] [Google Scholar]
- 62. Wang P, Gao J, Wan C, Zhang F, Xu Z et al. (2010) Divinyl chlorophyll (ide) a can be converted to monovinyl chlorophyll (ide) a by a divinyl reductase in rice. Plant Physiol 153: 994-1003. doi: 10.1104/pp.110.158477. PubMed: 20484022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. McCormac AC, Terry MJ (2002) Light-signalling pathways leading to the co-ordinated expression of HEMA1 and Lhcb during chloroplast development in Arabidopsis thaliana. Plant J 32: 549-559. doi: 10.1046/j.1365-313X.2002.01443.x. PubMed: 12445126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic analysis of ELM2 and the previously reported HO enzymes. The neighbor-joining method designed in the MEGA 4.0 software program was used to construct the phylogenetic tree. The branch length indicates the extent of divergence according to the scale at the bottom.
(TIF)
RT-PCR analysis of the 14 candidate genes. Z58-D and elm2-D are seedlings (above the mesocotyl) from the Zheng58 wild-type and the elm2 mutant, respectively, under constant darkness for 7 d; Z58-W and elm2-W are seedlings (above the mesocotyl) from the Zheng58 wild type and the elm2 mutant, respectively, under continuous white light for 7 d. The 14 candidate genes are labeled G1-G14. β-actin was amplified as a control.
(TIF)
Genomic sequence comparisons of the HO1-like gene on chromosome 9 in elm2 and B73. The predicted exons are underlined. The start codon and termination codon are indicated by black arrows. The predicted CDS in elm2 terminated prematurely; the length in elm2 is 552 bp whereas its counterpart in B73 is 729 bp.
(TIF)
Amino acid sequence comparison of HO1 paralogs in maize, sorgum, and rice. The black triangles denote the amino acid mutations in Zheng58-Chr6. The white triangle marks the premature termination in elm2-Chr6. The black arrow indicates the frame shift in elm2-Chr9.
(TIF)
De-etiolation responses in transgenic lines. The seedlings are, from left to right, the Columbia wild-type, the hy1-100 mutant, the transgenic lines hy1-100/ZmHO1-ox (with HO1 from Zheng58), and hy1-100/Zmho1-ox (with ho1 from elm2). A, constant darkness; B, white light; C, red light; D, far-red light; E, blue light. The sample size is 15-18 seedlings per treatment/genotype. Scale bar=1 mm.
(TIF)
Statistical measurement of hypocotyl length in transgenic lines. hy1-100/ZmHO1-ox and hy1-100/Zmho1-ox are transgenic lines with the HO1 sequence from Zheng58 and ho1 from elm2, respectively. The sample size is 15-18 seedlings per treatment/genotype. Bars denoted by the different letters were different significantly at P<0.01 according to Tukey's multiple range test.
(TIF)
The PCR-based molecular markers designed for fine mapping.
(PDF)
Gene annotation within the identified region.
(PDF)




