Analysis of Odf2 deletion mutants reveals regions important for the formation of basal body transition fibers and centriole distal appendages and distinct regions required for basal feet and subdistal appendages.
Abstract
Ciliogenesis is regulated by context-dependent cellular cues, including some transduced through appendage-like structures on ciliary basal bodies called transition fibers and basal feet. However, the molecular basis for this regulation is not fully understood. The Odf2 gene product, ODF2/cenexin, is essential for both ciliogenesis and the formation of the distal and subdistal appendages on centrioles, which become basal bodies. We examined the effects of Odf2 deletion constructs on ciliogenesis in Odf2-knockout F9 cells. Electron microscopy revealed that ciliogenesis and transition fiber formation required the ODF2/cenexin fragment containing amino acids (aa) 188–806, whereas basal foot formation required aa 1–59 and 188–806. These sequences also formed distal and subdistal appendages, respectively, indicating that the centriole appendages are molecularly analogous to those on basal bodies. We used the differential formation of appendages by Odf2 deletion constructs to study the incorporation and function of molecules associated with each appendage type. We found that transition fibers and distal appendages were required for ciliogenesis and subdistal appendages stabilized the centrosomal microtubules.
Introduction
Both ciliary basal bodies and centrioles display two kinds of appendage-like structures: transition fibers (TFs) and basal feet (BF) in ciliary basal bodies, and distal and subdistal appendages (DAs and SAs) in centrioles (Gibbons, 1961; Anderson, 1972; Vorobjev and Chentsov, 1982; Bornens et al., 1987; Paintrand et al., 1992). The presence of these appendages depends on the product of the Odf2 gene, ODF2/cenexin. This protein was first identified as a specific component of the sperm-tail outer dense fiber, and later as a ubiquitous component of centrioles, in which it is localized to the DAs and SAs (Oko and Clermont, 1988; Lange and Gull, 1995; Brohmann et al., 1997; Nakagawa et al., 2001; Ishikawa et al., 2005; Kunimoto et al., 2012). The targeted recombination of Odf2 in F9 cells showed that the Odf2 gene has essential roles in ciliogenesis and in forming the DAs and SAs of centrioles, which become basal bodies.
Among other characterized centrosome proteins, CEP164, CCDC123/CEP89/CEP123, CCDC41, SCLT1, and FBF1 were identified as DA components (Graser et al., 2007; Sillibourne et al., 2011; Tanos et al., 2013). In contrast, ninein, centriolin, ε-tubulin, and CEP170 are associated with SAs, suggesting that the two types of appendages have different roles (Bouckson-Castaing et al., 1996; Mogensen et al., 2000; Chang et al., 2003; Gromley et al., 2003; Guarguaglini et al., 2005). At least some of these components associate with ODF2/cenexin to form Odf2-based multifunctional molecular platforms in the centriole appendages, including platforms for Rab8 and Rab11, which are associated with DAs and SAs (Gromley et al., 2005; Graser et al., 2007; Ibi et al., 2011; Hehnly et al., 2012; Schmidt et al., 2012; Tanos et al., 2013).
The Odf2 gene expression can be completely suppressed by deleting exons 6–9 (Ex-6–Ex-9), and this deletion in F9 cells completely abrogates ciliogenesis. However, a recent report showed that the deletion of Odf2 Ex-6 and Ex-7 in mice permits the generation of cilia, due to the expression of Odf2’s C-terminal domains, but the BF of the ciliary basal bodies are lost (Kunimoto et al., 2012). Furthermore, in the trachea of these BF-lacking Odf2 mutant mice, the normally regular apical microtubule networks show aberrant organization. These findings shed light on the role of BF, especially in planar cell polarity (PCP) and the arrangement of apical microtubules. In addition, because centriolin is a component of BF, and the centriolin-based Rab11 pathway functions in vesicular trafficking, the BF are thought to play roles in a variety of biological processes (Ullrich et al., 1996; Ren et al., 1998; Knödler et al., 2010; Westlake et al., 2011; Hehnly et al., 2012).
In this study, we used deletion constructs of Odf2 lacking Ex-6 and Ex-7 (Δ6/7) or Ex-4 and Ex-5 (Δ4/5), as well as a deletion series of Odf2’s N-terminal and C-terminal domains, and examined ciliogenesis and the reconstitution of TFs and/or BF of basal bodies, as well as DAs and SAs of centrioles, in an Odf2-KO F9 cell line (Ishikawa et al., 2005). The appendage-modified cells enabled us to identify the Odf2 sequences responsible for ciliogenesis and for reconstituting each of the appendages.
Results and discussion
The N terminus of Odf2 is dispensable for ciliogenesis
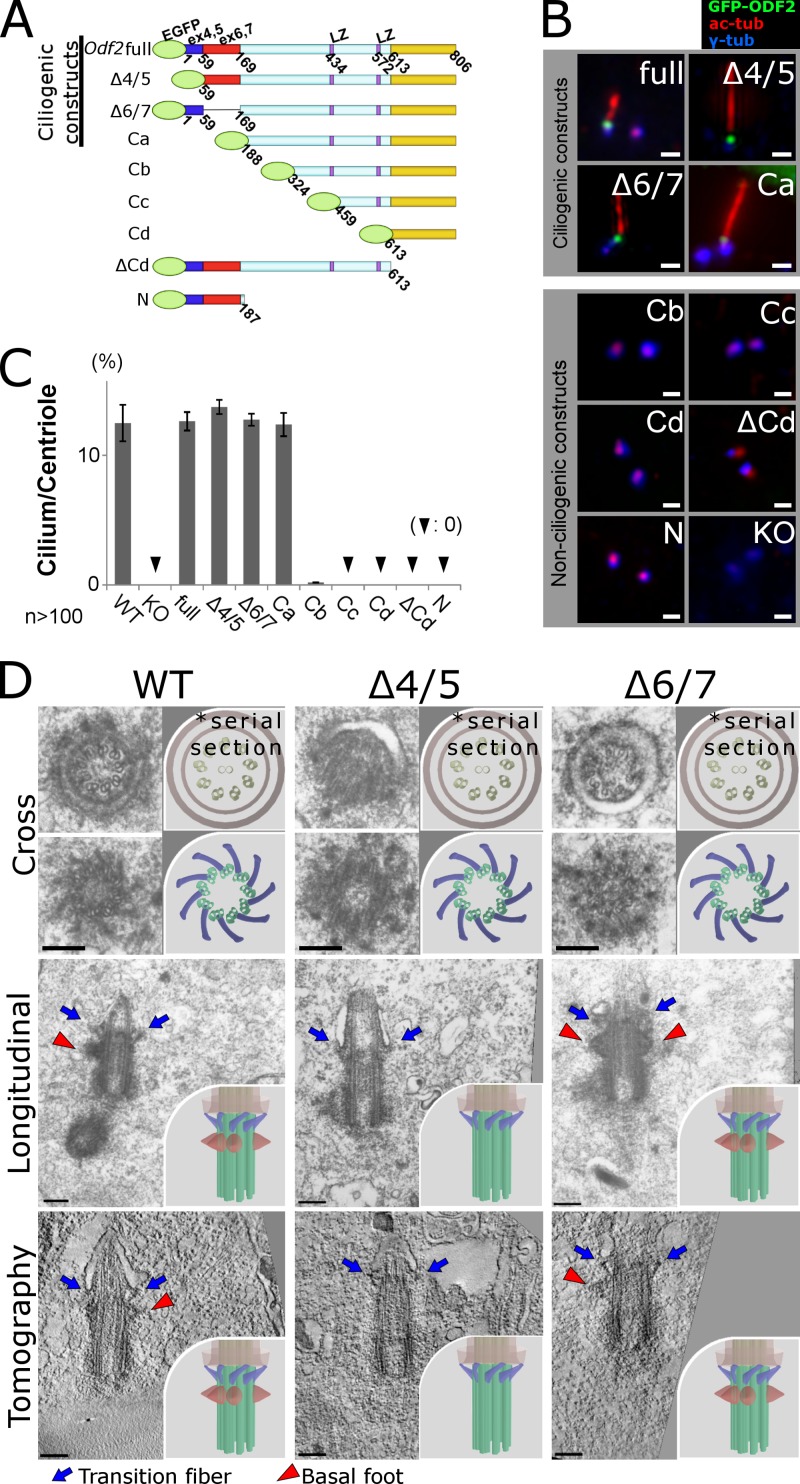
We first transfected full-length GFP-tagged Odf2 into Odf2-KO F9 cells, and immunostained for GFP (to detect GFP-ODF2/cenexin), acetylated α-tubulin (to detect primary cilia), and γ-tubulin (to detect basal bodies and centrioles; Fig. 1, A and B). The full-length Odf2 gene product was recruited to centrioles and ciliary basal bodies, and ciliogenesis was restored in Odf2-KO F9 cells by the full-length GFP-tagged Odf2 construct (Fig. 1, B and C).
