Abstract
Stomata play significant roles in plant evolution. A trio of closely related basic Helix-Loop-Helix (bHLH) subgroup Ia genes, SPCH, MUTE and FAMA, mediate sequential steps of stomatal development, and their functions may be conserved in land plants. However, the evolutionary history of the putative SPCH/MUTE/FAMA genes is still greatly controversial, especially the phylogenetic positions of the bHLH Ia members from basal land plants. To better understand the evolutionary pattern and functional diversity of the bHLH genes involved in stomatal development, we made a comprehensive evolutionary analysis of the homologous genes from 54 species representing the major lineages of green plants. The phylogenetic analysis indicated: (1) All bHLH Ia genes from the two basal land plants Physcomitrella and Selaginella were closely related to the FAMA genes of seed plants; and (2) the gymnosperm ‘SPCH’ genes were sister to a clade comprising the angiosperm SPCH and MUTE genes, while the FAMA genes of gymnosperms and angiosperms had a sister relationship. The revealed phylogenetic relationships are also supported by the distribution of gene structures and previous functional studies. Therefore, we deduce that the function of FAMA might be ancestral in the bHLH Ia subgroup. In addition, the gymnosperm “SPCH” genes may represent an ancestral state and have a dual function of SPCH and MUTE, two genes that could have originated from a duplication event in the common ancestor of angiosperms. Moreover, in angiosperms, SPCHs have experienced more duplications and harbor more copies than MUTEs and FAMAs, which, together with variation of the stomatal development in the entry division, implies that SPCH might have contributed greatly to the diversity of stomatal development. Based on the above, we proposed a model for the correlation between the evolution of stomatal development and the genes involved in this developmental process in land plants.
Introduction
The origin of terrestrial plants was a key event in the evolutionary history of life on earth [1], [2]. During the transition to a terrestrial habitat, ancestral land plants overcame several challenges, including growing in an environment with a limited water and mineral supply, surviving the harmful effects of enhanced ultraviolet and cosmic rays and defending against attack from a new and diversified set of microbes [2]. Stomata were a key evolutionary innovation that contributed to overcoming many of these challenges. Stomata first appeared more than 410 million years ago [3], [4] and generally consist of two guard cells (GCs) surrounding a pore in the epidermis except that a single guard cell that encircles the pore was found in the moss Funaria [5]. They play significant roles in protecting plants from a non-aqueous atmosphere by mediating gas exchange integral to photosynthesis and water transportation [4], [6]. Some of the earliest branching land plants, including liverworts, hornworts and some mosses, lack stomata, but all other land plants possess them [3]. Fossil evidence shows that the structure of stomata has remained more or less unchanged ever since its origin [3], [7], [8], but the developmental processes that lead to stomata have become more complex, such as the occurrence of amplifying divisions, and also more efficient throughout the land plant phylogeny [4], [9], [10]. These efficiencies in water use, gas exchange and photosynthetic rates are likely key to driving the adaptation of higher plants to a wide array of ecological niches although little is known about possible correlations between stomatal patterning and physiological function [9]–[19].
To date, most investigations of stomata have focused on their development and their reactions to environmental stressors (e.g., [3], [9], [13], [20]–[23]). However, in the last decade some of the genes that regulate stomatal development and patterning have been isolated. In Arabidopsis, these genes are a trio of closely related basic Helix-Loop-Helix (bHLH) genes, known as SPCH (SPEECHLESS), MUTE, and FAMA (here termed the SMF genes). These three genes work together with their heterodimeric partners - SCRM (SCREAM) and SCRM2 - to mediate sequential steps of the cell-state transitions that lead to stomatal formation. These steps include: i) asymmetric cell division that leads to ii) the acquisition of guard mother cell (GMC) identity and, finally, iii) the differentiation of guard cells (GCs) [24]–[28].
The bHLH family, which is typified by the highly conserved bHLH domain, is a large family of transcription factors that is distributed throughout the major eukaryotic lineages [29]. Phylogenetic analysis indicates that the bHLH genes from Arabidopsis are divided into 26 subgroups [27], [29], [30]. The three genes that contribute to stomatal development - i.e., SPCH, MUTE, and FAMA - belong to bHLH subgroup Ia, which also includes seven genes of unknown function. In contrast, SCRM and SCRM2 are members of subgroup IIIb [29].
The function of Ia and IIIb genes may be conserved over evolutionary time. For example, functional analyses have shown that PpSMF1 and PpSMF2, two bHLH group Ia members from the bryophyte Physcomitrella patens, can recapitulate elements of the SPCH, MUTE and FAMA overexpression phenotypes of Arabidopsis thaliana [31]. PpSMF1 can also partially complement mute and fama mutants in A. thaliana [31]. In addition, the partnership between the subgroup Ia SPCH/MUTE/FAMA and the subgroup IIIb SCRM/SCRM2 might have an ancient origin, because both of these two subgroups occur in the basal land plant, P. patens [32].
The evolutionary history of SPCH/MUTE/FAMA genes is nonetheless controversial [31], [33] because there is conflicting information about their origins and the relationships of their paralogs. At least two analyses found that the SPCH and MUTE genes are closest paralogs [29], [34], [35], whereas others suggest that SPCH and FAMA genes are closest paralogs [31], [33]. There are also discrepancies as to whether the homologs from basal land plants fall within the SPCH, MUTE and FAMA clades or represent sister-lineages to these clades [31]. It is important to note that the resolution of these phylogenetic issues will yield insight into the evolution of stomata, because each of the bHLH Ia genes performs a defined role in stomatal development.
Here we investigate the distribution and evolutionary relationships of bHLH Ia genes among land plants to better understand the evolution of genes involved in stomatal development. Thus far, evolutionary analyses of the members of Ia subgroup have been based on a small sample (n <12) of angiosperm species; here we survey a total of 51 species of land plants (and one green algae and two multicellular algae species) for the presence and distribution of subgroup Ia genes. Based on both phylogenetic and structural analyses of the bHLH Ia genes, we interpret their evolution in land plants, and we also predict a model for the correlation between the evolution of stomatal development and the genes involved in their development.
Materials and Methods
Ethics Statement
No specific permits were required for the sampling.
Identification of bHLH Ia Homologs
We surveyed a number of plant databases – such as Phytozome, NCBI, PGDD, PlantTFDB, EST, SRA and FLcDNA databases and other genome databases (Tables 1, 2, S1) - to identify bHLH Ia homologs from plant species. To retrieve Ia homologs from these databases, we performed tBLASTn and BLAST+ (ncbi-blast-2.2.27+) searches using the A. thaliana FAMA [Phytozome:AT3G24140] amino acid sequence as a query.
