The E3 ubiquitin ligase RNF168 recruits LSD1 to DNA damage sites, where it reduces histone methylation upstream of 53BP1 recruitment during the DNA damage response.
Abstract
Histone demethylation is known to regulate transcription, but its role in other processes is largely unknown. We report a role for the histone demethylase LSD1/KDM1A in the DNA damage response (DDR). We show that LSD1 is recruited directly to sites of DNA damage. H3K4 dimethylation, a major substrate for LSD1, is reduced at sites of DNA damage in an LSD1-dependent manner. The E3 ubiquitin ligase RNF168 physically interacts with LSD1 and we find this interaction to be important for LSD1 recruitment to DNA damage sites. Although loss of LSD1 did not affect the initial formation of pH2A.X foci, 53BP1 and BRCA1 complex recruitment were reduced upon LSD1 knockdown. Mechanistically, this was likely a result of compromised histone ubiquitylation preferentially in late S/G2. Consistent with a role in the DDR, knockdown of LSD1 resulted in moderate hypersensitivity to γ-irradiation and increased homologous recombination. Our findings uncover a direct role for LSD1 in the DDR and place LSD1 downstream of RNF168 in the DDR pathway.
Introduction
A double-stranded DNA break (DSB) represents a challenging problem for the cell, and its proper repair is critical for cell survival and the prevention of oncogenic transformation. In eukaryotes, DSBs initiate a signaling cascade on chromatin that coordinates ordered recruitment of specific factors to the damaged region, promotes cell cycle arrest, and effects DNA repair (Jackson and Bartek, 2009; Ciccia and Elledge, 2010). Studies in recent years have provided significant insight into the signaling cascade that mediates the DNA damage response (DDR), but the chromatin modifications important for DDR regulation remain incompletely understood.
The DDR cascade begins with the detection of DSBs by the MRN (MRE11–RAD50–NBS1) complex, which recruits and activates the ataxia telangiectasia mutated (ATM) kinase at DSBs to phosphorylate the variant histone H2A.X (Jackson and Bartek, 2009; Ciccia and Elledge, 2010). The formation of this phosphorylated histone (also known as γH2A.X) recruits the large scaffold phosphoprotein MDC1 to irradiation-induced foci (IRIF; Stewart et al., 2003; Stucki et al., 2005). MDC1 recruits the E3 ubiquitin ligase RNF8, which promotes ubiquitylation events near DSBs (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007). This ubiquitylation is further amplified by a second E3 ligase, RNF168, although recent evidence also suggests that RNF8 functions both upstream as well as downstream of RNF168 (Mattiroli et al., 2012). RNF168-mediated ubiquitylation promotes the recruitment of various downstream effector complexes (Doil et al., 2009; Stewart et al., 2009). One such complex includes BRCA1, which promotes repair primarily by homologous recombination (HR; Huen et al., 2010). A second effector is 53BP1, which promotes XRCC4-dependent nonhomologous end joining (Xie et al., 2007). Both BRCA1 and 53BP1 serve as tumor suppressors, at least partly because of their roles in DNA repair. It is well known that loss of BRCA1 significantly increases the risk of human breast and ovarian tumors, and mice bearing BRCA1 hypomorphic alleles are tumor prone (Huen et al., 2010). 53BP1 knockout mice are also prone to developing tumors in a variety of organ systems (Ward et al., 2005). Consistently, cells lacking 53BP1 harbor many signs of genomic instability, including hypersensitivity to genotoxic agents, increased aneuploidy, and loss of DNA damage–induced cell cycle arrest (FitzGerald et al., 2009). Localization of 53BP1 to IRIF is crucial for these functions, and the aforementioned upstream factors, particularly H2A/H2A.X ubiquitylation as well as dimethylation of histone H4 at lysine 20 (H4K20), are required for its recruitment (FitzGerald et al., 2009). Additional chromatin modifications associated with the DDR include Tip60-mediated histone acetylation (van Attikum and Gasser, 2009), as well as deacetylation of H3K56 (Miller et al., 2010). However, the entire spectrum of chromatin modifications and the associated enzymes required for the recruitment of 53BP1 or other IRIF factors to DNA damage sites are far from being completely understood.
Posttranslational modifications of histones, including methylation, acetylation, phosphorylation, and ubiquitylation, among others (Strahl and Allis, 2000), represent an important aspect of epigenetic regulation. Histone methylation, which occurs on both lysine and arginine residues, plays important roles in transcriptional activation and repression (Bedford and Clarke, 2009; Mosammaparast and Shi, 2010). The histone demethylase LSD1 (lysine-specific demethylase 1) mediates demethylation of histone H3K4me1/2 (dimethylated histone H3 lysine 4) and in so doing functions to repress transcription (Shi et al., 2004). Consistently, LSD1 is a component of transcriptional corepressor complexes containing histone deacetylases (You et al., 2001; Hakimi et al., 2002; Hakimi et al., 2003). LSD1 has also been shown to associate with the NuRD chromatin-remodeling corepressor complex (Wang et al., 2009). LSD1 also participates in transcriptional activation when associated with nuclear hormone receptors, i.e., androgen and estrogen receptors (Metzger et al., 2005; Garcia-Bassets et al., 2007), but the underlying molecular mechanism in this context is unclear. Notably, whether or not LSD1 plays a role outside of transcription remains unknown.
Here, we provide evidence that LSD1 plays a direct role in the DDR. We show that LSD1 is recruited to sites of DSBs in vivo, concomitant with a reduction of H3K4 dimethylation (H3K4me2), which occurs primarily in late S/G2 cells and is dependent on LSD1. Furthermore, recruitment of the downstream effector protein 53BP1 to IRIF is dependent on LSD1 in a subset of cells, which are also in the late S/G2 phase of the cell cycle. The mechanism of this appears to be reduced H2A/H2A.X ubiquitylation in the absence of LSD1, and, consistently, BRCA1 and Rap80 foci are also reduced upon LSD1 knockdown. We demonstrate that the E3 ubiquitin ligase RNF168 physically interacts with LSD1 and is important for recruitment of LSD1 to sites of DNA damage. Our findings suggest that LSD1 may play a direct role, possibly downstream of RNF168 but upstream of 53BP1 and BRCA1, in the DDR. Our findings further suggest a potential cross talk between H3K4 demethylation mediated by LSD1 and H2A/H2A.X ubiquitylation in the DDR.
Results
LSD1 is recruited to sites of DNA damage
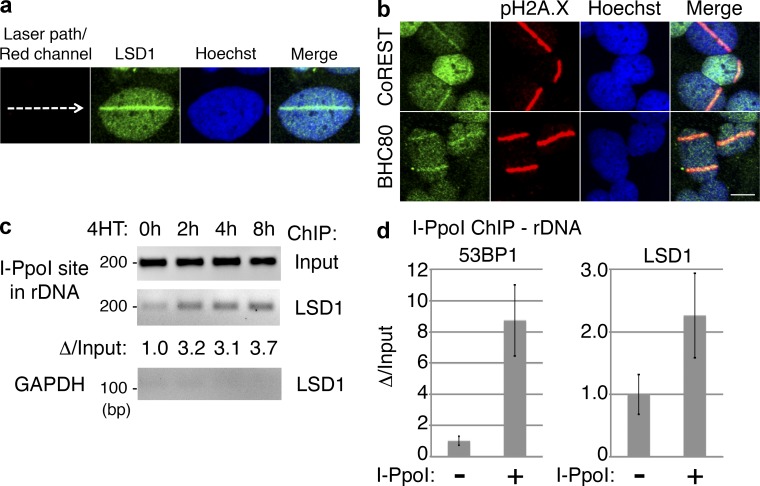
Recent studies have suggested that H3K4 methylation plays a role in controlling the DDR and recombination events (Cheung et al., 2010; Daniel and Nussenzweig, 2012). Specifically, H3K4 methylation appears to be tightly associated with regions of increased HR and crossovers during meiosis (Borde et al., 2009; Sommermeyer et al., 2013). We therefore wished to determine whether LSD1, an H3K4 demethylase, plays a role in the DDR by being recruited to sites of DNA damage. We used immunostaining to determine whether LSD1 concentrates at sites of DNA damage induced by UV laser microirradiation. Strikingly, 10 min after irradiation, endogenous LSD1 was found at the laser path, colocalizing with pH2A.X (Fig. 1 a and Fig. S1). Knockdown of LSD1 by shRNA resulted in the loss of LSD1 signal at the laser stripe, demonstrating specificity (Fig. S1, a and b). We also found two LSD1-associated proteins, CoREST and BHC80 (Lee et al., 2005; Shi et al., 2005), to colocalize with pH2A.X under the same conditions (Fig. 1 b). Importantly, neither the histone H3K9/K36 demethylase JMJD2A nor the H3K27 demethylase JMJD3 were colocalized with pH2A.X using laser microirradiation (Fig. S1 c).
Figure 1.
LSD1 is recruited to sites of DNA damage. (a) UV laser microirradiation was performed on U2OS cells, incubated at 37°C for 10 min, stained for endogenous LSD1, and analyzed by confocal microscopy. (b) U2OS cells were microirradiated as in panel a, and then stained for CoREST or BHC80 and pH2A.X as indicated. Bar, 10 µm. (c) PCR was performed using rDNA or GAPDH primers with LSD1 ChIP material after DSBs were induced with retroviral I-PpoI in U2OS cells. The indicated time points represent the length of 4-OH-tamoxifen (4HT) treatment to induce DSBs. Shown below is the change of LSD1 rDNA ChIP quantified and normalized to input. (d) U2OS cells transduced with control or I-PpoI–expressing virus and were treated with 4HT for 4 h. ChIP was then performed using antibodies against 53BP1 or LSD1, and real-time qPCR was done on the ChIP material using rDNA primers.
To confirm the association of LSD1 with sites of DNA damage, we turned to chromatin immunoprecipitation (ChIP) after DNA damage induction using an endonuclease. In this approach, a fusion protein consisting of the homing endonuclease I-PpoI, coupled to a hormone-responsive fragment of estrogen receptor, is targeted to the nucleus in the presence of tamoxifen, where DSBs are produced, primarily at ribosomal DNA (rDNA) repeats (Berkovich et al., 2007, 2008). Indeed, I-PpoI–induced DSBs recruited LSD1 to rDNA repeats (Fig. 1, c and d). As expected, I-PpoI–induced DSBs also recruited ATM to rDNA repeats, but a control IgG antibody did not produce significant ChIP signal (Fig. S1 d).