Figure 1.
Odf2 gene sequences required to generate cilia and reconstitute the appendages of ciliary basal bodies. (A) Schematic representation of the Odf2 deletion constructs, which were transfected into Odf2-KO F9 cells. LZ, leucine zipper motif. (B) Immunofluorescence for GFP (Odf2), γ-tubulin (centrioles/basal bodies), and acetylated tubulin (primary cilia) to examine the generation of cilia. Bars, 1 µm. (C) Percentage of cilia on centrioles in cells expressing the indicated construct (n > 100 in more than three independent experiments). (D) Electron micrographs showing transition fibers (TFs) and/or basal feet (BF) on ciliary basal bodies. Thin-section electron microscopic images show cross sections (Cross) and longitudinal sections (Longitudinal) of primary cilia. Tomography, UHVEMT images of basal bodies; TFs, blue arrows; BF, red arrowheads; WT, (TF+BF+) basal bodies in WT F9 cells; Δ4/5, (TF+BF−) basal bodies in Δ4/5 construct–expressing Odf2-KO F9 cells; Δ6/7, (TF+BF+) basal bodies in Δ6/7 construct–expressing Odf2-KO F9 cells. Bars, 0.2 µm. More than five samples were analyzed in each case. The cross section electron micrographs in the top two rows are the same images as in the top two rows of Fig. S3 A, where they are annotated for TFs and BF. Insets, schematic drawings of electron microscopic images of basal bodies.
Next, to reveal which Odf2 gene sequences are required for ciliogenesis, we exogenously expressed GFP-tagged Odf2 deletion constructs in the Odf2-KO cells, and examined the immunofluorescent signals for GFP, γ-tubulin, and acetylated α-tubulin (Fig. 1, A and B). The set of Odf2 deletion constructs was based on the previous finding that the C-terminal regions of ODF2/cenexin are critical for ciliogenesis (Kunimoto et al., 2012). When we examined Odf2 constructs that lacked certain N-terminal regions, the cells expressing the Ca construct (aa 188–806), but not the Cb (324–806), Cc (459–806), or Cd (613–806) construct, formed stable primary cilia (Fig. 1, A–C). The frequency of primary cilia among Odf2-KO F9 cells expressing the Ca construct was comparable to that in cells expressing the full-length construct; in contrast, cilia were quite infrequent in cells expressing Cb, and no cilia were observed in those expressing Cc or Cd. Thus, the N terminus of ODF2/cenexin was dispensable for primary ciliogenesis, and a sequence that was critical for ciliogenesis lay between the Ca and Cb constructs. Consistent with this finding, primary cilia were generated by cells expressing an Odf2 construct lacking Ex-4 and Ex-5 (Δ4/5) or one lacking Ex-6 and Ex-7 (Δ6/7) without the frame-shift (Fig. 1, B and C). These sequences included the region deleted from the Ca construct.
Furthermore, a nearly full-length construct, containing the two leucine zipper motifs but lacking the C-terminal domain (ΔCd), was not recruited to basal bodies and did not generate primary cilia. The same result was obtained for the N construct. Thus, the 188–806 aa fragment of ODF2/cenexin, which includes the two leucine motifs and the most C-terminal region, was indispensable for stable ciliogenesis; the N terminus was dispensable for it (Fig. 1, A–C; and Fig. S2). In previous reports on ODF2 knockdown experiments, DAs still formed (Graser et al., 2007; Schmidt et al., 2012; Tanos et al., 2013). We expect that low-level, residual ODF2 was sufficient for DA formation in those reports. In support of this idea, as mentioned above, the low-level expression of ODF2 C-terminal domains in mutant mice allowed DAs (and eventually cilia) to be generated (Kunimoto et al., 2012), which was not the case in Odf2-KO cells. Together, these findings support the idea that the C-terminal ODF2 domains are required to form DAs, and even low levels of these domains are sufficient for appendage formation.
Ciliogenic constructs of Odf2 reconstitute transition fibers
To learn the roles of specific Odf2 domains in the formation of basal body appendages, we transfected Odf2 deletion constructs, including the ciliogenic and nonciliogenic ones, into Odf2-KO F9 cells, and then examined the TFs and BF by electron microscopy (Fig. 1 D, Fig. S1, and Fig. S3 A). To make sure we examined the entire basal body, we used serial thin-section electron microscopy (EM) and ultra-high voltage electron microscopic tomography (UHVEMT). In cells expressing the ciliogenic constructs, i.e., the wild-type (full-length), Δ4/5, Δ6/7, and Ca constructs, TFs were associated with basal bodies at the base of the cilia, connecting them to the plasma membrane (Fig. 1 D, Fig. 3, Fig. S1, and Videos 1–4). These results indicated that the N terminus of Odf2 is dispensable for forming the molecular platform required to generate TFs and cilia.
