Abstract
Persistence of leukemic stem cells (LSC) after chemotherapy is thought to be responsible for relapse and prevents the curative treatment of acute myeloid leukemia (AML) patients. LSC and normal hematopoietic stem cells (HSC) share many characteristics and co-exist in the bone marrow of AML patients. For the development of successful LSC-targeted therapy, enabling eradication of LSC while sparing HSC, the identification of differences between LSC and HSC residing within the AML bone marrow is crucial. For identification of these LSC targets, as well as for AML LSC characterization, discrimination between LSC and HSC within the AML bone marrow is imperative. Here we show that normal CD34+CD38– HSC present in AML bone marrow, identified by their lack of aberrant immunophenotypic and molecular marker expression and low scatter properties, are a distinct sub-population of cells with high ALDH activity (ALDHbright). The ALDHbright compartment contains, besides normal HSC, more differentiated, normal CD34+CD38+ progenitors. Furthermore, we show that in CD34-negative AML, containing solely normal CD34+ cells, LSC are CD34– and ALDHlow. In CD34-positive AML, LSC are also ALDHlow but can be either CD34+ or CD34–. In conclusion, although malignant AML blasts have varying ALDH activity, a common feature of all AML cases is that LSC have lower ALDH activity than the CD34+CD38– HSC that co-exist with these LSC in the AML bone marrow. Our findings form the basis for combined functionally and immunophenotypically based identification and purification of LSC and HSC within the AML bone marrow, aiming at development of highly specific anti-LSC therapy.
Introduction
Only a small subpopulation of cells within acute myeloid leukemia (AML) is responsible for sustaining the leukemia [1]. This small subpopulation of leukemia-maintaining cells share cell surface markers with normal hematopoietic stem cells (HSC) and are capable of self-renewal and differentiation which has given them the name “leukemic stem cells” (LSC). Despite high remission rates after chemotherapy, only 30–40% of AML patients survive five years after diagnosis [2]. The main cause of present treatment failure is thought to be the insufficient eradication and survival of chemotherapy resistant LSC [3], [4]. Indeed, we have shown that high frequencies of CD34+CD38– LSC at diagnosis and after treatment predict relapse in AML [3], [5]. Beside LSC frequency, the capacity for an AML sample to give leukemic engraftment as well as recently identified LSC and HSC gene expression signatures have been linked to clinical outcome [6], [7]. Thus, LSC are prognostic and clinically important. The eradication of LSC may prevent relapse and therefore significantly improve long-term AML outcome.
LSC capable of initiating human AML in NOD/SCID mice were thought to be solely of the CD34+CD38– phenotype, similar to the normal HSC [1]. However, the LSC phenotype is more heterogeneous than initially realized and can even vary within a single AML patient [8]. Apart from the CD34+CD38– immunophenotype, other phenotypes have been associated with LSC properties such as the CD34+CD38+ and CD34– immunophenotypes [8], [9]. The CD34+CD38– cell compartment within the AML bone marrow (BM) includes both normal HSC and LSC. Selective anti-LSC therapies will be targeted to eradicate LSC while sparing HSC. Given that LSC and HSC share many features, the extent to which they differ will be a critical issue in development of LSC-targeted therapies with minimal toxicity. The search for these differences will be most relevant in HSC and LSC both obtained from the AML BM, thereby taking into account the effects of the AML microenvironment on both cell populations [10].
Discrimination and purification of CD34+CD38– LSC and CD34+CD38– HSC have been performed by using leukemia-associated proteins identified by us and others [11]–[15]. These include CLL-1, CD123 and several lineage markers [12]–[14]. CLL-1 is expressed on part of normal progenitors and in a portion of AML cases on LSC, but is absent on HSC [13]. Also lineage markers are absent on HSC, while in part of the AML cases, are expressed on both leukemic stem and progenitor cells [11], [12]. In general, immunophenotypic leukemia-associated markers are not expressed on all leukemic cells and not in all AML patients resulting in an inability to use a single marker for LSC identification in the whole AML patient population. This emphasizes the need for identification of additional markers of malignancy to detect LSC and discriminate these from HSC in all AML patients.
Recently, we discovered additional differences between HSC and LSC in both AML and chronic myeloid leukemia (CML) [5], [16]. We showed that normal and malignant stem cells can be identified by differences in light scatter properties (forward scatter, FSC and side scatter, SSC) and CD34 and/or CD45 expression. HSC values were 1–1.4 times (FSC) and 1–1.7 times (SSC) that of lymphocytes and defined as FSC/SSClow. LSC values are more than 1.4 times (FSC) and 1.7 times (SSC) that of the lymphocytes and defined as FSC/SSChigh. However, still in a considerable part of AML cases neither marker expression nor the combination with scatter properties is able to distinguish LSC from HSC. Therefore, we searched for a functional difference between normal HSC and LSC within the AML BM.
Aldehyde dehydrogenases (ALDHs) are cytosolic enzymes involved in the conversion of retinol (Vitamin A) to retinoic acids and important in HSC maturation, differentiation and loss of quiescence [17], [18]. These enzymes play a major role in the protection of BM progenitors from the cytotoxic effects of cyclophosphamide [19], [20] and increased the resistance to various other cytotoxic agents [20], [21]. The elevated expression of ALDH has been demonstrated in progenitors as compared to other hematopoietic cells like lymphocytes. Hematopoietic cells with high ALDH activity were enriched in the CD34+Lin- compartment indicating that ALDH activity is a marker for primitive hematopoietic stem/progenitor cells [22]–[27]. In AML, ALDH activity in the blast cells have been shown to correlate with clinical outcome [28], [29].
We investigated the ALDH activity in LSC as compared to normal CD34+CD38– HSC, both present in the BM of AML patients. We used discriminative aberrant cell surface markers and scatter aberrancies for the identification of HSC and LSC and analysed AML samples both from CD34-positive and CD34-negative AML. In this paper, we have demonstrated that, in both AML subtypes, high ALDH activity is an unique marker distinguishing CD34+CD38– HSC from CD34+ and CD34– LSC present within the BM of all the AML cases studied. In CD34-negative AML, CD34+CD38– stem cells are all normal (HSC) and have high ALDH activity. The LSC within this AML subtype are ALDHlow and of the CD34– immunophenotype. In CD34-positive AML, CD34+CD38– HSC are, like in CD34-negative AML, ALDHbright while the LSC are ALDHlow and either of the CD34+ or CD34– immunophenotype. This marked difference in ALDH activity between HSC and LSC offers an opportunity for identification and purification of LSC and CD34+CD38– HSC in every AML case.
Results
AML can be Divided in Two Subgroups Based on the Frequency and Pattern of CD34 Expression
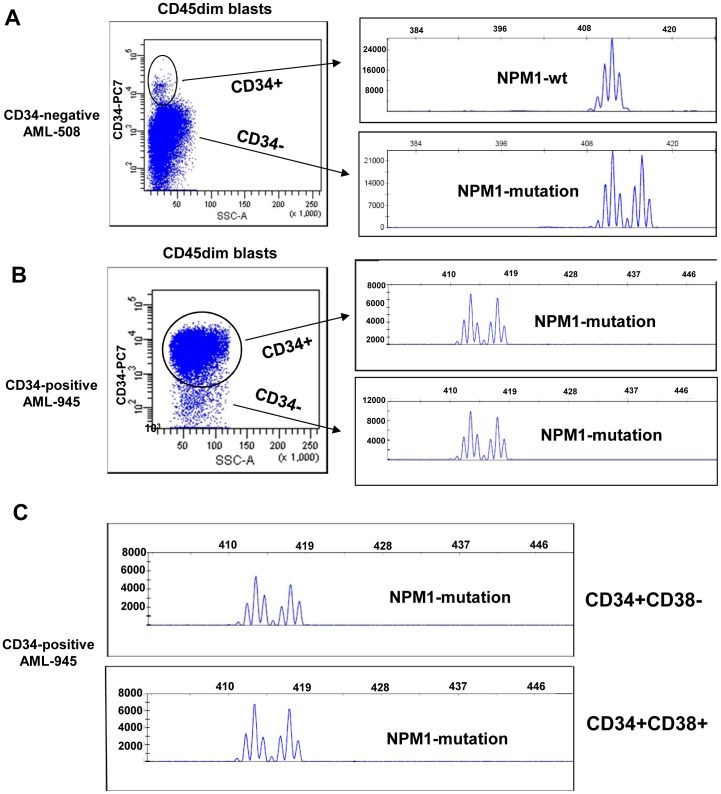
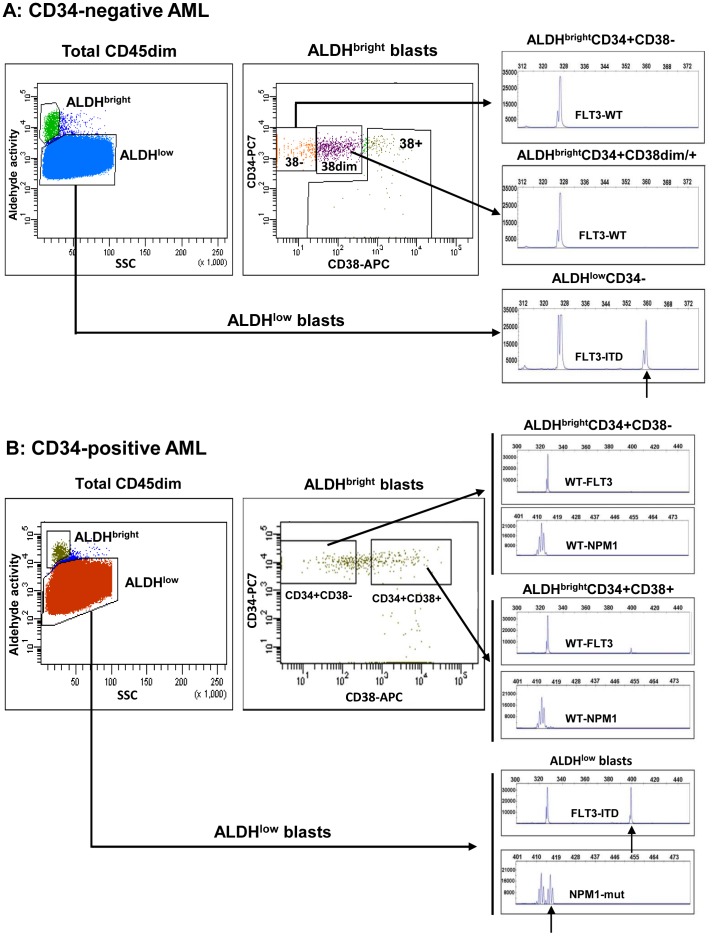
We have defined two groups of AML cases based on frequency and pattern of CD34 expression in the blast compartment. One of the subtypes, which we call CD34-negative AML, has a small, usually less than 1%, population of CD34+ cells which are cytogenetically and molecularly normal, as assessed by FISH analyses [30] and PCR (Figure 1A, AML-508). Thus, CD34-negative AML lacks CD34+ leukemic cells and consequently lacks also CD34+CD38– LSC. LSC in CD34-negative AML cases are of the CD34– immunophenotype. CD34-positive AML cases contain both leukemic CD34+ and CD34– cells, shown by the presence of molecular aberrancies in the CD34+ as well as the CD34– cell compartments (Figure 1B, AML-945), Within the CD34+ compartment, CD34+CD38– and CD34+CD38+ fractions contain mutated NPM1. Thus, LSC within CD34-positive AML can be either CD34+ or CD34– [8].