Table 1. Information of the bHLH Ia genes in the sampled plant species with whole genome sequences.
| Species | Abbr. | Copy number | Database* | |||
| FAMA | MUTE | SPCH | Other Ias | |||
| Volvox carteri | Vca | 0 | 0 | 0 | 0 | Phytozome |
| Physcomitrella patens | Ppa | 2 | 0 | 0 | 0 | Phytozome/PGDD |
| Selaginella moellendorffii | Smo | 3 | 0 | 0 | 0 | Phytozome/PGDD |
| Amborella trichopoda | Atr | 1 | 1 | 1 | 2 | Amborella GD |
| Aquilegia coerulea | Aco | 1 | 1 | 1 | 2 | Phytozome |
| Arabidopsis lyrata | Aly | 1 | 1 | 1 | 7 | Phytozome/PGDD |
| Arabidopsis thaliana | Ath | 1 | 1 | 1 | 7 | Phytozome/PGDD |
| Brachypodium distachyon | Bdi | 1 | 1 | 2 | 7 | Phytozome/PGDD |
| Brassica rapa | Bra | 3 | 3 | 3 | 14 | Phytozome/PGDD |
| Cajanus cajan | Cca | 1 | 1 | 1 | 7 | IIPG/PGDD |
| Capsella rubella | Cru | 1 | 1 | 1 | 9 | Phytozome |
| Carica papaya | Cpa | 1 | 0 | 1 | 4 | Phytozome/PGDD |
| Citrus clementina | Ccl | 1 | 1 | 1 | 3 | Phytozome |
| Citrus sinensis | Csi | 1 | 0 | 1 | 4 | Phytozome |
| Cucumis sativus | Csa | 1 | 1 | 1 | 5 | Phytozome/PGDD |
| Eucalyptus grandis | Egr | 1 | 1 | 1 | 5 | Phytozome |
| Fragaria vesca | Fve | 0 | 1 | 0 | 6 | PFR/PGDD |
| Glycine max | Gma | 2 | 2 | 4 | 12 | Phytozome/PGDD |
| Linum usitatissimum | Lus | 1 | 1 | 4 | 7 | Phytozome |
| Lotus japonicus | Lja | 0 | 1 | 0 | 6 | Kazusa/PGDD |
| Malus domestica | Mdo | 1 | 2 | 2 | 8 | Phytozome/PGDD |
| Manihot esculenta | Mes | 2 | 2 | 2 | 7 | Phytozome |
| Medicago truncatula | Mtr | 0 | 0 | 1 | 5 | Phytozome/PGDD |
| Mimulus guttatus | Mgu | 0 | 0 | 2 | 2 | Phytozome |
| Musa acuminata | Mac | 2 | 2 | 3 | 16 | Banana Genome/PGDD |
| Oryza sativa | Osa | 1 | 1 | 2 | 8 | Phytozome/PGDD |
| Phaseolus vulgaris | Pvu | 1 | 1 | 2 | 6 | Phytozome |
| Phoenix dactylifera | Pda | 1 | 2 | 1 | 5 | Date Palm Draft Sequence |
| Populus trichocarpa | Ptr | 2 | 1 | 2 | 9 | Phytozome/PGDD |
| Prunus persica | Per | 1 | 1 | 1 | 5 | Phytozome/PGDD |
| Ricinus communis | Rco | 1 | 1 | 1 | 4 | Phytozome/PGDD |
| Setaria italica | Sit | 1 | 1 | 2 | 8 | Phytozome |
| Solanum lycopersicum | Sly | 2 | 1 | 0 | 4 | SGN/PGDD |
| Solanum tuberosum | Stu | 2 | 0 | 0 | 4 | PGSC/PGDD |
| Sorghum bicolor | Sbi | 1 | 1 | 2 | 7 | Phytozome/PGDD |
| Thellungiella halophila | Tha | 2 | 1 | 1 | 8 | Phytozome |
| Theobroma cacao | Tca | 1 | 1 | 1 | 5 | CIRAD/PGDD |
| Vitis vinifera | Vvi | 1 | 1 | 1 | 5 | Phytozome/PGDD |
| Zea mays | Zma | 1 | 1 | 3 | 10 | Phytozome/PGDD |
Abbr., Abbreviation.*, database websites: Amborella GD, http://www.amborella.org/; Banana Genome, http://banana-genome.cirad.fr/; CIRAD, http://cocoagendb.cirad.fr/gbrowse/download.html; Date Palm Draft Sequence, http://qatar-weill.cornell.edu/research/datepalmGenome/download.html; IIPG, http://www.icrisat.org/gt-bt/iipg/Home.html; Kazusa, ftp://ftp.kazusa.or.jp/pub/lotus/; NCBI, http://www.ncbi.nlm.nih.gov/; Phytozome, http://www.phytozome.net; PFR, http://www.strawberrygenome.org/; PGSC, http://potatogenomics.plantbiology.msu.edu; PlantTFDB, http://planttfdb.cbi.edu.cn/.
Table 2. Information of the bHLH Ia genes in the plant species with EST or SRA databases.
| Species | Copy Number | EST & cDNA | SRA (spots) | SRA Submission | ||||
| SPCH | MUTE | FAMA | Other Ias | |||||
| Green algae | Nitella hyalina | – | – | – | – | 88,280 | 949,065 | SRA023590 |
| Nitella mirabilis | – | – | – | – | 83,526 | – | – | |
| Liverwort | Marchantia polymorpha | – | – | – | – | 33,722 | 22,854,396 | SRA026315 |
| Fern | Adiantum capillus-veneris | – | – | – | – | 30,544 | – | – |
| Gymnosperms | Cephalotaxus harringtonia | – | – | – | 1 | – | 695,559 | SRA023613 |
| Ginkgo biloba | – | – | 1 | 3 | 21,709 | 64,057 | SRA030487 | |
| Gnetum gnemon | – | – | – | 1 | 10,756 | 432,517 | SRA023615 | |
| Picea glauca# | 1 | – | – | 1 | 321,713 | 10,922,903 | SRA023921 | |
| Picea breweriana* | 1 | – | 1 | – | – | – | – | |
| Picea jezoensis* | 1 | – | 1 | – | – | – | – | |
| Picea smithiana* | 1 | – | 1 | – | – | – | – | |
| Picea sitchensis | – | – | – | 1 | 206,402 | – | – | |
| Pinus banksiana | – | – | 1 | – | 36,387 | 1,397,993 | SRA048732 | |
| Pinus taeda | – | – | 1 | 7 | 329,066 | 4,331,325 | SRA023533 | |
| Sciadopitys verticilliata | – | – | – | 1 | – | 484,806 | SRA023758 | |
#, Sequences from FLcDNA database of Arborea; *, sequences from PCR amplification; -, missing information.
We subjected the identified sequences to three additional filters. First, we discarded sequences that had lower sequence similarities to AT3G24140 than did the first non-subgroup-Ia member from A. thaliana [Phytozome: AT2G22750, a member of IVa]. Second, we examined individual sequences for two characteristic features of subgroup Ia genes: the bHLH domain and the SMF domain [30], [31]. These two domains were identified by Heim et al. [30]. Any sequence that lacked one of the domains was removed from further analysis. In addition, 36 putative Ia genes were modified by hand for a better alignment with the bHLH or SMF domain (Table S1). Finally, we culled redundant transcripts, sequences with incomplete bHLH or sequences with unalignable C-terminal domains from the final dataset.
DNA and RNA Extraction, PCR and RT-PCR Amplification, Cloning and Sequencing
Two types of bHLH Ia genes that grouped with SPCH/MUTE/FAMA genes were found in eight gymnosperm species, but we found no species that had both types. To assess whether these two types existed in one species, we amplified these Ia genes from the DNA and cDNA of three Picea species (Picea breweriana, P. jezoensis, and P. smithiana). Genomic DNAs were extracted from leaves using the modified cetyltrimethylammonium bromide (CTAB) method [36], [37]. Total RNA extraction, purification and first-strand cDNA synthesis followed the protocols of Guo et al. [38]. The primers used to amplify the Ia genes were designed to the EST sequence [GenBank:GW768467] of Pinus banksiana and the predicted unigene [Arborea:GQ04006] from Picea glauca, including FAMA-U5F3 and FAMA-U3R as well as bHLH2-U5F and bHLH2-U3R1 (Table S2). Polymerase chain reaction (PCR), product purification, cloning and sequencing followed the protocols of Ran et al. [39] except an annealing and sequencing reaction temperature of 60°C. In addition, some internal primers were used in sequencing (Table S2).