H3K4me2 demethylation marks sites of DNA damage and occurs primarily in late S/G2
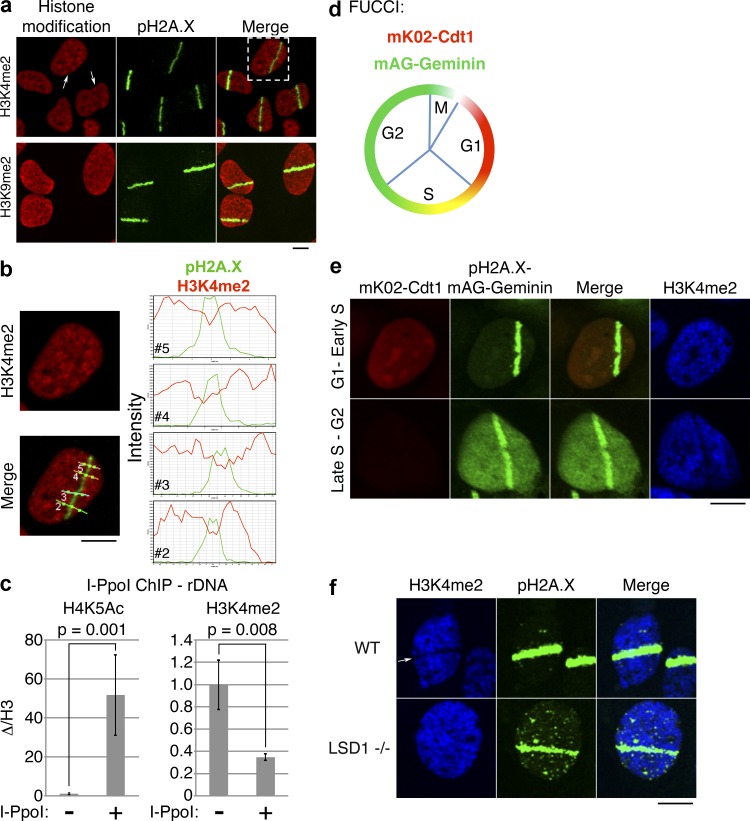
The fact that LSD1 is specifically recruited to sites of DNA damage suggested that a reduction of H3K4 methylation may be observed at sites of DNA damage. Consistent with this prediction, we found that H3K4me2 is lost in a significant population of cells upon laser microirradiation (Fig. 2 a, top). Profiling of the immunofluorescent signal confirmed an inverse correlation between pH2A.X and H3K4me2 (Fig. 2 b). We termed this loss of H3K4me2 at laser stripes as H3K4me2 antistripes. LSD1 has also been reported to demethylate H3K9me2 (Metzger et al., 2005), but we did not observe loss of H3K9me2 signal at pH2A.X stripes (Fig. 2 a, bottom). Furthermore, we did not observe loss of total H3 upon microirradiation (unpublished data). Collectively, these findings suggest that LSD1 may be responsible for the H3K4me2 reduction at DNA damage sites.
Figure 2.
H3K4me2 demethylation marks sites of DNA damage and is cell cycle and LSD1 dependent. (a) UV laser microirradiation was performed on U2OS cells, and the cells were stained with antibodies against the indicated histone modifications. Arrows indicate nuclei with apparent loss of H3K4me2 signal. (b) Magnified view of the cell from top right of panel a, showing H3K4me2 or merged H3K4me2/pH2A.X signals. Intensity profiles of H3K4me2 (red) and pH2A.X (green) signals through the four indicated lines (2–5) are shown on the right. (c) I-PpoI ChIP/qPCR was performed using the indicated histone modification-specific antibodies and primers specific for rDNA. Error bars represent ± standard error. (d) Schematic of FUCCI cells. FUCCI cells coexpress a fragment of Cdt1 linked to the fluorescent protein mK02 (monomeric Kusabira orange 2), as well as a fragment of Geminin linked to the fluorescent protein mAG (monomeric Azami green). (e) UV laser microirradiation was performed on U2OS-FUCCI cells and subsequently stained for pH2A.X and H3K4me2. (f) UV laser microirradiation was performed on wild-type (WT) and LSD1−/− MEFs and subsequently stained for the indicated histone modifications. Bars, 10 µm.
To confirm the aforementioned finding, we again turned to the I-PpoI system discussed earlier. As a positive control, we initially assessed acetylation at H4K5 after I-PpoI induced damage and found this mark dramatically increased (Fig. 2 c), consistent with the previously described recruitment of the histone acetyltransferase Tip60 to DNA damage foci, which acetylates H4K5 (Murr et al., 2006). Importantly, we observed a significant decrease in H3K4me2 (Fig. 2 c), consistent with the microirradiation results. Thus, LSD1 and H3K4me2 demethylation appear to mark sites of DNA damage in vivo.
We noticed that only about half of the cells that were positive for pH2A.X stripes had H3K4me2 antistripes (Fig. 2 a and see the quantitation in the following section). We hypothesized that this may be the result of a cell cycle–dependent process. To address this, we turned to fluorescence ubiquitylation cell cycle indicator (FUCCI) cells, which express fragments of two proteins, Cdt1 and geminin, whose levels vary in accordance with the cell cycle stage because of degradation by distinct E3 ubiquitin ligases (Sakaue-Sawano et al., 2008; Fig. 2 d). Fusion of these two fragments to red and green fluorescent proteins, respectively, allows the direct visualization of the cell cycle stage. Microirradiation of U2OS-FUCCI cells revealed a clear cell cycle dependence of H3K4me2 antistripes. 67% of late S/G2 cells had H3K4me2 antistripes, whereas only 21% of G1 cells had visible H3K4me2 antistripes (Fig. 2 e). Thus, H3K4me2 demethylation occurs at sites of DNA damage in a cell cycle–dependent fashion.
Why does H3K4me2 demethylation at sites of DNA damage occur primarily during late S/G2? One explanation could be that LSD1 expression varies accordingly as the cell cycle progresses. A previous study demonstrated that LSD1 levels are relatively low in G1/early S and are increased during cell cycle progression (Lv et al., 2010). We confirmed that LSD1 protein levels are in fact higher during G2 and fall when cells enter G1 (Fig. S1 e).
H3K4me2 demethylation at sites of DNA damage is LSD1 dependent
Is the loss of H3K4me2 at sites of DNA damage dependent on LSD1? We found that although loss of the H3K4me2 signal was readily observed upon UV laser microirradiation in wild-type mouse embryonic fibroblast (MEF) cells, the H3K4me2 signal was still present at pH2A.X laser stripes in LSD1−/− MEFs (Fig. 2 f and Fig. S1 f). Quantitation revealed that 54% of the wild-type MEFs had visible loss of H3K4me2 at pH2A.X stripes, whereas only 14% of LSD1−/− MEFs had H3K4me2 antistripes. These data suggest that LSD1 is a major player that mediates H3K4me2 demethylation at sites of DNA damage, although involvement of another H3K4 demethylase in this process cannot be excluded.
Recruitment of LSD1 to sites of DNA damage is dependent on RNF168
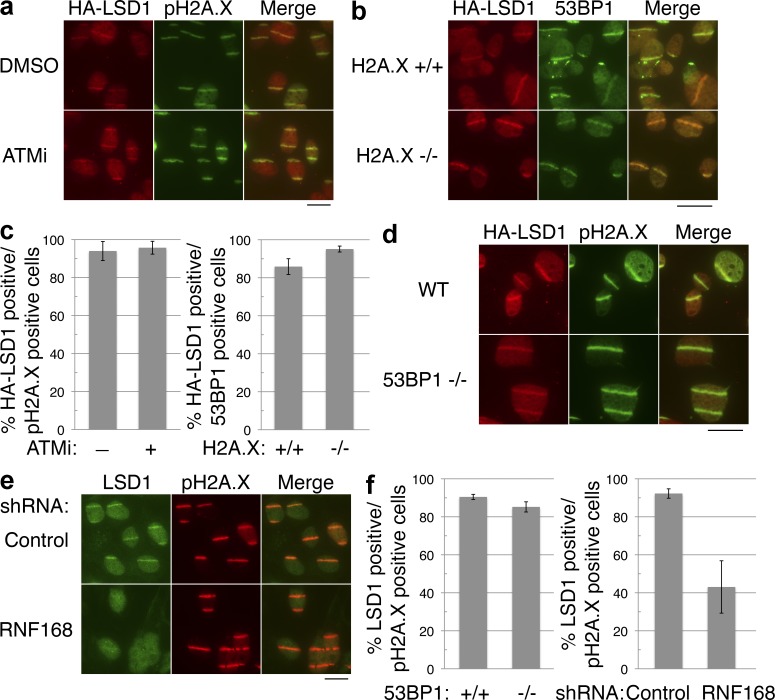
We next wished to determine the mechanism by which LSD1 is recruited to sites of DNA damage. We first considered whether LSD1 recruitment is dependent on the ATM or ataxia telangiectasia and Rad3 related (ATR) kinase pathways. Comparable numbers of cells treated with either the ATM inhibitor KU55933 or DMSO control displayed visible HA-LSD1 stripes overlapping with the pH2AX signal upon DNA damage (Fig. 3, a and c). There was also no change in LSD1 recruitment using an ATR inhibitor (Toledo et al., 2011; Fig. S1 g). At the concentrations used, both inhibitors were indeed functional (Fig. S2 a). Similarly, the loss of H2A.X did not affect LSD1 recruitment (Fig. 3, b and c). We considered the possibility that LSD1 recruitment may be PARP dependent because certain chromatin-modifying factors that are rapidly recruited to sites of damage are also PARP dependent (Chou et al., 2010; Polo et al., 2010). However, inhibition of PARP also did not affect LSD1 recruitment (Fig. S2, a–c). Interestingly, certain chromatin-associated factors, such as 53BP1, have rapid recruitment to sites of DNA damage even in the absence of H2A.X, but are not retained without H2A.X (Celeste et al., 2003). We then asked whether 53BP1 or a key factor responsible for its recruitment, RNF168, are important for LSD1 recruitment. Laser microirradiation experiments did not reveal a difference in the recruitment of LSD1 in 53BP1−/− MEFs (Fig. 3, d and f). However, knockdown of RNF168 (Fig. S2 d) caused a significant reduction in the recruitment of LSD1 to sites of DNA damage (Fig. 3 e). Quantitation revealed only ∼40% of pH2A.X-positive stripes were positive for LSD1 stripes upon RNF168 knockdown, as opposed to >90% in control knockdown cells (Fig. 3 f). RNF168 knockdown also reduced recruitment of HA-tagged LSD1 to microirradiation sites (unpublished data).
Figure 3.
LSD1 recruitment to sites of DNA damage is dependent on RNF168. (a) Flag-HA-LSD1 was stably expressed in U2OS cells, and then treated with the ATM inhibitor KU55933 (15 µM) or DMSO 1 h before laser microirradiation. The cells were then stained for HA and pH2A.X. (b) Wild-type (H2A.X+/+) or H2A.X-deficient (H2A.X−/−) MEFs stably expressing HA-tagged LSD1 were laser microirradiated and stained as in panel a. (c) Quantitation of a and b, with error bars representing the SD of duplicate experiments. (d) Wild-type or 53BP1-deficient (53BP1−/−) MEFs stably expressing Flag-HA–tagged LSD1 were laser microirradiated and stained as in panel a. (e) U2OS cells were treated with the indicated shRNAs, microirradiated, and stained for endogenous LSD1 and pH2A.X. (f) Quantitation of d and e, with error bars representing the SD of duplicate experiments. Bars, 20 µm.
