Erlins are cholesterol-binding proteins that restrict the activation of SREBPs, key transcription factors for cholesterol and fatty acid biosynthetic genes, under conditions of cholesterol sufficiency.
Abstract
Cellular cholesterol levels are controlled by endoplasmic reticulum (ER) sterol sensing proteins, which include Scap and Insig-1. With cholesterol sufficiency, Insig inhibits the activation of sterol regulatory element binding proteins (SREBPs), key transcription factors for cholesterol and fatty acid biosynthetic genes, by associating with Scap–SREBP complexes to promote their ER retention. Here we show that the multimeric ER proteins erlins-1 and -2 are additional SREBP regulators. Depletion of erlins from cells grown with sterol sufficiency led to canonical activation of SREBPs and their target genes. Moreover, SREBPs, Scap, and Insig-1 were physically associated with erlins. Erlins bound cholesterol with specificity and strong cooperativity and responded to ER cholesterol changes with altered diffusional mobility, suggesting that erlins themselves may be regulated by cholesterol. Together, our results define erlins as novel cholesterol-binding proteins that are directly involved in regulating the SREBP machinery. We speculate that erlins promote stability of the SREBP–Scap–Insig complex and may contribute to the highly cooperative control of this system.
Introduction
Cholesterol is an essential lipid of mammalian cells, serving as a structural component of membranes and a biosynthetic precursor to steroid hormones (Chang et al., 2006; Ikonen, 2008). Cholesterol increases stiffness and reduces permeability of membranes (Bretscher and Munro, 1993; Ikonen, 2008). It also is thought to participate in membrane-associated signaling and protein sorting (Simons and Gerl, 2010). Cholesterol has a central role in the etiology of many cardiovascular diseases, and pharmacological inhibitors of its biosynthesis are widely used as therapeutic agents.
Cholesterol biosynthesis and uptake are regulated by the sterol regulatory element binding protein (SREBP) transcription factors (Goldstein et al., 2006). SREBPs are synthesized as part of transmembrane precursors that are integrated in the ER membrane (Brown and Goldstein, 1999). The SREBP precursors are bound to the ER transmembrane chaperone Scap, which contains a sterol-sensing domain (Radhakrishnan et al., 2004; Motamed et al., 2011). Under conditions of cholesterol sufficiency, cholesterol-bound Scap associates with Insig, which promotes ER retention of the SREBP–Scap complex (Goldstein et al., 2006). However, when ER cholesterol decreases below a critical threshold, Scap undergoes a conformational change that allows packaging of SREBP–Scap in COPII-coated vesicles for subsequent transport to the Golgi. This is accompanied by increased ER-associated degradation (ERAD) of Insig (Gong et al., 2006). In the Golgi, site-specific proteases release the cytosolic transcription factor domain of SREBPs that activates genes for cholesterol and fatty acid biosynthesis (Goldstein et al., 2006). When cholesterol levels are restored, the SREBP–Scap–Insig complex again accumulates in the ER. SREBP retention in the ER involves a highly cooperative, switch-like response to cholesterol (Radhakrishnan et al., 2008). The molecular basis for this effect is not understood.
Erlins are ∼40-kD proteins that originally were characterized by their fractionation in cholesterol-enriched, detergent-resistant membrane derivatives (Browman et al., 2006). Erlins contain a membrane-inserted segment at their N terminus and are localized to the ER lumen in ∼1,000-kD heteromultimeric complexes (Pearce et al., 2007, 2009; Hoegg et al., 2009). They contain an ∼180-residue stomatin, prohibitin, flotillin, HflK/C homology domain, found in more than 6,500 proteins from Archaea to higher eukaryotes, which is proposed to organize membrane microdomains (Langhorst et al., 2005; Browman et al., 2007). Erlins promote ERAD of the activated IP3 receptor (Pearce et al., 2007, 2009) and of 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR; Jo et al., 2011). We now show that erlins are cholesterol-binding proteins that interact with SREBP–Scap–Insig and are needed for restricting SREBP activation under conditions of cholesterol sufficiency. Our results reveal a new complexity of the machinery that regulates SREBPs and identify erlins as components that contribute to cellular cholesterol homeostasis.
Results and discussion
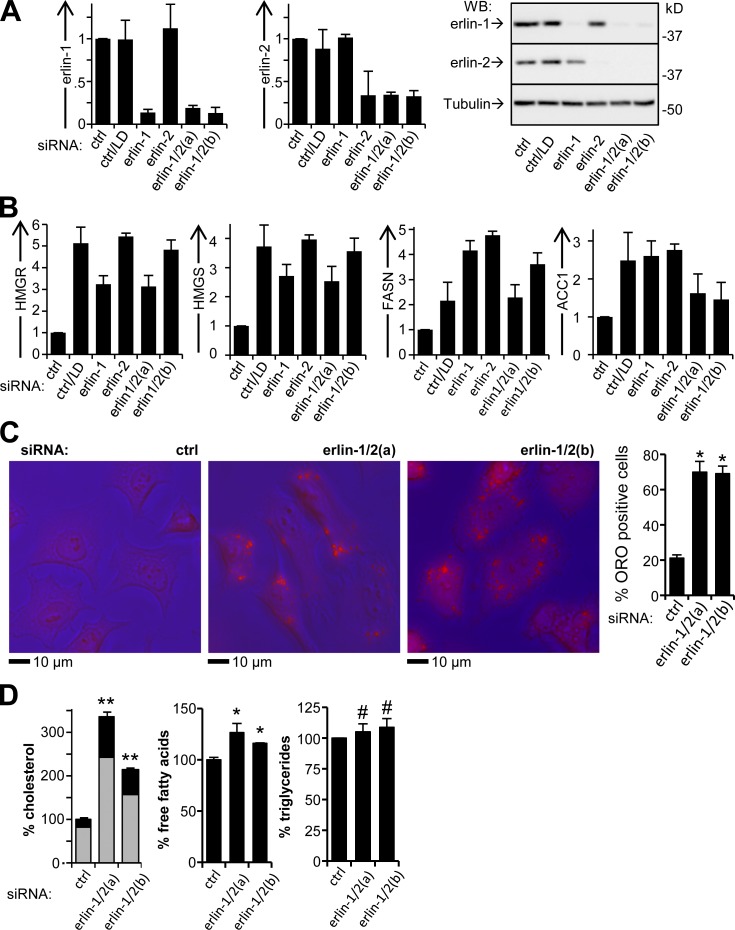
We analyzed whether siRNA-mediated knockdown of erlin-1 and/or erlin-2 in HeLa cells affects the activity of SREBPs under conditions of cholesterol sufficiency (Fig. 1). As a positive control for SREBP activation, lipid-depleted cell cultures were analyzed in parallel (Sakai et al., 1996). This condition strongly activates both SREBP-2, which controls genes governing cholesterol biosynthesis, and SREBP-1a, which regulates genes for both cholesterol and fatty acid biosynthesis (Hannah et al., 2001). Whereas erlin levels were strongly reduced with targeting siRNAs, they were not affected by lipid depletion (LD; Fig. 1 A).
Figure 1.
SREBP target gene activation and lipid accumulation in cells with erlin depletion. (A) Silencing of erlin-1 and -2 with siRNA. HeLa cells were transfected with erlin-targeting or control (ctrl) siRNAs and incubated in complete medium or with LD, and were analyzed by q-RT-PCR (left and middle) and Western blotting (right) for erlin mRNA and protein levels. (B) q-RT-PCR of SREBP target genes. P-values for all comparisons to control <0.031; n = 5. (C) Oil Red O (ORO) staining of siRNA-transfected cells. Overlays of phase contrast (blue) and red fluorescence (left) and quantification of Oil Red O staining (right). *, P < 10−4; n = 3. (D) Quantification of cellular lipids in erlin-depleted cells. (left) Esterified cholesterol (gray sections) is the difference between total cholesterol and free cholesterol (black). Values were normalized to phosphoglyceride levels and expressed relative to control. **, P < 10−4; *, P < 4 × 10−4; #, P > 0.36; n = 3. Error bars indicate standard deviations.
To functionally assess SREBP activation, we used quantitative RT-PCR (q-RT-PCR) to determine mRNA levels for well-established SREBP transcriptional targets (Fig. 1 B). These included two genes involved in cholesterol biosynthesis that are activated by both SREBP-2 and SREBP-1a, HMGR and 3-hydroxy-3-methylglutaryl-CoA synthase (HMGS), and two genes involved in fatty acid synthesis that are preferentially activated by SREBP-1a, fatty acid synthase (FASN) and acetyl-CoA carboxylase 1 (ACC1; Horton et al., 2002). The transcripts for HMGR and HMGS were elevated to ∼300–500% of the control level by siRNAs that silenced either one or both erlins. Similarly, the levels of FASN and ACC1 mRNAs were increased to ∼200–300% of the control. These changes closely paralleled the effects seen with LD (Fig. 1 B). Erlin silencing in HepG2 hepatoma cells grown with cholesterol sufficiency led to a similar up-regulation of the genes reporting activation of the SREBP-1a and SREBP-2 pathways (Fig. S1 A).
We verified that erlin depletion did not induce an ER stress response that might activate SREBPs indirectly (Lee and Ye, 2004) by examining the splicing of XBP1 mRNA (Walter and Ron, 2011). Whereas treatment with the ER stressor tunicamycin resulted in the shift of all detectable XBP1 mRNA to the spliced form (Fig. S1 B), only the unspliced form of XBP1 was detectable in erlin-silenced cells, similar to control cells (Fig. S1 B). Moreover, the tubuloreticular morphology of the ER seen in live HeLa cells did not detectably change with erlin loss (Fig. S1 C).