Figure 1. AML can be divided in two subtypes based on frequency and pattern of CD34 expression.
Representative flow cytometric staining patterns of CD34 expression (CD34 versus SCC) are shown for (A) a CD34-negative AML case (0.03% CD34+ cells within the blast compartment) and (B and C) a CD34-positive AML case (72% CD34+ cells within the blast compartment). The CD34-negative AML cases contain a discrete, usually less than 1%, CD34+ cell population. This CD34+ population is completely devoid of molecular aberrancies (in this case mutated NPM1)(A). The CD34-positive AML case contains a large CD34+ cell population, usually more than 1%, which contains the leukemia-associated mutated NPM1 protein (B). The CD34+CD38– and CD34+CD38– fractions from a CD34-positive case contain the FLT3-ITD and NPM1 mutation (C).
In Healthy BM, CD34+CD38− HSC are ALDHbright while ALDH Activity Decreases upon Differentiation
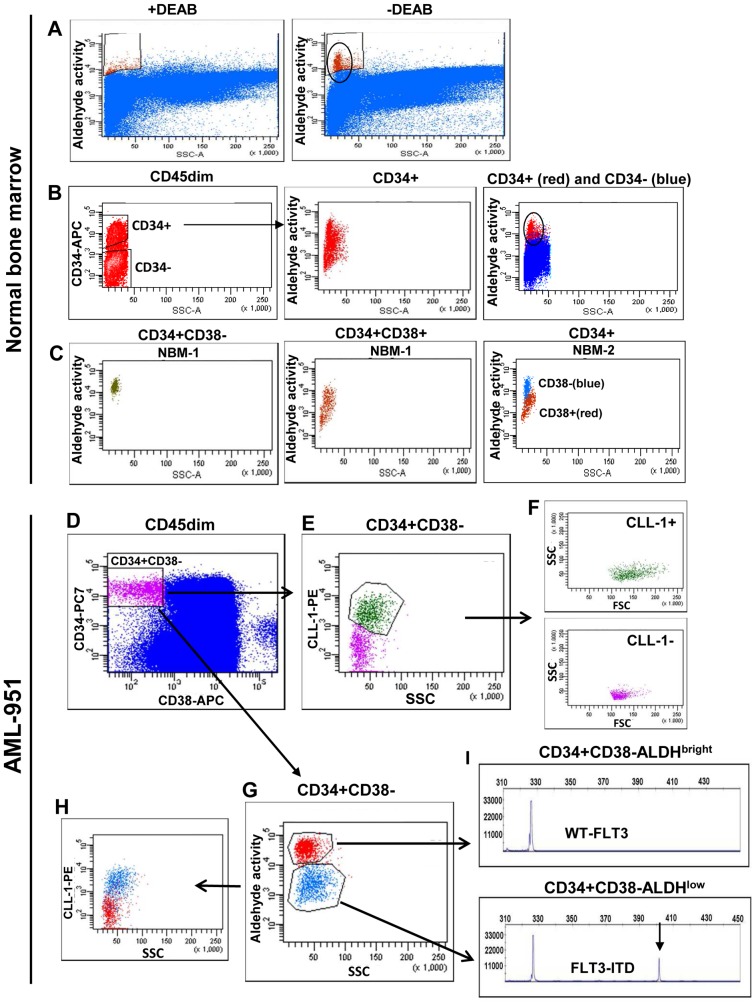
BM from a healthy donor contains a small population of cells with high ALDH activity (Figure 2A). This ALDH activity can be inhibited by incubation with diethylaminobenzaldehyde (DEAB), an inhibitor of the enzymatic activity of the ALDH proteins (Figure 2A). The CD34+ population within normal BM contained this same population of high ALDH activity cells (Figure 2B, circle), which we called ALDHbright. The CD34– cell population within normal BM is ALDHlow (Figure 2B, right panel, blue). The stem cell compartment within normal BM, defined as the CD34+CD38– cells, was completely retained in this high ALDH activity compartment (two examples in Figure 2C, left panel, green and right panel, blue) and visible as a single population, while the CD34+CD38+ more differentiated progenitor cells were ALDHlow (Figure 2C, middle and right panels, red). Thus, ALDH activity is high in CD34+CD38– HSC and decreases when cells become more differentiated. The normal BM CD34+CD38–ALDHbright cells comprised the complete (>99%) CD34+CD38– population of cells (n = 5).
Figure 2. CD34+CD38– HSC have higher ALDH activity than co-existing CD34+CD38– LSC.
(A–C) CD34+CD38– HSC within normal bone marrow are ALDHbright and the level of ALDH activity decreases upon differentiation to CD38+ progenitors. Representative flow cytometric ALDH activity patterns (ALDH versus SSC) are shown for (A) total CD45dim normal bone marrow cells treated with or without DEAB and (B) CD45dimCD34+ cells (middle panel) and both CD45dimCD34+ cells (red) and CD45dimCD34– cells (blue)(right panel). In C, the ALDH activity versus SCC of CD34+CD38– stem cells (panel 1 in green and panel 3 in bleu) and CD34+CD38+ progenitor cells (red in panel 2 and panel 3) from the normal BM is shown. (D–I) In CD34-positive AML, the ALDH activity of CD34+CD38– HSC is higher than that of the CD34+CD38– LSC. The aldefluor assay was performed on cells of a CD34-positive AML case (AML-951) and cells were subsequently labeled with anti-CD45 PERCP, anti-CD34 PC7, anti-CD38 APC and anti-CLL1 PE. The CD34+CD38– stem cells (D, purple) showed to be partly CLL-1+ and partly CLL-1– (E). The CLL-1+ stem cells (green) are FSC/SSChigh as compared to the CLL-1– cells (purple)(F). Investigation of the ALDH activity of the CD34+CD38– compartment showed that the CD34+CD38– stem cell population (D) segregates into an ALDHbright (red) and an ALDHlow (blue) population (G). The ALDHbright cells (red) are CLL-1 negative (H) and contain only wild type FLT3 kinase (I, upper panel). The ALDHlow cells (blue) are largely CLL-1 positive (H) and contain FLT3-ITD+ cells (I, lower panel). The arrow indicates the FLT3-ITD.
In CD34-positive AML, the CD34+CD38− ALDHbright Cells Retain the HSC while CD34+CD38− ALDHlow Cells are LSC
In the CD34+CD38– stem cell compartment of CD34-positive AML cases (n = 19) two separate ALDH activity populations, which we refer to as ALDHbright and ALDHlow (Table 1, Figure 2D and 2G), were seen. The CD34+CD38– ALDHlow compartment was not observed in normal donor BM as outlined in the left panel of Figure 2C. In CD34-positive AML cases, CD34+CD38– cells that express aberrant leukemia-associated immunophenotypic markers are neoplastic [12], [31]. Expression of these leukemia-associated markers is often not on all leukemic cells and consequently not on all LSC. Therefore, CD34+CD38– cells that lack aberrant marker expression can be either HSC or LSC. In the marker-negative CD34+CD38– population we can, in many cases, discriminate between HSC and LSC using additional characteristics, such as scatter properties. HSC have a lower FSC and/or SSC than the LSC population [5], [16]. On the basis of expression of aberrant markers and scatter properties we showed that CD34+CD38– ALDHbright cells in CD34-positive AML (n = 19) had no expression of immunophenotypic leukemia-associated markers (median 0.03% marker expression on CD34+CD38– ALDHbright cells, Table 1) and are lower in FSC and/or SSC than ALDHlow cells (data not shown), strongly suggesting that ALDHbright cells are CD34+CD38– HSC. The level of ALDH activity corresponds with that of normal BM CD34+CD38– HSC (Figure 2C, right and middle panel). The ALDHlow cells had aberrant marker expression (frequency corresponds with that of marker expression on leukemic CD45dim blasts of the corresponding AML, median is 25% marker expression on CD34+CD38– ALDHlow cells, Table 1) and a higher FSC/SSC, suggesting to be CD34+CD38– LSC. Thus, putative normal HSC are contained within the ALDHbright compartment while LSC are ALDHlow.