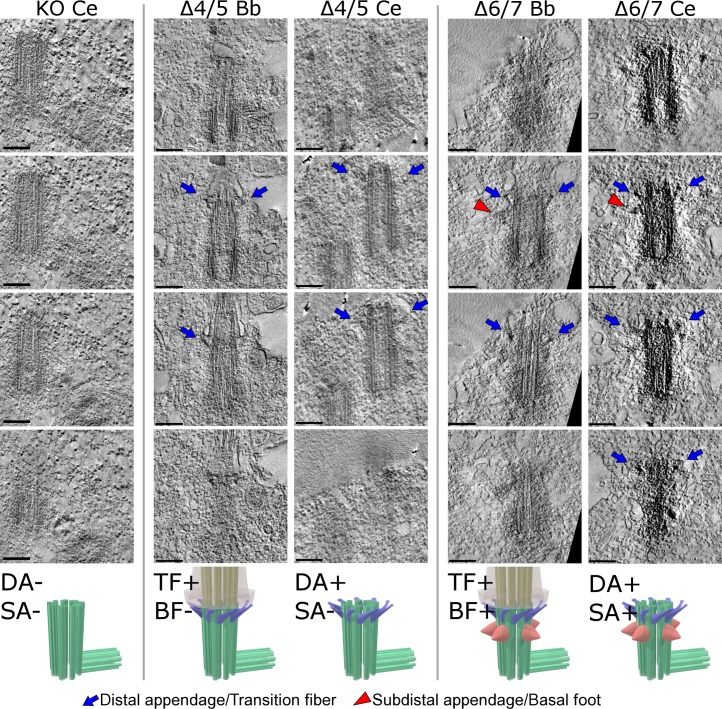
Figure 3.
Ultra-high voltage electron microscopic tomographic (UHVEMT) images of basal bodies/centrioles. Four slices are shown (see Videos 1–9). Bb, basal body; Ce, centriole; Δ4/5 and Δ6/7, deletion mutants of Odf2 that were exogenously expressed in Odf2-KO F9 cells; TFs/DAs, blue arrows; BF/SAs, red arrowheads. Bars, 0.2 µm.
ODF2/cenexin exons 4/5 are required to reconstitute the basal feet in ciliogenic constructs
In the cells of Odf2 mutant mice that expressed only the C-terminal Odf2 sequences, the TFs formed, but the BF did not (Kunimoto et al., 2012). We therefore examined BF formation in wild-type F9 cells or in Odf2-KO F9 cells transfected with the ciliogenic Odf2-deletion constructs (Δ4/5, Δ6/7, and Ca) by serial thin-section EM and UHVEMT. We found that the Δ4/5 and Ca constructs did not form BF, whereas the full-length and Δ6/7 constructs did (Fig. 1 D, Fig. 3, Fig. S1, Fig. S3 A, and Videos 1–4), suggesting that Ex-4/5 was required for the formation of BF on basal bodies. The large C-terminal region in the Ca construct (aa 188–806) was required and sufficient for the formation of TFs and for ciliogenesis. Together, these findings demonstrated the domains of Odf2 required to form TFs and BF on ciliary basal bodies.
The same respective domains of Odf2 reconstitute the homologous appendage types in ciliary basal bodies and centrioles
It is generally accepted that centrioles become basal bodies, depending on context-dependent cellular cues (Vorobjev and Chentsov, 1982; Bornens, 2002; Dawe et al., 2007; Hoyer-Fender, 2010; Kobayashi and Dynlacht, 2011; Nigg and Stearns, 2011). The similar morphological characteristics of the appendages for basal bodies and centrosomes was first noted in early EM studies, which identified two kinds of protrusions on the distal sides of the microtubule barrels (Gibbons, 1961; Sorokin, 1962; Anderson, 1972; Dirksen and Satir, 1972; Reed et al., 1984). That is, the TFs of the basal body morphologically resembled the DAs of the centriole, and the BF of the basal body resembled the centriole SAs. Later, the TFs and DAs were found to contain CEP164, CCDC123/CEP89/CEP123, CCDC41, SCLT1, and FBF1 (Graser et al., 2007; Tanos et al., 2013), and the BF and SAs to contain ninein and centriolin (Ishikawa et al., 2005; Vladar and Stearns, 2007; Kunimoto et al., 2012), supporting the structural homology of these appendages. Here we examined this similarity using specific domains of the Odf2 gene to reconstitute the appendages in ciliary basal bodies and centrioles.
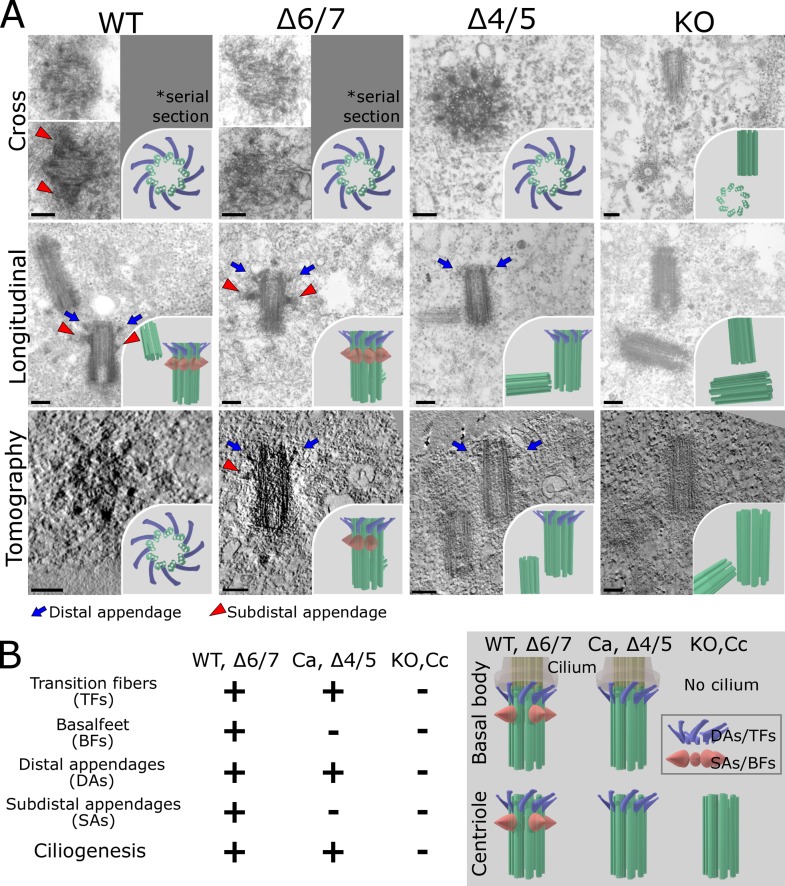
We first examined the centrosomes in cells expressing the Odf2 ciliogenic constructs that formed TFs, and found that these constructs also formed the centriole DAs. In contrast, the nonciliogenic constructs did not reconstitute the DAs (Fig. 2, Fig. 3, Fig. S1, Fig. S3 B, and Videos 5–9). Thus, the same domains of Odf2 were required to form ciliary TFs and centriole DAs, suggesting that these structures had some molecular architecture and function in common.
Figure 2.