Consistent with the transcriptional activation of SREBP target genes, erlin silencing caused a substantial increase in the frequency of cells containing cytoplasmic lipid inclusions stained with Oil Red O (Fig. 1 C), sites for accumulation of excess cholesterol such as cholesteryl esters (Chang et al., 2006; Walther and Farese, 2009). Likewise, the end products of the biosynthetic pathways activated by SREBPs were increased by erlin silencing (Fig. 1 D). Cellular cholesterol was increased to ∼200–230% of the control level (Fig. 1 D, left) with little change in the proportion of free and esterified cholesterol. Free fatty acid was increased to 115–125% of the control (Fig. 1 D, middle), although triglyceride levels were unchanged (Fig. 1 D, right).
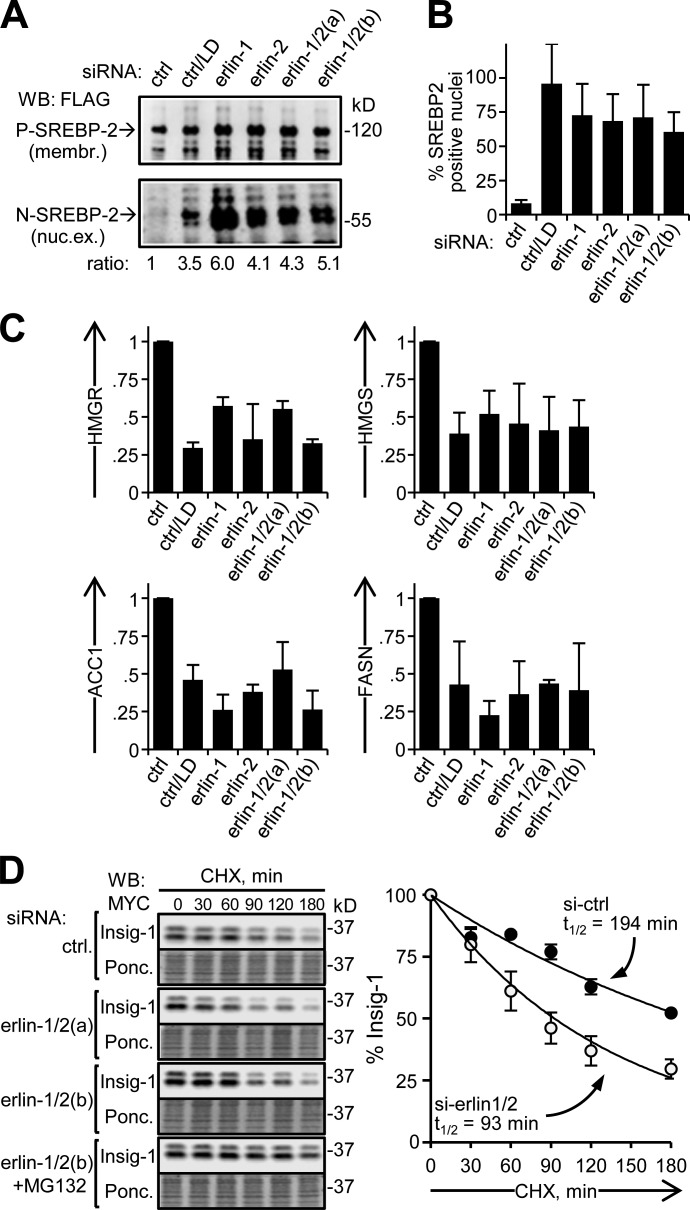
We next investigated whether the activation of SREBP target genes by erlin silencing under cholesterol sufficiency involves the canonical pathway that is normally induced by cholesterol depletion (Goldstein et al., 2006; Fig. 2). First, we found that silencing of erlin-1 and/or erlin-2 led to the appearance of proteolytically cleaved SREBP-2 in nuclear extracts, as seen with LD (Fig. 2 A). Correspondingly, SREBP-2 accumulated in the nucleus (Fig. 2 B and Fig. S2). Second, we determined that SREBP target gene activation with erlin silencing involves Scap. Fatostatin, a small molecule that binds to Scap and inhibits ER to Golgi transport of SREBP (Kamisuki et al., 2009), reduced SREBP target gene activation to similar extents for both erlin knockdown and LD (Fig. 2 C). Finally, knockdown of erlins increased the rate of proteasomal degradation of Insig-1 (Fig. 2 D), similar to the destabilization of Insig induced by cholesterol depletion (Gong et al., 2006). Together these data indicate that activation of SREBP target genes with erlin silencing occurs by the canonical Scap-mediated pathway. This implies that erlins inhibit the production of cholesterol by restraining SREBP activation. These cholesterol-limiting effects could be amplified by the ability of erlin-2 to promote ERAD of HMGR (Jo et al., 2011).
Figure 2.
Validation of SREBP activation with erlin depletion. (A) SREBP-2 processing. HEK293 cells were transfected with FLAG-SREBP-2 and siRNA, and membrane (membr.) and nuclear extract (nuc. ex.) fractions were analyzed by Western blotting. Lipid-depleted control, second lane. Membrane fractions showing SREBP-2 precursor (P-SREBP-2; ∼120 kD; top) and nuclear extracts showing the processed form of SREBP-2 (N-SREBP-2; ∼60 kD; bottom). N-SREBP-2/P-SREBP-2 ratios, relative to the control, are indicated. (B) Nuclear accumulation of SREPB-2. HeLa cells were examined by immunofluorescence with an antibody to the N-terminal domain of endogenous SREBP-2 and scored for nuclear SREBP-2 enrichment (nuclear/cytoplasmic ratio >1.8; see Fig. S2). P-values for all comparisons to control ≤1.2 × 10−6; n = 3. (C) Inhibition of SREBP target gene activation with fatostatin. Sets of siRNA-transfected HeLa cultures were treated with either vehicle (DMSO) or fatostatin. Lipid-depleted control, second lane. Values shown are average ratios of relative mRNA levels in fatostatin- versus DMSO-treated samples. P-values for all comparisons to control <0.027; n = 3. (D) Insig turnover in erlin-depleted cells. HEK293 cells transfected with Insig-1-Myc and siRNAs were treated with cycloheximide (CHX) for the times indicated (and, where specified, with MG132) and analyzed by Western blotting. Shown are Western blots (top panels) and corresponding Ponceau S–stained membranes (Ponc.; bottom panels) of a typical experiment. Insig-1 signals were quantified, normalized to protein loaded, and plotted to determine t1/2 values for Insig-1 turnover. (right) Closed circles, control; open circles, erlin depleted. P-values for si-erlin versus si-control data points at 60–180 min are <0.003; n = 4. Error bars indicate standard deviations.
To determine whether the SREBP regulatory machinery is physically associated with erlins, we analyzed immunoprecipitates (IPs) of endogenous erlins from cell lysates (Fig. 3 A). Under conditions of cholesterol sufficiency, we found that Insig-1, Scap, SREBP-1a, and SREBP-2 all coimmunoprecipitated (coIPed) with erlin-2 (Fig. 3 A) and the associated erlin-1 (Fig. S3 B). We also detected coIP of the ubiquitin E3 ligase gp78 with erlin-2 as reported previously (Pearce et al., 2007; Jo et al., 2011). The coIP of these proteins with erlins was specific because three other transmembrane proteins of the ER (derlin-1, Sec61α, and calnexin) as well as the peripheral ER protein p97 were not detectable in erlin IPs (Fig. 3 A). Moreover, after cholesterol depletion, Insig-1, Scap, SREBP-1a, and SREBP-2 were undetectable or strongly diminished in erlin IPs (Fig. 3 A). However, gp78 association was unchanged as seen before (Pearce et al., 2007; Jo et al., 2011). This supports the model that a physical association of erlins with SREBP–Scap–Insig is functionally relevant to regulation of the SREBP pathway.
Figure 3.
Association of erlins with the SREBP regulatory machinery. (A) CoIP of SREBP pathway proteins with endogenous erlin-2 in lysates of HeLa cells grown with complete (F) or LD medium. Samples were probed by Western blotting as indicated. CANX, calnexin. 10 times more cell equivalents were loaded in IP lanes compared with input, except for erlin-2 blot (1 equivalent). Western blot images are typical of three separate repeats. Numbers below the panel indicate LD/F ratios of IP target proteins averaged from three experiments. P-values for LD versus complete (F): Scap, Srebp-1, and Srebp-2, <3.5 × 10−4; erlin-2 and gp78, >0.61. (B) Diagram of the inducible erlin clustering system. (C) Fluorescence images showing Insig localization to induced erlin clusters. Cells expressing the indicated combinations of recombinant proteins were incubated without (left column) or with (all other columns) rapamycin. Corresponding GFP and red mCherry fluorescent images of a representative cell from each condition are shown in grayscale. Arrows indicate some Insig-1-mCherry foci colocalized with erlin-GFP foci. (D) CoIP of recombinant erlin and SREBP–Scap–Insig. Lysates of 293T cells expressing various combinations of erlin-2-V5, Insig-1-Myc, hamster Scap (cgScap), and FLAG-SREBP-2 were IP with anti-V5 antibody. Input (Inp.; 1 equivalent) and IPed material (IP; 10 equivalents, except for erlin-2 blot [1 equivalent]) was analyzed by Western blotting. Certain transfections (asterisk) received four times more Insig-1-Myc cDNA to compensate for the instability of Insig in the absence of overexpressed Scap (Gong et al., 2006). (E) CoIP of recombinant erlin and Scap with Insig-1 depletion. Experiments were performed as in D with the indicated sample depleted of endogenous Insig-1 with siRNA. Insig-1 mRNA levels determined by q-RT-PCR are shown in the bar graph (bottom). Error bars indicate standard deviations.
