A biosensor of local kinesin activity demonstrates that PKC and EB1 promote the activation of KIF17 on dynamic microtubules, where it contributes to microtubule stabilization in epithelia.
Abstract
We showed previously that the kinesin-2 motor KIF17 regulates microtubule (MT) dynamics and organization to promote epithelial differentiation. How KIF17 activity is regulated during this process remains unclear. Several kinesins, including KIF17, adopt compact and extended conformations that reflect autoinhibited and active states, respectively. We designed biosensors of KIF17 to monitor its activity directly in single cells using fluorescence lifetime imaging to detect Förster resonance energy transfer. Lifetime data are mapped on a phasor plot, allowing us to resolve populations of active and inactive motors in individual cells. Using this biosensor, we demonstrate that PKC contributes to the activation of KIF17 and that this is required for KIF17 to stabilize MTs in epithelia. Furthermore, we show that EB1 recruits KIF17 to dynamic MTs, enabling its accumulation at MT ends and thus promoting MT stabilization at discrete cellular domains.
Introduction
Modulation of microtubule (MT) dynamics and reorganization of the MT cytoskeleton are key events accompanying cellular morphogenesis during differentiation and tissue remodeling (Gierke and Wittmann, 2012). This change in cytoskeletal organization and dynamics is often mediated by an evolutionarily conserved mechanism involving capture of MT plus ends by cortical factors that favor local MT stabilization (Gundersen, 2002; Wu et al., 2006). We showed that, in epithelial cells, the kinesin-2 family motor KIF17 associates with MT plus ends via an interaction with the EB1 (end-binding protein 1). We also demonstrated that KIF17 dampens MT dynamics, contributes to MT stabilization, and is necessary for polarization of epithelial cells in 3D matrices. We proposed that active KIF17 could participate in regulating interactions of MT plus ends and cortical proteins during MT capture and stabilization (Jaulin and Kreitzer, 2010). However, how KIF17 activity is regulated temporally and spatially to contribute to MT dynamics and epithelial polarization is not known.
Kinesins are MT-stimulated mechanoenzymes that use ATP hydrolysis to generate motile forces (Schliwa and Woehlke, 2003; Vale, 2003). Several kinesins, including KIF17, are regulated by an autoinhibitory mechanism wherein the motor and tail domains interact, resulting in reduced MT binding and/or ADP release (Coy et al., 1999; Hackney and Stock, 2000; Imanishi et al., 2006; Dietrich et al., 2008; Verhey and Hammond, 2009; Hammond et al., 2010; Jaulin and Kreitzer, 2010). To understand how KIF17 is regulated in epithelial cells for MT stabilization, we designed kinesin biosensor constructs that are monitored using fluorescence lifetime imaging microscopy (FLIM). These biosensors provide a readout of kinesin conformation based on measurements of intramolecular Förster resonance energy transfer (FRET) efficiency (Wallrabe and Periasamy, 2005); inactive motors in a compact conformation generate FRET, whereas active motors in an extended conformation do not. FRET-based, sensitized emission approaches have been used in live cells to detect kinesin-1 and kinesin-3 in compact and extended conformations (Cai et al., 2007; Hammond et al., 2009). However, quantitative determination of active and inactive kinesin populations and their spatial distributions cannot be resolved with this approach. By comparison, FLIM improves sensitivity, is quantitative, and allows independent determinations of FRET efficiency and the fraction of interacting donor molecules (Piston and Kremers, 2007; Padilla-Parra and Tramier, 2012). Here, we apply phasor analysis to FLIM (Clayton et al., 2004; Redford and Clegg, 2005; Caiolfa et al., 2007; Digman et al., 2008), allowing us to localize active and inactive kinesin populations in single cells for the first time across large datasets.
Using a KIF17 biosensor, we provide direct evidence that KIF17 conformation and activity are regulated by EB1 and PKC. Our data suggest that PKC activates KIF17 for binding to dynamic MTs and that EB1 promotes KIF17 accumulation in an active form at the ends of dynamic MTs. Both EB1 and active PKC affect KIF17 conformation in cells and are likely to contribute to selective MT stabilization by KIF17 in epithelia. The data presented here provide the first direct visualization of extended, active and compact, inactive kinesin populations in living cells and demonstrate that conformational biosensors monitored by FLIM, combined with phasor analysis of lifetime data, provide a significant technical advance over current approaches to study kinesin regulation in living cells.