Reconstitution of centriole appendages by Odf2 deletion constructs. (A) Electron micrographs showing distal appendages (DAs) and/or subdistal appendages (SAs) on centrioles. Thin-section electron microscopic images show cross sections (Cross) and longitudinal sections (Longitudinal) of centrioles. More than five samples were analyzed in each case. Tomography, UHVEMT images of centrioles; DAs, blue arrows; SAs, red arrowheads; WT, (DA+SA+) centriole in WT F9 cells; Δ4/5, (DA+SA−) centriole in Δ4/5 construct–expressing Odf2-KO F9 cells; Δ6/7, (DA+SA+) centriole in Δ6/7 construct–expressing Odf2-KO F9 cells; KO, (DA−SA−) centriole; TFs, blue arrows; BF, red arrowheads. Bars, 0.2 µm. Insets, schematic drawings of electron microscopic images of centrioles. (B) Summary of findings on the reconstitution of the basal body/centriole appendages.
Next, because Ex-4/5 was required for BF to form in the basal bodies of primary cilia in Odf2-KO F9 cells, we examined the formation of centriole SAs by Ex-4/5–containing constructs by serial thin-section EM and UHVEM. We found that all of the Ex-4/5–containing ciliogenic constructs formed SAs in centrioles. In contrast, the ciliogenic constructs lacking the Ex-4/5 sequence did not form SAs (Fig. 2 A, Fig. 3, Fig. S3 B, and Videos 6–9). Thus, the same domains of ODF2/cenexin were needed to form the basal body BF and centriole SAs, indicating that they have common molecular bases (Fig. 2 B). Together, these results supported the idea that the appendages of basal bodies and centrioles are homologous (Azimzadeh and Marshall, 2010; Seeley and Nachury, 2010; Kunimoto et al., 2012).
Immunofluorescence analysis of the molecular components in appendages reconstituted by Odf2
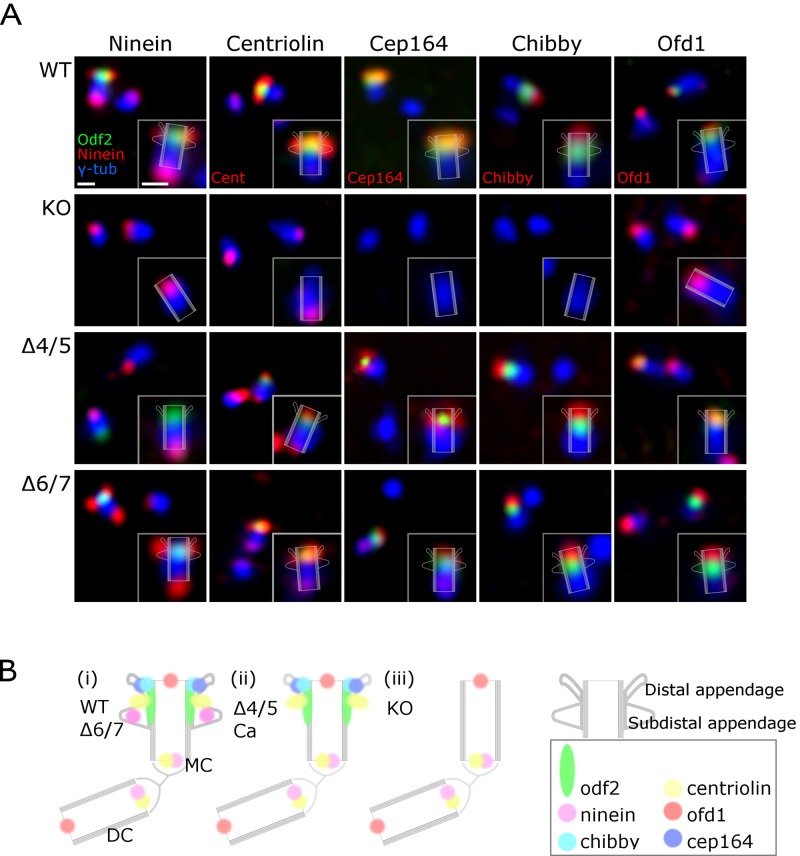
The two sets of homologous appendages on basal bodies and centrioles appear to be molecularly distinct. Their dependence on different domains of the Odf2 gene product could result in the distinct sets of associated molecules incorporated into the different appendage types. The three patterns of reconstitution (i.e., centrioles bearing no appendages, only DAs, or DAs and SAs, depending on the sequences contained in the Odf2 deletion construct) provided a powerful tool for confirming which molecular components are associated with DAs or SAs, respectively. We therefore examined the immunofluorescence signals for DA- and SA-associated proteins (ninein, centriolin, and CEP164) and centriole proteins (chibby and OFD1) in their respective reconstituted appendages. To examine the relative spatial relationships of the appendages in centrosomes, we stained the centrosomes with anti-GFP to visualize GFP-tagged ODF2/cenexin and with anti–γ-tubulin to label centrosomes (Fig. 4).
Figure 4.
Immunofluorescence microscopic images of centriole appendages in the DA+SA+, DA-SA−, and DA+SA− patterns. (A) Immunofluorescence for ninein, centriolin, CEP164, and OFD1. WT, (DA+SA+) centriole in WT F9 cells; KO, (DA-SA−) centriole in Odf2-KO F9 cells; Δ4/5, (DA+SA−) centriole in Δ4/5 construct–expressing Odf2-KO F9 cells; Δ6/7, (DA+SA+) centriole in Δ6/7 construct–expressing Odf2-KO F9 cells. (B) Schematic drawing showing the localization of centrosomal components. MC, mother centriole; DC, daughter centriole. Bars, 500 nm.
We first found that the distal ends of the Odf2-KO centrioles were negative for all the antigens except OFD1. The ninein and centriolin signals were detected at the proximal ends of mother and daughter centrioles regardless of Odf2 expression, as previously reported (Mogensen et al., 2000; Ishikawa et al., 2005). Next, we found that centrioles associated with DAs and SAs (due to exogenous transfection of the full-length or Δ6/7 Odf2 constructs) were positive for CEP164, ninein, centriolin, chibby, and OFD1 at their distal ends. These results strongly suggested that the DA and SA structures represent supramolecular complexes that require Odf2 expression to form.
We then examined the appendage proteins in DA+SA− centrioles by immunofluorescence, using the Δ4/5 Odf2 construct in Odf2-KO cells. We found that CEP164, a DA component, was restored on the distal end of centrioles. Chibby, an appendage component that had not been assigned to a particular appendage (Voronina et al., 2009; Steere et al., 2012), was also restored on the distal end of centrioles, although it remains to be studied whether chibby is a DA component and/or a component associated with the ciliary vesicle or transition zone, the assembly of which depends on DA-mediated membrane docking.