We extended this analysis by examining cells that ectopically overexpressed erlin-2 and Insig-1, Scap, and SREBP-2 in various combinations (Fig. 3 D). IPs of erlin-2 contained similar levels of ectopic Insig-1, whether or not Scap and/or SREBP-2 were coexpressed. When ectopic erlin-2 and Scap were expressed without Insig-1, a low level of Scap coIPed with erlins. Endogenous Insig-1 was responsible for at least most of this coIPed Scap because it diminished approximately threefold after knockdown of Insig-1 (Fig. 3 E). In contrast, the amount of coIPed Scap increased approximately fourfold when Insig-1 was expressed together with Scap (Fig. 3, D and E). Finally, ectopic SREBP-2 was found in erlin-2 IPs only when Scap–Insig was coexpressed. These results suggest that Insig-1 is a more proximal binding partner of erlins that stimulates binding of SREBP–Scap to the erlin–Insig complex. However, decisive resolution of this issue, and whether the interactions of erlins with the SREBP–Scap–Insig complex is direct or involves bridging proteins, awaits further analysis.
We used wide-field fluorescence microscopy of erlin-GFP and Insig-mCherry to examine whether erlins and Insig colocalize in live cells. We were unable to visualize discrete erlin complexes by conventional imaging. Therefore, we developed a rapamycin-inducible system (Klemm et al., 1997) to cluster erlin multimers into resolvable units (Fig. 3 B). We monitored Insig association with these foci (Fig. 3 C) and used quantitative methods to characterize the sample set (Fig. S3).
Both erlin-GFP and Insig-mCherry were distributed in an ER-like pattern in untreated cells. However, 20 min after rapamycin addition numerous distinctive foci of GFP appeared (Figs. 3 C and S3 D). Insig-1-mCherry foci also were observed after the addition of rapamycin (Fig. 3 C). We found that 64% (±5.5%) of the Insig-mCherry foci colocalized with conspicuous erlin-GFP foci (three experiments, n = 55 cells). When the assay contained calnexin-mCherry instead of Insig-1-mCherry, we never observed mCherry foci in cells with erlin-GFP foci (Fig. 3 C; n = 28 cells). Finally, no rapamycin-induced GFP or mCherry foci were obtained in cells transfected with erlin-2-GFP and Insig-1-mCherry only (Fig. 3 C; n = 38 cells). The specific coclustering of Insig-1 and the erlin complex in this live-cell assay provides further evidence for a physical interaction between these components.
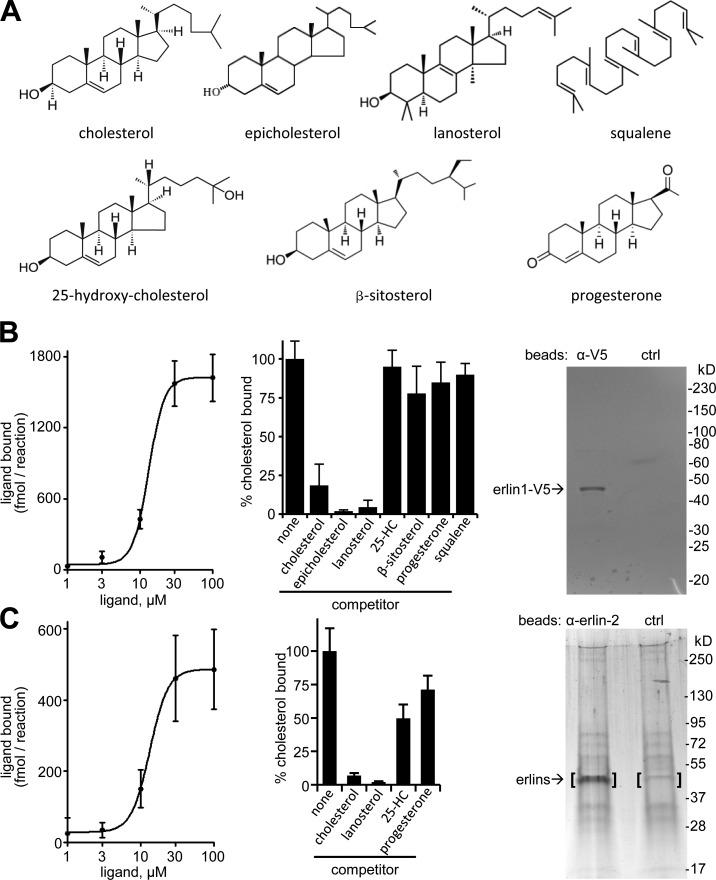
Because SREBP activation is modulated by binding of cholesterol to Scap (Goldstein et al., 2006), and because erlin interactions with the SREBP machinery appear to be sensitive to changes in cholesterol (Fig. 3 A), we examined whether cholesterol binds to erlins (Fig. 4; see Materials and methods). We found that cholesterol bound to recombinant erlin-1 with an apparent Kd of 13.2 µM and approximately six molecules of cholesterol per erlin monomer at saturation (Fig. 4 B). The binding curve fit best to a sigmoidal function (r2 = 0.98) with a Hill coefficient of 4.05, indicating a high degree of binding cooperativity. The binding of radiolabeled cholesterol to erlin-1 was specific, as it was strongly diminished by a 10-fold excess of unlabeled cholesterol, epicholesterol, or lanosterol, but was not substantially reduced by progesterone, 25-hydroxycholesterol, β-sitosterol, or squalene (Fig. 4 B). We also found that the native erlin-1/2 complex isolated from murine liver microsomal membranes saturably bound cholesterol (Fig. 4 C; apparent Kd = 13.4 µM; approximately four molecules of cholesterol bound per molecule of erlin monomer at saturation). Here the binding curve was best described by a sigmoidal function (r2 = 0.91) with a Hill coefficient of 3.47. Competition assays showed sterol binding specificity similar to recombinant erlin-1 (Fig. 4 C). Supporting our results, erlin-2 was specifically labeled with a photoreactive cholesterol analogue in a proteome-wide screen (Hulce et al., 2013). Cholesterol binding by erlins may occur outside the lipid bilayer as suggested for NPC1 (Infante et al., 2008a) and Scap (Motamed et al., 2011) or may involve a segment of erlins with close bilayer interaction.
Figure 4.
Cholesterol binding by erlins. (A) Structures of cholesterol and related compounds used. (B) Analysis of purified recombinant erlin-1 with cholesterol binding and competition. Cholesterol binding curve (left), competition of 5 µM of radiolabeled cholesterol by 50 µM of the indicated nonlabeled compounds (middle), and analysis of purified erlin-1-V5 by SDS-PAGE and Coomassie blue staining (right). Shown is representative material eluted from anti-V5 beads (left lane) and control IgG beads (right lane). Densitometry established >90% purity for erlin-1-V5. Error bars indicate standard deviations. (C) Binding and competition of cholesterol to the erlin complex from mouse liver microsomal membranes. Left and middle panels are the same as in B. (right) SDS-PAGE with silver staining of material eluted from anti–erlin-2 beads (left lane) and control beads (right lane). The bracketed gel areas were analyzed by mass spectrometry. Erlin-1 (14.2% sequence coverage) and erlin-2 (18.2% sequence coverage) were found only in the anti–erlin-2 sample.
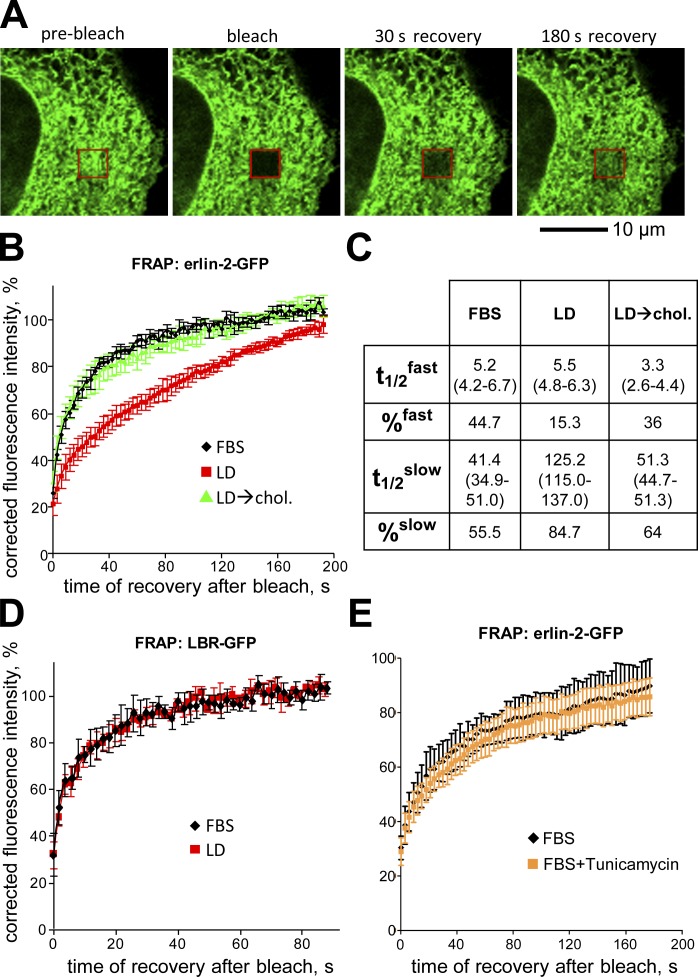
We used FRAP to examine whether the physical state of the erlin complex is sensitive to ER cholesterol levels (Fig. 5, A and B). We compared the diffusional mobility of erlin-2-GFP in cells grown in cholesterol replete (FBS) versus lipid-depleted conditions. In both cases, the FRAP recovery curves fit a two-phase exponential equation (r2 > 0.99), describing erlin-2 pools with “fast” and “slow” recovery kinetics (Fig. 5 C). In FBS, approximately half of the erlin-2-GFP was in a slow pool. After cholesterol depletion nearly all of it was in a slow pool, albeit with substantially slower recovery kinetics than the FBS slow pool. After restoration of cholesterol, both fast and slow pools of erlin-2-GFP had FRAP recovery times similar to cholesterol-replete cultures. Cholesterol-dependent changes in diffusional mobility were not observed for the model ER transmembrane protein laminin B receptor (LBR)–GFP (Ellenberg et al., 1997), showing that the effect on erlin-2 is selective (Fig. 5 D). Moreover, the effect cannot be ascribed to an ER stress response because there was no significant change in the mobility of erlin-2 in tunicamycin-treated cells (Fig. 5 E). Although we have not yet determined the molecular basis for the changes in erlin mobility with cholesterol depletion, our data nonetheless show that the physical properties of erlins are altered by standard conditions that induce SREBP pathway activation.