Table 1. CD34+CD38–stem cell frequencies and their marker expression in the ALDHbright and ALDHlow compartment in CD34-positive AML.
| CD34-pos AML de novo | Marker | % CD34+CD38– ALDHbright | Marker+ %& # |
| 575 | CD33 | 51.3 | 1.7 (2c) ‡ |
| 670 | CLL-1 | 54.4 | 0.3 (1c) |
| 726 | CD7 | 34.5 | 0.3 (3c) |
| 808 | none | 22.7 | – |
| 945 | CLL-1/CD19 | 44.6 | 1.1 (2c) |
| 951 | CLL-1 | 51.9 | 1.5 (23c) |
| 966 | CD7 | 58.9 | – |
| 1013 | CLL-1 | 22 | 0.67 (15c) |
| 1016 | CD11b | 0.25 | 0 |
| 1022 | CLL-1 | 38 | 0 |
| 1030 | CD22 | 10 | 3 (2c) |
| 1034 | CLL-1 | 3 | 0 |
| 1036 | CLL-1 | 20 | 0 |
| 1047 | CD7 | 5.5 | 0 |
| 1048 | CD7 | 92 | 0.03 |
| 1057 | CD19 | 7 | 0 |
| 1263 | CD33/CD123 | 84 | 0 |
| 1305 | CLL-1 | 58 | 3.5 (6c) |
| 1320 | CD2 | 92.2 | 0 |
| Median | 38 | 0.03 | |
| CD34-pos AML de novo | Marker | % CD34+CD38– ALDHlow | Marker+ % & # |
| 575 | CD33 | 0.01 | 100 |
| 670 | CLL-1 | 0.1 | 4.5 |
| 726 | CD7 | 0 | – |
| 808 | none | 0.2 | – |
| 945 | CLL-1/CD19 | 0.1 | 86.4 |
| 951 | CLL-1 | 0.1 | 76.7 |
| 966 | CD7 | 0.5 | 24.4 |
| 1013 | CLL-1 | 2.36 | 15 |
| 1016 | CD11b | 0.28 | 13 |
| 1022 | CLL-1 | 0 | – |
| 1030 | CD22 | 3.5 | 6 |
| 1034 | CLL-1 | 0.12 | 40 |
| 1036 | CLL-1 | 15 | 25 |
| 1047 | CD7 | 0.18 | 12 |
| 1048 | CD7 | 34 | 12 |
| 1057 | CD19 | 0.15 | 67 |
| 1263 | CD33/CD123 | 1.7 | 99 |
| 1305 | CLL-1 | 0.7 | 84 |
| 1320 | CD2 | 0.6 | 7.2 |
| Median | 0.2 | 24.7 |
Detection of immunophenotypic aberrancies in CD34+CD38– ALDHbright and ALDHlow compartments of CD34-positive AML.
The percentage of CD34+CD38– cells in the total ALDHbright or ALDHlow compartments is indicated.
in the ALDHbright CD34+CD38– compartment the aberrant marker expression, both CLL-1 and lineage markers, is indicative for the neoplastic character of the cells.
Percentage expression does not quantitatively correlate with % of neoplastic cells: a shift in fluorescence of a total cell population may result e.g. in 50% marker expression based on normalization by isotype controls of negative cell populations.
data in between parenthesis represent number of cells in cases where there is <25 cells positive for a marker.
This putative normal HSC, CD34+CD38– ALDHbright, cell population of 7 of these 19 CD34-positive AML cases was essentially devoid of cells with the leukemia-specific cytogenetic abnormalities FLT3-ITD and/or mutated NPM1 (Table 2). An ALDH activity analysis of such a CD34-positive AML, in this case FLT3-ITD-positive, is shown in Figure 2D–I (AML-951). The CD34+CD38– compartment contains both CLL-1+ and CLL-1– cells (Figure 2E), whereby the CLL-1+ stem cells are in general FSC/SSChigh as compared to the CLL-1– cells (Figure 2F). Analysis of the ALDH activity of the CD34+CD38– compartment showed that it segregates into an ALDHbright and an ALDHlow population (Figure 2G). The ALDHbright cells are CLL-1– (Figure 2H), FSC/SSClow (Figure 2F) and contained only wild type FLT3 kinase (Figure 2I, upper panel), indicating HSC. The ALDHlow cells are for a major part CLL-1+ (Figure 2H), FSC/SSChigh (Figure 2F) and contain FLT3-ITD+ cells (Figure 2I, lower panel), indicating LSC.
Table 2. Presence of FLT3-ITD and mutated NPM1 in ALDHbright and ALDHlow compartments in CD34-positive AML.
| Patient # FLT3-ITD | CD34– cells | CD34+CD38 ALDHlow | CD34+CD38– ALDHbright |
| 575 | pos | 50% ITD | wt |
| 808 | pos | 50% ITD | wt |
| 951 | pos | 42% ITD | wt |
| 966 | pos | 43% ITD | wt |
| 1263 | pos | 60% ITD | wt |
| Patient # NPM1 | CD34– cells | CD34+CD38 ALDHlow | CD34+CD38 ALDHbright |
| 575 | mut | mut | wt |
| 808 | mut | mut | wt |
| 945 | mut | mut | wt (<5%) |
| 670 | mut | mut | wt |
Detection of molecular aberrancies in CD34+CD38– ALDHbright and ALDHlow compartments of CD34-positive AML. The FLT3-ITD percentage is determined in the total leukemic blast population (data not shown), the CD34– cell population and the CD34+CD38-ALDHlow and CD34+CD38-ALDHbright compartments. The NPM1 mutation analysis is not quantative. # the ALDHbright compartment contained in one case a small mutant peak (AML-945), likely caused by relatively poor separation of ALDHbright and ALDHlow populations due to overlap in the boundary region.
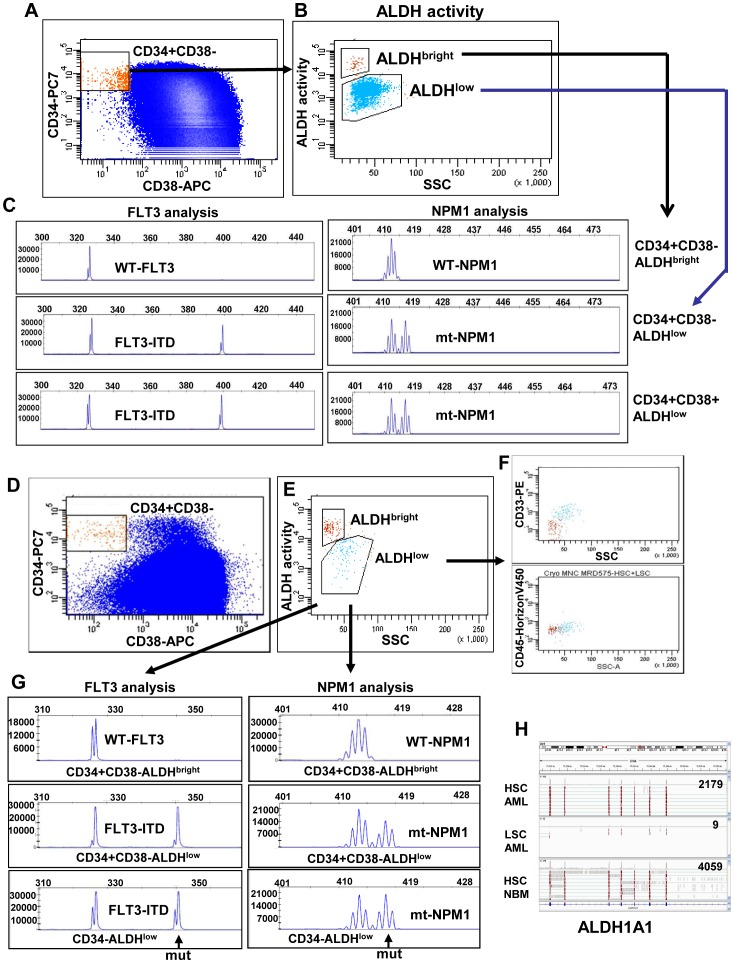
Development of acute leukemia follows the rules of the two-hit model; cells have to acquire a mutation interfering with differentiation and a mutation conferring a proliferative advantage to become neoplastic [32]. Consequently, there is a possibility that hematopoietic cells with only one detectable aberrant leukemia-associated molecular mutation still have a normal phenotype. To confirm that the ALDHbright CD34+CD38– cells are normal HSC and the ALDHlow CD34+CD38– cells are LSC we analysed the presence of molecular aberrancies in the ALDH compartments from CD34-positive AML cases with two molecular aberrancies, FLT3-ITD and mutated NPM1 (AML-575 and AML-808). CD34+CD38– AML cells can be divided in an ALDHbright and ALDHlow compartment (Figure 3A,B, AML-808 and 3D,E, AML-575). In case of AML 575, the ALDHbright cells are negative for CD33 and low in SSC strongly suggesting normal HSC (Figure 3F). In both these AML cases the ALDHbright compartment contains neither an FLT3-ITD nor an NPM1 mutation (Figure 3C, AML-808 and 3G, AML-575 upper panels), indicating HSC, while the ALDHlow compartment has both these leukemia-associated mutations (Figure 3C, AML-808 and 3G, AML-575 middle panels), indicating LSC.