Retention of LSD1 at microirradiation sites is transient
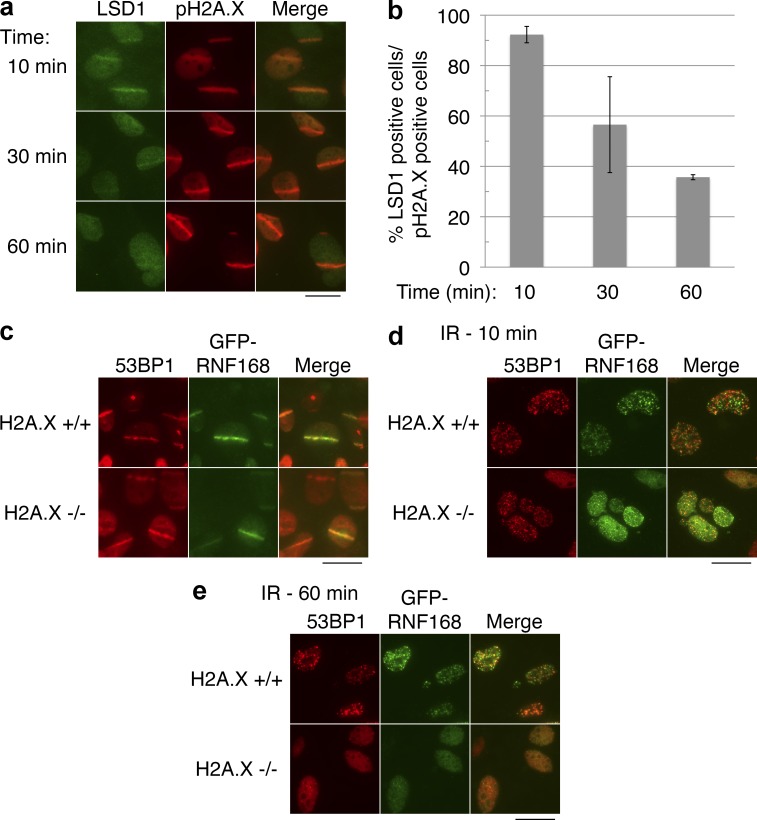
Many other chromatin-associated factors, such as KAP1 and CHD4, are recruited rapidly to sites of DNA damage but become dissociated quickly after their recruitment (Ziv et al., 2006; Polo et al., 2010), whereas other factors, such as 53BP1, are retained for hours (Celeste et al., 2003). We wished to determine the retention kinetics of LSD1 at sites of DNA damage. Although over 90% of cells had visible LSD1 stripes 10 min after microirradiation, this was reduced significantly 60 min after microirradiation (Fig. 4, a and b). We conclude that LSD1 is recruited to sites of DNA damage but that its retention is relatively transient.
Figure 4.
Transient retention of LSD1 and a rapid, H2A.X-independent recruitment of RNF168 to sites of DNA damage. (a) U2OS cells were laser microirradiated, incubated for the indicated time at 37°C, and fixed and stained for LSD1 and pH2A.X. (b) Quantitation of panel a. Error bars indicate ± SD of duplicate experiments. (c) Wild-type (H2A.X+/+) or H2A.X-deficient (H2A.X−/−) MEFs expressing GFP-tagged RNF168 were laser microirradiated, incubated for 10 min at 37°C, and fixed and stained as indicated. (d and e) Cells from panel c were exposed to 5 Gy IR, incubated at 37°C for the indicated times, and stained as in c. Bars, 20 µm.
Rapid, transient recruitment of RNF168 to damage sites is H2A.X independent
The aforementioned results were surprising because localization of RNF168 to sites of DNA damage is thought to be H2A.X dependent, but LSD1 recruitment is H2A.X independent although apparently RNF168 dependent. We noticed that most studies on RNF168 and the requirement of H2A.X for its recruitment to sites of damage were done using longer time scales, typically 1–4 h after irradiation (Doil et al., 2009; Stewart et al., 2009), whereas our microirradiation studies on LSD1 were done on a shorter timescale because of its transient retention described in the previous section. Therefore it was possible that RNF168 is rapidly recruited to sites of damage in a manner independent of H2A.X, but that its retention is H2A.X dependent, as previously described for 53BP1 (Celeste et al., 2003). Indeed, we found that GFP-RNF168 recruitment to sites of laser-induced damage is clearly seen in the absence of H2A.X 10 min after irradiation (Fig. 4 c). Although 100% of H2A.X+/+ cells had visible GFP-RNF168 recruitment at microirradiation sites, 100% of H2A.X−/− cells also had GFP-RNF168 recruitment (using 53BP1 as the microirradiation site indicator). We could also observe the recruitment of RNF168 to IR-induced foci (IRIF) in the same cells 10 min after irradiation (Fig. 4 d). We should note that GFP-RNF168 formed very distinct foci in H2A.X+/+ cells, whereas some background GFP-RNF168 nucleoplasmic staining was noticeable in H2A.X−/− cells, whose foci were somewhat smaller. Consistent with the previously published data, RNF168 was not retained at sites of DNA damage in the absence of H2A.X 1 h after irradiation (Fig. 4 e; Doil et al., 2009; Stewart et al., 2009). Therefore, like 53BP1, an H2A.X-independent mechanism exists for the initial recruitment of RNF168 to sites of DNA damage. We should note that a recent study suggested that the H2A.X-independent rapid recruitment of 53BP1 to IRIF is dependent on RNF168 (Bohgaki et al., 2011). Thus, it appears that both LSD1 and 53BP1 may depend on RNF168 for this initial rapid recruitment to DNA damage sites.
LSD1 interacts with RNF168
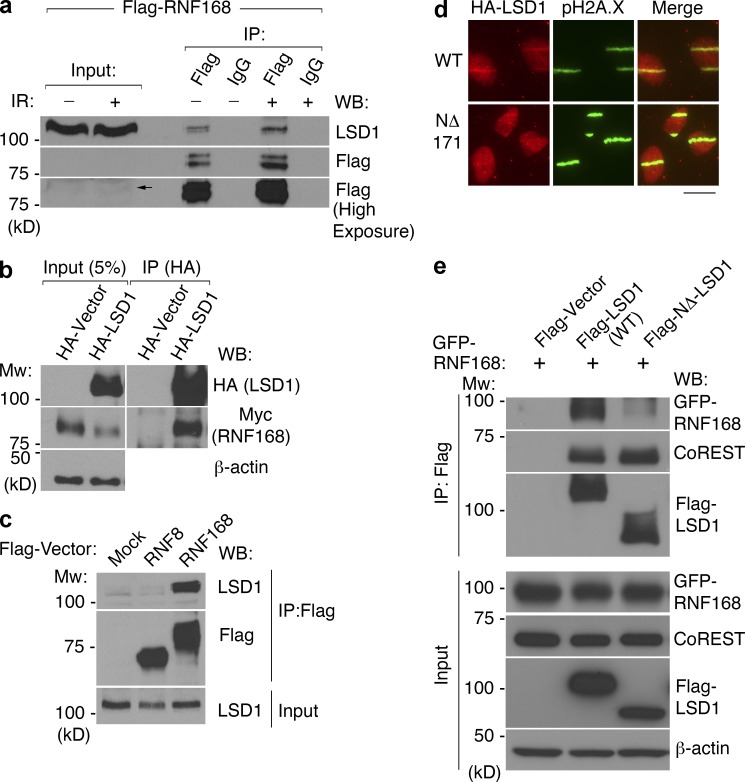
Reminiscent of CHD4 and its dependency on physical interaction with RNF8 for recruitment to damage sites (Luijsterburg et al., 2012), it was possible that the recruitment of LSD1 to sites of DNA damage was dependent on physical interaction with RNF168. To test this, we first took an unbiased approach by generating a stable cell line expressing Flag-tagged RNF168 and immunoprecipitating the tagged RNF168 and its associated proteins from irradiated and nonirradiated cells for mass spectrometry analysis. Indeed, LSD1 was present in both samples, along with many other interactors (Table S1). We then confirmed the presence of LSD1 in the Flag-purified RNF168 complex by Western blotting (Fig. 5 a). We further demonstrated by reciprocal immunoprecipitation that HA-LSD1 effectively coimmunoprecipitated myc-tagged RNF168 (Fig. 5 b). Although immunoprecipitation of Flag–RNF168 from asynchronous cells demonstrated its association with LSD1, this interaction seems to be enhanced in nocodazole-arrested cells, at least partly because there was greater RNF168 present (Fig. S2 e). Whether the endogenous, untagged RNF168 is similarly enriched at the G2 phase of the cell cycle remains to be determined. As a control, we failed to identify an association between LSD1 and the other damage-associated E3 ligase RNF8 under the same conditions (Fig. 5 c). Using coimmunoprecipitation, we also confirmed the physical interaction between endogenous LSD1 and RNF168 (Fig. S2 f).
Figure 5.
RNF168 interacts with LSD1. (a) RNF168 was immunoprecipitated using Flag or control antibody from nuclear extract prepared from 293T cells stably expressing Flag-HA-RNF168, with or without irradiation as indicated. Western blotting was then performed against Flag or endogenous LSD1. Arrow indicates the Flag-HA-RNF168, which is expressed at relatively low levels. (b) Myc-RNF168 and HA-LSD1 were coexpressed, and LSD1 was immunoprecipitated using an HA antibody, followed by Western blotting as indicated. (c) Immunprecipitation was performed as in panel a from nocodazole-arrested 293T cells mock infected or stably expressing Flag-HA-RNF8 or Flag-HA-RNF168 as indicated. (d) Tagged wild-type LSD1 or NΔ-171 LSD1 were stably expressed in U2OS cells, microirradiated, incubated for 15 min, and then stained for HA and pH2A.X. Bar, 20 µm. (e) Immunprecipitation was performed from 293T cells expressing the indicated vectors, followed by Western blotting as indicated.
Is the catalytic activity of RNF168 important for the recruitment of LSD1 to sites of DNA damage? We first asked whether LSD1 may associate with a catalytically inactive RNF168. In fact, immunoprecipitation of Flag-LSD1 effectively coimmunoprecipitated both wild type and a catalytically inactive form of RNF168 (Fig. S2 g). Expression of an shRNA-resistant, wild-type form of RNF168 rescued the ability of LSD1 to be recruited to laser-induced DNA damage sites after endogenous RNF168 knockdown (Fig. S3, a and b). Interestingly, a catalytically inactive form of RNF168 was also capable of rescuing LSD1 recruitment to sites of DNA damage (Fig. S3, a and b). These experiments strongly suggested that the catalytic activity of RNF168 is not required for the recruitment of LSD1 to DNA damage sites.