Figure 5.
Cholesterol-dependent changes in diffusional mobility of erlin-2. (A) Confocal images during FRAP of a HeLa cell expressing erlin-2-GFP. Shown are images before fluorescence bleaching (pre-bleach) or at the indicated times after bleaching (red box). (B) Plot showing FRAP of erlin-2-GFP under conditions of cholesterol sufficiency (FBS; black), cholesterol depletion (LD; red), or cholesterol depletion followed by incubation with 50 µM cholesterol–β-MCD complex for 3 h (LD→chol.; green). Shown are averaged data from n = 7 cells. (C) Table summarizing erlin-2-GFP FRAP data. t1/2 and percentage values were calculated from two-phase association functions best describing the FRAP curves shown in B. (D) Plot of FRAP of LBR-GFP under lipid-sufficient (FBS; black) and lipid-depleted (LD; red) conditions; n = 5 cells. (E) Plot of FRAP of erlin-2-GFP under lipid-replete conditions without (FBS; black) or with tunicamycin (FBS+Tunicamycin; orange) to induce ER stress; n = 6 cells. Error bars indicate standard deviations.
In summary, our work has identified erlins as novel ER regulators of SREBPs that are important for cholesterol homeostasis. Because erlins bind cholesterol and physically interact with SREBP–Scap–Insig, we suggest that they directly promote ER retention of SREBP–Scap. Several nonmutually exclusive models could explain this effect. First, erlin multimers could provide a scaffold for maintaining the integrity of the SREBP–Scap–Insig complex. Second, erlins could catalytically promote the structural states of Scap and/or Insig that are competent to retain SREBPs in the ER. Third, erlins might negatively regulate proteasomal degradation of Insig. Alternatively, cholesterol association with erlins might nucleate the formation of cholesterol-rich microdomains in the ER that stimulate the interaction of Insig with SREBP–Scap. This would be analogous to the concept of cholesterol-rich “rafts” in the plasma membrane as regulators of membrane protein functions and signaling (Lingwood and Simons, 2010). In any case, the highly cooperative binding of cholesterol to erlins might stabilize an erlin conformation that could support the functional output we have defined and contribute to the switch-like cooperative regulation of the SREBP system by cholesterol (Radhakrishnan et al., 2008).
Materials and methods
DNA constructs
pLBR-GFP for the expression of the 238–amino acid N-terminal portion of LBR fused to GFP (Ellenberg et al., 1997) was obtained from H. Worman (Columbia University, New York, NY). pSec61-GFP for the expression of Sec61α-GFP under control of a CMV promoter (Greenfield and High, 1999) was obtained from T. Rapoport (Harvard Medical School, Boston, MA). pCMV-Insig-1-Myc for the expression of C-terminally MYC epitope–tagged human Insig-1 was obtained from the American Type Culture Collection (deposited by J.L. Goldstein, University of Texas Southwestern Medical Center, Dallas, TX). pcDNA-HSV-cgScap-T7 was constructed by lifting the desired coding sequence from pTK-HSV-SCAP-T7 (Hua et al., 1996; American Type Culture Collection) using PCR with specific primers, adding the four nontranslated nucleotides (CACC) in front of the start codon to make the PCR product compatible with directional TOPO cloning into pcDNA3.1/V5-His/D-TOPO (Invitrogen). The resulting construct allowed expression of the tagged hamster Scap protein (cgScap) under control of a CMV promoter. pcDNA-FLAG-SREBP-2 was constructed by PCR amplifying the coding sequence for human SREBP-2 from template pTK-HSV-SREBP-2 (Hua et al., 1996; provided by J. Ye, University of Texas Southwestern Medical Center, Dallas, TX) with specific primers adding an N-terminal FLAG tag, a stop codon immediately downstream of the ORF, and four nontranslated nucleotides (CACC) in front of the start codon to make the PCR product compatible with directional TOPO cloning into pcDNA3.1/V5-His/D-TOPO. This construct allowed expression of the tagged protein FLAG-Srebp-2 under control of a CMV promoter. Plasmid pErlin-2-GFP was constructed as follows. Human erlin-2 cDNA was amplified from IMAGE clone 5296776 (Thermo Fisher Scientific) by PCR and cloned into pCR2.1-TOPO (Invitrogen) to result in pCR-erlin-2. From there, the SalI–BamHI fragment containing the erlin-2 coding sequence was transferred into pEGFP-N1 (Takara Bio Inc.) to yield a C-terminal GFP fusion protein under control of a CMV promoter. pcDNA-erlin-1-FKBP-V5 was constructed by first PCR amplifying the ORF for human erlin-1 from clone #4214940 (Thermo Fisher Scientific) using specific oligonucleotides and adding a glycine-serine (GSGSG) linker at the C terminus of erlin-1 and four nontranslated nucleotides (CACC) in front of the erlin-1 start codon to make the PCR product compatible with directional TOPO cloning into pcDNA3.1/V5-His/D-TOPO. This created pcDNA-erlin-1. To produce pcDNA-erlin-1-FKBP, a PCR fragment encoding three copies of FKBP flanked by unique XbaI and XhoI sites was PCR amplified from a plasmid DNA template (Klemm et al., 1997) (Ohba et al., 2004). This PCR product was digested with restriction enzymes XbaI and XhoI and cloned into pcDNA-erlin-1 via identical restriction sites. Construct pcDNA-erlin-1-FKBP-V5 was created by linearizing pcDNA-erlin-1-FKBP with XhoI, filling in the resulting 3′ recessed ends, and religating the DNA resulting DNA. This adjusts the sequence so that erlin-1-FKBP was brought in frame with a C-terminal V5 epitope tag for the expression of erlin-1-FKBP/V5 under control of a CMV promoter. pcDNA-erlin-2-GFP-FRB was constructed by PCR amplifying the ORF encoding erlin-2-GFP of pErlin-2-GFP with specific primers, adding a GSGSG linker sequence to the C terminus of erlin-2-GFP and four nontranslated nucleotides (CACC) in front of the erlin-2 start codon to make the PCR product compatible with directional TOPO cloning. This PCR product was cloned into pcDNA3.1/V5-His/D-TOPO to produce pcDNA-erlin-2-GFP. To fuse erlin-2-GFP to the FRB domain of mTOR (Klemm et al., 1997), a PCR product encoding FRB was amplified from a DNA template (Ohba et al., 2004) using primers that introduced unique NotI and XbaI sites 5′ and 3′ of the FRB coding sequence, respectively. This PCR product was digested with NotI and XbaI and cloned into the identical restriction sites in pcDNA-Erlin-2-GFP to create pcDNA-erlin-2-GFP-FRB for the expression of erlin-2-GFP-FRB under control of a CMV promoter. pcDNA-Insig-1-mCherry was constructed as follows. First, the region of clone #3794 (Thermo Fisher Scientific) harboring the full-length human Insig-1 coding sequence was amplified by PCR with specific primers. The reverse primer added a SalI restriction site immediately 3′ of the last codon of Insig-1. This fragment was cloned into pCR2.1 (Invitrogen) to result in pCR-Insig-1. Second, the region of plasmid pmCherry-N1 (Takara Bio Inc.) encoding the red fluorescent protein mCherry was amplified by PCR with specific primers, adding a SalI restriction site and a sequence encoding a GSGSG linker immediately upstream and in frame with the CDS for mCherry. This PCR product was cloned into pCR2.1 to create pCR-mCherry. Next, the SacI–SalI fragment of pCR-Insig-1 harboring the ORF for human Insig-1 was ligated into pCR-mCherry via identical restriction sites, fusing Insig-1 in frame with mCherry in the resulting construct, pCR-Insig-1-mCherry. This construct was used as a template to PCR amplify the Insig-1-mCherry ORF with flanking primers, adding a sequence encoding three consecutive FLAG tags followed by a translational stop codon after the last codon of mCherry sequence and the four nontranslated nucleotides (CACC) in front of the start codon for Insig-1 to allow directional TOPO cloning of the PCR product into pcDNA3.1/V5-His/D-TOPO. The resulting construct, pcDNA-Insig-1-mCherry, allowed expression of the red fluorescent fusion protein under control of a CMV promoter. pcDNA-CANX-mCherry was constructed by PCR amplifying full-length human calnexin (CANX) coding sequence from clone #4342723 (Thermo Fisher Scientific) with specific primers, introducing a SalI restriction site immediately after the last codon of the ORF for CANX. The PCR product was cloned into pCR4 (Invitrogen) to result in pCR4-CANX. The SalI–SpeI insert of pCR-mCherry (see above) was ligated into the SalI–SpeI–digested pCR4-CANX to create an in-frame fusion of CANX with mCherry in pCR4-CANX-mCherry. From here, a 2.2-kb EcoRI–SbfI restriction fragment was purified and the EcoRI site was filled in to generate a blunt DNA end. This fragment was ligated with the SbfI–HindIII backbone portion of pcDNA-INSIG-1-mCherry after blunting the HindIII site of the vector fragment to make it ligatable with the blunted EcoRI site of the CANX-mCherry insert. This created pcDNA-CANX-mCherry for the expression of the red fluorescent fusion protein under control of a CMV promoter. All constructs were verified by DNA sequencing (Genewiz).