Phylogenetic Reconstruction
Coding sequences of bHLH Ia genes were aligned using the program Clustal X version 2.0 [40]. The conserved bHLH and SMF domains (a total of 468 bp or 156 aa) [31] were used for the phylogenetic analyses. Substitution saturation was tested using DAMBE 5.3.00 [41], which showed that the third positions were saturated. jModeltest 2 [42] and ProtTest 3.2 [43] were used to identify the best-fit models of nucleotide and amino acid (AA) sequence evolution, respectively. Using both Akaike Information Criterion (AIC) and Bayesian Information Criterion (BIC), the GTR+I+G model was selected for nucleotide sequence analyses and the JTT+I+G model for AA analyses. Bayesian and maximum-likelihood (ML) trees were constructed with MrBayes version 3.1.2 [44] and PhyML version 2.4.4 [45], respectively.
Selection Test and Nucleotide Diversity Analysis
ML analyses were performed to identify regions or clades of the bHLH Ia genes that may have been subject to diversifying selection, using the Fitmodel program version 0.5.3 [46] and the codeml program of PAML version 4.6 [47], respectively. Because the Ia gene dataset was very large, we limited these analyses to a subset of subgroup Ia sequences. SPCH/MUTE/FAMA homologs, all bHLH other Ias from gymnosperms, all bHLH Ias from Physcomitrella and Selaginella, and some of bHLH other Ias from four angiosperm species (Arabidopsis thaliana, Glycine max, Linum usitatissimum and Oryza sativa) were analyzed with Fitmodel, in which the M0, M3, M3+S1 and M3+S2 models were applied. Three genic classes, including clade A, and Ang-MUTE and Ang-SPCH in clade B (Fig. 1A), were analyzed with the branch-site and site models (codeml program), respectively. The parameter settings followed Guo et al. [38]. To test the evolutionary rate variation, DnaSP version 5.10.00 [48] was used to calculate the nucleotide diversity (Pi), the number of nonsynonmous substitutions per nonsynonymous site (Ka), the number of synonymous substitutions per synonymous site (Ks), and the ω (Ka/Ks) value for the sequences of the angiosperm FAMA, MUTE, SPCH and other Ia genes (Fig. 1) because some clades lack members from non-flowering plants.
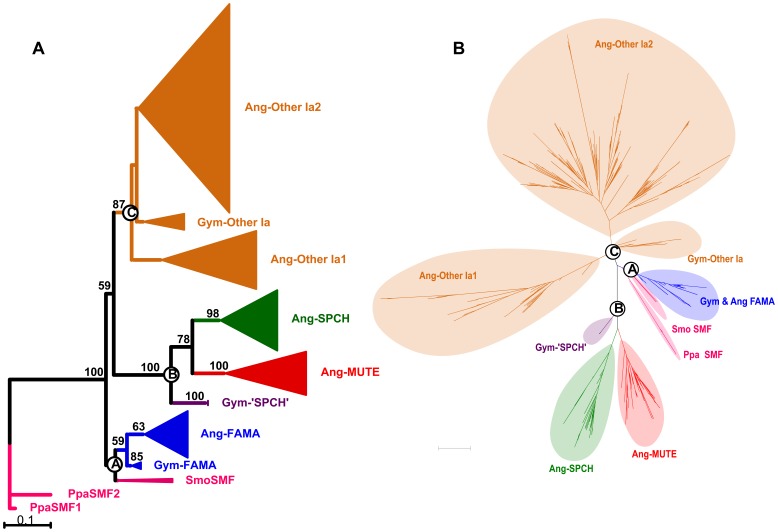
Figure 1. Maximum likelihood (ML) trees of the bHLH Ia genes constructed based on the nucleotide sequences.
A. The tree was rooted with the Ia genes from Physcomitrella patens; B. The tree was not rooted. Numbers above branches indicate bootstrap values higher than 50%. Ang, angiosperm; Gym, gymnosperm; Smo, Selaginella moellendorffii; Ppa, Physcomitrella patens.
Results
Sequence Characterization
We searched complete genomes as well as EST and SRA resources to identify bHLH Ia homologs in basal (non-angiosperm) land plants. Using Blast-based strategies (see Materials and Methods) with a loose searching criteria (E value = 1), we characterized two and three SMF genes in the bryophyte Physcomitrella patens and the lycopsid Selaginella moellendorffii, respectively. However, using similar search methods, we were unable to identify bHLH Ia homologs from the complete genome of a green algae (Volvox carteri), and the EST+cDNA databases of two multicellular green alga (Nitella hyalina and Nitella mirabilis) (88,280 and 88,526 sequences, respectively), a liverwort (Marchantia polymorpha) (33722 sequences) and a fern Adiatum capillus-veneris (30,544 sequences) (Table 2). The inability to identify Ia homologs from EST databases of the liverwort and fern could reflect reality, or it could be caused by the size of EST databases. We note, however, that other non-Ia bHLH genes were successfully retrieved from these species. In addition, we identified 19 Ia homologs from the EST or SRA databases of eight gymnosperms (Table S1), and PCR-amplified six sequences from three spruce species (Table 2). Of the 25 sequences from gymnosperms, 4, 6 and 15 showed similar gene structures with SPCH, FAMA and Other Ias, respectively.
Within angiosperms, SMF (SPCH, MUTE and FAMA) homologs were identified in all 36 species sampled, but homologs of individual genes were not found in five species for FAMA, four species for SPCH and five species for MUTE. However, 14, 6, and 8 species harbored more than one SPCH, MUTE and FAMA homolog (Table 1). In addition, 2–16 copies of other (i.e., non-SPCH, non-MUTE and non-FAMA) Ia members were found throughout the 36 angiosperm species. In total, 395 bHLH Ia sequences from 49 species of land plants were used in the final analyses (Table S1). These coding sequences ranged from 543 (AcoMUTE) to 3114 bp (FveIa_1).
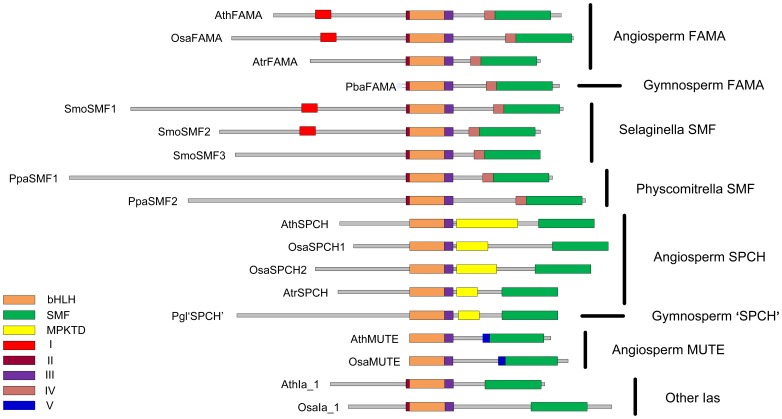
All of the sequences in the final dataset contained both bHLH and SMF domains, but additional domains were also detected (Fig. 2), as suggested by MacAlister and Bergmann [31]. These domains helped to differentiate gene structure among putative SPCH, MUTE and FAMA genes and also to corroborate phylogenetic inferences based on the bHLH and SMF domain sequences (see below). In angiosperms, the FAMA homologs have a pre-bHLH domain (II) and two unique and highly conserved regions; one of these (I) is in the N-terminus between the start codon and the bHLH domain and the other (IV) is immediately N-terminal to the SMF domain. However, four putative FAMA homologs (AtrFAMA CpaFAMA, EgrFAMA, and StuFAMA) lacked region I. All MUTE homologs lacked the pre-bHLH domain (II) and had a unique pre-SMF domain (V). Finally, all SPCH homologs had a MPKTD domain, but it differed in length.