These experiments suggested that physical association between LSD1 and RNF168 may be important for the recruitment of LSD1 to damaged chromatin. We wished to identify the domain of LSD1 important for its recruitment to sites of DNA damage. We made several mutations in LSD1 in regions of the protein that are predicted to not affect its catalytic activity or its interaction with the CoREST complex in the hope of isolating a mutant that would selectively abrogate its recruitment to sites of DNA damage (see examples of mutants tested in Fig. S3 c). One mutant form of LSD1 that we constructed was an N-terminal deletion removing the first 171 amino acids of LSD1 (NΔ-LSD1). This deletion mutant retains catalytic activity and was previously used for cocrystallization with CoREST (Yang et al., 2006). Strikingly, in comparison to wild-type LSD1, NΔ-LSD1 was recruited to sites of DNA damage relatively poorly (Fig. 5 d). 15 min after laser microirradiation, 77% of cells had visible recruitment of wild-type LSD1, compared with only 35% of cells expressing NΔ-LSD1. This suggested a requirement of the N-terminal region for effective LSD1 recruitment to DNA damage sites. Interestingly, the same N-terminal region also appears to be required for physical interaction with RNF168, as NΔ-LSD1 displayed a significantly reduced interaction with RNF168 (Fig. 5 e). Importantly, this LSD1 mutant immunoprecipitated an equal amount of CoREST compared with wild-type LSD1 (Fig. 5 e), suggesting that the loss of interaction with RNF168 is most likely not because of improper folding of the truncated LSD1, which is consistent with a previous structural study (Yang et al., 2006). Collectively, these results strongly suggest that the recruitment of LSD1 to sites of DNA damage is mediated primarily by physical interaction with RNF168 and is independent of RNF168 activity.
Loss of LSD1 causes γ-irradiation (γ-IR) hypersensitivity and increased HR
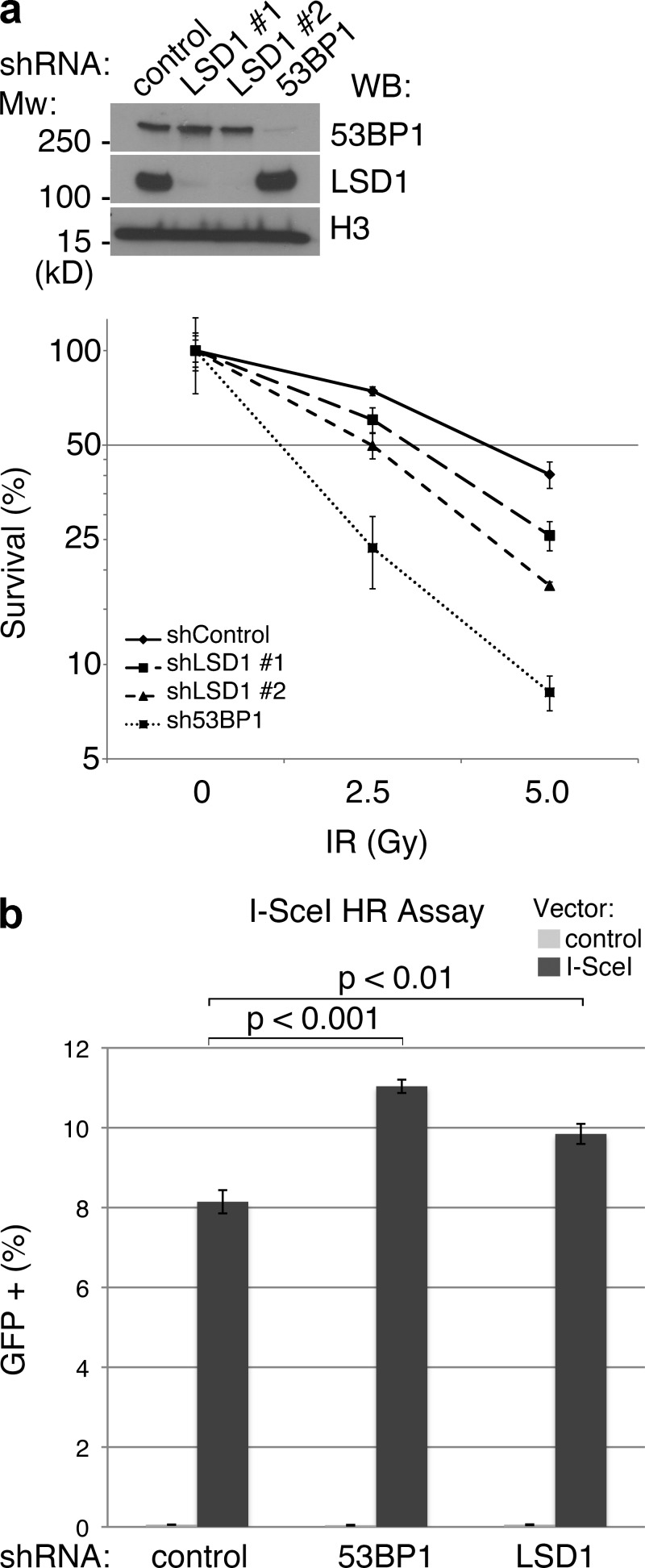
Cells lacking factors directly involved in the DDR, such as 53BP1 and MDC1, are hypersensitive to γ-IR (FitzGerald et al., 2009; Coster and Goldberg, 2010). Therefore, we tested whether this was the case with LSD1. LSD1 was knocked down using lentiviral-mediated shRNA, and knockdown of 53BP1 was also performed in parallel as a positive control (Fig. 6 a). These cells were then used in a colony formation assay, which showed that knockdown of LSD1 caused a moderate sensitivity to γ-IR (approximately two- to threefold at 5 Gy; Fig. 6 a). This degree of γ-IR hypersensitivity, although modest, is similar to what is observed for certain other chromatin-modifying factors that have recently been shown to play a role in the DDR, such as components of the NuRD–CHD4 chromatin remodeling complex (Chou et al., 2010; Larsen et al., 2010; Polo et al., 2010; Smeenk et al., 2010).
Figure 6.
LSD1 promotes the cellular response to DNA damage. (a) Lentiviral knockdown was performed in HeLa cells using the indicated shRNAs, followed by Western blotting as indicated (top). Colony formation assay was then performed in quadruplicate using the knockdown cells. Y axis represents colony survival normalized to control. (b) U2OS-DR-GFP reporter cells were treated with the indicated shRNAs, and frequency of HR was determined by flow cytometry after transfection with control or I-SceI vector. Error bars represent ± SD.
As suggested earlier, H3K4 methylation has been shown to mark recombination hotspots (Cheung et al., 2010). Whether or not this modification is causative in promoting HR is unknown. Our evidence suggested that LSD1 can demethylate H3K4 at sites of DNA damage, and if in fact H3K4 methylation promotes HR, LSD1 may act to inhibit HR. We therefore tested whether knockdown of LSD1 affects HR using the established I-SceI HR reporter assay (Weinstock et al., 2006). Indeed, we found that loss of LSD1 modestly increased HR in these cells (Fig. 6 b). As a positive control we showed that loss of 53BP1 also increased HR, consistent with previous findings (Xie et al., 2007; Bunting et al., 2010).
LSD1 promotes 53BP1 foci formation primarily in late S/G2 cells
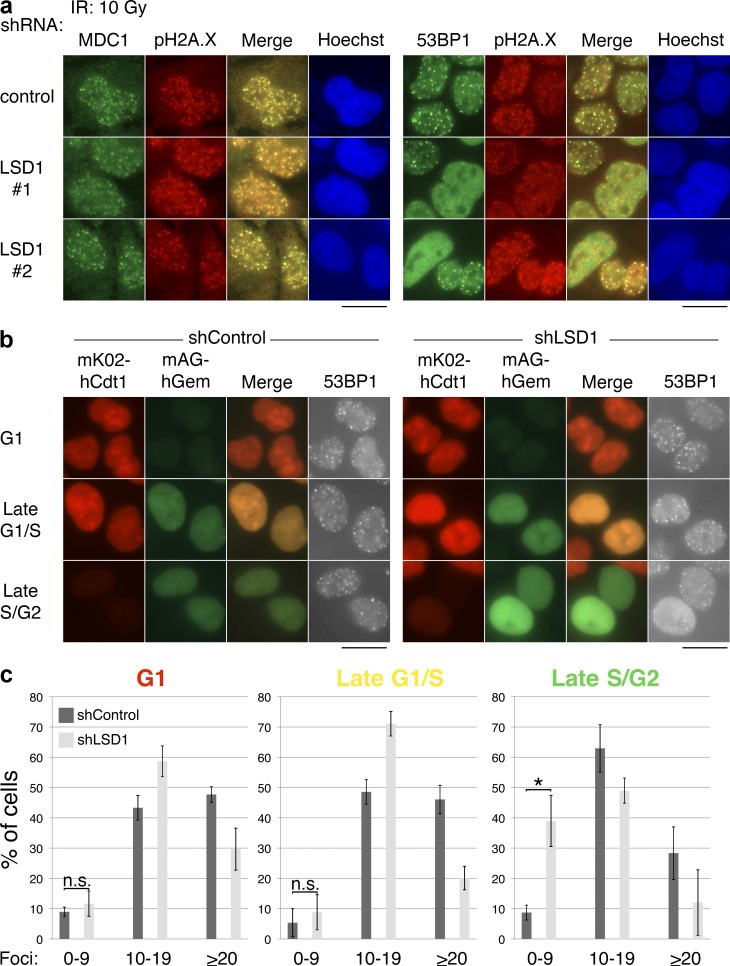
Numerous studies have suggested that RNF168 plays an important role in the recruitment of 53BP1 to sites of DNA damage (Doil et al., 2009; Stewart et al., 2009). Because LSD1 can interact with RNF168, is recruited to sites of damage in an RNF168-dependent manner, and regulates HR like 53BP1, we investigated whether LSD1 plays a role in the recruitment of 53BP1 to IRIF. We used lentiviral shRNA constructs to knock down LSD1 in U2OS cells (Fig. S1 a). These cells were then treated with γ-IR (10 Gy) and IRIF was assessed by immunofluorescence microscopy. Knockdown of LSD1 did not affect induction of pH2A.X or MDC1 foci in response to γ-IR (Fig. 7 a, left), suggesting that the initial signaling events related to the DDR are intact and not LSD1 dependent. In contrast, knockdown of LSD1 with three different shRNAs, but not two control shRNAs, had a noticeable effect on the formation of 53BP1 IRIF in a subset of cells (Fig. 7 a and not depicted). Some LSD1 knockdown cells had significantly more diffuse nucleoplasmic 53BP1 signal, while other cells lacked 53BP1 foci. These data suggest that LSD1 plays a role in the recruitment of 53BP1 to IRIF downstream of MDC1 and pH2A.X.
Figure 7.