Cell culture and RNAi
HeLa, HEK293, HEK293T, and HepG2 cells (American Type Culture Collection) were maintained in complete medium (DMEM, 10% FBS, 2 mM glutamine, 100 µg/ml penicillin, and 100 µg/ml streptomycin). Unless stated otherwise, for acute LD, cells were cultured in LD medium (same as above, but with 5% LPDS instead of FBS) for 24 h and then treated with 50 µM lovastatin for 4 h.
siRNAs targeting human Insig-1 (5′-CCCACAAAUUUAAAGAGA-3′; Jo et al., 2011), erlin-1 (siRNA erlin-1, 5′-CCACAAATAGGAGCAGCAT-3′) or -2 (siRNA erlin-2, 5′-GCCTCTCCGGTACTAACAT-3′) individually, or erlin-1 and -2 simultaneously (siRNA erlin-1/2(a); 5′-AGAAGCAATGGCCTGGTAC-3′) and the nontargeting control siRNA (control, 5′-ACTGTCACAAGTACCTACA-3′; Pearce et al., 2007) were purchased from Integrated DNA Technologies. The second reagent for double depletion of erlin-1 and -2, termed siRNA erlin-1/2(b), was an equimolar mix of siRNAs erlin-1 and 2. Unless stated otherwise, siRNA and DNA transfections were performed on cells resuspended in complete medium after trypsinization (reverse transfection). In a typical knockdown experiment. 0.4 ml of a HeLa cell suspension (3 × 105 cells) was added to a preformed complex of 40 pmol siRNA and 4 µl Dharmafect-3 transfection reagent, incubated for 10 min, and seeded on 6-well plates prepared with 2 ml of complete medium. Fresh medium was applied after overnight incubation, and cells were assayed 48 h after transfection.
q-RT-PCR
cDNA was synthesized from 1 µg of total RNA (RNeasy Plus kit; QIAGEN) using the Transcriptor First Strand cDNA Synthesis kit (Roche) and analyzed by q-RT-PCR with the use of 2× Power SYBR Green PCR Master Mix (Applied Biosystems) on a Prism 7900HT (Applied Biosystems) under default instrument settings. Transcripts of interest (RefSeq accession numbers are given in parentheses) were detected using the following oligonucleotides primer pairs (forward/reverse): erlin-1 (NM_001100626.1), 5′-GAAAGCTCACTCCCCTCTAAG-3′/5′-TGTTCCCACTTAACCCCTTG-3′; erlin-2 (NM_007175.6), 5′-ACGCTTCAAGAGGTCTACATTG-3′/5′-ATTGCCTCTGGTATGTTGGG-3′; Insig-1 (NM_005542.4), 5′-CTTCCCCGAGGAGGTGAT-3′/5′-GGCCCATTCTCTCTTAAATTTGTG-3′; HMGR (NM_000859.2), 5′-TGACCTTTCCAGAGCAAGC-3′/5′-CCAACTCCAATCACAAGACATTC-3′; HMGS (NM_001098272.1), 5′-CATTAGACCGCTGCTATTCT-3′/5′-AGCCAAAATCATTCAAGGTA-3′; ACC1 (NM_198834.1), 5′-GAGGGAAGGGAATTAGAAAA-3′/5′-CTTGAACCTGTCTGAAGAG-3′; FASN (NM_004104.4), 5′-CAGAGTCGGAGAACTTGCAG-3′/5′-GGAGGCATCAAACCTAGACAG-3′; HPRT1 (NM_000194), 5′-TTCCAGACAAGTTTGTTGTAGGAT-3′/5′-GCAGATGGCCACAGAACTAG-3′. Target gene expression was normalized to HPRT1 expression and is shown relative to control samples (ΔΔCt method [Livak and Schmittgen, 2001]).
Western blot analysis
Western blotting was done following a standard procedure (Sambrook et al., 1989) using NuPAGE gels (Invitrogen) and nitrocellulose membranes (Whatman). Antibodies detecting tubulin (Abcam), calnexin (BD), derlin-1 (Cell Signaling Technology), SREBP-1 (BD), SREBP-2 (BD), gp78 (Cell Signaling Technology), p97 (Cell Signaling Technology), Sec61 (Abcam), FLAG epitope (Flag-M2; Sigma-Aldrich), Myc epitope (Sigma-Aldrich), hamster Scap (clone 9D5; Santa Cruz Biotechnology, Inc.), human Scap (Bethyl Laboratories, Inc.), or the V5 epitope tags (Invitrogen) were purchased from commercial sources. Erlin-2 was detected with our in-house generated rabbit polyclonal antibody that was generated and affinity purified against the full-length immunogen. The erlin-1 7D3 monoclonal antibody was a gift of S. Robbins (University of Calgary, Alberta, Canada). Blots were developed by chemilumiscence. Detection and quantitative analyis was performed using digital imaging systems (UVP; Alpha Innotech) and their native software applications.
Cellular lipid determination
Lipid droplets were detected by Oil Red O staining adapted from Goodpaster et al. (2000) and Koopman et al. (2001). In brief, the control and erlin targeting siRNA transfected HeLa cultures were grown on 6-well cell culture dishes, washed with PBS, and fixed with 10% paraformaldehyde in PBS (diluted from a 16% methanol-free paraformaldehye stock solution; Electron Microscopy Sciences). The wells were washed with water, equilibrated into 60% isopropanol, and stained with Oil Red O solution (0.12% Oil Red O in 60% isopropanol; Electron Microscopy Sciences) for 10 min. Samples were washed briefly with water and covered with 50% glycerol in PBS. Images were acquired on a DM-IRE2 epifluorescence microscope (Leica) equipped with a CC4742-95 digital CCD camera (Hamamatsu Photonics). Phase-contrast images were taken to identify cells. Oil red O was visualized by fluorescence imaging in the Texas red channel.
To quantify cellular lipids, cells were extracted with chloroform/methanol (2:1), evaporated to dryness, and dissolved in 50 µl methanol. This extract was used for fluorimetric assays to determine levels of total cholesterol (Amplex red cholesterol assay kit; Invitrogen), free fatty acids (Free fatty acid assay kit; Cayman), and triglycerides (Triglyceride assay kit; Cayman). For determination of unesterified cholesterol, the Amplex red cholesterol assay was performed as prescribed, but with omission of cholesterol esterase from the reaction. Total phospholipid content was quantified by ammonium ferrothiocyanate complex assay (Stewart, 1980; Chen et al., 2006). Here, small samples of the methanol extracts (see above) were diluted with chloroform to a total volume of 150 µl and mixed with an equal volume of aqueous ammonium ferrothiocyanate solution formed from 27 g/l FeCl3 × 6 H2O and 30.4 g/l NH4SCN. The amount of water-insoluble complex drawn into the organic phase of the reaction was determined spectrophotometrically by measuring absorbance at a wavelength of 460 nm. Phospholipid concentrations were determined by comparison to a standard curve produced with L-α-phosphatidylcholine dissolved in chloroform (Sigma-Aldrich).