Figure 2. Structural diagram of the bHLH Ia genes from some representative species of land plants.
The regions that are unique to FAMA, MUTE, and SPCH are marked with different colors.
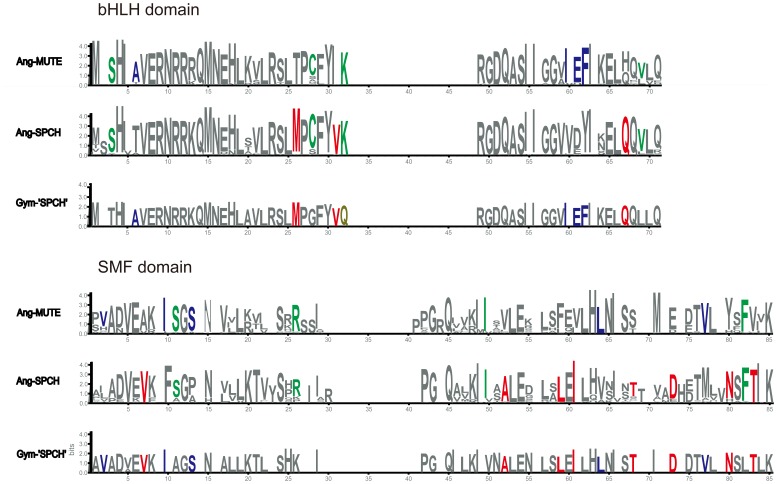
The gene structures in non-angiosperm taxa merit special attention. In gymnosperms, two structures were found, one similar to angiosperm FAMA genes and the other similar to angiosperm SPCH genes. In Selaginella moellendorffii, the three bHLH Ia genes are structurally similar to the angiosperm FAMA genes, especially in domains II and IV, and the domain I was also found in SmoSMF1 and SmoSMF2. In Physcomitrella patens, the two Ia genes are also similar to the angiosperm FAMA genes due to the presence of domains II and IV. Moreover, the other Ia genes also have domain II (Fig. 2). In brief, according to the gene structure analysis, the basal land plants (bryophyte and lycopsid) only harbored FAMA Ia genes, while both FAMA and SPCH Ia genes occurred in gymnosperms. This was consistent with the reconstruction of ancestral gene structure using Mesquite version 2.75 [49], which is shown in Fig. S1. The conserved sequence patterns of the angiosperm SPCH and MUTE genes as well as the gymnosperm ‘SPCH’ genes were generated with WebLogo3 (http://weblogo.berkeley.edu/) and shown in Fig. 3.
Figure 3. Graphical sequence logo representation of bHLH and SMF domains of seed-plant SPCH/MUTE genes.
The conserved sequence pattern was generated using WebLogo3 (http://weblogo.berkeley.edu/). Bits represent the relative frequency of amino acids.
Phylogenetic Analyses
The ML trees generated by nucleotide and AA sequences of the bHLH Ia genes are topologically identical except some branches with low bootstrap support (Figs. 1, S2). Because most Bayesian prosterior probabilities were lower than 0.90, we did not consider the Bayesian tree. The ML trees of the nucleotide sequences with and without outgroups are shown in Figure 1A and 1B, respectively. From the rooted trees, we found that the Ia genes of vascular plants are monophyletic with high bootstrap support (100%) and could be divided into three main clades (A, B and C) (Figs. 1A, S2A, S2B). Clade A included the SMF genes from Selaginella moellendorffii and the FAMA homologs from seed plants (gymnosperm and angiosperm). Clade B comprised two sister subclades, i.e., the gymnosperm ‘SPCH’ genes and the angiosperm MUTE and SPCH genes. Clade C comprised all the other Ia genes from gymnosperms and angiosperms. From the unrooted tree, the bHLH Ia members from Physcomitrella patens grouped with Clade A (Figs. 1B, S2C).
Selection Test and Nucleotide Diversity
The likelihood ratio test (LRT) of the nested models in the Fitmodel program showed that the M3+S1 model was significantly better than the other ones. In the M3+S1 model, the switching rates between ω values (ω1 to ω2, ω1 to ω3, and ω2 to ω3) are equally imposed. Under this model, no codon or branch was inferred to be under relaxed selection (ω3 = 0.32) (Table 3).
Table 3. Results of the LRT test of the models in Fitmodel for the bHLH Ia genes.
| M0 | M3 | M3+S1 | M3+S2 | |
| lnL | −26059.06 | −25848.39 | −25656.58 | −25707.45 |
| ω1 ω2 ω3 | 0.08 | 0.01 0.07 0.20 | 0.00 0.07 0.32 | 0.01 0.19 58.10 |
| p 1 p 2 p 3 | 1.00 | 0.31 0.51 0.17 | 0.40 0.42 0.18 | 0.65 0.34 0.00 |
| R 01 R 02 R 12 | 1.59 1.59 1.59 | 1.66 9.02 434.75 |
The site model test indicated that no site in the three classes A, Ang-MUTE and Ang-SPCH (Fig. 1) experienced positive selection. Using the branch-site model, no positively selected site was found in the angiosperm SPCH or MUTE genes when they acted as the background clade to each other. For the FAMA homologs, four branches (vascular plants, seed plants, angiosperms, and eudicots) were tested, and one positively selected site was detected in the branches of seed plants and angiosperms, respectively, but the positive selection was not supported by the LRT test due to a significance much higher than 0.05 (Table S3).
For angiosperms, the FAMA genes showed the lowest Pi, Ka and ω values, while the SPCH and MUTE genes had similar Pi, Ka, Ks and ω values. In addition, the highest Pi, Ka, Ks and ω values were found in the other Ia genes (Table 4).
Table 4. Sequence information of angiosperm SPCH, MUTE, FAMA and Other Ias.
| Dataset | N | Pi | Ka | Ks | ω (Ka/Ks) |
| SPCH | 53 | 0.25742±0.00454 | 0.11749 | 0.69853 | 0.168 |
| MUTE | 38 | 0.25526±0.00887 | 0.11350 | 0.73050 | 0.155 |
| FAMA | 41 | 0.21573±0.00868 | 0.05844 | 0.72591 | 0.091 |
| Other Ias | 233 | 0.29867±0.00363 | 0.16091 | 0.75129 | 0.214 |
N, Number of sequences; Pi, Nucleotide diversity; Ka, The number of nonsynonymous substitutions per nonsynonymous site; Ks: The number of synonymous substitutions per synonymous site; ω: Ka/Ks.