LSD1 promotes 53BP1 IRIF formation in late S/G2 cells. (a) LSD1 affects 53BP1 foci formation in a subset of cells but does not affect pH2A.X or MDC1 foci formation. U2OS cells were infected with the indicated lentiviral shRNAs, exposed to 10 Gy IR, incubated at 37°C for 1 h, and processed for immunofluorescence using the indicated antibodies. (b) U2OS-FUCCI cells were infected with the indicated lentiviral shRNAs, exposed to 10 Gy IR, incubated at 37°C for 1 h, and processed for immunofluorescence using an antibody against 53BP1. Bars, 20 µm. (c) The experiment in panel b was performed in triplicate, and 53BP1 foci were quantified as shown. Error bars represent ± SD. *, P < 0.01; n.s., not significant.
Because of the cell cycle–specific nature of LSD1 expression and H3K4me2 demethylation during DNA damage, we wished to determine whether the 53BP1 foci defect was also cell cycle specific. We knocked down LSD1 in U2OS-FUCCI cells and analyzed 53BP1 foci formation after γ-IR. Loss of LSD1 affected 53BP1 foci formation primarily in cells that were in late S/G2 (Fig. 7, b and c). With LSD1 knockdown, ∼40% of cells in late S/G2 had <10 53BP1 foci, whereas <10% of control knockdown cells had the same phenotype (Fig. 7 c). A more modest reduction in foci (i.e., from >20 to the 10–19-foci/cell nucleus range) was observed in other phases of the cell cycle upon LSD1 knockdown. In contrast, knockdown of RNF8 affected 53BP1 foci formation in all stages of the cell cycle (Fig. S3, d and e). This cell cycle–specific phenotype associated with LSD1 was not likely to be caused by a major difference in the cell cycle distribution of cells upon LSD1 knockdown, as this was unchanged relative to control knockdown cells, consistent with previously published data (Scoumanne and Chen, 2007; unpublished data).
Is the catalytic activity of LSD1 important for its role in promoting 53BP1 foci formation in late S/G2? To address this issue, we stably expressed tagged, shRNA-resistant wild-type or catalytically inactive LSD1 (K661A; Lee et al., 2005) in U2OS-FUCCI cells, followed by knockdown of the endogenous LSD1 (Fig. S4 a). Importantly, the wild-type and catalytically inactive versions of LSD1 were both recruited to laser-induced DNA damage sites (Fig. S4 b). These cells were then treated with γ-IR, and 53BP1 foci formation was assessed in late S/G2 cells. Wild-type LSD1, but not the catalytically inactive form, improved the percentage of late S/G2 cells that had 53BP1 foci, although not to the same degree as control knockdown (Fig. S4, c and d). This may be because of the N-terminal tag of the exogenously expressed LSD1 or the presence of multiple LSD1 isoforms (Zibetti et al., 2010). Nevertheless, the difference between control rescue and wild-type LSD1 rescue was statistically significant. These results suggest that the catalytic activity of LSD1 promotes 53BP1 foci formation in late S/G2 cells.
LSD1 promotes H2A/H2A.X ubiquitylation downstream of RNF168 recruitment
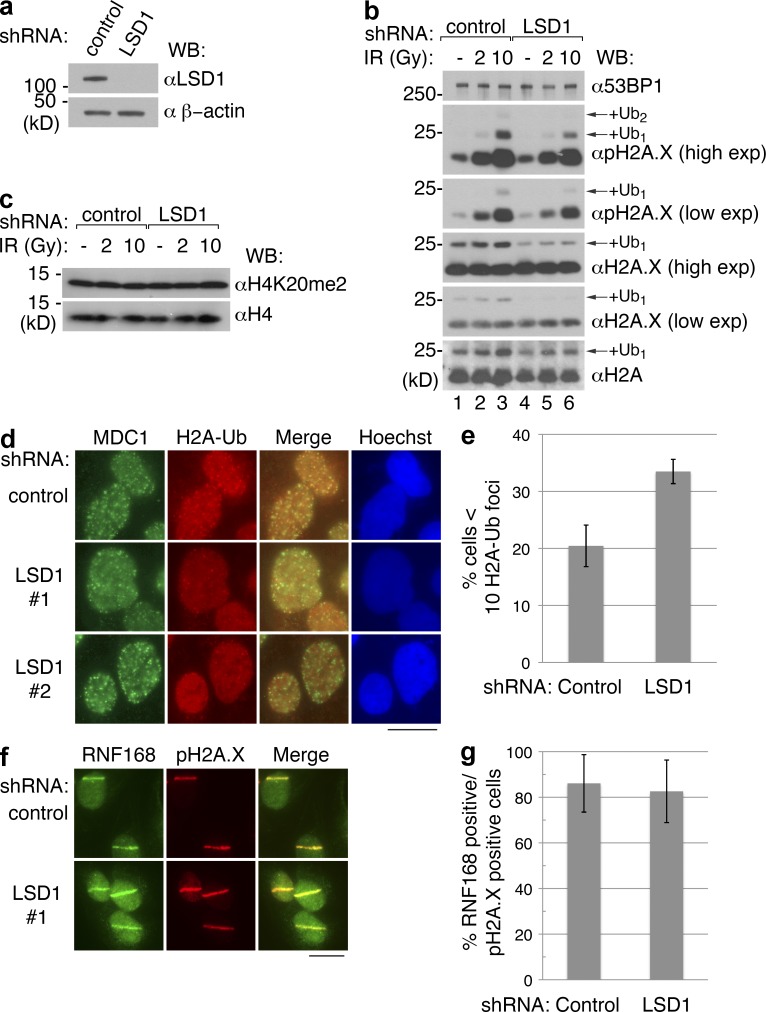
RNF168 is an E3 ligase that promotes H2A/H2A.X ubiquitylation, and this activity is important for the recruitment of 53BP1 to sites of damage (Doil et al., 2009; Stewart et al., 2009). Our data suggested that LSD1 is associated with RNF168 and promotes 53BP1 recruitment. Therefore, we next determined whether IR-induced H2A/H2A.X ubiquitylation is affected in the absence by LSD1 late in the cell cycle. Cells were synchronized in late G2 using sequential thymidine-nocodazole block after transduction with control or LSD1-specific shRNA, followed by γ-IR treatment (Fig. 8, a and b). LSD1 knockdown did not significantly alter IR-induced pH2A.X, consistent with the lack of observable difference in pH2A.X foci formation (Fig. 7 a). However, there was a reduced signal corresponding to ubiquitylated H2A.X and ubiquitylated pH2A.X, particularly upon γ-IR (Fig. 8 b). Global levels of H2A ubiquitylation appeared to be significantly affected only with γ-IR (Fig. 8 b, bottom, compare lanes 1 and 4), suggesting that LSD1 does not play a significant role in H2A ubiquitylation in the absence of DNA damage. 53BP1 recruitment may also be affected by dimethylation of H4K20, but we did not observe a difference in the global methylation status of H4K20me2 before or after γ-IR (Fig. 8 c).
Figure 8.
LSD1 promotes H2A/H2A.X ubiquitylation upon DNA damage. (a) 293T cells were infected with control or LSD1-specific shRNA, and then Western blotted with the indicated antibodies. (b and c) Cells from panel a were synchronized in late G2 using thymidine-nocodazole and were harvested 4 h after the indicated doses of γ-IR. Whole cell extracts were analyzed by Western blot using the indicated antibodies. (d) After knockdown and selection, U2OS cells were irradiated and processed for immunofluorescence using antibodies against MDC1 and ubiquitylated H2A. (e) Quantitation of panel d. (f) U2OS cells treated with control or LSD1-specific shRNA were laser microirradiated and stained with the indicated antibodies. (g) Quantitation of panel f. Bars, 20 µm.
To determine whether histone ubiquitylation was reduced at DNA damage foci, we analyzed IRIF formation using a monoclonal antibody specific for ubiquitylated H2A (H2A-Ub). Indeed, H2A-Ub foci were reduced in upon LSD1 knockdown (Fig. 8, d and e). Other DNA damage factors, particularly BRCA1 and Rap80, also depend on ubiquitylation at damage sites for their recruitment (Huen et al., 2010). Consistently, loss of LSD1 also affected the recruitment of these two factors to IRIFs, albeit modestly (Fig. S5, a and c). We also observed a modest reduction in ubiquitin conjugate formation at IRIFs upon LSD1 knockdown, which was similarly cell cycle dependent (Fig. S5 d).
Finally, we asked whether LSD1 affects histone ubiquitylation by promoting recruitment of RNF168 or RNF8. We did not observe a change in the localization of RNF168 to laser stripes upon LSD1 knockdown (Fig. 8, f and g). Furthermore, we did not observe a reduction in RNF8 IRIF formation upon LSD1 knockdown (Fig. S5 e). Collectively, our results suggest that LSD1 promotes histone ubiquitylation at sites of DNA damage downstream of E3 ligase recruitment.
Discussion
LSD1 was discovered in 2004 as the first histone demethylase that mediates H3K4me1/2 demethylation and promotes transcriptional repression (Shi et al., 2004). Since then, it has become clear from work done by various groups that LSD1 has a multitude of functions associated with transcription (Klose and Zhang, 2007; Cloos et al., 2008; Mosammaparast and Shi, 2010). However, besides the recently described noncatalytic role of JMJD2A (Mallette et al., 2012), there is little evidence in the literature that any histone demethylase plays a direct role in the DDR. Previous work had suggested that the Caenorhabditis elegans LSD1 homologue SPR-5 plays a role in meiotic DSB repair, but the precise molecular mechanism or its functional evolutionary conservation were not clear (Nottke et al., 2011). In this study, we provide numerous lines of evidence supporting the notion that human LSD1 plays a direct role in the DDR. First, endogenous LSD1 colocalizes with pH2A.X at sites of UV laser microirradiation, coincident with H3K4me2 demethylation (Figs. 1 and 2). We did not observe LSD1 at IR-induced foci, possibly because most of the cellular LSD1 has functions associated with transcription and therefore the background binding to chromatin does not allow a sufficient difference between the signal and the noise to allow LSD1 to be seen at these foci. Consistent with the laser microirradiation data, DSBs induced by I-PpoI endonuclease recruit LSD1 and reduce H3K4me2 (Figs. 1 and 2). Furthermore, H3K4me2 demethylation at sites of DNA damage is promoted by LSD1 (Fig. 2). Finally, LSD1 interacts with RNF168 and its recruitment to sites of DNA damage is RNF168 dependent (Figs. 3–5). Collectively, these results strongly suggest that LSD1 plays a direct role in the DDR pathway.
Our results suggest that LSD1 is recruited to sites of DNA damage in a manner dependent on RNF168 but independent of H2A.X and ATM signaling (Fig. 3). This recruitment is also independent of RNF168 catalytic activity (Fig. S3), reminiscent of the mechanism by which NuRD–CHD4 is recruited to sites of DNA damage, which is also H2A.X independent and relies at least partially on physical association with RNF8 (Luijsterburg et al., 2012). Thus it appears that these E3 ligases, like 53BP1, may be recruited transiently to sites of DNA damage in a manner independent of H2A.X, but their retention therein requires H2A.X. Further work is necessary to determine the mechanism behind the rapid transient recruitment of RNF168 to DNA damage sites.