Centriolin was also observed at the distal end of (DA+SA−) centrioles in Δ4/5 Odf2-expressing Odf2-KO cells. This finding was unexpected because centriolin is reportedly an SA component (Gromley et al., 2003), and our data suggest that ninein and centriolin are differentially associated with SAs and that centriolin is associated with the product of the Δ4/5 Odf2 construct (Fig. 4 B). Centriolin is reported to interact with Rab11 GTPase and to regulate its activity through Evi5 in vesicular trafficking (Gromley et al., 2005; Hehnly et al., 2012). Thus, it is most likely that centriolin, as a complex with the product of the Δ4/5 Odf2 construct, forms the platform for Rab11-related vesicular trafficking, possibly in a different way from the case in (DA+SA+) centrioles.
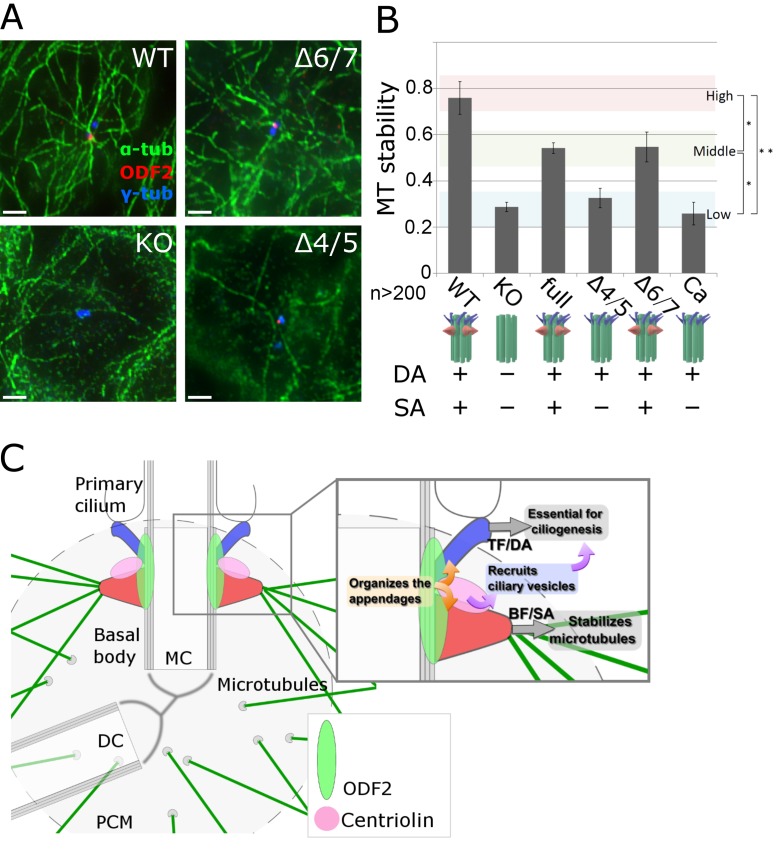
Centrosomal subdistal appendages help stabilize the centrosomal microtubules
We next examined the stabilization of centrosomal microtubules in transfected cells treated with nocodazole, which interferes with microtubule polymerization. We found that under nocodazole treatment, the number of microtubules associated with centrosomes was significantly greater in wild-type (DA+SA+) cells than in Odf2-KO (DA−SA−) cells, based on the number of microtubules in the asters surrounding the centrioles (Fig. 5, A and B). In a previous study, we showed that the cell cycle and centrosomal microtubule-organizing center (MTOC) activity are unaffected by the loss of both appendages (Ishikawa et al., 2005). Consistent with this finding, our results of a microtubule regrowth assay (Brandt and Lee, 1993; Gaglio et al., 1996; Ishikawa et al., 2005) using centrosomes depleted of microtubules by prolonged nocodazole/cold treatment showed that the MTOC activity in the wild-type and Odf2-KO cells was the same (Fig. S3, C and D).
Figure 5.
Specific role of centrosomal subdistal appendages (SAs) in stabilizing centriole microtubules (MTs). (A) Immunofluorescence images of MTs in DA+SA+, DA+SA−, and DA−SA− centrioles under nocodazole treatment, an MT-destabilizing condition. WT, (DA+SA+) centriole in WT F9 cells; Δ4/5, (DA+SA−) centriole in Δ4/5 construct–expressing Odf2-KO F9 cells; Δ6/7, (DA+SA+) centriole in Δ6/7 construct–expressing Odf2-KO F9 cells. (B) Quantification of MT stability. The relative MT stability is shown for the indicated Odf2 construct (*, P = 0.01; **, P = 0.005; n > 200 in more than three independent experiments). (C) Schematic drawing of the specific roles of the appendages of ciliary basal bodies/centrioles. In the proposed model, separate domains in Odf2 serve as the molecular platform on which the appendages are constructed. Note that centriolin associates with ODF2/cenexin at the base of the SA (or where the base would be in the absence of SAs), and recruits vesicles whose cargoes support ciliogenesis. The BF/SAs (red) stabilize MTs, whereas the TFs/DAs (blue) are essential for ciliogenesis. MC, mother centriole; DC, daughter centriole.
Similar differences were detected between SA− centrioles and those with reconstituted SAs. The number of centrosomal microtubules was not significantly higher on the DA+SA− centrioles in cells transfected with the Δ4/5 or Ca construct, compared with Odf2-KO (DA−SA−) cells, suggesting that the resistance to nocodazole was not increased by the presence of DAs. In contrast, the full-length or Δ6/7 Odf2 constructs, which reconstituted both the DA and SA in Odf2-KO cells, increased the cells’ resistance to nocodazole. These findings suggested that SA stabilizes the centrosomal microtubules against depolymerization, consistent with the idea that the DA and SA centriole appendages have distinct functions. That is, DAs are required and sufficient for ciliogenesis, whereas SAs stabilize the centrosomal microtubules.
Conclusion
We here showed that different Odf2 sequences were required to reconstitute the basal body appendages as TF+BF− or TF+BF+, and the same sequences were required to reconstitute, respectively, the DA and SA centriole appendages. A comparison of the structural characteristics of the three appendage patterns on basal bodies (TF+BF+, TF+BF−, and TF−BF−) and on centrioles (DA+SA+, DA+SA−, DA−SA−), respectively, showed that the TFs and BF are structurally and possibly functionally homologous to the DAs and SAs, respectively. The N-terminal domain reconstituted the BF/SA on basal bodies or centrioles only when the TF/DA was formed by the C-terminal domains of Odf2. Alternatively, the assembly of BF/SAs could require both the N-terminal and C-terminal domains of the ODF2 protein, whether the TFs/DAs are present or not. Thus, the location of the TF/DA and BF/SA appendages at the distal end of the nine microtubule triplets might be determined primarily by the TF/DA. Given that the distinct appendage structures both depended on the Odf2 gene product, it is likely that they are integrated in some cooperative way into a unified sophisticated system (Fig. 5 C).
In this respect, the functional relationship between the two appendage types is not simple because the SA component centriolin was found, unexpectedly, associated with SA− centrioles. Although centriolin may not be directly involved in SA formation, it may be involved in some function of the SA. In this respect, regulators of Rab8 and Rab11, such as Rabin8 and Evi5, were recently implicated in the SA/BF and DA/TF systems through centriolin and CEP164, respectively (Knödler et al., 2010; Westlake et al., 2011; Hehnly et al., 2012). Thus, ciliogenesis may represent the actions of functionally integrated systems of the TF/DA and BF/SA.