Figure 3. ALDHbright CD34+CD38– cells are devoid of both the FLT3 and NPM1 mutation.
AML-808 contains both FLT3-ITD and mutated NPM1 and has no immunophenotypic aberrancy present. However, AML-808 can be segregated into a normal and a leukemic CD34+CD38– compartment by measuring ALDH activity (A–C). The CD34+CD38– stem cells (A, orange) segregate into two separate ALDH compartments (B). The CD34+CD38–ALDHbright cells of AML-808 do neither contain FLT3-ITD nor mutated NPM1 (C, upper panels), while the CD34+CD38-ALDHlow cells (blue) do (C, middle panel). The ALDHlow cells have both mutations (C, lower panel). AML-575 contains both FLT3-ITD and mutated NPM1. The CD34+CD38– cells (red) segregate into two separate ALDH compartments (D, middle panel). The ALDHbright (red) cells within AML-575 lack the immunophenotypic aberrancy CD33 and are low in SSC compared to the ALDHlow cells (blue)(F). The CD34+CD38-ALDHbright cells do neither contain FLT3-ITD nor mutated NPM1 (G, upper panels), while the CD34+CD38-ALDHlow cells do (G, middle panel). The ALDHlow cells have both mutations (G, lower panel). (H) CD34+CD38– HSC, CD34+CD38– LSC (purification based on CLL1+, CD34 expression and scatter properties) from AML-598, and CD34+CD38– HSC from normal BM were purified. RNA was isolated and RNA sequencing was performed. The most abundant ALDH mRNA expressed within both normal HSC fractions (AML and normal BM) was the ALDH1A1 enzyme. LSC had hardly any expression of the ALDH1A1 form. Numbers indicate the amount of ALDH1A1 reads within that fraction.
In the Absence of Immunophenotypic Marker Aberrancies, CD34+CD38− HSC can be Discriminated from CD34+CD38− LSC Solely on ALDH Activity
In part of the AML cases (1/19 in our CD34-positive AML population, AML-808), the CD34+CD38– compartment can not be segregated into HSC and LSC by expression of aberrant immunophenotypic markers or by scatter properties. In such cases, nevertheless ALDH activity identified two separate populations within the CD34+CD38– compartment (Figure 3A and 3B). The ALDHbright CD34+CD38– cells lacked both FLT3-ITD and mutated NPM1 (Figure 3C, upper panels), indicating normal HSC while the ALDHlow CD34+CD38– cells contained the FLT3-ITD and mutated NPM1 (Figure 3C, lower panels), indicating LSC. Thus, even in the absence of marker aberrancies, CD34+CD38– HSC can be discriminated from CD34+CD38– LSC based on ALDH activity.
Different isoforms of ALDH might contribute to the ALDH activity profile seen. To search for the ALDH enzyme(s) responsible for the high ALDH activity seen in HSC as compared to LSC, the HSC from healthy donor BM and CD34+CD38– HSC and LSC from the AML BM were purified. HSC and LSC purified from the AML BM were defined as CLL-1-, FSC/SSClow and CLL-1+, FSC/SSChigh., respectively. These stem cell fractions were analysed for ALDH isoform expression by RNA-sequencing. The putative HSC contained within the AML BM have high expression of the ALDH1A1 enzyme as compared to almost no expression in the LSC (Figure 3H, AML-598). Moreover, in HSC from the BM of a healthy donor, like in HSC from the AML BM, the ALDH1A1 isoform is highly expressed (Figure 3H). In HSC and LSC within this AML BM other ALDH enzymes are expressed be it at much lower level than ALDH1A1 in the HSC. ALDH3B1 is the only ALDH member that is higher expressed in LSC as compared to HSC (5 fold).
CD34-positive AML: Inter-Individual Variation in the Difference in ALDH Activity between LSC and HSC
With flow cytometry analysis, the relative ALDH activity can be measured as mean fluorescence intensity (MFI) level of the population. We standardized the ALDH-MFI values of normal and neoplastic stem cell candidates by dividing these by the ALDH-MFI value of lymphocytes present within the same sample. Lymphocytes are negative for ALDH, resulting in MFI values that can be considered as background signal and as a stable characteristic of the individual sample. Treatment with DEAB was used in 9/19 samples as an extra control and showed similar basement ALDH activity levels in most samples used (Table S1). From these 9 CD34-positive AML cases the CD34+CD38– HSC and CD34+CD38– LSC, as defined by marker expression and scatter properties, were analysed for their ALDH activity MFI. The marker-negative compartment (containing HSC) had median 6.9-fold (range 1.7–28.9) higher ALDH activity than the marker-positive LSC compartment (Table S1). This way of calculation showed marked variations in the fold difference in ALDH activity of HSC as compared to LSC in individual samples. There are cases where LSC ALDH activity levels come close to levels in HSC (Figure S1, AML-1048) while in others, LSC and HSC ALDH activity levels are far apart (Figure S1, AML-1036), indicating inter-individual variation in the difference between ALDH activity in LSC and HSC. Inhibition of the high activity of ALDHbright CD34+CD38– cells by DEAB was not in all cases till the same level as the LSC present in the same sample (Figure S1, AML-1036 and AML-1013). The ALDHlow CD34+CD38– cell compartment segregated, in some AML cases, in a population which could be inhibited by DEAB and a population which could not (Figure S1, AML-1048), suggesting the existence of LSC populations with various levels of ALDH activity within one CD34+CD38– ALDHlow compartment. Some AML cases had mainly normal ALDHbright CD34+CD38– cells within the BM (Figure S1, AML-1013).
In CD34-negative AML, CD34+CD38– Cells have High ALDH Activity and are Normal while LSC are CD34– and ALDHlow
In CD34-negative AML, all the CD34+CD38– cells have a normal phenotype and consequently LSC are CD34– (Figure 1A) [30]. In all CD34-negative AML cases that we analysed (n = 13), the CD34+CD38– cells were retained in the ALDHbright compartment while there were essentially no CD34+CD38– cells in the ALDHlow compartment (Table 3). The few CD34+CD38– cells that were ALDHlow meet the criteria FSC/SSClow, confirming the absence of CD34+ LSC in CD34-negative AML cases (Table 3, median 48% CD34+CD38– in ALDHbright and median 0% CD34+CD38– in ALDHlow). The bulk of the AML (CD34– cells) were ALDHlow (Table 3, example in Figure 4A and B).
Table 3. CD34+CD38– frequency and aberrant marker expression in ALDHbright and ALDHlow compartments in CD34-negative AML.
| CD34-neg AML de novo | Marker | % CD34+CD38− ALDHbright * | Marker+ % & |
| 464 | CD56 | 83.8 | 0 |
| 508 | CLL-1 | 35 | 0 |
| 813 | CLL-1 | 24.6 | 0.1 (2c) ‡ |
| 822 | CLL-1 | 50.4 | 0 |
| 1021 | CLL-1 | 75 | 0 |
| 1024 | CD7 | 9 | 0 |
| 1027 | CLL-1 | 48 | – |
| 1028 | CD7 | 60 | 0 |
| 1035 | CLL-1 | 70 | 0 |
| 1045 | CLL-1 | 14 | 0 |
| 1054 | CD56 | 3 | 0 |
| 1063 | CLL-1 | 57 | 0 |
| 1276 | CLL-1 | 7.9 | 0.1 |
| Median | 48 | 0 | |
| CD34-neg AML de novo | Marker | % CD34+CD38− ALDHlow * | Marker+ % & |
| 464 | CD56 | 0 | – |
| 508 | CLL-1 | 0 | – |
| 813 | CLL-1 | 0 | – |
| 822 | CLL-1 | 0 | – |
| 1021 | CLL-1 | 0.89 | 24 |
| 1024 | CD7 | 2.7 (10c) | 0 |
| 1027 | CLL-1 | 1.9 (5c) | – |
| 1028 | CD7 | 0.05 | 0.44 (1c) |
| 1035 | CLL-1 | 0.02 (14c) | 20 (9c) |
| 1045 | CLL-1 | 0 | – |
| 1054 | CD56 | 0 | – |
| 1063 | CLL-1 | 0.01 (8c) | – |
| 1276 | CLL-1 | 0 | – |
| Median | 0 | 10.22 |
Detection of immunophenotypic aberrancies in CD34+CD38– ALDHbright and ALDHlow compartments of CD34-negative AML.
Percentage of CD34+CD38– cells within the ALDHbright or ALDHlow compartments.
in the ALDHbright CD34+CD38– compartment, the aberrant marker expression, both CLL-1 and lineage markers, is indicative for the neoplastic character of the cells.
data in between parenthesis represent number of cells in cases where there is <25 cells with a marker. The ALDHlow compartment in CD34-negative AML cases is essentially devoid of CD34+CD38– stem cells.
Figure 4. The ALDH activity of CD34+CD38– HSC is higher than the CD34– bulk in CD34-negative AML.
The aldefluor assay was performed on the bulk of cells (AML-464). Cells were subsequently labeled with anti-CD45 PERCP, anti-CD34 PC7, anti-CD38 APC and anti-CD56 PE. (A) The CD34+CD38– stem cells (A, left panel, orange) were negative for CD56 (A, middle panel, CD34+CD38– in orange) and low in FSC/SCC (A, right panel). (B) The ALDH activity of these CD34+CD38– cells is high and shows a separate SCClowALDHbright population of cells (B, CD34+CD38– cells in orange). (C) Detection of FLT3-ITD by length fragment analysis showed that the ALDHbright compartment was devoid of FLT3-ITD (top) while the ALDHlow compartment has the FLT-ITD (bottom); arrow indicate the mutation.