Previous work had suggested that LSD1 plays at least an indirect role in the DDR by demethylating p53, thereby inhibiting the activation function of p53 (Huang et al., 2007). However, other work suggested that p53-independent mechanisms play a role in the altered cell cycle response to DNA damaging agents in the absence of LSD1, although transcriptional mechanisms could not be ruled out (Scoumanne and Chen, 2007). The work described here may help explain at least some of the p53-independent functions of LSD1 in the DDR. As suggested earlier, recent studies have identified H3K4 methylation as a mark of meiotic DSB formation and crossovers in yeast and mice, respectively (Borde et al., 2009; Cheung et al., 2010). Furthermore, the H3K4 methyltransferase PRDM9 has been suggested to regulate the methylation status at these sites to promote meiotic recombination (Cheung et al., 2010). Our finding that loss of LSD1 causes an increase in HR is consistent with these data. Thus, the status of H3K4 methylation may serve as a regulator in determining whether HR is deployed for DNA repair.
Our work suggests that LSD1 participates in the DDR by promoting H2A/H2A.X ubiquitylation events at sites of damage during late S/G2. This suggests a possible cross talk between H3K4 demethylation and H2A/H2A.X ubiquitylation. What could be the mechanism behind this potential cross talk? Recent evidence has suggested that two distinct histone ubiquitylation events occur at sites of DNA damage. These include H2A K119 ubiquitylation, which is mediated by polycomb repressive complex 1 (PRC1; Ismail et al., 2010; Ginjala et al., 2011), as well as H2A K13/15 ubiquitylation, which is mediated by RNF168 (Gatti et al., 2012; Mattiroli et al., 2012). Very recent work has demonstrated that the latter modification acts in concert with methylation at H4K20 to promote recruitment of 53BP1 (Fradet-Turcotte et al., 2013). Our data, which demonstrates that LSD1 interacts with RNF168, and that 53BP1 recruitment is reduced in the absence of LSD1 in late S/G2, suggests that RNF168 ubiquitylation events are affected by LSD1. However, our antibody-based approach in the analysis of H2A ubiquitylation events in vivo is limited, as it does not discriminate between H2A-K15 and H2A-K119 ubiquitylation events. Given that PRC1-mediated ubiquitylation of H2A-K119 has been suggested to affect 53BP1 recruitment (Ismail et al., 2010), it is formally possible that LSD1 may affect ubiquitylation events through mechanisms that are both RNF168 dependent and independent (i.e., through PRC1). Lastly, we note that LSD1 appears to affect other ubiquitylation events at sites of DNA damage as well because BRCA1/Rap80 and ubiquitin chain (FK2) formation are also reduced in the absence of LSD1 (Fig. S5). Recent work has also suggested that removal of ubiquitylated proteins by p97/VCP (such as L3MBTL1) promotes 53BP1 recruitment to sites of DNA damage (Acs et al., 2011; Meerang et al., 2011). It is formally possible that LSD1 functions to promote 53BP1 recruitment via such an alternative mechanism. However, the reduction of H2A/H2A.X ubiquitylation, as well as reduction of other damage foci proteins that depend on these ubiquitylation events, make these alternative mechanisms appear less likely.
It should be noted that PRC1-mediated H2A K119 ubiquitylation has been known for some time to be a mark of gene silencing (Wang et al., 2004). This modification, and indeed the presence and spreading of a transcriptionally silent region of chromatin, occurs near sites of DSBs in mammalian cells and is at least partially dependent on RNF168 (Shanbhag et al., 2010). Formation of silent chromatin marks, such as methylation of H3K9, and recruitment of the NuRD–CHD4 complex are two other examples of a transient, repressive chromatin state that appear to be critical for the DDR (Chou et al., 2010; Larsen et al., 2010; Polo et al., 2010; Smeenk et al., 2010; Chiolo et al., 2011). Indeed, several other groups have observed that loss of the NuRD–CHD4 also reduces H2A/H2A.X ubiquitylation during DNA damage (Larsen et al., 2010; Smeenk et al., 2010). It is likely that one function of LSD1-mediated demethylation in this pathway is to facilitate such a repressive chromatin environment near DSBs, which may repress transcription near these sites. Further work is necessary to elucidate the molecular mechanisms of cross talk between the formation of transcriptionally repressive chromatin and the recruitment of chromatin-associated factors, such as 53BP1 and BRCA1, which are thought to be critical in regulating the DDR.
Materials and methods
Plasmids
LSD1 cDNA was cloned into pMSCV-Flag-HA for retroviral expression or pHAGE-CMV-Flag-HA for lentiviral expression. RNF168 cDNA (provided by R. Greenberg, University of Pennsylvania, Philadelphia, PA) was cloned into pHAGE-CMV-Flag-HA, pHAGE-CMV-GFP, and pHAGE-CMV-myc. RNF8 cDNA (provided by S. Elledge, Harvard Medical School, Boston, MA) was cloned into pHAGE-CMV-Flag-HA. The retroviral vector pBABE-HA-ER-I-PpoI was provided by M. Kastan (Duke University, Durham, NC; Berkovich et al., 2007). Lentiviral shRNA constructs for LSD1, 53BP1, RNF168, and RNF8 were obtained from Thermo Fisher Scientific (clones used were as follows: shLSD1: TRCN0000046072 [#1] and TRCN0000046068 [#2]; sh53BP1: TRCN0000018865; shRNF168: TRCN0000034134; shRNF8: TRCN0000003441). Mutations in LSD1 and RNF168 were produced by site-directed mutagenesis and confirmed by sequencing.
Cell culture and viral transduction
HeLa, 293T, MEFs, and U2OS cells were maintained as previously described (Mulligan et al., 2008). The U2OS-FUCCI cells were made by lentiviral cotransduction of mKO2-hCdt1 (30/12) and mAG-hGem (1/110) into U2OS cells and were provided by S. Elledge. The U2OS-DR-GFP cells were originally constructed by integration of the DR-GFP reporter construct into the HPRT gene on the X chromosome and have been previously described (Weinstock et al., 2006). The LSD1−/− MEFs (provided by M.G. Rosenfeld, University of California, San Diego, La Jolla, CA) were originally made by targeted deletion of exon 6 of LSD1, resulting in an alteration of the reading frame and loss of detectable LSD1 (Wang et al., 2007). The H2A.X−/− MEFs (provided by A. Nussenzweig, National Cancer Institute, Bethesda, MD) were originally made by inserting the Neo cassette into the H2A.X locus (Celeste et al., 2003). Preparation of viruses and cell transduction were performed as previously described (Mulligan et al., 2008). In brief, lentiviruses were produced by cotransfection of the lentiviral plasmid with helper vectors (pHDM-VSV-G, pHDM-tat1b, pHDM-HgPM2, and pRC-CMV-RaII) into 293T cells, and viral supernatants were collected after 60–72 h. Cells infected with lentiviral shRNAs were selected after transduction with 1 µg/ml puromycin. The ATM inhibitor was provided by B. Sleckman (Washington University in St. Louis, St. Louis, MO) and the ATR inhibitor was provided by O. Fernandez-Capetillo (Spanish National Cancer Research Center, Madrid, Spain).
Laser microirradiation and immunofluorescent microscopy
For laser microirradiation, U2OS cells were grown on LabTek II chamber slides (Thermo Fisher Scientific) in the presence of 10 µM BrdU for 24–48 h before induction of DNA damage by a UV-A laser using an inverted microscope (Observer.Z1; Carl Zeiss) with a Palm microbeam laser microdissection workstation or with an inverted microscope (IX-81; Olympus) with an MMI CellCut plus microdissection workstation (Chou et al., 2010). After incubation at 37°C (typically 10 min for LSD1 and histone modifications, unless otherwise indicated), cells were washed once with cold PBS, extracted with CSK buffer plus 0.5% Triton X-100 for 2–5 min, washed again with cold PBS, and fixed with PBS containing 3.2% paraformaldehyde for 20 min. CSK/Triton buffer extraction step was omitted when using U2OS-FUCCI cells. For IRIF analysis, U2OS cells were infected with the indicated retroviral or lentiviral vectors and plated onto coverslips. The cells were then irradiated (10 Gy or as indicated) and incubated at 37°C for 1 h. After washing with PBS, cells were fixed with PBS containing 3.2% paraformaldehyde for 20 min, and then washed extensively with IF wash buffer (PBS containing 0.5% NP-40 and 0.02% NaN3). The fixed cells were then incubated with blocking buffer (IF wash buffer with 10% FBS) and stained with the indicated antibodies diluted in blocking buffer at room temperature for 1–2 h or at 4°C overnight. After staining with secondary antibodies (conjugated with Alexa Fluor 350, 488, or 594 or Pacific Blue; EMD Millipore) and Hoechst 33342 (Sigma-Aldrich), where indicated, samples were mounted using Prolong Gold mounting medium (Invitrogen). Confocal imaging was performed on a FluoView FV1000 laser scanning confocal system (Olympus) connected to an inverted microscope (IX-81) equipped with PLAPON 60×O/NA 1.42 objective. FV10-ASW version 2.1c software was used to acquire images and to quantify H3K4me2 antistripes. Epifluorescent microscopy was performed on a fluorescent microscope (Eclipse E600; Nikon) using a 60×/1.4 Plan-Apo oil immersion lens and ProgRes CapturePro 2.7.7 software or a microscope (BX-53; Olympus) using an ApoN 60×/1.49 NA oil immersion lens and cellSens Dimension software. Raw images were exported into Adobe Photoshop, and for any adjustments in image contrast or brightness, the levels function was applied in Photoshop using identical settings for all images of a given experiment. For quantification of laser stripes, ∼30–100 microirradiated cells were analyzed. For foci quantitation in U2OS-FUCCI cells, images of 150–300 total cells were analyzed in triplicate or as otherwise indicated.
I-PpoI ChIP
ChIP studies on DSBs were performed using the previously described method (Berkovich et al., 2008) except that the ChIP-IT Express kit (Active Motif) was used for the ChIP procedure. The antibodies used for ChIP are described in Table S2. Primers used for amplification of ChIP products are as follows: rRNA forward, 5′-TGGATCAGAAGGGCAAAAGC-3′; rRNA reverse, 5-’TAGGAAGAGCCGACATCGAAGG-3′; GAPDH forward, 5′-TCGGTTCTTGCCTCTTGTC-3′; GAPDH reverse, 5′-CTTCCATTCTGTCTTCCACTC-3′. For real-time analysis, the material from one ChIP was amplified and quantified using SYBR green in triplicate and expressed as fold change ± standard error.