Here we showed that the distinct molecular constitutions of the appendages, which are located close to one another near the distal end of basal bodies and centrioles, are based on different Odf2 domain usage. The transfection experiments used here provided a good tool for dissecting the molecular components of the different appendages. Furthermore, defining the different Odf2 domains used in the formation of each appendage type provided a helpful tool for identifying new components required for building the separate appendage structures. Further studies along these lines will illuminate the molecular composition and functions of these homologous structures: the basal body–associated TFs and BF and the centriole-associated DAs and SAs. Another important future goal is to elucidate how the functional platforms associated with distinct appendages are integrated into centrosome/basal body– and microtubule-related cellular events.
Materials and methods
Antibodies
Rabbit anti-CEP164 polyclonal antibodies (pAbs) were a gift from G. Pereira (DKFZ-ZMBH Alliance, German Cancer Research Center, Heidelberg, Germany). The rat anti-ODF2 monoclonal antibody (mAb) was newly generated in our laboratory. Synthetic peptides corresponding to the C-terminal ODF2/cenexin protein sequence were used to immunize rats, and hybridoma cells were cloned by a standard method, as described previously (Tsukita et al., 1994). In short, hybridoma cells were generated by fusing immunized rat lymphocytes and P3 mouse myeloma cells. We obtained two clones, MAT159 and MAT195, named after the people who generated them (Murakami/Aihara/Tateishi). The specificities of these antibodies were examined using Odf2-KO F9 cells by immunofluorescence. The rat anti-centriolin mAb and rabbit anti-ninein pAb were also generated previously (Ishikawa et al., 2005). The cDNAs encoding amino acids 953–1228 of mouse ninein and 1–261 of mouse centriolin (N-centriolin) were subcloned into pGEX-4T-1 (GE Healthcare), and their GST fusion proteins were expressed in Escherichia coli. They were purified on glutathione Sepharose 4B columns (GE Healthcare), and used as antigens in rabbits or rats. In addition, two other anti-centriolin antibodies were used, and yielded the same results: rat anti-centriolin mAb that we generated but have not described previously, and commercially available rabbit anti-centriolin pAb (Sigma-Aldrich). The following antibodies were purchased: rabbit anti-ODF2 pAb (Abcam), rabbit anti-chibby pAb (EMD Millipore), mouse anti–α-tubulin mAb (Sigma-Aldrich), rat anti–α-tubulin mAb (Abcam), rabbit anti–γ-tubulin pAb (Sigma-Aldrich), mouse anti–γ-tubulin mAb (Sigma-Aldrich), and mouse anti-acetylated α-tubulin mAb (Sigma-Aldrich). All primary antibodies were used at a dilution of 100× to ∼1,000×. Alexa Fluor 488–, 568–, and 647–labeled secondary antibodies were commercially obtained (Invitrogen). All secondary antibodies were used at a dilution of 1,000×.
Constructs and cell culture
The product of Odf2 (ENSMUST00000113757) was considered to be full length. The Odf2 deletion constructs were subcloned into the pCAGGS expression vector, which is designed to express N-terminally tagged EGFP-fused proteins. F9 cells are derived from mouse embryonic teratocarcinoma, and were cultured on gelatin-coated dishes in DMEM with 10% FBS. To establish stable F9 cell lines expressing each Odf2 construct, each expression vector was transfected into Odf2-KO F9 cells, which were described previously (Ishikawa et al., 2005). In F9 cells, exons 6–9 were deleted in the 22 exons of Odf2.
Microtubule stability assay
Confluent cells were transfected, cultured on coverslips, and treated with 500 nM nocodazole for 5 min at 4°C. The cells were then fixed and processed for immunofluorescence microscopy. Microtubule stability was defined as the proportion of cells showing an aster-like microtubule arrangement.
Microtubule regrowth assay
Confluent stably transfected cells were cultured on coverslips, and treated with 10 µM nocodazole for 1 h at 4°C. The cells were then washed with DMEM and incubated for another 25 min to allow microtubule regrowth (Tanaka et al., 2012). The cells were then fixed and processed for immunofluorescence microscopy.
Electron microscopy
All samples were prepared using a classical method (Ishikawa et al., 1969). F9 cells were cultured in a 6-well plate. After reaching confluence, the cells were incubated in IEM buffer (30 mM Hepes, pH 7.3, 1 mM EGTA, 0.1 mM EDTA, 25 mM NaCl, 5 mM MgSO4, 1 mM DTT, and 1% volume of protease inhibitor mixture [Sigma-Aldrich]) at 37°C for 3 min. After three washes in MT-stabilizing buffer with 1 mM taxol, the cells were fixed in 2.5% glutaraldehyde, 2% formaldehyde, and 0.1% tannic acid in 100 mM Hepes buffer (pH 7.5), followed by post-fixation with 1% OsO4 in 100 mM Hepes buffer (pH 7.5). The samples were dehydrated and embedded in Epon 812 (Polysciences).
For the TEM analysis, samples were serially thin-sectioned at 50 or 70 nm, and then 15–30 serial sections were analyzed by TEM (JEM-1010; JEOL). For the ultra-high acceleration voltage electron microscopy (UHVEM) analysis, the samples were thick-sectioned at 700 nm and mounted on formvar-coated 50/75-mesh molybdenum grids. Colloidal gold particles (20-nm diameter) were deposited on both sides of each section, and the samples were observed by UHVEM at 1 or 2 MeV acceleration voltage (H-3000; Hitachi). UHVEM images were taken at 25,000×, from −60 to +60 degrees at 2-degree intervals around a single tilt axis, and images were acquired with an SSCCD camera (model F415S; TVIPS GmbH). Image calibration and 3D reconstructions of each series were performed using IMOD software (Kremer et al., 1996; The Boulder Laboratory For 3-D Electron Microscopy of Cells, Boulder, CO).
Immunofluorescence
Cells were cultured on gelatin-coated coverslips. To observe the primary cilia, the cells were incubated at 4°C for 30 min before fixation to decrease the amount of cytoplasmic acetylated microtubules. In other cases, this step was skipped. The cells were fixed in ice-cold methanol for 10 min, washed with PBS three times, and permeabilized with 0.5% Triton X-100 in PBS for 15 min. After being washed with PBS, the cells were soaked in 1% BSA in PBS for 30 min at room temperature. The cells were then incubated with primary antibodies for 1 h in a humidified chamber. The cells were then washed with PBS and incubated with Alexa Fluor 488–, 555–, or 647–conjugated secondary antibodies (Molecular Probes) for 30 min. The samples were then washed with PBS, and mounted in fluorescence mounting medium (Dako). Images were obtained with a DeltaVision system (Applied Precision) equipped with a microscope (model IX70; Olympus) and observed using 40× Plan Apo NA 1.4 and 63× Plan Apo NA 1.4 oil immersion objectives. Images of cytoplasmic microtubules were obtained using an ELYRA S.1 (SR-SIM) system (Carl Zeiss) and a 100× α-Plan Apo NA 1.46 oil immersion objective. To quantify cilia, cells displaying primary cilia were identified by immunofluorescence with an anti-acetylated α-tubulin mAb. All images were acquired at 25°C. Images were prepared using Adobe Photoshop, Illustrator (Adobe systems), and ImageJ (National Institutes of Health).