Proteolytic SREBP-2 activation assay
For SREBP-2 cleavage studies (Fig. 2 A), ∼1.5 × 107 HEK293 cells were transfected with 50 µg pcDNA-FLAG-SREBP-2 and 50 µg pcDNA using Optifect reagent (Invitrogen) and seeded in two individual T164 cm2 cell culture flasks. After 38 h cells were trypsinized, reverse transfected with 200 pmol siRNA and Dharmafect-1 reagent (Thermo Fisher Scientific), and seeded on six individual 10-cm dishes. After another 24 h, this procedure was repeated with 150 pmol siRNA and Dharmafect-3. 6 h after this last transfection the cell culture medium was changed to fresh DMEM with either 10% FBS, 5% LPDS, or 50 µM compactin (mevastatin; Sigma-Aldrich) and 50 µM sodium mevalonate (Sigma-Aldrich) for LD. 20 h later, 25 µg/ml ALLN (Sigma-Aldrich) was added to each dish. Cells were incubated for another 3 h and subjected to subcellular fractionation, essentially as described previously (Hua et al., 1996). Each 10-cm culture dish was washed with PBS and subsequently with hypotonic buffer B (10 mM Hepes-KOH, pH 7.4, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM EDTA, 0.5 mM EGTA, 1 mM DTT, 25 µg/ml ALLN, and protease inhibitor cocktail [Roche]). Next, cells were swollen on the dish with 1 ml buffer B for 10 min at 4°C. Thereafter, cells were scraped off the dish, collected, and disrupted by forcing the suspension through a 22-gauge needle attached to a small syringe (10 repetitions). Intact nuclei were collected by centrifugation at 1000 g for 5 min, extracted by incubation with 150 µl buffer C (10 mM Hepes-KOH, pH 7.4, 0.45 M NaCl, 2.5% vol/vol glycerol, 1.5 mM MgCl2, 0.5 mM EDTA, 0.5 mM EGTA, 1 mM DTT, 25 µg/ml ALLN, and protease inhibitor cocktail) at 4°C for 30 min, and separated from insoluble nuclear material by centrifugation at 20,000 g for 10 min. Microsomal membranes were isolated from post-nuclear supernatants by centrifugation at 100,000 g for 30 min at 4°C. Membrane pellets and nuclear extracts were adjusted with SDS-PAGE loading dye to contain 2% SDS. Equal amounts of protein from each fraction and experimental conditions were analyzed by Western blotting with anti-FLAG antibody to detect the N terminus of FLAG-SREBP-2.
Cycloheximide chase analysis
For Insig-1 turnover experiments (Fig. 2 D), ∼2.5 × 107 HEK293 cells were transfected with 100 µg pCMV-Insig-1-MYC using Optifect (Invitrogen) and cultured on 500-cm2 dishes. 38 and 54 h later the cells received control or erlin targeting siRNAs (10 pmol/105 cells) by reverse transfection with Dharmafect-1 reagent. At the time of the second siRNA transfection, aliquots of 3 × 105 cells were seeded in wells of 6-well plates. After 21 h, 100 µg/ml cycloheximide (Sigma-Aldrich) was added. Where desired, 20 µM of the proteasomal inhibitor MG132 (Sigma-Aldrich) was added to a set of wells 3 h before CHX treatment. Individual wells were washed with PBS and harvested into SDS-PAGE loading dye at the time points indicated. Samples were analyzed by Western blotting with anti-Myc antibody.
IP
For IP of endogenous erlin complexes, we used nearly confluent cultures of HeLa cells that had been transfected with a small quantity of pCMV-Insig-1-Myc to express trace amounts of the tagged protein. This allowed unambiguous identification of Insig-1 with anti-Myc antibodies because commercial anti–Insig-1 antibodies showed lot-to-lot variations in quality. Cells were lysed at ∼107 cells/ml in 50 mM Tris-HCl, pH 7.8, 100 mM NaCl, 1% Triton X-100, and EDTA-free Protease Inhibitor Cocktail; cleared of insoluble components by centrifugation; and incubated with 3 µg of either α–erlin-2 for 2 h at 4°C. Immune complexes were captured on protein G–coupled Dynabeads (Invitrogen) and washed three times with ice-cold lysis buffer. Proteins were eluted with SDS dye and heating to 100°C for 5 min. Interacting proteins were detected directly by Western blotting with specific antibodies.
For IP experiments with recombinant proteins (Fig. 3) the expression constructs were mixed at the combinations indicated (Fig. 3 B) and the amount of DNA per sample was adjusted to 35 µg with a pcDNA vector carrying no cDNA insert (empty vector). The DNA was diluted into 400 µl OptiMEM (Invitrogen) and complexed with 150 µl LF2000 for reverse transfection of ∼2 × 106 293T cells. Cells were seeded on 10-cm culture dishes prepared with 10 ml complete medium and were incubated for 24 h before analysis. Where applicable (Fig. 3 E), nontargeting or Insig-1–specific siRNA (600 pmol per 10-cm dish; ∼50 nM) was introduced into 293T cells 24 h before transfection with the indicated expression constructs. Dishes were washed once with ice-cold IP buffer (50 mM Tris-HCl and 100 mM NaCl, pH 7.8) and cells were collected by scraping. The cell material was adjusted to a total volume of 400 µl with IP buffer, to which was added proteinase inhibitor cocktail and Triton X-100 to a final concentration of 1%. Cells were lysed by brief sonication (5 s) and incubation on ice for 5 min. Lysates were cleared by centrifugation at 16,000 g for 10 min. The resulting supernatant, with protein concentrations ranging from 6 to 10 mg/ml, served as input material for IP. Erlin-2-V5 and associated proteins were precipitated by incubation with 5 µg anti-V5 antibody on ice for 2 h. Formed immune complexes were bound to protein G magnetic beads (Invitrogen) for 45 min on ice, collected, washed thrice with IP buffer containing 0.2% Triton X-100, and eluted into 70 µl of SDS-PAGE loading buffer containing 50 mM DTT by heating in a boiling water bath for 5 min. A 50-µl sample of the input material was carried through the same incubation steps as the IP reaction for accurate comparison of the starting and IPed material. Recombinant erlin-2-V5, FLAG-SREBP-2, and Insig-1-Myc were detected in Western blotting by antibodies against the epitope tags. Hamster (Cricetus griseus) Scap was analyzed using an antibody specific for Scap from this species (labeled cgScap throughout this paper).
Microscopy and image analysis
To evaluate activation of SREBP-2 by immunofluorescence, cells were grown on 35-mm glass bottom cell culture dishes for 36 h, fixed, and labeled with an antibody recognizing the N-terminal transcription factor portion of SREBP-2 (Cayman) followed by anti–rabbit Alexa 568 (Invitrogen) and the DNA dye Hoechst 33342 (Invitrogen). Samples were covered with ProLong Gold (Invitrogen) before imaging. Images were acquired at room temperature using a 40× plan apo objective (1.4 NA) on a DM-IRE2 fluorescent microscope (Leica), and analyzed using ImageJ software v1.43u (Schneider et al., 2012) as follows. To assess nuclear levels of the N-terminal transcription factor portion of SREBP-2, nuclei were identified according to their staining with Hoechst. The nuclear SREBP-2 fluorescence signal was quantified and normalized to the fluorescent DNA signal in the same nucleus (SREBP-2nuc./DNA). To determine nuclear enrichment of SREBP-2, the SREBP-2 fluorescence signal was measured along a 5-pixel-wide line drawn through a cell. This line was split into nuclear and cytoplasmic segments by reference to Hoechst staining. The average pixel intensities of SREBP-2 in the nuclear (SREBP-2nuc.) and cytoplasmic segments (SREBP-2cyt.) were computed and the nuclear/cytoplasmic ratio calculated as SREBP-2nuc./SREBP-2cyt..
Inducible erlin focus formation experiments exploited the rapamycin-dependent interaction between FKBP12 and the FRB domain of mTOR (Klemm et al., 1997). HeLa cells were transfected as described (Fig. 3, C and D) with constructs for the expression of erlin-1-FKBP, erlin-2-GFP-FRB, and Insig-1-mCherry mixed at a 1:1:3 ratio (10 µg DNA total) using 10 µl Lipofectamine 2000 for reverse transfection of 3 × 105 HeLa cells. Cells were seeded and incubated in phenol red–free complete medium on 35-mm glass bottom dishes (MatTek Corporation) for ∼20 h. Where indicated, 200 ng/ml rapamycin was added for 20 min before imaging live cells at room temperature in normal atmosphere. Images were acquired with a 60× Plan-Apochromat (1.4 N.A.) oil immersion objective on an Eclipse TE2000-T fluorescence microscope (Nikon), using a dual excitation light source and a TwinCam dual emission camera system (Cairn Research) for simultaneous acquisition of images in the FITC (erlin-2-GFP-FRB) and Texas red (Insig-1-mCherry and CANX-mCherry) fluorescent channels. Imaging was concluded within 60 min of rapamycin addition. Colocalization between erlin-GFP and Insig-1-mCherry foci was evaluated using NIH ImageJ v1.43u software as follows. Red (Insig-1-mCherry) and green (erlin-2-GFP-FRB) channels of an image were separated into individual grayscale images. Typically ∼6–12 easily identifiable foci in the Insig-1-mCherry image were chosen and outlined by circles 3–6 pixels in diameter. A mask was created with these regions of interest (ROIs) and transferred to the identical position on the erlin-GFP image. If the circles from the red channel surrounded foci in the green channel, this indicated that a corresponding erlin-GFP focus existed and foci were scored as colocalized. Further characterization of this inducible clustering system and fluorescence intensity characterization of erlin-GFP foci is described in Fig. S3.
To assess ER morphology, pSec61-GFP transfected cells grown on 35-mm glass bottom dishes in complete medium (DMEM with 10% FBS) were labeled for 15 min with Hoechst 33342 and imaged live through a 60× plan apo objective (1.4 NA) on a 710 laser scanning confocal microscope (Carl Zeiss) equipped with a live-cell chamber (37°C, humidified 5% CO2 atmosphere). A pinhole setting of 1 Airy Unit was used. Images created by Zen2008 software (Carl Zeiss) were viewed, converted, and cropped in ImageJ.
All images shown are representative of the situation observed during microscopy. Adjustments to brightness and contrast, as far as they were necessary for publication purposes, were minimal, linear, and applied evenly to the entire image.