Discussion
Evolutionary History of the bHLH Ia Genes Involved in Stomatal Development
Genome-wide analyses of the bHLH transcription factor family have found that SPCH, MUTE and FAMA, together with some other proteins of unknown functions, form a well supported clade (subgroup Ia) [27], [29], [30], [35], which is characterized by a conserved region of 18 amino acids immediately N-terminal to the bHLH domain and a domain at the C-terminus of the protein [30], [31]. Some previous phylogenetic analyses found that the SPCH and MUTE genes were the closest sister paralogs (The SPCH genes were not monophyletic) and the FAMA genes grouped with some other subgroup Ia members [29], [34], [35]. However, other studies that focused on the SMF (SPCH, MUTE, FAMA) genes did not obtain the same phylogenetic relationships. For example, Liu et al. [50] found that the SPCH/MUTE/FAMA genes from Arabidopsis thaliana, Oryza sativa and Zea mays formed three monophyletic groups, respectively, although the functions of SPCH and MUTE from Oryza are somewhat divergent from their homologs in Arabidopsis. In contrast, by sampling Arabidopsis, Populus, Oryza, and Physcomitrella, Peterson et al. [33] found that the angiosperm SPCH and MUTE genes formed a clade sister to a clade comprising the angiosperm FAMA genes and the Physcomitrella bHLH Ia members. In addition, MacAlister and Bergmann [31] constructed a phylogeny of the SMF genes based on a sampling of nine angiosperms plus Selaginella moellendorffii and Physcomitrella patens with complete genome sequences and a EST sequence (Pgl‘SPCH’ [PgSPCH]) from the conifer Picea glauca. They found that the angiosperm SPCH, MUTE and FAMA genes formed monophyletic groups, respectively, and SPCH was closer to FAMA than MUTE. Particularly, SmoSMF2 (SmSMF2) and SmoSMF3 (SmSMF3) (Ia members of Selaginella) were sister to the angiosperm FAMA genes, while SmoSMF1 (SmSMF1) (also an Ia member of Selaginella), PpaSMF1 (PpSMF2) and PpaSMF2 (PpSMF2) (Ia members of Physcomitrella) were sister to the angiosperm MUTE genes with strong bootstrap supports, and PglSPCH was sister to the angiosperm MUTE genes with a weak support (49%). This phylogeny also seems to be supported by the domain architectures of the genes that they designated [31]. Therefore, as discussed above, the evolutionary history and patterns of the bHLH Ia genes involved in stomatal development are still greatly controversial, especially the phylogenetic positions of the Ia genes from basal land plants and gymnosperms.
In the present study, putative homologs of the bHLH Ia genes were identified from 49 species representing four major lineages of land plants (bryophyte, lycophyte, gymnosperm, and angiosperm). Because previous phylogenetic studies indicate that the bHLH Ia genes are monophyletic [26], [29], [34], [35] and non-bHLH amino acid motifs are highly conserved in each bHLH subgroup [29], [30], we only include the bHLH Ia members in this study in order to avoid incorrect phylogenetic topology that could be caused by improper alignment between different subgroups. Also, considering that the Ia genes first appeared in Physcomitrella patens, the two Ia members from this species (PpaSMF1/2) were used as functional outgroups in the phylogenetic analysis. The ML trees based respectively on the amino acid sequences and the first plus second codon positions both support that the Ia genes from vascular plants could be divided into three clades. That is, Clade A comprises the SMF genes from Selaginella moellendorffii and the FAMA homologs from seed plants (gymnosperm and angiosperm); Clade B includes two monophyletic sister groups, i.e., the gymnosperm “SPCH” genes and the angiosperm MUTE and SPCH genes; and Clade C includes other Ia members from gymnosperms and angiosperms that could not be grouped with SPCH/MUTE/FAMA (Fig. 1A). The unrooted ML tree also supports the three clades and a close relationship between the Ia genes from Physcomitrella patens and Clade A (Fig. 1B).
The SPCH, MUTE and FAMA genes of angiosperms form monophyletic groups, respectively, which is corroborated by their different gene structures (Figs. 1, 2). All SPCH genes have the unique and conserved MPKTD domain, although with different lengths, and this domain is important for regulating the SPCH activity in response to phosphorylation by the MAP kinases [51]. In addition, except PvuSPCH1 and LusSPCH3, all other SPCHs have a conserved position of stop codon. The MUTE genes have a unique conserved region (V) [31], and lack some residues preceding the bHLH domain that are present in all the other bHLH Ia members with various lengths. In contrast, the FAMA genes have high AA sequence similarity (Table 4), and harbor three unique domains (I, II and IV) (Fig. 2).
For gymnosperms, the FAMA and ‘SPCH’ genes were found in the EST databases of Ginkgo biloba, Picea glauca, Pinus banksiana and Pinus taeda (Tables 2, S1), and were successfully PCR-amplified from Picea breweriana, P. jezoensis and P. sitchensis. We also tried to amplify the ‘MUTE’ genes from gymnosperms using different primers, but failed. Moreover, we blasted the draft genome sequence of Picea abies just released, which could have covered 96% of the protein-coding genes of this species [52], and found four bHLH Ia genes [ConGenIE:MA_57244g0010, ConGenIE:MA_686524g0010, ConGenIE:MA_120602g0010 and ConGenIE:MA_130776g0010] (http://congenie.org). When the four genes were added into our dataset, the generated tree is topologically identical to Figure 1 and indicates that MA_57244g0010 is a FAMA homolog and MA_120602g0010 is a ‘SPCH’ homolog. The other two genes belong to other Ias (Tree not shown). Therefore, all available evidence strongly suggests that the MUTE gene does not occur in gymnosperms.
Undoubtedly, as suggested by MacAlister and Bergmann [31], the gymnosperm FAMA genes are sister to the angiosperm FAMA genes (Fig. 1), and the sister relationship is also supported by the high similarities of gene structures (Fig. 2). However, the gymnosperm ‘SPCH’ genes are sister to a clade comprising the angiosperm SPCH and MUTE genes with 100% bootstrap support, although they are very similar to the angiosperm SPCH genes in structure, especially the presence of the N-terminal sequence before the bHLH domain and the partial MPKTD domain (Fig. 2). By comparing the AA sequences of the bHLH and SMF domains, we found that the gymnosperm ‘SPCH’ genes are similar to the angiosperm SPCH sequences in some sites, but are identical to the angiosperm MUTE sequences in some other sites (Fig. 3). Additionally, deletion of the MPKTD domain of SPCH can generate proteins with functions similar to MUTE [27], [51]. Therefore, we deduce that the gymnosperm ‘SPCH’ genes may be closely related to and function as the common ancestor of the angiosperm SPCH/MUTE genes, and the MUTE genes could be derived from the ancestral copy by losing the MPKTD domain and the N-terminal sequences before the bHLH domain. This inference is also consistent with the reconstruction of ancestral gene structure (Fig. S1).
All bHLH Ia genes from S. moellendorffii are located in Clade A (FAMA), which is in line with Pires and Dolan [29] and Peterson et al. [33] but different from MacAlister and Bergmann [31]. In MacAlister and Bergmann [31], SmoSMF1 was placed sister to the angiosperm MUTE genes whereas SmoSMF2–3 were sister to the angiosperm FAMA genes. Comparing sequences of the conserved domains and structures of these genes, we found that all members from Selaginella have the pre-SMF domain (IV) and the unique region prior to the bHLH domain (II) (Fig. 2). Meanwhile, SmoSMF1 and SmoSMF2 have the conserved region in the N-terminus (I) that occurs in most angiosperm FAMA genes. This information, together with the high AA sequence similarity among SmoSMFs and the FAMA genes of gymnosperms and angiosperms (data not shown) and the phylogenetic reconstruction (Fig. 1), strongly suggests that all bHLH Ia members of Selaginella have close relationships with the FAMA genes. In addition, the function of the FAMA genes might be ancestral in bHLH Ia subgroup because PpaSMFs and SmoSMFs from the basal land plants are structurally identical and closely related to the FAMA genes of gymnosperms and angiosperms (Figs. 1, 2, S1).