Purification of RNF168 complex, MS/MS identification, and immunoprecipitation
The complex purification was performed as previously described (Nakatani and Ogryzko, 2003; Mulligan et al., 2008), with the following modifications. Lentiviral Flag-HA–tagged RNF168 was stably expressed in 293T cells. Nuclei were prepared by incubation of cells in buffer A (10 mM Hepes, pH 7.6, 3 mM MgCl2, 10 mM KCl, 0.5% NP-40, and 5% glycerol) with phosphatase and protease inhibitors (Roche) for 10 min. After centrifugation, nuclear pellets were resuspended in buffer A containing 300 mM KCl. The nuclear extract was then diluted 1:1 with buffer A, incubated with anti-Flag (M2) beads (Sigma-Aldrich), washed extensively with buffer A, and eluted with Flag peptide (Sigma-Aldrich). The complex components were digested with trypsin, vacuum centrifuged to dryness, and resuspended in sample loading buffer (5% formic acid/5% acetonitrile). Samples were analyzed by HPLC (Eksigent NanoLC; AB Sciex) into PicoTips (New Objective) coupled to a LTQ ion trap mass spectrometer (Thermo Fisher Scientific) operated in positive ion mode. After a survey scan the six most abundant precursors were selected for fragmentation in CID mode. For identification, raw files were processed to mgf files using a user-written script, and ProteinPilot Software (version 4.5; AB Sciex) was used to search the data against a UniProt target decoy human database appended with common contaminants (using the common Repository of Adventitious Proteins). A 1% false discovery rate for protein identifications was determined using the Posterior Error Probability algorithm integrated into ProteinPilot. Immunoprecipitation of Flag-tagged proteins in 293T cells was performed by lysis of cells in 50 mM Tris, pH 7.9, 150 mM NaCl, 1% Triton X-100, 5% glycerol, and protease and phosphatase inhibitor cocktails (Thermo Fisher Scientific). The lysate was cleared by centrifugation and incubated overnight at 4°C with M2-agarose beads. After extensive washing in the same buffer, bound material was eluted using Laemmli sample buffer and analyzed by Western blotting. Endogenous immunoprecipitation of RNF168 was performed using HeLa nuclear extracts essentially as previously described (Dango et al., 2011). In brief, nuclear extract (∼2 mg) was precleared with PrA-Sepharose (Thermo Fisher Scientific), and then incubated overnight at 4°C in the presence of 5 µg of antibody and 2.5 mg of BSA. The extract was then incubated with PrA-Sepharase for 1 h at 4°C, centrifuged, and washed extensively in TAP buffer (50 mM Tris, pH 7.9, 100 mM KCl, 5 mM MgCl2, 0.2 mM EDTA, 0.1% NP-40, and 10% glycerol), and bound material was eluted using Laemmli sample buffer and analyzed by Western blotting.
Colony formation assay and I-SceI recombination assay
For colony formation assays, HeLa cells were infected with control shRNA or shRNA targeting LSD1 or 53BP1, selected with puromycin, plated at low density, and irradiated with the indicated doses of IR. The cells were incubated for 12–14 d, fixed, and stained with crystal violet. The experiment was performed in quadruplicate for each shRNA and IR dose. The I-SceI recombination assay was performed using the U2OS-DR-GFP cells as previously described (Weinstock et al., 2006). In brief, the U2OS-DR-GFP cells were infected with the indicated lentiviral shRNAs. After selection, the cells were transfected with the pCBA-Sce plasmid (Weinstock et al., 2006) using Lipofectamine 2000 (Invitrogen). After incubation for 72 h, the cells were analyzed by FACS.
Antibodies
Antibodies used in this study are listed in Table S2.
Online supplemental material
Fig. S1 describes additional experiments demonstrating the recruitment of LSD1 to sites of DNA damage. Figs. S2 and S3 refer to experiments regarding the mechanism of LSD1 recruitment to sites of damage and its interaction with RNF168. Fig. S4 describes the role of the catalytic activity of LSD1 in 53BP1 recruitment. Fig. S5 relates to the mechanism of LSD1 in the DDR. Table S1 contains the proteomic data for the Flag-purified RNF168 complex. Table S2 contains information regarding the antibodies used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201302092/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201302092.dv.
Supplementary Material
Acknowledgments
We wish to thank Barry Sleckman and members of the Shi laboratory for helpful discussions. In particular, we wish to thank Hank Qi for help with complex purification and Shigeki Iwase for help with immunofluorescence microscopy. We thank Steve Elledge for the RNF8 cDNA, the U2OS-FUCCI cells, and assistance with the laser microirradiation experiments, as well as Daniel Durocher for the anti-RNF168 antibody and Roger Greenberg for the RNF168 cDNA. We thank Andre Nussenzweig for the H2A.X−/− MEFs, M. Geoff Rosenfeld for the LSD1−/− MEFs, Barry Sleckman for the ATM inhibitor, and Oscar Fernandez-Capetillo for the ATR inhibitor. We wish to thank the Harvard Medical School Department of Pathology flow cytometry and confocal microscopy core facilties.
N. Mosammaparast was supported by a Ruth L. Kirschstein Institutional National Research Service Award (T32-HL007627) and a Clinical Scientist Research Career Development Award (K08 CA158133). This work was supported by grants from the National Institutes of Health to Y. Shi (CA118487) and to J.W. Harper (AG011085 and GM054137). Y. Shi is an American Cancer Society Research Professor.
Y. Shi is a cofounder of Constellation Pharmaceuticals, Inc., and is a member of its scientific advisory board.
Footnotes
Abbreviations used in this paper:
- ATM
- ataxia telangiectasia mutated
- ATR
- ataxia telangiectasia and Rad3 related
- ChIP
- chromatin immunoprecipitation
- DDR
- DNA damage response
- DSB
- double-stranded DNA break
- FUCCI
- fluorescence ubiquitylation cell cycle indicator
- γ-IR
- γ-irradiation
- HR
- homologous recombination
- IRIF
- irradiation-induced foci
- MEF
- mouse embryonic fibroblast
- PRC1
- polycomb repressive complex 1
- rDNA
- ribosomal DNA
References
- Acs K., Luijsterburg M.S., Ackermann L., Salomons F.A., Hoppe T., Dantuma N.P. 2011. The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat. Struct. Mol. Biol. 18:1345–1350 10.1038/nsmb.2188 [DOI] [PubMed] [Google Scholar]
- Bedford M.T., Clarke S.G. 2009. Protein arginine methylation in mammals: who, what, and why. Mol. Cell. 33:1–13 10.1016/j.molcel.2008.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovich E., Monnat R.J., Jr, Kastan M.B. 2007. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat. Cell Biol. 9:683–690 10.1038/ncb1599 [DOI] [PubMed] [Google Scholar]
- Berkovich E., Monnat R.J., Jr, Kastan M.B. 2008. Assessment of protein dynamics and DNA repair following generation of DNA double-strand breaks at defined genomic sites. Nat. Protoc. 3:915–922 10.1038/nprot.2008.54 [DOI] [PubMed] [Google Scholar]
- Bohgaki T., Bohgaki M., Cardoso R., Panier S., Zeegers D., Li L., Stewart G.S., Sanchez O., Hande M.P., Durocher D., et al. 2011. Genomic instability, defective spermatogenesis, immunodeficiency, and cancer in a mouse model of the RIDDLE syndrome. PLoS Genet. 7:e1001381 10.1371/journal.pgen.1001381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde V., Robine N., Lin W., Bonfils S., Géli V., Nicolas A. 2009. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 28:99–111 10.1038/emboj.2008.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting S.F., Callén E., Wong N., Chen H.T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L., et al. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 141:243–254 10.1016/j.cell.2010.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A., Fernandez-Capetillo O., Kruhlak M.J., Pilch D.R., Staudt D.W., Lee A., Bonner R.F., Bonner W.M., Nussenzweig A. 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5:675–679 10.1038/ncb1004 [DOI] [PubMed] [Google Scholar]
- Cheung V.G., Sherman S.L., Feingold E. 2010. Genetics. Genetic control of hotspots. Science. 327:791–792 10.1126/science.1187155 [DOI] [PubMed] [Google Scholar]
- Chiolo I., Minoda A., Colmenares S.U., Polyzos A., Costes S.V., Karpen G.H. 2011. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 144:732–744 10.1016/j.cell.2011.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou D.M., Adamson B., Dephoure N.E., Tan X., Nottke A.C., Hurov K.E., Gygi S.P., Colaiácovo M.P., Elledge S.J. 2010. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. USA. 107:18475–18480 10.1073/pnas.1012946107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A., Elledge S.J. 2010. The DNA damage response: making it safe to play with knives. Mol. Cell. 40:179–204 10.1016/j.molcel.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloos P.A., Christensen J., Agger K., Helin K. 2008. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 22:1115–1140 10.1101/gad.1652908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coster G., Goldberg M. 2010. The cellular response to DNA damage: a focus on MDC1 and its interacting proteins. Nucleus. 1:166–178 10.4161/nucl.1.2.11176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dango S., Mosammaparast N., Sowa M.E., Xiong L.J., Wu F., Park K., Rubin M., Gygi S., Harper J.W., Shi Y. 2011. DNA unwinding by ASCC3 helicase is coupled to ALKBH3-dependent DNA alkylation repair and cancer cell proliferation. Mol. Cell. 44:373–384 10.1016/j.molcel.2011.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J.A., Nussenzweig A. 2012. Roles for histone H3K4 methyltransferase activities during immunoglobulin class-switch recombination. Biochim. Biophys. Acta. 1819:733–738 10.1016/j.bbagrm.2012.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D.H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J., et al. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 136:435–446 10.1016/j.cell.2008.12.041 [DOI] [PubMed] [Google Scholar]
- FitzGerald J.E., Grenon M., Lowndes N.F. 2009. 53BP1: function and mechanisms of focal recruitment. Biochem. Soc. Trans. 37:897–904 10.1042/BST0370897 [DOI] [PubMed] [Google Scholar]
- Fradet-Turcotte A., Canny M.D., Escribano-Díaz C., Orthwein A., Leung C.C., Huang H., Landry M.C., Kitevski-LeBlanc J., Noordermeer S.M., Sicheri F., Durocher D. 2013. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 499:50–54 10.1038/nature12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bassets I., Kwon Y.S., Telese F., Prefontaine G.G., Hutt K.R., Cheng C.S., Ju B.G., Ohgi K.A., Wang J., Escoubet-Lozach L., et al. 2007. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 128:505–518 10.1016/j.cell.2006.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti M., Pinato S., Maspero E., Soffientini P., Polo S., Penengo L. 2012. A novel ubiquitin mark at the N-terminal tail of histone H2As targeted by RNF168 ubiquitin ligase. Cell Cycle. 11:2538–2544 10.4161/cc.20919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginjala V., Nacerddine K., Kulkarni A., Oza J., Hill S.J., Yao M., Citterio E., van Lohuizen M., Ganesan S. 2011. BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Mol. Cell. Biol. 31:1972–1982 10.1128/MCB.00981-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi M.A., Bochar D.A., Chenoweth J., Lane W.S., Mandel G., Shiekhattar R. 2002. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc. Natl. Acad. Sci. USA. 99:7420–7425 10.1073/pnas.112008599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi M.A., Dong Y., Lane W.S., Speicher D.W., Shiekhattar R. 2003. A candidate X-linked mental retardation gene is a component of a new family of histone deacetylase-containing complexes. J. Biol. Chem. 278:7234–7239 10.1074/jbc.M208992200 [DOI] [PubMed] [Google Scholar]