Online supplemental material
Fig. S1 provides data on the electron micrographs of appendages reconstituted on centrioles and basal bodies by the transfected Ca construct. Fig. S2 shows Western blotting analysis of Odf2-KO F9 cells in which GFP-tagged Odf2 deletion constructs, used in this study, were expressed. Fig. S3 shows the serial cross sections of basal bodies and centrioles with two or one appendages, reconstituted by expression of deleted series of Odf2, or with no appendages. Also in Fig. S3, MTOC activity on the centrioles in wild-type and Odf2-KO F9 cells is shown. Video 1 shows the UHVEM tomographic images of the ciliary basal body of an Odf2-WT F9 cell and its schematic drawing. Video 2 shows the UHVEM tomographic images of the ciliary basal body of an Odf2-KO F9 cell expressing the Δ4/5 construct and its schematic drawing. Video 3 shows the UHVEM tomographic images of the ciliary basal body of an Odf2-KO F9 cell expressing the Δ6/7 construct and its schematic drawing. Video 4 shows the UHVEM tomographic images of the ciliary basal body of an Odf2-KO F9 cell expressing the Ca construct and its schematic drawing. Video 5 shows the UHVEM tomographic images of a centriole in an Odf2-WT F9 cell and its schematic drawing. Video 6 shows the UHVEM tomographic images of a centriole in an Odf2-KO cell and its schematic drawing. Video 7 shows the UHVEM tomographic images of a centriole in an Odf2-KO F9 cell expressing the Δ4/5 construct and its schematic drawing. Video 8 shows the UHVEM tomographic images of a centriole in an Odf2-KO F9 cell expressing the Δ6/7 construct and its schematic drawing. Video 9 shows the UHVEM tomographic images of a centriole in an Odf2-KO F9 cell expressing the Ca construct and its schematic drawing. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201303071/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201303071.dv.
Supplementary Material
Acknowledgments
The authors thank M. Uji, A. Hagiwara-Yano, and F. Takenaga for technical assistance, and members of our laboratories for discussion. We also thank the undergraduate students, H. Murakami and C. Aihara, for their collaboration in generating the rat anti-ODF2/cenexin mAbs (MAT159 and MAT195); and M. Sudol, as well as G. Gray and L. Miglietta, for reading our manuscript. The anti-CEP164 pAb was a gift from G. Pereira.
This work was supported in part by a Grant-in-Aid for Scientific Research on Innovative Areas and for Scientific Research (A) to S. Tsukita and by the “Nanotechnology Platform” project (no. 12024046) of the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
Footnotes
Abbreviations used in this paper:
- BF
- basal feet
- DA
- distal appendage
- MTOC
- microtubule-organizing center
- SA
- subdistal appendage
- TF
- transition fiber
- UHVEMT
- ultra-high voltage electron microscopic tomography
References
- Anderson R.G. 1972. The three-dimensional structure of the basal body from the rhesus monkey oviduct. J. Cell Biol. 54:246–265 10.1083/jcb.54.2.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh J., Marshall W.F. 2010. Building the centriole. Curr. Biol. 20:R816–R825 10.1016/j.cub.2010.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M. 2002. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 14:25–34 10.1016/S0955-0674(01)00290-3 [DOI] [PubMed] [Google Scholar]
- Bornens M., Paintrand M., Berges J., Marty M.C., Karsenti E. 1987. Structural and chemical characterization of isolated centrosomes. Cell Motil. Cytoskeleton. 8:238–249 10.1002/cm.970080305 [DOI] [PubMed] [Google Scholar]
- Bouckson-Castaing V., Moudjou M., Ferguson D.J., Mucklow S., Belkaid Y., Milon G., Crocker P.R. 1996. Molecular characterisation of ninein, a new coiled-coil protein of the centrosome. J. Cell Sci. 109:179–190 [DOI] [PubMed] [Google Scholar]
- Brandt R., Lee G. 1993. Functional organization of microtubule-associated protein tau. Identification of regions which affect microtubule growth, nucleation, and bundle formation in vitro. J. Biol. Chem. 268:3414–3419 [PubMed] [Google Scholar]
- Brohmann H., Pinnecke S., Hoyer-Fender S. 1997. Identification and characterization of new cDNAs encoding outer dense fiber proteins of rat sperm. J. Biol. Chem. 272:10327–10332 10.1074/jbc.272.15.10327 [DOI] [PubMed] [Google Scholar]
- Chang P., Giddings T.H., Jr, Winey M., Stearns T. 2003. Epsilon-tubulin is required for centriole duplication and microtubule organization. Nat. Cell Biol. 5:71–76 10.1038/ncb900 [DOI] [PubMed] [Google Scholar]
- Dawe H.R., Farr H., Gull K. 2007. Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J. Cell Sci. 120:7–15 10.1242/jcs.03305 [DOI] [PubMed] [Google Scholar]
- Dirksen E.R., Satir P. 1972. Ciliary activity in the mouse oviduct as studied by transmission and scanning electron microscopy. Tissue Cell. 4:389–403 10.1016/S0040-8166(72)80017-X [DOI] [PubMed] [Google Scholar]
- Gaglio T., Saredi A., Bingham J.B., Hasbani M.J., Gill S.R., Schroer T.A., Compton D.A. 1996. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J. Cell Biol. 135:399–414 10.1083/jcb.135.2.399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons I.R. 1961. The relationship between the fine structure and direction of beat in gill cilia of a lamellibranch mollusc. J. Biophys. Biochem. Cytol. 11:179–205 10.1083/jcb.11.1.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S., Stierhof Y.-D., Lavoie S.B., Gassner O.S., Lamla S., Le Clech M., Nigg E.A. 2007. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 179:321–330 10.1083/jcb.200707181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromley A., Jurczyk A., Sillibourne J., Halilovic E., Mogensen M., Groisman I., Blomberg M., Doxsey S. 2003. A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J. Cell Biol. 161:535–545 10.1083/jcb.200301105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromley A., Yeaman C., Rosa J., Redick S., Chen C.-T., Mirabelle S., Guha M., Sillibourne J., Doxsey S.J. 2005. Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell. 123:75–87 10.1016/j.cell.2005.07.027 [DOI] [PubMed] [Google Scholar]
- Guarguaglini G., Duncan P.I., Stierhof Y.D., Holmström T., Duensing S., Nigg E.A. 2005. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol. Biol. Cell. 16:1095–1107 10.1091/mbc.E04-10-0939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehnly H., Chen C.T., Powers C.M., Liu H.L., Doxsey S. 2012. The centrosome regulates the Rab11- dependent recycling endosome pathway at appendages of the mother centriole. Curr. Biol. 22:1944–1950 10.1016/j.cub.2012.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer-Fender S. 2010. Centriole maturation and transformation to basal body. Semin. Cell Dev. Biol. 21:142–147 10.1016/j.semcdb.2009.07.002 [DOI] [PubMed] [Google Scholar]