Among the bHLH Ia genes from angiosperms, the FAMA genes are more conserved than the other ones, showing not only the lowest nucleotide diversity (Pi = 0.21573), but also the lowest nonsynonymous substitution rate (Ka = 0.05844) and Ka/Ks ratio (ω = 0.091). The SPCH and MUTE genes have similar values of nucleotide diversity, nonsynonymous and synonymous substitution rates, and Ka/Ks (Table 4). Therefore, the SPCH/MUTE/FAMA genes might have experienced different selective pressures, although the fact that no site or branch was detected to be under positive or relaxed selection using Fitmodel and codeml suggests conserved functions of all of them (Table 3, Table S3). Nevertheless, the functional shift could have occurred repeatedly in the SPCH genes, considering that more copies of them are maintained in some angiosperm species (Table 1), and that the functional divergence of different SPCH copies has been reported in Oryza sativa and Zea mays [50]. It cannot be ruled out that the multiple conspecific copies of the bHLH Ia subgroup, especially for the SPCH/MUTE/FAMA genes, were caused by ancient or recent genome duplications (http://chibba.agtec.uga.edu/duplication/). For example, some species that have experienced recent whole-genome duplication (WGD), such as Brassica rapa, Glycine max, Musa acuminate, Populus trichocarpa, Solanum lycopersicum, Solanum tuberosum and Zea mays, harbor more bHLH Ia genes. However, recent WGD cannot explain why more SPCH genes are retained than the MUTE and FAMA genes in some species (Table 1). Until now, no functional studies have been performed for other bHLH Ia genes (Fig. 1A, Clade C). These genes experienced frequent duplication and extinction events, both ancient and recent, and thus it would be interesting to investigate their functional differentiation in the future (Fig. S2).
Evolution of Stomatal Development in Land Plants
The developmental process of stomata has evolved and become complex with the evolution of land plants, especially the occurrence of amplifying divisions [14]. This process has been well studied in Arabidopsis, which includes three main steps, i.e., asymmetric entry divisions of the meristemoid mother cells (MMCs) to create meristemoids (M), self-renew via amplifying divisions or direct differentiation of meristemoids into GMCs, and symmetrical divisions of GMCs to form GCs [53]. However, in mosses and lycophytes, stomata develop by a simple process, in which a single asymmetric or symmetric division is followed by differentiation of a GMC, then GCs ( [54],reviewed by [55]). For ferns, an epidermal cell may go through one or two asymmetric divisions, then differentiates into a GMC, and finally into GCs [56]. In gymnosperms, different stomatal development patterns have been reported. The meristemoid divides once symmetrically to generate a GMC that then develops into GCs in Pinus [57], but it is interesting that Ginkgo biloba, a living fossil of gymnosperms, possesses both asymmetric divisions in perigenous neighbor cells like grasses and amplifying divisions within the stomatal lineage like Arabidopsis [58]. The basal angiosperms and dicots are basically similar to A. thaliana in stomatal development, although many of them show some differences in amplifying divisions [59]. In monocots, the formation of two guard cells needs two steps if not considering the subsidiary cells, i.e., one asymmetric division to generate a GMC, and a symmetric division to form two guard cells [60], [61]. The presence or absence of at least one asymmetric division in the cell lineage leading to the guard mother cell is a potential key factor in land-plant evolution. However, it is unclear which kind of division (symmetric or asymmetric) is ancestral in land plants [55]. More works are needed on the stomatal development of early land plants.
Based on the above information, it is obvious that the basic developmental process of stomata has not greatly changed in land plants if we do not consider the formation of the neighbor cells. At least two steps are necessary and conserved. One is the formation of GMC by one or two symmetric or asymmetric divisions, and the other is the formation of two GCs by a symmetric division. The two steps are directly mediated by the MUTE and FAMA genes, which has been proved by the experiments that PpaSMF1 and PpaSMF2 can partially rescue the fama and mute phenotypes [31], and by the fact that the MUTE and FAMA genes are functionally conserved in the monocots Oryza sativa and Zea mays [50], and the eudicot A. thaliana (Reviewed by [53]).
The SPCH gene controls the first asymmetric division of protodermal cells to initiate the stomatal lineage in Arabidopsis. However, functions of the two SPCH homologs from Oryza sativa (OsaSPCH1, OsaSPCH2) have somewhat diverged. The OsaSPCH2 plays a role in promoting the early events of the stomatal development whereas OsaSPCH1 does not [50]. In addition, the SPCH genes of angiosperms have experienced more duplication events and harbor more copies than the MUTE and FAMA genes (Table 1). All this information, together with the variation of the stomatal development in the entry division [33], [58], implies that the SPCH genes could have contributed greatly to the diversity of stomatal development in angiosperms.
MacAlister and Bergmann [31] constructed a model to explain the developmental complexity of the stomatal lineage. In their model, a single multifunctional bHLH Ia gene was originally responsible for both specification of GMC identity and GC differentiation in early land plants. Then, the ancestral Ia gene was duplicated into two genes, MUTE for GMC identity and FAMA for GC differentiation. Finally, a third Ia member, SPCH, originated from another gene duplication, which allows for the typical stomatal lineage in angiosperms, including amplifying divisions. This model was based on the phylogeny of the SPCH/MUTE/FAMA genes constructed in their study that sampled nine angiosperms, Picea glauca, Physcomitrella patens and Selaginella moellendorffii, and particularly based on their finding that PpaSMF1, PpaSMF2 and SmoSMF1 grouped with the angiosperm MUTEs. However, in that phylogeny, it is difficult to understand that PglSPCH, one Ia member from the gymnosperm P. glauca, was grouped with the angiosperm SPCH genes, although with only 49% bootstrap support, while no MUTE gene was found in gymnosperms. Furthermore, that phylogeny is topologically different from the phylogenies constructed in the present and all other previous studies [29], [33], [35], [50]. Therefore, it seems incredible that MUTE originated before SPCH if the MUTE gene does not occur in gymnosperms.
In the present study, all bHLH Ia genes from the two basal land plants Physcomitrella and Selaginella show close relationships with the FAMA genes of gymnosperms and angiosperms, and the angiosperm SPCH and MUTE genes form a clade sister to the gymnosperm ‘SPCH’ genes (Fig. 1). Hence, we agree with MacAlister and Bergmann [31] that an ancestral Ia gene with multifunction could control the specification of GMC identity and GC differentiation in basal land plants. However, we argue that SPCH originated earlier than MUTE based on the gene phylogeny and structure. The SPCH and MUTE genes are easily to be identified because they have a MPKTD domain and a truncated N-terminus, respectively. Theoretically, it is easier for MUTE to lose the N-terminus than for SPCH to gain the MPKTD domain [31]. However, the MPKTD domain could have originated before the divergence of gymnosperms and angiosperms, given the fact that the gymnosperm ‘SPCH’ genes have a truncated MPKTD domain whereas the MUTE genes are only found in angiosperms. Furthermore, the partial deletion of the MPKTD in SPCH generates a protein that more resembles MUTE in function [51], also supporting that the MUTE gene might have derived from the ancestor of the SPCH gene by loss of the MPKTD domain. To reveal the evolutionary history of the MPKTD domain, more species of lycophytes and ferns need to be studied.