In CD34-negative AML cases, ALDHbright cells lack expression of aberrant immunophenotypic markers (Table 3, median 0%), have low scatter properties (not shown) but most importantly are devoid of the molecular aberrancies FLT3-ITD and mutated NPM1 (Table 4, example Figure 4C AML-464). In AML-464, ALDHbright CD34+CD38– cells lack the aberrant marker CD56 present in the leukemic blasts of this patient (Figure 4A , middle panel) and have low FSC/SSC (Figure 4A, right panel). The ALDHbright cells are devoid of FLT3-ITD (Figure 4C, upper panel) and the NPM1 mutation (data not shown) while the ALDHlow CD34– compartment contained FLT3-ITD positive cells (Figure 4C lower panel) and mutated NPM1 (not shown).
Table 4. The presence of FLT3-ITD and mutated NPM1 in ALDHbright and ALDHlow compartments in CD34-negative AML.
| Patient # FLT3-ITD | CD34− ALDHlow | CD34+CD38− ALDHbright |
| 1027 | 48% ITD | 0% ITD |
| 464 | 32% ITD | 0% ITD |
| Patient # NPM1 | CD34− ALDHlow | CD34+CD38 ALDHbright |
| 822 | mut | wt |
| 1276 | mut | wt |
| 464 | mut | wt |
| 508 | mut | wt |
Detection of molecular aberrancies in CD34+CD38– ALDHbright and ALDHlow compartments of CD34-negative AML. The molecular nature of the CD34+CD38-ALDHbright and CD34– compartments in CD34-negative AML cases. Detection of FLT3-ITD and mutated NPM1 was performed in the various cell fractions. The FLT3-ITD percentage is determined in the total leukemic blast population (data not shown), the CD34+CD38-ALDHbright and CD34– compartments. The NPM1 mutation analysis is not quantative.
We have studied the presence of molecular aberrancies in the ALDH compartments in five CD34-negative AML cases (Table 4). The ALDHbright CD34+CD38– compartment is devoid of molecular mutations (Table 4) in all the five AML patients. The ALDHlow compartment contained almost exclusively CD34– cells having aberrant marker expression and molecular mutations (Table 3 and 4, Figure 4).
CD34-negative AML: Inter-individual Variation in the Difference in ALDH Activity between CD34+CD38-HSC and CD34– cells
In CD34-negative AML, we standardized the ALDH-MFI values of normal stem cell candidates and CD34– neoplastic cells by dividing these by the ALDH-MFI value of lymphocytes present within the same AML sample (Table S2). Eight CD34-negative AML cases were treated with and without DEAB and in these we analysed the ALDH activity MFI level of the CD34+CD38– HSC, defined by lack of immunophenotypic marker expression, and the ALDH activity MFI level of the CD34– cells. The marker negative CD34+CD38– compartment (HSC) had a median 4.3-fold (range 2.2–18.7) higher ALDH activity than the CD34– cells (Table S2).
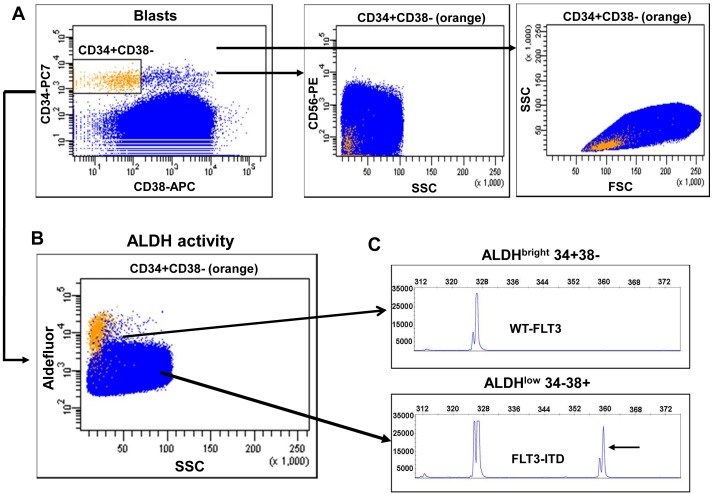
The ALDHbright Compartment in Both CD34-positive and CD34-negative AML Contained CD34+CD38+ Progenitors with a Normal Phenotype
The ALDHbright compartment in the BM of both CD34-positive and CD34-negative AML cases contained CD34+CD38– HSC, more differentiated CD34+CD38+ progenitors and CD34– cells (Figure 5A and 5B, Table S3). The CD34+CD38+ ALDHbright cells in CD34-negative AML cases neither expressed an aberrant lineage marker, nor had a molecular aberrancy as tested for four CD34-negative AML cases (Table S3A, Figure 5A, AML-464). In CD34-positive AML, the CD34+CD38+ ALDHbright cells had solely the wt-FLT3 protein present in two of the five cases tested. In 3/5 CD34-positive AML cases that we analyzed a very small amount of ALDHbright progenitors contained FLT3-ITD and/or NPM1 mutations and might therefore be leukemic (Table S3B, Figure 5B).
Figure 5. The presence of FLT3-ITD and mutated NPM1 in ALDHbright CD34+CD38− progenitor populations.
(A) In CD34-negative AML cases, the ALDHbright population of AML cells consists, besides CD34+CD38− stem cells of CD38dim and CD38+ progenitors (A, middle panel, AML-464) (Table S3). The complete ALDHbright compartment in CD34-negative AML has a normal phenotype as proven by the absence of FLT3-ITD in the CD34+CD38− (A, right upper panel) as well as the CD38dim (A, right middle panel) and CD38+ (data not shown) population of cells. The ALDHlow compartment contains the FLT3-ITD (arrow in right lower panel), indicating leukemic cells. (B) In CD34-positive AML cases, the ALDHbright population contains, apart from normal CD34+CD38− HSC, CD34+CD38+ progenitors and CD34− cells (Figure 5B middle panel)(Table S3). The ALDHbright CD34+CD38− stem cells lack molecular aberrancies (B, right upper two panels) and are therefore normal. The CD34+CD38+ progenitor compartment (B, right middle two panels) in this case has very small FLT3-ITD and NPM1 mutant peaks likely originating from purification of ALDHlow cells within the ALDHbright compartment. The ALDHlow compartment is largely neoplastic (B, right lower 2 panels).
An ALDH Positive Population can be Identified in AML Patients
In AML, ALDH activity in leukemic blasts has been shown to define a subgroup with adverse prognosis and superior NOD/SCID engrafting potential [28], [29]. We focussed on differences in ALDH activity between HSC and LSC, whereby we identified a small population of high ALDH activity normal HSC (designated as ALDHbright). The ALDHlow compartment contained cells with various levels of ALDH activity within one AML patient but also inter-individual ALDH activity differences, relative to the ALDHbright compartment. These differences were also seen in earlier studies [28], [33], [34] and we therefore compared our results with those. Seventeen AML samples (both CD34-positive and CD34-negative AML cases) were treated with DEAB in order to define an ALDH negative and positive population similarly as done by others [28], [29], [33](Figure S2 and Table S4). Similar to Cheung et al [28], we defined samples as ALDH-positive when the proportion of ALDH+ cells, defined by DEAB treatment, in the total sample was more than 5%. Three of our 17 cases (18%) were ALDH-positive (8.2%, 9.5%, 12.6%). In a study by Pearce et al, ALDH activity in AML was classified, based on the shape, level and scatter properties of the ALDH positive compartment, as rare, numerous or negative [33]. In our study 18/32 samples (56%) matched the ‘rare’ ALDH activity pattern and 14 matched the ‘numerous’ pattern (Figure S2A and S2B and Table S4).
Discussion
In the present study, we have identified the activity of ALDH enzymes as a consistent functional discrimination between LSC and HSC concomitantly present in the BM of both CD34-negative and CD34-positive AML patients. Although HSC and LSC can, in a considerable part of AML cases, be distinguished using aberrancies of marker expression [12]–[14] and scatter properties [5], [16], assessment of ALDH activity enables such discrimination in all AML cases even in the absence of aberrancies. To our knowledge, there is no single parameter that allows identification of both normal and neoplastic stem cells in every AML patient. Association of high ALDH activity and ALDH1A1 expression solely with normal HSC and not with LSC within the AML BM may have important implications for treatment options, relapse prediction, and identification of HSC and LSC aiming at LSC-specific therapeutic target identification.
First, the relapse that occurs in 50% of AML patients after a seemingly successful chemotherapeutic treatment is likely caused by persistent LSC. This is reflected by the fact that frequency of LSC both at diagnosis and after treatment as well as expression of LSC signatures are tightly linked to the survival of AML patients [3], [5], [7]. For this reason, establishing LSC frequency at diagnosis and monitoring after treatment may become very important for clinical risk assessment and treatment decision making. Since cell surface markers are less stable due to interactions with the microenvironment or chemotherapy treatment, marker expression used nowadays for LSC detection might be less valuable than using a cytoplasmic functional maker like ALDH. In addition, heterogeneity of aberrant marker expression on LSC, i.e. either absent or present on part of the LSC compartment, and consequently the difficulty in identification of all LSC, will be circumvented by using ALDH activity as a marker.
Second, it is thought that LSC need to be eradicated in order to prevent relapse and to cure AML. The optimal anti-LSC therapy will spare the normal HSC. ALDH activity as a strong discriminative marker allows purification of LSC and HSC paving the way for LSC-specific target identification.