Erlin purification and lipid binding assay
Recombinant human erlin-1-V5 was expressed in Schneider S2 insect cells using the DES TOPO TA Expression Kit (Invitrogen). In brief, S2 cells stably transfected with a plasmid encoding human erlin-1 and labeled at the C-terminal end with the V5 epitope and a hexahistidine tag, which was under the control of the copper inducible metallothionein promoter (pMT-erlin-1-V5). Cells were grown in suspension to a density of 107 cells/ml before induction of protein expression with 0.5 mM CuSO4 for 48 h. Cells were pelleted and lysed in a buffer of 50 mM Tris-HCl, pH 8, 100 mM NaCl, and 1% Triton X-100. This crude lysate was clarified by centrifugation at 20,000 g for 20 min. The resulting cleared lysate was processed by FPLC purification of the recombinant protein using metal-chelate affinity chromatography. The recombinant protein was highly enriched in a single fraction eluting at 500 mM imidazole.
Erlin-1-V5 was further purified and prepared for lipid binding assays from the imidazole eluate by immunoadsorption. In brief, anti-V5 antibody was cross-linked to protein G dynabeads at 0.1 µg antibody/µl beads, using BS3 (Invitrogen). Non-specific control beads were prepared identically except that normal mouse IgG (Invitrogen) was used. Anti-V5 and control beads were incubated overnight at 4°C with aliquots of the erlin-1-V5–containing imidazole fraction. Beads were then washed twice with buffer (25 mM Hepes, pH 7.1, 1 mM MgCl2, and 0.2% Triton X-100) containing 250 mM NaCl, followed by two washes with buffer containing 500 mM NaCl. The washed beads were pulled down again and resuspended in storage buffer (10 mM Hepes, pH 7.4, 150 mM NaCl, 1 mM MgCl2, and 0.01% Triton X-100) before use in lipid binding reactions. Each sample for a binding experiment contained ∼400 ng of highly purified erlin-1-V5 (>90% pure; Fig. 4 A).
Native erlin complex was purified from microsomal preparations isolated from the livers of young adult mice by differential centrifugation (Walter and Blobel, 1983) as follows. Livers from 10 C57B6/J mice (five male and five female, ∼3 mo old) fed ad libitum with normal chow were collected at the end of the dark cycle. Livers were washed in PBS, followed by hypotonic buffer (5 mM Hepes, pH 7.0, and 10 mM KCl). Typically 5–6 g of tissue were minced, transferred to a Potter homogenizer, and mixed with 3.5 weight volumes (∼20 ml) of hypotonic buffer supplemented with proteinase inhibitor cocktail. The tissue was homogenized on ice with seven strokes from a motorized pestle revolving at 1,500 rpm. The homogenate was transferred to a 50-ml conical tube and subjected to a first centrifugation for 10 min at 1,000 g. The resulting post-nuclear supernatant was collected, adjusted to contain 0.25 M sucrose, and centrifuged for 10 min at 6,600 g in a 45Ti rotor to pellet mitochondria. The post-mitochondrial supernatant was loaded onto a discontinuous gradient made from layers of (bottom to top) 2 M sucrose, 1.3 M sucrose, and the 0.25-M sucrose containing post-mitochondrial material. The step gradients were centrifuged for 4 h at 27,000 rpm (∼131,300 g) in a SW28 rotor. The flocculent layer at the interface between 1.3 and 2 M sucrose, identified by Western blotting to be most highly enriched for erlin protein, was collected and used for the preparation of magnetic beads for cholesterol binding reactions.
Magnetic beads for binding reactions containing endogenous erlins were produced by coupling 6 µg of anti–erlin-2310 antibody, affinity purified from our rabbit polyclonal anti–erlin-2 serum against the synthetic peptide CGGLGKQFEGLSDDKLG (representing amino acids 310–325 of erlin-2), to protein G Dynabeads (see above). The resulting anti–erlin-2 beads were incubated with a lysate of 200 µl microsomes (0.65 mg protein/ml) in 30 mM Hepes, pH 7.1, 200 mM NaCl, 1.2 mM MgCl2, and 0.3% Triton X-100 overnight at 4°C. Control beads were produced by incubating the microsomal lysate with anti–erlin-2310 beads together with 0.1 mg/ml antigenic peptide. Beads were washed four times with 500 mM NaCl and 0.2% Triton X-100 and resuspended in 100 µl of storage buffer immediately before use in binding reactions. The erlin-specific bead preparations contained on average a total of ∼200 ng of strongly enriched erlin complex.
The lipid binding analysis was adapted from the procedure in Infante et al. (2008b). Lipid binding reactions contained 3 µl beads (erlin-1-V5, native erlin, or matching control beads), lipids at the indicated concentrations, and 10 mM Hepes, pH 7.4, 150 mM NaCl, 1 mM MgCl2, and Triton X-100 at the subcritical micellar concentration of 0.005% (wt/vol) in a total volume of 50 µl. For binding saturation and competition experiments, cholesterol and other lipids, including a defined trace amount of 3H-cholesterol (5 pmol/reaction), were dissolved and/or mixed in ethanol at 40× final concentration before diluting the lipids into a reaction master mix. Reactions were started by addition of beads, incubated at room temperature for 1 h, washed three times with buffer, and eluted with 200 mM glycine, pH 1.9, 0.005% Triton X-100, and eluates were analyzed by scintillation counting. To determine erlin-specific binding, the amount of eluted radioactivity from control reactions was subtracted from that of erlin-containing reactions. Binding data were analyzed mathematically as described in the Statistical analysis section.
FRAP analysis
Living pErlin-2-GFP– or pLBR-GFP–transfected HeLa cells, kept in DMEM supplemented as indicated in the legend to Fig. 5 (lipid complete, depletion conditions, with or without tunicamycin), were visualized on a 710 scanning laser confocal microscope (Carl Zeiss) equipped with a live-cell chamber (37°C, humidified 5% CO2 atmosphere) and controlled by Zen2008 software. Imaging used a 63× plan apo objective (1.4 NA) and a pinhole setting of 1 Airy unit. Pre- and post-bleach images were recorded using the 488-nm emission of a 40-mW argon laser at 7% power and a fast line speed of 10. After five prebleach scans, a 4-µm2 bleach spot was generated in the cytoplasmic/ER region of a selected cell using 80% laser power with a slower line speed setting of 5 for eight iterations. Post-bleach fluorescence intensity was monitored by recording an image every 3 s for 65 cycles using the original laser and line speed settings. Simultaneously, fluorescence data from an unbleached, unrelated 4-µm2 area within the same cell was recorded for normalization purposes. Numerical fluorescence intensity data from bleached and control regions, generated by the Zen2008 software, were exported into Excel 2007 (Microsoft). Data from the bleached ROI were corrected for the loss of fluorescence owing to post-bleach imaging by normalization to the nonbleached region in the same cell as described (Ellenberg et al., 1997). Data from at least three separate experiments (5–10 cells/condition) were averaged and further evaluated as described in the Statistical analysis section. Selected images shown in Fig. 5 were exported from Zen2008 in TIFF format and processed and labeled in ImageJ v1.43u.
Statistical analysis
Values presented are averages from multiple biological repeats of an experiment. Error bars indicate SD. Significance of results was determined by unpaired two-tailed t test between control and experimental samples. Functions that best describe lipid binding and FRAP data were determined by F-test, and best-fit values were calculated based on averages of at least three samples per data point. Covariation of nuclear SREBP-2 concentration and the SREBP-2 nuclear/cytoplasmic ratio was determined by Pearson correlation. All statistical analysis was performed using GraphPad Prism versions 4.03 and 5.04 (GraphPad Software) and Excel 2007.
Online supplemental material
This section contains supplemental figures showing q-RT-PCR data for activation of SREBP target genes in HepG2 cells, PCR analysis of the UPR status in erlin-silenced cells, and micrographs illustrating the ER morphology in erlin-silenced cells (Fig. S1); SREBP-2 activation after erlin silencing determined by IF and image analysis (Fig. S2); and a biochemical and microscopic characterization of the rapamycin-inducible erlin/Insig-1 clustering system (Fig. S3). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201305076/DC1.
Supplementary Material
Acknowledgments
We thank V. DeLorme-Walker, C. DerMardirossian, and A. Liu for help with light microscopy; S. Robbins, H. Worman, T. Rapoport, and J. Ye for reagents; and members of our laboratory for help and stimulating discussions.
This work was supported by National Institutes of Health grants GM28521 and HL82545 to L. Gerace and postdoctoral fellowships to P.W. Vesely from the Austrian Science Fund (J2922-P12) and the American Heart Association (11POST7590202).