- Ibi M., Zou P., Inoko A., Shiromizu T., Matsuyama M., Hayashi Y., Enomoto M., Mori D., Hirotsune S., Kiyono T., et al. 2011. Trichoplein controls microtubule anchoring at the centrosome by binding to Odf2 and ninein. J. Cell Sci. 124:857–864 10.1242/jcs.075705 [DOI] [PubMed] [Google Scholar]
- Ishikawa H., Bischoff R., Holtzer H. 1969. Formation of arrowhead complexes with heavy meromyosin in a variety of cell types. J. Cell Biol. 43:312–328 10.1083/jcb.43.2.312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Kubo A., Tsukita S., Tsukita S. 2005. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat. Cell Biol. 7:517–524 10.1038/ncb1251 [DOI] [PubMed] [Google Scholar]
- Knödler A., Feng S., Zhang J., Zhang X., Das A., Peränen J., Guo W. 2010. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc. Natl. Acad. Sci. USA. 107:6346–6351 10.1073/pnas.1002401107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Dynlacht B.D. 2011. Regulating the transition from centriole to basal body. J. Cell Biol. 193:435–444 10.1083/jcb.201101005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., McIntosh J.R. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Kunimoto K., Yamazaki Y., Nishida T., Shinohara K., Ishikawa H., Hasegawa T., Okanoue T., Hamada H., Noda T., Tamura A., et al. 2012. Coordinated ciliary beating requires Odf2-mediated polarization of basal bodies via basal feet. Cell. 148:189–200 10.1016/j.cell.2011.10.052 [DOI] [PubMed] [Google Scholar]
- Lange B.M., Gull K. 1995. A molecular marker for centriole maturation in the mammalian cell cycle. J. Cell Biol. 130:919–927 10.1083/jcb.130.4.919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen M.M., Malik A., Piel M., Bouckson-Castaing V., Bornens M. 2000. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J. Cell Sci. 113:3013–3023 [DOI] [PubMed] [Google Scholar]
- Nakagawa Y., Yamane Y., Okanoue T., Tsukita S., Tsukita S. 2001. Outer dense fiber 2 is a widespread centrosome scaffold component preferentially associated with mother centrioles: its identification from isolated centrosomes. Mol. Biol. Cell. 12:1687–1697 10.1091/mbc.12.6.1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A., Stearns T. 2011. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat. Cell Biol. 13:1154–1160 10.1038/ncb2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oko R., Clermont Y. 1988. Isolation, structure and protein composition of the perforatorium of rat spermatozoa. Biol. Reprod. 39:673–687 10.1095/biolreprod39.3.673 [DOI] [PubMed] [Google Scholar]
- Paintrand M., Moudjou M., Delacroix H., Bornens M. 1992. Centrosome organization and centriole architecture: their sensitivity to divalent cations. J. Struct. Biol. 108:107–128 10.1016/1047-8477(92)90011-X [DOI] [PubMed] [Google Scholar]
- Reed W., Avolio J., Satir P. 1984. The cytoskeleton of the apical border of the lateral cells of freshwater mussel gill: structural integration of microtubule and actin filament-based organelles. J. Cell Sci. 68:1–33 [DOI] [PubMed] [Google Scholar]
- Ren M., Xu G., Zeng J., De Lemos-Chiarandini C., Adesnik M., Sabatini D.D. 1998. Hydrolysis of GTP on rab11 is required for the direct delivery of transferrin from the pericentriolar recycling compartment to the cell surface but not from sorting endosomes. Proc. Natl. Acad. Sci. USA. 95:6187–6192 10.1073/pnas.95.11.6187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K.N., Kuhns S., Neuner A., Hub B., Zentgraf H., Pereira G. 2012. Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis. J. Cell Biol. 199:1083–1101 10.1083/jcb.201202126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley E.S., Nachury M.V. 2010. The perennial organelle: assembly and disassembly of the primary cilium. J. Cell Sci. 123:511–518 10.1242/jcs.061093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillibourne J.E., Specht C.G., Izeddin I., Hurbain I., Tran P., Triller A., Darzacq X., Dahan M., Bornens M. 2011. Assessing the localization of centrosomal proteins by PALM/STORM nanoscopy. Cytoskeleton (Hoboken). 68:619–627 10.1002/cm.20536 [DOI] [PubMed] [Google Scholar]
- Sorokin S. 1962. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J. Cell Biol. 15:363–377 10.1083/jcb.15.2.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steere N., Chae V., Burke M., Li F.Q., Takemaru K., Kuriyama R. 2012. A Wnt/beta-catenin pathway antagonist Chibby binds Cenexin at the distal end of mother centrioles and functions in primary cilia formation. PLoS ONE. 7:e41077 10.1371/journal.pone.0041077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N., Meng W., Nagae S., Takeichi M. 2012. Nezha/CAMSAP3 and CAMSAP2 cooperate in epithelial-specific organization of noncentrosomal microtubules. Proc. Natl. Acad. Sci. USA. 109:20029–20034 10.1073/pnas.1218017109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos B.E., Yang H.J., Soni R., Wang W.J., Macaluso F.P., Asara J.M., Tsou M.F.B. 2013. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 27:163–168 10.1101/gad.207043.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita S., Oishi K., Sato N., Sagara J., Kawai A., Tsukita S. 1994. ERM family members as molecular linkers between the cell surface glycoprotein CD44 and actin-based cytoskeletons. J. Cell Biol. 126:391–401 10.1083/jcb.126.2.391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich O., Reinsch S., Urbé S., Zerial M., Parton R.G. 1996. Rab11 regulates recycling through the pericentriolar recycling endosome. J. Cell Biol. 135:913–924 10.1083/jcb.135.4.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar E.K., Stearns T. 2007. Molecular characterization of centriole assembly in ciliated epithelial cells. J. Cell Biol. 178:31–42 10.1083/jcb.200703064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev I.A., Chentsov Y.S. 1982. Centrioles in the cell cycle. I. Epithelial cells. J. Cell Biol. 93:938–949 10.1083/jcb.93.3.938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina V.A., Takemaru K., Treuting P., Love D., Grubb B.R., Hajjar A.M., Adams A., Li F.Q., Moon R.T. 2009. Inactivation of Chibby affects function of motile airway cilia. J. Cell Biol. 185:225–233 10.1083/jcb.200809144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlake C.J., Baye L.M., Nachury M.V., Wright K.J., Ervin K.E., Phu L., Chalouni C., Beck J.S., Kirkpatrick D.S., Slusarski D.C., et al. 2011. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc. Natl. Acad. Sci. USA. 108:2759–2764 10.1073/pnas.1018823108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.