Third, it could be hypothesized that firm differences in ALDH activity between HSC and LSC in the BM of AML patients might result in consequent resistance of HSC for particular drugs subjected to ALDH-dependent detoxification. The high ALDH activity in HSC, compared to LSC, provided to be effective for LSC, predicts lower toxicity of alkylating agents, such as cyclophosphamide. The difference between the ALDH activity in LSC and HSC defines here the therapeutic window. We are currently testing drugs, known to be dependent on low ALDH activity for proper activity, i.e. LSC-specific killing. Besides this, we are using drug libraries to identify novel drugs that can be detoxified via high ALDH activity. We hypothesize that AML patients with a large difference in ALDH activity between HSC and LSC might benefit from ALDH activity dependent treatment strategies.
Lately, ALDH has received considerable attention as a functional marker for identification of cells with enhanced tumorigenic/metastatic potential and elevated therapeutic resistance in several cancers of epithelial origin [35]–[37]. The relative functional contribution of ALDH activity to tumor-initiating potential is not clear and has not been the subject of this study. It might be that leukemic cells with considerable higher ALDH activity than other leukemic cells have enrichment of leukemia-initiating potential or might be more resistant to therapy. As in the study of Gerber et al. [34], we observed various populations of LSC within the ALDHlow compartment (defined by the inhibition with DEAB) which might represent LSC populations with different levels of ALDH activity. In the study by Pearce et al [33], as well as in the study by Berger et al [34], ALDH+ cells with a normal genotype were found in part of AML cases. The low incidence of positive cases, as compared to the study of Pearce et al. [33], and the high number of ‘rare’ pattern cases, as compared to the study of Cheung et al. [28], that we found is possibly related to the high number of CD34-negative AML cases that we have included in our study.
The difference in ALDH activity level as seen between normal and neoplastic stem cells may also add to the discussion and controversies regarding the cell of origin for AML, i.e. derived from transformation of HSC or from more committed progenitors [38], [39]. The fact that AML LSC do not exhibit ALDH activity at levels as high as those of HSC suggests that the cell giving rise to AML LSC was a progenitor endowed only in part with stem cell features, with the exclusion of an enhanced ALDH activity.
Overall, we show a marked difference between ALDH activity of HSC and LSC within the AML BM indicating the importance of ALDH activity as a functional stem cell biomarker and its potential usefulness in identification and purification of HSC and LSC, with the aim of treatment decision making, relapse prediction and development of LSC-specific therapies.
Methods
Patient Samples, AML Blast Phenotype, and Purification
BM samples were collected after informed consent from 32 AML patients at diagnosis. Normal BM was obtained after informed consent from healthy donors or patients undergoing cardiac surgery. The patients we used in this study gave informed consent following the procedure that was approved by the Medical Ethical Committee of our institute, the METC-VUmc. We (the Hematology department of the VUMC) have formal approval from the METC-VUmc for all our studies in acute myeloid leukemia patients. The patients gave written informed consent and the material of patients that did not gave this consent is not used for research purposes. AML phenotyping, cytogenetic, and molecular genetic analysis were carried out in the context of routine diagnostics at our facility. Cells were analyzed freshly or after thawing of samples frozen in liquid nitrogen. Mononuclear cells were isolated by Ficoll gradient (1.077 g/ml Amersham Biosciences, Freiburg Germany). Red blood cells were lysed by 10 minutes incubation on ice, using 10 ml lysing solution containing 155 mM NH4Cl, 10 mM KHCO3, 0.1 mM Na2EDTA pH7.4 applied directly to the cell pellet. AML cells were frozen in RPMI (Gibco, Paisley, Uk) with 20% heat-inactivated fetal bovine serum (FBS, Greiner, Alphen a/d Rijn, The Netherlands) and 10% dimethylsulfoxide (Riedel-de Haen, Seelze, Germany) in isopropanol-filled containers and subsequently stored in liquid nitrogen. When needed for analysis, cells were thawed and suspended in pre-warmed RPMI with 40% FBS at 37°C. Cells were washed in PBS with 0.1% HSA.
Aldehyde Dehydrogenase Activity Assay
Primary AML cells (1×106/ml) were resuspended in the aldefluor assay buffer at a concentration of 2×106 cells/ml. Cells were incubated with the ALDH-substrate BAAA (Stem cell Technologies, Aldefluor assay, 1 µl/ml) with or without the ALDH inhibitor, diethylamino-benzaldehyde (DEAB, 1.5 mM in 95% ethanol) and incubated at 37°C for 60 minutes according to the manufacturer protocol. After performing the Aldefluor assay, cells were resuspended in icecold PBS/0.1%HSA and washed once. The BAAA substrate can be detected in the FITC channel of the flow cytometer.
Cell Labeling, Flow Cytometry and Cell Purification
AML cells were incubated for 30 minutes on ice with monoclonal antibody (MoAb) combinations consisting of phycoerythrin (PE), allophycocyanin (APC), R-phycoerythrin-cyanine 7 (PC7), peridinin chlorophyll protein (PERCP) labeled MoAbs, anti-CD45 PerCp, Anti-CD34 PC7, Anti-CD38 APC, and anti-CD7 PE, Anti-CD56 PE, Anti-CLL1 PE, Anti-CD19 PE, Anti-CD33 PE, Anti-CD22 PE, Anti-CD123 PE. The antibodies were all from BD Biosciences (BD Biosciences, San Jose, CA, USA). After antibody staining, cells were washed with icecold PBS/0.1%HSA, resuspended in 250 µl cold PBS/0.1%HSA and stained for 5 minutes with Sytox Blue (Invitrogen, Molecular Probes) enabling to exclude dead cells. Cells were kept on ice before FACS analysis. Labeled samples were analyzed and cells were sorted using a flow cytometer (BD FACSAria, equipped with red, blue, ultra-violet and infrared solid-state lasers; BD Biosciences, San Jose, CA, USA). Prior to molecular analysis, subfractions were sorted. Data acquisition was performed using either a FACS Calibur or a FACS CantoII, both from BD Biosciences. Analysis was performed using Cellquest and FACS Diva software (BD Biosciences). For analysis of the presence molecular aberrancies, cells were sorted directed into cold culture medium. Purity of sorted population was in most cases >98%.
Length Fragment Analysis of FLT3 and NPM1 Mutations
To determine mutational status of flow cytometer-sorted subpopulations, only cryo-preserved cells were used. Genomic DNA (gDNA) was isolated from pellets of cells according to manufacturer’s instructions using the Quiagen Allprep kit (Quiagen Benelux B.V., Venlo, The Netherlands). Mutation profiling was performed on isolated gDNA as previously described for FLT3/ITD and NPM1. Direct PCR was applied to cell sorted subfractions under similar reaction conditions, except for the use of lower reaction volumes (always 10 µl). Mutations in NPM1exon12 were analyzed with PCR with the following primers: NPM1forward: 5′-TTAACTCTCTGGTGGTAGAATGA-3′; NPM1 reverse: 5-CTGACCACCGCTACTATGT-3, located in intron 11 and exon12 of the NPM1 genomic DNA, respectively. FLT3 was amplified using the primers spanning the entire transmembrane and JM domains of FLT3. Subsequent fragment analysis was performed with a tetrachlorofluorescein phosphoramidite-labeled forward primer (Biolegio, Nijmegen, The Netherlands). For both FLT3 and NPM1 analysis, lymphocytes served as an internal negative control.
RNA Sequencing
Total RNA from cells (normal BM HSC, AML HSC and LSC) was extracted with TRIzol reagent (Invitrogen, Leek, The Netherlands) according to the manufacturer’s protocol. The quality of RNA samples was measured on a 2100 Bioanalyzer (Agilent, Amstelveen, Netherlands). The sequencing libraries, each with individual Illumina indexes, were constructed using the TruSeq™ RNA Sample Prep Kit v2 (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. The resulting library DNA concentration and molarity was checked on a 2100 Bioanalyzer. Next, a mixture of three 10 pM libraries were pooled equimolar and the resulting DNA was clustered onto a V3 flowcell lane using a c-Bot cluster station and subsequently sequenced in single read fashion for 50 bp using the Illumina HiSeq2000 and V3 SBS Chemistry. Sequence reads were aligned to the human reference genome. In order to determine whether the aldehyde dehydrogenases are differentially expressed between samples, the R package DEGseq was used [40].
Supporting Information
CD34-positive AML cases; variation in the difference in ALDH activity between ALDHbright and ALDHlow compartments. ALDH versus SCC of the CD34+CD38– compartment of three CD34-positive AML cases. ALDH activity segregates the CD34+CD38– compartment in CD34+CD38– ALDHbright and CD34+CD38– ALDHlow cells. One AML case, AML-1048, with a relatively small difference between HSC and LSC ALDH activity levels (panel 1). An AML case, AML-1036, with a large difference between ALDH activity levels of HSC and LSC (panel 2). One AML case, AML-1013, with mainly HSC (panel 3).
(TIF)
Defining the ALDH compartments as has been done by others, amount of ALDH positive cells and pattern (29,34). The amount of ALDH positive cells is determined (normalized with the DEAB inhibitor) and the samples are defined as ALDH- (<5% ALDH positive cells from the total AML, A) or ALDH+ (>5% ALDH positive cells, B) With this method used by Cheung et al. (28) AML patients are classified based on percentage of ALDH positive cells defined by DEAB treatment. Our classification shows that 18% of AML patients are positive for ALDH (more than 5% of cells are ALDH+). The pattern of ALDH activity is determined by the shape, level and scatters properties of the ALDH activity as defined by Pearce et al. (33) The “rare” pattern ALDH activity is seen in (A) and the numerous pattern of ALDH activity is seen in (B). All the positive AML cases have the numerous pattern.