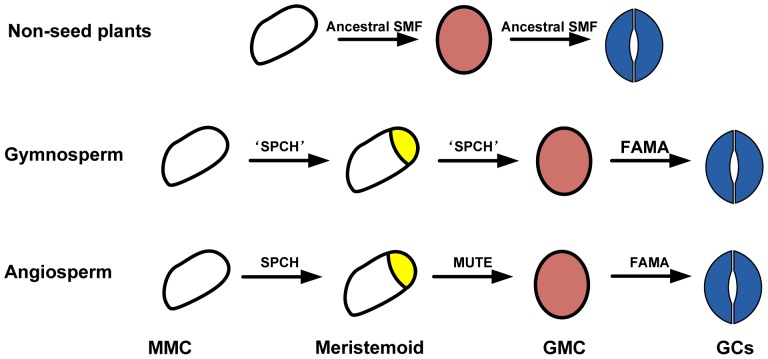
Based on the phylogeny and structure of the bHLH Ia genes (Figs. 1, 2), we deduce that a single multifunctional Ia gene responsible for the stomatal development originated in early land plants. Then, the ancestral Ia gene gave rise to two genes by a duplication in the ancestor of seed plants, one keeping the function of FAMA while the other (like the gymnosperm ‘SPCH’ genes) acquiring the MPKTD domain and having a dual function of SPCH and MUTE. Finally, the ‘SPCH’ gene evolved into the two genes SPCH and MUTE by another duplication in the ancestor of angiosperms. The SPCH retained the MPKTD domain, whereas the MUTE lost it, and got a later start codon at the beginning of the bHLH domain due to the disruption of the initial start codon (Fig. 4). One may argue that the gymnosperm FAMA genes could have multifunction as the FAMA genes in basal land plants. However, it is clear that the gymnosperm ‘SPCH’ genes are sister to the angiosperm SPCH-MUTE genes (Fig. 1). For a better understanding of functions of the SPCH/MUTE/FAMA genes, more functional studies should be done in non-flowering vascular plants in the future.
Figure 4. A predictive model of the stomatal development with the evolution of land plants.
(A) A single, multifunctional Ia gene responsible for the formation of both guard mother cell (GMC) and guard cells (GCs). (B) An ancestral ‘SPCH’ that originally occurred in gymnosperms by a gene duplication has a dual function of MUTE and SPCH and allows for the divergence between FAMA and ‘SPCH’. (C) The ‘SPCH’ gene evolved into the two genes SPCH and MUTE by another duplication in the ancestor of angiosperms.
Conclusions
In this study, we revealed the evolutionary relationships of three bHLH Ia genes (SPCH, MUTE and FAMA) of land plants that mediate sequential steps of stomatal development based on a wide sampling. The most interesting findings include the close relationship between the Ia genes from basal land plants (Physcomitrella and Selaginella) and the FAMA genes of seed plants, and the sister relationship between the gymnosperm ‘SPCH’ genes and the angiosperm SPCH and MUTE genes. Also, we proposed a model for the correlation between the evolution of stomatal development and the genes involved in this developmental process in land plants. However, only a few of bHLH Ia genes of gymnosperms that are available were included in this study, and almost nothing is known about the functions of these genes. It would be of great interest to investigate evolution and function of the ‘SPCH’ and FAMA genes in more gymnosperms in the future.
Supporting Information
Phylogeny of the bHLH Ia genes with a reconstruction of ancestral gene structure.
(PDF)
Maximum-likelihood (ML) trees of the bHLH Ia genes. A, A rooted ML tree based on amino acid sequences with the bHLH Ia genes from Physcomitrella patens as outgroups. B, A rooted tree based on nucleotide sequences with the bHLH Ia genes from P. patens as outgroups. C, An unrooted ML tree of the Ia genes constructed based on amino acid sequences. Numbers above branches refer to bootstrap values higher than 50%. The stars denote some inferred duplication events. Gene names are shown in Table S1. Ang, angiosperms; Gym, gymnosperms; Smo, moellendorffii; Ppa, Physcomitrella patens.
(PDF)
The sources of the Ia members of the bHLH transcriptional factors we analyzed.
(XLS)
The primers used in this study. * represent the primers used in the gene amplification.
(DOC)
Results of the site and branch-site analyses of the SPCH/MUTE/SPCH genes.
(XLS)
Acknowledgments
The authors thank Prof. Brandon S. Gaut for his careful reading and insightful comments that greatly improved the manuscript, Dr. Dong-Mei Guo for her assistance in the Fitmodel analysis, Ming-Ming Wang for his assistance in retrieving the bHLH Ia genes in gymnosperms and Ms. Wan-Wing Jin in the DNA sequencing.
Funding Statement
This work was supported by the National Natural Science Foundation of China (Grant numbers 30870166, 30990240 and 30425028). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bateman RM, Crane PR, DiMichele WA, Kenrick PR, Rowe NP, et al. (1998) Early evolution of land plants: Phylogeny, physiology, and ecology of the primary terrestrial radiation. Annu Rev Ecol Syst 29: 263–292. [Google Scholar]
- 2. Delaux PM, Nanda AK, Mathe C, Sejalon-Delmas N, Dunand C (2012) Molecular and biochemical aspects of plant terrestrialization. Perspect Plant Ecol Evol Syst 14: 49–59. [Google Scholar]
- 3. Edwards D, Kerp H, Hass H (1998) Stomata in early land plants: an anatomical and ecophysiological approach. J Exp Bot 49: 255–278. [Google Scholar]
- 4. Raven JA (2002) Selection pressures on stomatal evolution. New Phytol 153: 371–386. [DOI] [PubMed] [Google Scholar]
- 5. Sack FD, Paolillo DJ (1985) Incomplete cytokinesis in Funaria stomata. Am J Bot 72: 1325–1333. [Google Scholar]
- 6. Torii KU (2007) Stomatal patterning and guard cell differentiation. Plant Cell Monogr 9: 343–359. [Google Scholar]
- 7. Beerling DJ, Franks PJ (2009) Evolution of stomatal function in 'lower' land plants. New Phytol 183: 921–925. [DOI] [PubMed] [Google Scholar]
- 8. Chater C, Kamisugi Y, Movahedi M, Fleming A, Cuming AC, et al. (2011) Regulatory mechanism controlling stomatal behavior conserved across 400 million years of land plant evolution. Curr Biol 21: 1025–1029. [DOI] [PubMed] [Google Scholar]
- 9. Haworth M, Elliott-Kingston C, McElwain JC (2011) Stomatal control as a driver of plant evolution. J Exp Bot 62: 2419–2423. [DOI] [PubMed] [Google Scholar]
- 10. McAdam SAM, Brodribb TJ, Ross JJ, Jordan GJ (2011) Augmentation of abscisic acid (ABA) levels by drought does not induce short-term stomatal sensitivity to CO2 in two divergent conifer species. J Exp Bot 62: 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Doi M, Shimazaki K (2008) The stomata of the fern Adiantum capillus-veneris do Not respond to CO2 in the dark and open by photosynthesis in guard cells. Plant Physiol 147: 922–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brodribb TJ, McAdam SAM, Jordan GJ, Feild TS (2009) Evolution of stomatal responsiveness to CO2 and optimization of water-use efficiency among land plants. New Phytol 183: 839–847. [DOI] [PubMed] [Google Scholar]
- 13. Brodribb TJ, McAdam SAM (2011) Passive origins of stomatal control in vascular plants. Science 331: 582–585. [DOI] [PubMed] [Google Scholar]
- 14. McAdam SA, Brodribb TJ (2012) Stomatal innovation and the rise of seed plants. Ecol Lett 15: 1–8. [DOI] [PubMed] [Google Scholar]
- 15. de Boer HJ, Eppinga MB, Wassen MJ, Dekker SC (2012) A critical transition in leaf evolution facilitated the Cretaceous angiosperm revolution. Nat Commun 3: 1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Robinson JM (1994) Speculations on carbon-dioxide starvation, Late Tertiary evolution of stomatal regulation and floristic modernization. Plant Cell Environ 17: 345–354. [Google Scholar]
- 17. Franks PJ, Beerling DJ (2009) CO2-forced evolution of plant gas exchange capacity and water-use efficiency over the Phanerozoic. Geobiology 7: 227–236. [DOI] [PubMed] [Google Scholar]
- 18. Doi M, Wada M, Shimazaki K (2006) The fern Adiantum capillus-veneris lacks stomatal responses to blue light. Plant Cell Physiol 47: 748–755. [DOI] [PubMed] [Google Scholar]
- 19. Croxdale JL (2000) Stomatal patterning in angiosperms. Am J Bot 87: 1069–1080. [PubMed] [Google Scholar]