- Huang J., Sengupta R., Espejo A.B., Lee M.G., Dorsey J.A., Richter M., Opravil S., Shiekhattar R., Bedford M.T., Jenuwein T., Berger S.L. 2007. p53 is regulated by the lysine demethylase LSD1. Nature. 449:105–108 10.1038/nature06092 [DOI] [PubMed] [Google Scholar]
- Huen M.S., Grant R., Manke I., Minn K., Yu X., Yaffe M.B., Chen J. 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 131:901–914 10.1016/j.cell.2007.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen M.S., Sy S.M., Chen J. 2010. BRCA1 and its toolbox for the maintenance of genome integrity. Nat. Rev. Mol. Cell Biol. 11:138–148 10.1038/nrm2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail I.H., Andrin C., McDonald D., Hendzel M.J. 2010. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J. Cell Biol. 191:45–60 10.1083/jcb.201003034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S.P., Bartek J. 2009. The DNA-damage response in human biology and disease. Nature. 461:1071–1078 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose R.J., Zhang Y. 2007. Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 8:307–318 10.1038/nrm2143 [DOI] [PubMed] [Google Scholar]
- Kolas N.K., Chapman J.R., Nakada S., Ylanko J., Chahwan R., Sweeney F.D., Panier S., Mendez M., Wildenhain J., Thomson T.M., et al. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 318:1637–1640 10.1126/science.1150034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen D.H., Poinsignon C., Gudjonsson T., Dinant C., Payne M.R., Hari F.J., Rendtlew Danielsen J.M., Menard P., Sand J.C., Stucki M., et al. 2010. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J. Cell Biol. 190:731–740 10.1083/jcb.200912135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.G., Wynder C., Cooch N., Shiekhattar R. 2005. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 437:432–435 [DOI] [PubMed] [Google Scholar]
- Luijsterburg M.S., Acs K., Ackermann L., Wiegant W.W., Bekker-Jensen S., Larsen D.H., Khanna K.K., van Attikum H., Mailand N., Dantuma N.P. 2012. A new non-catalytic role for ubiquitin ligase RNF8 in unfolding higher-order chromatin structure. EMBO J. 31:2511–2527 10.1038/emboj.2012.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv S., Bu W., Jiao H., Liu B., Zhu L., Zhao H., Liao J., Li J., Xu X. 2010. LSD1 is required for chromosome segregation during mitosis. Eur. J. Cell Biol. 89:557–563 10.1016/j.ejcb.2010.01.004 [DOI] [PubMed] [Google Scholar]
- Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 131:887–900 10.1016/j.cell.2007.09.040 [DOI] [PubMed] [Google Scholar]
- Mallette F.A., Mattiroli F., Cui G., Young L.C., Hendzel M.J., Mer G., Sixma T.K., Richard S. 2012. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 31:1865–1878 10.1038/emboj.2012.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiroli F., Vissers J.H., van Dijk W.J., Ikpa P., Citterio E., Vermeulen W., Marteijn J.A., Sixma T.K. 2012. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell. 150:1182–1195 10.1016/j.cell.2012.08.005 [DOI] [PubMed] [Google Scholar]
- Meerang M., Ritz D., Paliwal S., Garajova Z., Bosshard M., Mailand N., Janscak P., Hübscher U., Meyer H., Ramadan K. 2011. The ubiquitin-selective segregase VCP/p97 orchestrates the response to DNA double-strand breaks. Nat. Cell Biol. 13:1376–1382 10.1038/ncb2367 [DOI] [PubMed] [Google Scholar]
- Metzger E., Wissmann M., Yin N., Müller J.M., Schneider R., Peters A.H., Günther T., Buettner R., Schüle R. 2005. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 437:436–439 [DOI] [PubMed] [Google Scholar]
- Miller K.M., Tjeertes J.V., Coates J., Legube G., Polo S.E., Britton S., Jackson S.P. 2010. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 17:1144–1151 10.1038/nsmb.1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast N., Shi Y. 2010. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 79:155–179 10.1146/annurev.biochem.78.070907.103946 [DOI] [PubMed] [Google Scholar]
- Mulligan P., Westbrook T.F., Ottinger M., Pavlova N., Chang B., Macia E., Shi Y.J., Barretina J., Liu J., Howley P.M., et al. 2008. CDYL bridges REST and histone methyltransferases for gene repression and suppression of cellular transformation. Mol. Cell. 32:718–726 10.1016/j.molcel.2008.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murr R., Loizou J.I., Yang Y.G., Cuenin C., Li H., Wang Z.Q., Herceg Z. 2006. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat. Cell Biol. 8:91–99 10.1038/ncb1343 [DOI] [PubMed] [Google Scholar]
- Nakatani Y., Ogryzko V. 2003. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 370:430–444 10.1016/S0076-6879(03)70037-8 [DOI] [PubMed] [Google Scholar]
- Nottke A.C., Beese-Sims S.E., Pantalena L.F., Reinke V., Shi Y., Colaiácovo M.P. 2011. SPR-5 is a histone H3K4 demethylase with a role in meiotic double-strand break repair. Proc. Natl. Acad. Sci. USA. 108:12805–12810 10.1073/pnas.1102298108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo S.E., Kaidi A., Baskcomb L., Galanty Y., Jackson S.P. 2010. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 29:3130–3139 10.1038/emboj.2010.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A., Kurokawa H., Morimura T., Hanyu A., Hama H., Osawa H., Kashiwagi S., Fukami K., Miyata T., Miyoshi H., et al. 2008. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 132:487–498 10.1016/j.cell.2007.12.033 [DOI] [PubMed] [Google Scholar]
- Scoumanne A., Chen X. 2007. The lysine-specific demethylase 1 is required for cell proliferation in both p53-dependent and -independent manners. J. Biol. Chem. 282:15471–15475 10.1074/jbc.M701023200 [DOI] [PubMed] [Google Scholar]
- Shanbhag N.M., Rafalska-Metcalf I.U., Balane-Bolivar C., Janicki S.M., Greenberg R.A. 2010. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 141:970–981 10.1016/j.cell.2010.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A., Shi Y. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 119:941–953 10.1016/j.cell.2004.12.012 [DOI] [PubMed] [Google Scholar]
- Shi Y.J., Matson C., Lan F., Iwase S., Baba T., Shi Y. 2005. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell. 19:857–864 10.1016/j.molcel.2005.08.027 [DOI] [PubMed] [Google Scholar]
- Smeenk G., Wiegant W.W., Vrolijk H., Solari A.P., Pastink A., van Attikum H. 2010. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J. Cell Biol. 190:741–749 10.1083/jcb.201001048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommermeyer V., Béneut C., Chaplais E., Serrentino M.E., Borde V. 2013. Spp1, a member of the Set1 complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Mol. Cell. 49:43–54 [DOI] [PubMed] [Google Scholar]
- Stewart G.S., Wang B., Bignell C.R., Taylor A.M., Elledge S.J. 2003. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 421:961–966 10.1038/nature01446 [DOI] [PubMed] [Google Scholar]
- Stewart G.S., Panier S., Townsend K., Al-Hakim A.K., Kolas N.K., Miller E.S., Nakada S., Ylanko J., Olivarius S., Mendez M., et al. 2009. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 136:420–434 10.1016/j.cell.2008.12.042 [DOI] [PubMed] [Google Scholar]
- Strahl B.D., Allis C.D. 2000. The language of covalent histone modifications. Nature. 403:41–45 10.1038/47412 [DOI] [PubMed] [Google Scholar]
- Stucki M., Clapperton J.A., Mohammad D., Yaffe M.B., Smerdon S.J., Jackson S.P. 2005. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 123:1213–1226 10.1016/j.cell.2005.09.038 [DOI] [PubMed] [Google Scholar]
- Toledo L.I., Murga M., Zur R., Soria R., Rodriguez A., Martinez S., Oyarzabal J., Pastor J., Bischoff J.R., Fernandez-Capetillo O. 2011. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat. Struct. Mol. Biol. 18:721–727 10.1038/nsmb.2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H., Gasser S.M. 2009. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 19:207–217 10.1016/j.tcb.2009.03.001 [DOI] [PubMed] [Google Scholar]
- Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R.S., Zhang Y. 2004. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 431:873–878 10.1038/nature02985 [DOI] [PubMed] [Google Scholar]
- Wang J., Scully K., Zhu X., Cai L., Zhang J., Prefontaine G.G., Krones A., Ohgi K.A., Zhu P., Garcia-Bassets I., et al. 2007. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 446:882–887 10.1038/nature05671 [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang H., Chen Y., Sun Y., Yang F., Yu W., Liang J., Sun L., Yang X., Shi L., et al. 2009. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 138:660–672 10.1016/j.cell.2009.05.050 [DOI] [PubMed] [Google Scholar]
- Ward I.M., Difilippantonio S., Minn K., Mueller M.D., Molina J.R., Yu X., Frisk C.S., Ried T., Nussenzweig A., Chen J. 2005. 53BP1 cooperates with p53 and functions as a haploinsufficient tumor suppressor in mice. Mol. Cell. Biol. 25:10079–10086 10.1128/MCB.25.22.10079-10086.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock D.M., Nakanishi K., Helgadottir H.R., Jasin M. 2006. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol. 409:524–540 10.1016/S0076-6879(05)09031-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie A., Hartlerode A., Stucki M., Odate S., Puget N., Kwok A., Nagaraju G., Yan C., Alt F.W., Chen J., et al. 2007. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol. Cell. 28:1045–1057 10.1016/j.molcel.2007.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Gocke C.B., Luo X., Borek D., Tomchick D.R., Machius M., Otwinowski Z., Yu H. 2006. Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol. Cell. 23:377–387 10.1016/j.molcel.2006.07.012 [DOI] [PubMed] [Google Scholar]
- You A., Tong J.K., Grozinger C.M., Schreiber S.L. 2001. CoREST is an integral component of the CoREST- human histone deacetylase complex. Proc. Natl. Acad. Sci. USA. 98:1454–1458 10.1073/pnas.98.4.1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zibetti C., Adamo A., Binda C., Forneris F., Toffolo E., Verpilli C., Ginellli E., Mattevi A., Sala C., Battaglioli E. 2010. Alternative splicing of the histone demethylase LSD1/KDM1 contributes to the modulation of neurite morphogenesis in the mammalian nervous system. J. Neurosci. 30:2521–2532 10.1523/JNEUROSCI.5500-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y., Bielopolski D., Galanty Y., Lukas C., Taya Y., Schultz D.C., Lukas J., Bekker-Jensen S., Bartek J., Shiloh Y. 2006. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol. 8:870–876 10.1038/ncb1446 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.