(TIF)
Quantification of ALDH activity in marker defined CD34+CD38– HSC and CD34+CD38– LSC compartments in CD34 positive AML. MFI values of CD34+CD38–marker- and CD34+CD38–marker+ were standardized by dividing them with the ALDH-MFI values from lymphocytes present in the same sample. AML samples were treated with diethylamino-benaldehyde (DEAB) to compare background MFI values in each cell population. Comparison of the median of the ALDH-MFI values of two populations; CD34+CD38– marker- cells and CD34+CD38– marker+ cells from 9 (9/19 CD34-positive AML cases were treated with DEAB) CD34-positive AML samples (median values are indicated) was done. In all CD34-positive AML cases, the MFI of CD34+CD38–marker- cells was divided with the MFI of the CD34+CD38–marker+ cells to obtain fold induction of ALDH activity in HSC compared to LSC within the AML. MFI is mean fluorescent intensity.
(TIF)
Quantification of ALDH activity in marker defined CD34+CD38– HSC and CD34– compartments in CD34 negative AML cases. *MFI is mean fluorescent intensity. MFI values of CD34+CD38–marker- cells and CD34– cells were standardized by dividing them with the ALDH-MFI values from lymphocytes present in the same sample. AML samples were treated with diethylamino-benaldehyde (DEAB) to compare background MFI values in each cell population. Comparison of the median of the ALDH-MFI values of two populations; CD34+CD38– marker- cells and CD34– cells from 8 (8/14 CD34-negative AML cases were treated with DEAB) CD34-negative AML samples (median values are indicated) was done. In all CD34-negative AML cases, the MFI of CD34+CD38–marker- cells was divided by the MFI of the CD34– cells to obtain fold induction of ALDH activity in HSC compared to the bulk of the AML. *MFI is mean fluorescent intensity.
(TIF)
The presence of FLT3-ITD and mutated NPM1 in ALDHbright CD34+CD38dim/+ progenitor populations. (A) CD34-negative AML cases. 4/4 CD34+CD38+/dim cells were negative. (B) CD34-positive AML cases. # In 3/5 CD34-positive AML cases, the CD34+CD38+ progenitor population has a tiny population of mutated cells present (likely contamination with ALDHlow cells). ne: not evaluable because CLL-1 (and sometimes CD33) is not a reliable aberrant marker for malignancy of CD34+CD38+ progenitors since part of normal progenitors and more mature CD34– cells can have CLL-1 and lineage marker expression (12). na: not applicable because no leukemia-associated aberrant marker present or expression to low.
(TIF)
Classification of ALDH defined AML cases. Percentage of ALDH positivity defined by DEAB treatment in the total AML (Panel 2). Percentage of ALDH positivity defined by DEAB treatment in the CD45dim blast population of cells (panel 3). AML cases that are defined as positive (more than 5% of the total AML population) are indicated as plus. AML cases that are defined as negative (less than 5% of the total AML population) are indicated as min (panel 4, Cheung et al.28). AML cases were defined as rare or numerous (panel 4, Pearce et al.33). In our cohort there are no AML cases with the negative pattern. The percentage of CD45dim blasts in the total AML (panel 6), the frequency of CD34+ cells within the total CD45dim population (panel 7), the total amount of CD34+CD38– cells within the CD34+ compartment (panel 8) and the total amount of CD34+CD38– cells within the CD45dim population of cells (panel 9) is indicated in this table.
(TIF)
Acknowledgments
We would like to thank Dr. R. Kerkhoven and Dr. I. de Rink (Netherlands Cancer Institute, Amsterdam, The Netherlands) for performing RNA sequencing and RNAseq data analysis. We thank the members of the Hematology laboratory at the VU University Medical Center for collecting and storing AML patient samples during the past years and are grateful to Jacqueline Cloos for helpful discussions and advice.
Funding Statement
This work was supported by the Association for International Cancer Research (AICR 08-0075). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bonnet D, Dick JE (1997) Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 3: 730–737. [DOI] [PubMed] [Google Scholar]
- 2.Lowenberg B, Griffin JD, Tallman MS (2003) Acute myeloid leukemia and acute promyelocytic leukemia. Hematology Am Soc Hematol Educ Program 82–101. [PubMed]
- 3. Van Rhenen A, Feller N, Kelder A, Westra AH, Rombouts E, et al. (2005) High stem cell frequency in acute myeloid leukemia at diagnosis predicts high minimal residual disease and poor survival. Clin Cancer Res 11: 6520–6527. [DOI] [PubMed] [Google Scholar]
- 4. Ishikawa F, Yoshida S, Saito Y, Hijikata A, Kitamura H, et al. (2007) Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat Biotech 11: 1315–1321. [DOI] [PubMed] [Google Scholar]
- 5.Terwijn M, Rutten AP, Kelder A, Snel AN, Scholten WJ, et al.. (2010) Accurate detection of residual leukemic stem cells in remission bone marrow predicts relapse in acute myeloid leukemia patients. Blood ASH Annu Meet Abstr 759.
- 6. Pearce DJ, Taussig D, Zibara K, Smith LL, Ridler CM, et al. (2006) AML engraftment in the NOD/SCID assay reflects the outcome of AML: implications for our understanding of the heterogeneity of AML. Blood 107: 1166–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, et al. (2011) Stem cell gene expression programs influence clinical outcome in human leukemia. Nature Med 17: 1086–1093. [DOI] [PubMed] [Google Scholar]
- 8. Taussig DC, Vargaftig J, Miraki-Moud F, Griessinger E, Sharrock K, et al. (2010) Leukemia-initiating cells from some acute myeloid leukemia patients with mutated nucleophosmin reside in the CD34(-) fraction. Blood 115: 1976–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taussig DC, Miraki-Moud F, Anjos-Afononso F, Pearce DJ, Allen K, et al.. (2008) Anti-CD38 antibody mediated clearance of human repopulating cells masks the heterogeneity of leukemia-initiating cells. Blood 112; 568–575. [DOI] [PubMed]
- 10. Buggins AG, Milojkovic D, Arno MJ, Lea NC, Mufti GJ, et al. (2001) Microenvironment produced by acute myeloid leukemia cells prevents T cell activation and proliferation by inhibition of NF-kappaB, c-Myc, and pRb pathways. J Immunol 167: 6021–6030. [DOI] [PubMed] [Google Scholar]
- 11. Feller N, van der Pol MA, van Stijn A, Weijers GW, Westra AH, et al. (2004) MRD parameters using immunophenotypic detection methods are highly reliable in predicting survival in acute myeloid leukaemia. Leukemia 18: 1380–1390. [DOI] [PubMed] [Google Scholar]
- 12. van Rhenen A, Moshaver B, Kelder A, Feller N, Nieuwint AW, et al. (2007) Aberrant marker expression patterns on the CD34+CD38– stem cell compartment in acute myeloid leukemia allows to distinguish the malignant from the normal stem cell compartment both at diagnosis and in remission. Leukemia 8: 1700–1707. [DOI] [PubMed] [Google Scholar]
- 13. van Rhenen A, van Dongen GA, Kelder A, Rombouts EJ, Feller N, et al. (2007) The novel AML stem cell associated antigen CLL-1 aids in discrimination between normal and leukemic stem cells. Blood 110: 2659–2666. [DOI] [PubMed] [Google Scholar]
- 14. Jordan CT, Upchurch D, Szilvassy SJ, Guzman ML, Howard DS, et al. (2000) The interleukin-3 receptor alpha chain is a unique marker for human acute myelogenous leukemia stem cells. Leukemia 10: 1777–1784. [DOI] [PubMed] [Google Scholar]
- 15. Bakker AB, van den Oudenrijn S, Bakker AQ, Feller N, van Meijer M, et al. (2004) C-type lectin-like molecule-1: a novel myeloid cell surface marker associated with acute myeloid leukemia. Cancer Res 64: 8443–8450. [DOI] [PubMed] [Google Scholar]
- 16.Janssen JJ, Deenik W, Smolders KG, van Kuijk BJ, Pouwels W, et al.. (2011) Residual normal stem cells can be detected in newly diagnosed chronic myeloid leukemia patients by a new flow cytometric approach and predict for optimal response to imatinib. Leukemia Dec 9. doi: 10.1038/leu.2011.347. [DOI] [PubMed]
- 17. Chute JP, Muramoto GG, Whitesides J, Colvin M, Safi R, et al. (2006) Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc Natl Acad Sci USA 103: 11707–11712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Duester G (2000) Families of retinoid dehydrogenases regulating vitamin A function: production of visual pigment and retinoic acid. Eur J Biochem 267: 4315–4324. [DOI] [PubMed] [Google Scholar]
- 19. Magni M, Shammah S, Schiró R, Mellado W, Dalla-Favera R, et al. (1996) Induction of cyclophosphamide-resistance by aldehyde-dehydrogenase gene transfer. Blood 87: 1097–1103. [PubMed] [Google Scholar]
- 20. Wang JS, Fang Q, Sun DJ, Chen J, Zhou XL, et al. (2001) Genetic modification of hematopoietic progenitor cells for combined resistance to 4-hydroperoxycyclophosphamide, vincristine, and daunorubicin. Acta Pharmacol Sin 10: 949–955. [PubMed] [Google Scholar]
- 21. Takebe N, Zhao SC, Adhikari D, Mineishi S, Sadelain M, et al. (2001) Generation of dual resistance to 4-hydroperoxycyclophosphamide and methotrexate by retroviral transfer of the human aldehyde dehydrogenase class 1 gene and a mutated dihydrofolate reductase gene. Mol Ther 1: 88–96. [DOI] [PubMed] [Google Scholar]