- 20. Darwin F (1898) Observations on stomata. Phil Trans R Soc Lond B 190: 531–621. [Google Scholar]
- 21. Berry JA, Beerling DJ, Franks PJ (2010) Stomata: key players in the earth system, past and present. Curr Opin Plant Biol 13: 232–239. [DOI] [PubMed] [Google Scholar]
- 22. Ruszala EM, Beerling DJ, Franks PJ, Chater C, Casson SA, et al. (2011) Land plants acquired active stomatal control early in their evolutionary history. Curr Biol 21: 1030–1035. [DOI] [PubMed] [Google Scholar]
- 23. McAdam SA, Brodribb TJ (2013) Ancestral stomatal control results in a canalization of fern and lycophyte adaptation to drought. New Phytol 198: 429–441. [DOI] [PubMed] [Google Scholar]
- 24. Kanaoka MM, Pillitteri LJ, Fujii H, Yoshida Y, Bogenschutz NL, et al. (2008) SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell 20: 1775–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pillitteri LJ, Torii KU (2007) Breaking the silence: three bHLH proteins direct cell-fate decisions during stomatal development. BioEssays 29: 861–870. [DOI] [PubMed] [Google Scholar]
- 26. MacAlister CA, Ohashi-Ito K, Bergmann DC (2007) Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445: 537–540. [DOI] [PubMed] [Google Scholar]
- 27. Ohashi-Ito K, Bergmann DC (2006) Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18: 2493–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gudesblat GE, Schneider-Pizon J, Betti C, Mayerhofer J, Vanhoutte I, et al. (2012) SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat Cell Biol 14: 548–554. [DOI] [PubMed] [Google Scholar]
- 29. Pires N, Dolan L (2010) Origin and diversification of basic-helix-loop-helix proteins in plants. Mol Biol Evol 27: 862–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, et al. (2003) The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20: 735–747. [DOI] [PubMed] [Google Scholar]
- 31. MacAlister CA, Bergmann DC (2011) Sequence and function of basic helix-loop-helix proteins required for stomatal development in Arabidopsis are deeply conserved in land plants. Evol Dev 13: 182–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vaten A, Bergmann DC (2012) Mechanisms of stomatal development: an evolutionary view. EvoDevo 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Peterson KM, Rychel AL, Torii KU (2010) Out of the mouths of plants: the molecular basis of the evolution and diversity of stomatal development. Plant Cell 22: 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li X, Duan X, Jiang H, Sun Y, Tang Y, et al. (2006) Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol 141: 1167–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Doyle JA, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19: 11–15. [Google Scholar]
- 37.Rogers SO, Bendich AJ (1988) Extraction of DNA from plant tissues. In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Biology Manual. Boston, MA: Kluwer Academic Publishers. 1–10.
- 38. Guo D–M, Ran J–H, Wang X-Q (2010) Evolution of the cinnamyl/sinapyl alcohol dehydrogenase (CAD/SAD) gene family: The emergence of real lignin is associated with the origin of bona fide CAD. J Mol Evol 71: 202–218. [DOI] [PubMed] [Google Scholar]
- 39. Ran J–H, Gao H, Wang X-Q (2010) Fast evolution of the retroprocessed mitochondrial rps3 gene in Conifer II and further evidence for the phylogeny of gymnosperms. Mol Phylogenet Evol 54: 136–149. [DOI] [PubMed] [Google Scholar]
- 40. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xia X, Xie Z (2001) DAMBE: software package for data analysis in molecular biology and evolution. J Hered 92: 371–373. [DOI] [PubMed] [Google Scholar]
- 42. Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Darriba D, Taboada GL, Doallo R, Posada D (2011) ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27: 1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- 45. Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704. [DOI] [PubMed] [Google Scholar]
- 46. Guindon S, Rodrigo AG, Dyer KA, Huelsenbeck JP (2004) Modeling the site-specific variation of selection patterns along lineages. Proc Natl Acad Sci U S A 101: 12957–12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- 48. Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- 49.Maddison WP, Maddison DR Mesquite: a modular system for evolutionary analysis. Version 2.75 [http://mesquiteproject.org].
- 50. Liu T, Ohashi-Ito K, Bergmann DC (2009) Orthologs of Arabidopsis thaliana stomatal bHLH genes and regulation of stomatal development in grasses. Development 136: 2265–2276. [DOI] [PubMed] [Google Scholar]
- 51. Lampard GR, MacAlister CA, Bergmann DC (2008) Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322: 1113–1116. [DOI] [PubMed] [Google Scholar]
- 52. Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin YC, et al. (2013) The Norway spruce genome sequence and conifer genome evolution. Nature 497: 579–584. [DOI] [PubMed] [Google Scholar]
- 53. Pillitteri LJ, Torii KU (2012) Mechanisms of stomatal development. Annu Rev Plant Biol 63: 591–614. [DOI] [PubMed] [Google Scholar]
- 54. Payne WW (1979) Stomatal patterns in embryophytes - their evolution, ontogeny and interpretation. Taxon 28: 117–132. [Google Scholar]
- 55.Rudall PJ, Hilton J, Bateman RM (2013) Several developmental and morphogenetic factors govern the evolution of stomatal patterning in land plants. New Phytol DOI: 10.1111/nph.12406. [DOI] [PubMed]
- 56. Apostolakos P, Panteris E, Galatis B (1997) Microtubule and actin filament organization during stomatal morphogenesis in the fern Asplenium nidus. 1. Guard cell mother cell. Protoplasma 198: 93–106. [DOI] [PubMed] [Google Scholar]
- 57. Johnson RW, Riding RT (1981) Structure and ontogeny of the stomatal complex in Pinus strobus L. and Pinus banksiana Lamb. Am J Bot 68: 260–268. [Google Scholar]
- 58. Rudall PJ, Rowland A, Bateman RM (2012) Ultrastructure of stomatal development in Ginkgo biloba . Int J Plant Sci 173: 849–860. [Google Scholar]
- 59. Carpenter KJ (2005) Stomatal architecture and evolution in basal angiosperms. Am J Bot 92: 1595–1615. [DOI] [PubMed] [Google Scholar]
- 60. Sack FD (1994) Structure of the stomatal complex of the monocot Flagellaria indica . Am J Bot 81: 339–344. [Google Scholar]
- 61. Stebbins GL, Jain SK (1960) Developmental studies of cell differentiation in the epidermis of monocotyledons: I. Allium, rhoeo, and commelina. Dev Biol 2: 409–426. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogeny of the bHLH Ia genes with a reconstruction of ancestral gene structure.
(PDF)
Maximum-likelihood (ML) trees of the bHLH Ia genes. A, A rooted ML tree based on amino acid sequences with the bHLH Ia genes from Physcomitrella patens as outgroups. B, A rooted tree based on nucleotide sequences with the bHLH Ia genes from P. patens as outgroups. C, An unrooted ML tree of the Ia genes constructed based on amino acid sequences. Numbers above branches refer to bootstrap values higher than 50%. The stars denote some inferred duplication events. Gene names are shown in Table S1. Ang, angiosperms; Gym, gymnosperms; Smo, moellendorffii; Ppa, Physcomitrella patens.
(PDF)
The sources of the Ia members of the bHLH transcriptional factors we analyzed.
(XLS)
The primers used in this study. * represent the primers used in the gene amplification.
(DOC)
Results of the site and branch-site analyses of the SPCH/MUTE/SPCH genes.
(XLS)