Footnotes
Abbreviations used in this paper:
- ACC1
- acetyl-CoA carboxylase 1
- coIPed
- coimmunoprecipitated
- ERAD
- ER-associated degradation
- FASN
- fatty acid synthase
- HMGR
- 3-hydroxy-3-methylglutaryl-CoA reductase
- HMGS
- 3-hydroxy-3-methylglutaryl-CoA synthase
- IP
- immunoprecipitate
- LBR
- laminin B receptor
- LD
- lipid depletion
- q-RT-PCR
- quantitative RT-PCR
- ROI
- region of interest
- SREBP
- sterol regulatory element binding protein
References
- Bretscher M.S., Munro S. 1993. Cholesterol and the Golgi apparatus. Science. 261:1280–1281 10.1126/science.8362242 [DOI] [PubMed] [Google Scholar]
- Browman D.T., Resek M.E., Zajchowski L.D., Robbins S.M. 2006. Erlin-1 and erlin-2 are novel members of the prohibitin family of proteins that define lipid-raft-like domains of the ER. J. Cell Sci. 119:3149–3160 10.1242/jcs.03060 [DOI] [PubMed] [Google Scholar]
- Browman D.T., Hoegg M.B., Robbins S.M. 2007. The SPFH domain-containing proteins: more than lipid raft markers. Trends Cell Biol. 17:394–402 10.1016/j.tcb.2007.06.005 [DOI] [PubMed] [Google Scholar]
- Brown M.S., Goldstein J.L. 1999. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc. Natl. Acad. Sci. USA. 96:11041–11048 10.1073/pnas.96.20.11041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.Y., Chang C.C., Ohgami N., Yamauchi Y. 2006. Cholesterol sensing, trafficking, and esterification. Annu. Rev. Cell Dev. Biol. 22:129–157 10.1146/annurev.cellbio.22.010305.104656 [DOI] [PubMed] [Google Scholar]
- Chen I.H., Huber M., Guan T., Bubeck A., Gerace L. 2006. Nuclear envelope transmembrane proteins (NETs) that are up-regulated during myogenesis. BMC Cell Biol. 7:38 10.1186/1471-2121-7-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberg J., Siggia E.D., Moreira J.E., Smith C.L., Presley J.F., Worman H.J., Lippincott-Schwartz J. 1997. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 138:1193–1206 10.1083/jcb.138.6.1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein J.L., DeBose-Boyd R.A., Brown M.S. 2006. Protein sensors for membrane sterols. Cell. 124:35–46 10.1016/j.cell.2005.12.022 [DOI] [PubMed] [Google Scholar]
- Gong Y., Lee J.N., Lee P.C., Goldstein J.L., Brown M.S., Ye J. 2006. Sterol-regulated ubiquitination and degradation of Insig-1 creates a convergent mechanism for feedback control of cholesterol synthesis and uptake. Cell Metab. 3:15–24 10.1016/j.cmet.2005.11.014 [DOI] [PubMed] [Google Scholar]
- Goodpaster B.H., Theriault R., Watkins S.C., Kelley D.E. 2000. Intramuscular lipid content is increased in obesity and decreased by weight loss. Metabolism. 49:467–472 10.1016/S0026-0495(00)80010-4 [DOI] [PubMed] [Google Scholar]
- Greenfield J.J., High S. 1999. The Sec61 complex is located in both the ER and the ER-Golgi intermediate compartment. J. Cell Sci. 112:1477–1486 [DOI] [PubMed] [Google Scholar]
- Hannah V.C., Ou J., Luong A., Goldstein J.L., Brown M.S. 2001. Unsaturated fatty acids down-regulate srebp isoforms 1a and 1c by two mechanisms in HEK-293 cells. J. Biol. Chem. 276:4365–4372 10.1074/jbc.M007273200 [DOI] [PubMed] [Google Scholar]
- Hoegg M.B., Browman D.T., Resek M.E., Robbins S.M. 2009. Distinct regions within the erlins are required for oligomerization and association with high molecular weight complexes. J. Biol. Chem. 284:7766–7776 10.1074/jbc.M809127200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton J.D., Goldstein J.L., Brown M.S. 2002. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109:1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X., Sakai J., Brown M.S., Goldstein J.L. 1996. Regulated cleavage of sterol regulatory element binding proteins requires sequences on both sides of the endoplasmic reticulum membrane. J. Biol. Chem. 271:10379–10384 10.1074/jbc.271.17.10379 [DOI] [PubMed] [Google Scholar]
- Hulce J.J., Cognetta A.B., Niphakis M.J., Tully S.E., Cravatt B.F. 2013. Proteome-wide mapping of cholesterol-interacting proteins in mammalian cells. Nat. Methods. 10:259–264 10.1038/nmeth.2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonen E. 2008. Cellular cholesterol trafficking and compartmentalization. Nat. Rev. Mol. Cell Biol. 9:125–138 10.1038/nrm2336 [DOI] [PubMed] [Google Scholar]
- Infante R.E., Radhakrishnan A., Abi-Mosleh L., Kinch L.N., Wang M.L., Grishin N.V., Goldstein J.L., Brown M.S. 2008a. Purified NPC1 protein: II. Localization of sterol binding to a 240-amino acid soluble luminal loop. J. Biol. Chem. 283:1064–1075 10.1074/jbc.M707944200 [DOI] [PubMed] [Google Scholar]
- Infante R.E., Abi-Mosleh L., Radhakrishnan A., Dale J.D., Brown M.S., Goldstein J.L. 2008b. Purified NPC1 protein. I. Binding of cholesterol and oxysterols to a 1278-amino acid membrane protein. J. Biol. Chem. 283:1052–1063 10.1074/jbc.M707943200 [DOI] [PubMed] [Google Scholar]
- Jo Y., Sguigna P.V., DeBose-Boyd R.A. 2011. Membrane-associated ubiquitin ligase complex containing gp78 mediates sterol-accelerated degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J. Biol. Chem. 286:15022–15031 10.1074/jbc.M110.211326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamisuki S., Mao Q., Abu-Elheiga L., Gu Z., Kugimiya A., Kwon Y., Shinohara T., Kawazoe Y., Sato S., Asakura K., et al. 2009. A small molecule that blocks fat synthesis by inhibiting the activation of SREBP. Chem. Biol. 16:882–892 10.1016/j.chembiol.2009.07.007 [DOI] [PubMed] [Google Scholar]
- Klemm J.D., Beals C.R., Crabtree G.R. 1997. Rapid targeting of nuclear proteins to the cytoplasm. Curr. Biol. 7:638–644 10.1016/S0960-9822(06)00290-9 [DOI] [PubMed] [Google Scholar]
- Koopman R., Schaart G., Hesselink M.K. 2001. Optimisation of oil red O staining permits combination with immunofluorescence and automated quantification of lipids. Histochem. Cell Biol. 116:63–68 [DOI] [PubMed] [Google Scholar]
- Langhorst M.F., Reuter A., Stuermer C.A. 2005. Scaffolding microdomains and beyond: the function of reggie/flotillin proteins. Cell. Mol. Life Sci. 62:2228–2240 10.1007/s00018-005-5166-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.N., Ye J. 2004. Proteolytic activation of sterol regulatory element-binding protein induced by cellular stress through depletion of Insig-1. J. Biol. Chem. 279:45257–45265 10.1074/jbc.M408235200 [DOI] [PubMed] [Google Scholar]
- Lingwood D., Simons K. 2010. Lipid rafts as a membrane-organizing principle. Science. 327:46–50 10.1126/science.1174621 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Motamed M., Zhang Y., Wang M.L., Seemann J., Kwon H.J., Goldstein J.L., Brown M.S. 2011. Identification of luminal Loop 1 of Scap protein as the sterol sensor that maintains cholesterol homeostasis. J. Biol. Chem. 286:18002–18012 10.1074/jbc.M111.238311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T., Schirmer E.C., Nishimoto T., Gerace L. 2004. Energy- and temperature-dependent transport of integral proteins to the inner nuclear membrane via the nuclear pore. J. Cell Biol. 167:1051–1062 10.1083/jcb.200409149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce M.M., Wang Y., Kelley G.G., Wojcikiewicz R.J. 2007. SPFH2 mediates the endoplasmic reticulum-associated degradation of inositol 1,4,5-trisphosphate receptors and other substrates in mammalian cells. J. Biol. Chem. 282:20104–20115 10.1074/jbc.M701862200 [DOI] [PubMed] [Google Scholar]
- Pearce M.M., Wormer D.B., Wilkens S., Wojcikiewicz R.J. 2009. An endoplasmic reticulum (ER) membrane complex composed of SPFH1 and SPFH2 mediates the ER-associated degradation of inositol 1,4,5-trisphosphate receptors. J. Biol. Chem. 284:10433–10445 10.1074/jbc.M809801200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A., Sun L.P., Kwon H.J., Brown M.S., Goldstein J.L. 2004. Direct binding of cholesterol to the purified membrane region of SCAP: mechanism for a sterol-sensing domain. Mol. Cell. 15:259–268 10.1016/j.molcel.2004.06.019 [DOI] [PubMed] [Google Scholar]
- Radhakrishnan A., Goldstein J.L., McDonald J.G., Brown M.S. 2008. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab. 8:512–521 10.1016/j.cmet.2008.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai J., Duncan E.A., Rawson R.B., Hua X., Brown M.S., Goldstein J.L. 1996. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell. 85:1037–1046 10.1016/S0092-8674(00)81304-5 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. 1989. Transfer of proteins from SDS-polyacrylamide gels to solid supports: immunological detection of immobilized proteins (Western blotting). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 18.60–18.75 [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K., Gerl M.J. 2010. Revitalizing membrane rafts: new tools and insights. Nat. Rev. Mol. Cell Biol. 11:688–699 10.1038/nrm2977 [DOI] [PubMed] [Google Scholar]
- Stewart J.C. 1980. Colorimetric determination of phospholipids with ammonium ferrothiocyanate. Anal. Biochem. 104:10–14 10.1016/0003-2697(80)90269-9 [DOI] [PubMed] [Google Scholar]
- Walter P., Blobel G. 1983. Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 96:84–93 10.1016/S0076-6879(83)96010-X [DOI] [PubMed] [Google Scholar]
- Walter P., Ron D. 2011. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 334:1081–1086 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]
- Walther T.C., Farese R.V., Jr 2009. The life of lipid droplets. Biochim. Biophys. Acta. 1791:459–466 10.1016/j.bbalip.2008.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.