- 22. Kastan MB, Schlaffer E, Russo JE, Colvin OM, Civin CI, et al. (1990) Direct demonstration of elevated aldehyde dehydrogenase in human hematopoietic progenitor cells. Blood 75: 1947–1950. [PubMed] [Google Scholar]
- 23. Armstrong L, Stojkovic M, Dimmick I, Ahmad S, Stojkovic P, et al. (2004) Phenotypic characterization of murine primitive hematopoietic progenitor cells isolated on basis of aldehyde dehydrogenase activity. Stem cells 22: 1142–1151. [DOI] [PubMed] [Google Scholar]
- 24. Gentry T, Deibert E, Foster SJ, Haley R, Kurtzberg J, et al. (2007) Isolation of early hematopoietic cells, including megakaryocyte progenitors, in the ALDH-bright cell population of cryopreserved, banked UC blood. Cytotherapy 9: 569–576. [DOI] [PubMed] [Google Scholar]
- 25. Storms RW, Trujillo AP, Springer JB, Shah L, Colvin OM, et al. (1999) Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc Natl Acad Sci USA 96: 9118–9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christ O, Lucke K, Imren S, Leung K, Hamilton M, et al.. (2007) Improved purification of hematopoietic stem cells based on their elevated aldehyde dehydrogenase activity. Haematologica 92: 1165–11 72. [DOI] [PubMed]
- 27. Hess DA, Meyerrose TE, Wirthlin L, Craft TP, Herrbrich PE, et al. (2004) Functional characterization of highly purified human hematopoietic repopulating cells isolated according to aldehyde dehydrogenase activity. Blood 104: 1648–1655. [DOI] [PubMed] [Google Scholar]
- 28. Cheung AM, Wan TS, Leung JC, Chan LY, Huang H, et al. (2007) Aldehyde dehydrogenase activity in leukemic blasts defines a subgroup of acute myeloid leukemia with adverse prognosis and superior NOD/SCID engrafting potential. Leukemia 21: 1423–30. [DOI] [PubMed] [Google Scholar]
- 29.Ran D, Schubert M, Pietsch L, Taubert I, Wuchter P, et al.. (2009) Aldehyde dehydrogenase activity among primary leukemia cells is associated with stem cell features and correlates with adverse clinical outcome. Exp Hematology 12;1423–1434. [DOI] [PubMed]
- 30. van der Pol MA, Feller N, Roseboom M, Moshaver B, Westra G, et al. (2003) Assessment of the normal or leukemic nature of CD34+ cells in acute myeloid leukemia with low percentages of CD34 cells. Haematologica 88: 983–993. [PubMed] [Google Scholar]
- 31. Taussig DC, Pearce DJ, Simpson C, Rohatiner AZ, Lister TA, et al. (2005) Hematopoietic stem cells express multiple myeloid markers: implications for the origin and targeted therapy of acute myeloid leukemia. Blood 106: 4086–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dash A, Gilliland DG (2001) Molecular genetics of acute myeloid leukaemia. Best Pract Res Clin Haematol 1: 49–84. [DOI] [PubMed] [Google Scholar]
- 33. Pearce DJ, Taussig D, Simpson C, Allen K, Rohatiner AZ, et al. (2005) Characterization of cells with a high aldehyde dehydrogenase activity from cord blood and acute myeloid leukemia samples. Stem cells 23: 752–760. [DOI] [PubMed] [Google Scholar]
- 34. Gerber JM, Smith BD, Ngwang B, Zhang H, Vala MS, et al. (2012) A clinically relevant population of leukemic CD34+CD38– cells in acute myeloid leukemia. Blood 119: 3571–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, et al. (2007) ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell 1: 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jiang F, Qiu Q, Khanna A, Todd NW, Deepak J, et al. (2009) Aldehyde Dehydrogenase 1 Is a Tumor Stem Cell-Associated Marker in Lung Cancer. Mol Cancer Res 7: 330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tanei T, Morimoto K, Shimazu K, Kim SJ, Tanji Y, et al. (2009) Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin Cancer Res 15: 4234–4241. [DOI] [PubMed] [Google Scholar]
- 38. Wang JC (2010) Good cells gone bad: the cellular origins of cancer. Trends Mol Med 16: 145–151. [DOI] [PubMed] [Google Scholar]
- 39. Goardon N, Marchi E, Atzberger A, Quek L, Schuh A, et al. (2011) Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell 19: 138–152. [DOI] [PubMed] [Google Scholar]
- 40. Wang L, Feng Z, Wang X, Wang X, Zhang X (2010) DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26: 136–138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CD34-positive AML cases; variation in the difference in ALDH activity between ALDHbright and ALDHlow compartments. ALDH versus SCC of the CD34+CD38– compartment of three CD34-positive AML cases. ALDH activity segregates the CD34+CD38– compartment in CD34+CD38– ALDHbright and CD34+CD38– ALDHlow cells. One AML case, AML-1048, with a relatively small difference between HSC and LSC ALDH activity levels (panel 1). An AML case, AML-1036, with a large difference between ALDH activity levels of HSC and LSC (panel 2). One AML case, AML-1013, with mainly HSC (panel 3).
(TIF)
Defining the ALDH compartments as has been done by others, amount of ALDH positive cells and pattern (29,34). The amount of ALDH positive cells is determined (normalized with the DEAB inhibitor) and the samples are defined as ALDH- (<5% ALDH positive cells from the total AML, A) or ALDH+ (>5% ALDH positive cells, B) With this method used by Cheung et al. (28) AML patients are classified based on percentage of ALDH positive cells defined by DEAB treatment. Our classification shows that 18% of AML patients are positive for ALDH (more than 5% of cells are ALDH+). The pattern of ALDH activity is determined by the shape, level and scatters properties of the ALDH activity as defined by Pearce et al. (33) The “rare” pattern ALDH activity is seen in (A) and the numerous pattern of ALDH activity is seen in (B). All the positive AML cases have the numerous pattern.
(TIF)
Quantification of ALDH activity in marker defined CD34+CD38– HSC and CD34+CD38– LSC compartments in CD34 positive AML. MFI values of CD34+CD38–marker- and CD34+CD38–marker+ were standardized by dividing them with the ALDH-MFI values from lymphocytes present in the same sample. AML samples were treated with diethylamino-benaldehyde (DEAB) to compare background MFI values in each cell population. Comparison of the median of the ALDH-MFI values of two populations; CD34+CD38– marker- cells and CD34+CD38– marker+ cells from 9 (9/19 CD34-positive AML cases were treated with DEAB) CD34-positive AML samples (median values are indicated) was done. In all CD34-positive AML cases, the MFI of CD34+CD38–marker- cells was divided with the MFI of the CD34+CD38–marker+ cells to obtain fold induction of ALDH activity in HSC compared to LSC within the AML. MFI is mean fluorescent intensity.
(TIF)
Quantification of ALDH activity in marker defined CD34+CD38– HSC and CD34– compartments in CD34 negative AML cases. *MFI is mean fluorescent intensity. MFI values of CD34+CD38–marker- cells and CD34– cells were standardized by dividing them with the ALDH-MFI values from lymphocytes present in the same sample. AML samples were treated with diethylamino-benaldehyde (DEAB) to compare background MFI values in each cell population. Comparison of the median of the ALDH-MFI values of two populations; CD34+CD38– marker- cells and CD34– cells from 8 (8/14 CD34-negative AML cases were treated with DEAB) CD34-negative AML samples (median values are indicated) was done. In all CD34-negative AML cases, the MFI of CD34+CD38–marker- cells was divided by the MFI of the CD34– cells to obtain fold induction of ALDH activity in HSC compared to the bulk of the AML. *MFI is mean fluorescent intensity.
(TIF)
The presence of FLT3-ITD and mutated NPM1 in ALDHbright CD34+CD38dim/+ progenitor populations. (A) CD34-negative AML cases. 4/4 CD34+CD38+/dim cells were negative. (B) CD34-positive AML cases. # In 3/5 CD34-positive AML cases, the CD34+CD38+ progenitor population has a tiny population of mutated cells present (likely contamination with ALDHlow cells). ne: not evaluable because CLL-1 (and sometimes CD33) is not a reliable aberrant marker for malignancy of CD34+CD38+ progenitors since part of normal progenitors and more mature CD34– cells can have CLL-1 and lineage marker expression (12). na: not applicable because no leukemia-associated aberrant marker present or expression to low.
(TIF)
Classification of ALDH defined AML cases. Percentage of ALDH positivity defined by DEAB treatment in the total AML (Panel 2). Percentage of ALDH positivity defined by DEAB treatment in the CD45dim blast population of cells (panel 3). AML cases that are defined as positive (more than 5% of the total AML population) are indicated as plus. AML cases that are defined as negative (less than 5% of the total AML population) are indicated as min (panel 4, Cheung et al.28). AML cases were defined as rare or numerous (panel 4, Pearce et al.33). In our cohort there are no AML cases with the negative pattern. The percentage of CD45dim blasts in the total AML (panel 6), the frequency of CD34+ cells within the total CD45dim population (panel 7), the total amount of CD34+CD38– cells within the CD34+ compartment (panel 8) and the total amount of CD34+CD38– cells within the CD45dim population of cells (panel 9) is indicated in this table.
(TIF)