Results and discussion
Active KIF17 in an extended conformation localizes at the cell periphery in MDCK epithelial cells
We and others have shown that KIF17 undergoes a salt-dependent change in conformation in vitro (Imanishi et al., 2006; Hammond et al., 2010; Jaulin and Kreitzer, 2010). To detect KIF17 conformations in cells directly, we designed intramolecular FRET expression constructs encoding full-length KIF17 tagged with mCherry (mCh) and Emerald GFP (EmGFP) at N and C termini, respectively. We used FLIM to measure the decay in EmGFP lifetime caused by quenching by mCh in live MDCK cells (Bastiaens and Squire, 1999). In all experiments, donor lifetimes in cells expressing FRET constructs (mCh-KIF17-EmGFP) were referenced to cells expressing donor alone (KIF17-EmGFP). Only cells with low to medium expression levels were chosen for analysis to avoid artifacts caused by overexpression. Lifetime data were analyzed using the phasor approach. This method is amenable to rapid analysis of a large number of cells and reports the localization of kinesin populations in compact and extended states in the cell as well as the relative amount of each population (see Materials and methods and Fig. S1 [B and C] for details).
We first determined that N- and C-terminal tags did not interfere with KIF17 function by testing whether FRET constructs stabilized MTs when expressed in MDCK cells, as described previously for the constitutively active, extended conformation hinge mutant GFP-KIF17G754E (Jaulin and Kreitzer, 2010). In cells expressing mCh-KIF17-EmGFP, KIF17-EmGFP, GFP-KIF17, or mCh-KIF17G754E-EmGFP, we observed an increase in nocodazole (NZ)-resistant, posttranslationally modified (detyrosinated or acetylated) stable MTs (Fig. 1, A and E; and not depicted). Thus, fluorescently tagged wild-type and mutant KIF17 are active for MT stabilization, demonstrating that these constructs are functional in cells.
Figure 1.
Localization of active and inactive populations of KIF17 in MDCK cells. (A) Immunostaining of tyrosinated and detyrosinated tubulin in cells microinjected with mCh-KIF17-EmGFP (outlined cells and inset) and treated for 45 min with 33 µM NZ. (B) Representative FLIM phasor analysis of mCh-KIF17-EmGFP in cells. Fluorescence image (inset) and the distribution of KIF17 in extended, active (red mask on image and red circle on phasor plot, FRETeff < 7%) and compact, inactive (green mask on image and green circle on phasor plot, FRETeff = 7–21%) forms determined from measured lifetimes. Inset on the phasor plot shows analysis of mCh-KIF17G754E-EmGFP. (C) Representative FLIM phasor analysis of mCh-KIF17G754E-EmGFP in cells as in B. (D) Box–whisker plots showing the distribution of FRETeff and populations of active and inactive mCh-KIF17-EmGFP and mCh-KIF17G754E-EmGFP in in MDCK cells. Data represent the indicated number of cells (n) obtained from three or more independent experiments ± SEM. Box–whisker plots show minimum, 25th percentile, median, 75th percentile, maximum, and mean FRET values. (E) Quantification of immunoblots probed for detyrosinated (Glu) and acetylated (Ace) tubulin in control or mCh-KIF17-EmGFP transfected cells. Ratio of tubulin/actin was normalized to 1 ± SD in controls. Data are derived from five experiments. Bars, 20 µm.
As shown previously for GFP-KIF17 (Jaulin and Kreitzer, 2010), exogenously expressed mCh-KIF17-EmGFP appeared primarily as diffuse cytoplasmic fluorescence (Fig. 1 B, inset grayscale image), suggesting that the majority of KIF17 is in a compact, inactive conformation as previously described (Hammond et al., 2010). Using FLIM, we measured a mean FRET efficiency (EAV) for mCh-KIF17-EmGFP of 6.8 ± 0.2% (Fig. 1 D and Table 1). However, this mean could reflect a mixture of both compact (FRET signal) and extended (non-FRET) populations of mCh-KIF17-EmGFP. To discriminate between these two populations, we mapped the spatial distribution of FRET and non-FRET species in cells using phasor analysis. Based on our FRET confidence limit (see Materials and methods), pixels in the image with FRETeff ≤ 7% represent KIF17 in an extended conformation, whereas pixels with FRETeff = 7–21% represent KIF17 in a compact form (Fig. 1 B, phasor plot). Phasor analysis revealed that 64.9 ± 2.0% of mCh-KIF17-EmGFP is in a compact state (Fig. 1 D). This pool of mCh-KIF17-EmGFP localizes throughout the cytoplasm but does not generally extend to the cell periphery (Fig. 1 B, green mask). A smaller but significant proportion (25.9 ± 1.4%) of mCh-KIF17-EmGFP was in the extended conformation (Fig. 1 D) and localized prominently near the cell periphery (Fig. 1 B, red mask). The cortical distribution of extended KIF17 is consistent with the EB1-dependent localization of endogenous protein to MT plus ends and a role in plus end–mediated MT stabilization (Jaulin and Kreitzer, 2010). It is likely that some extended KIF17 is also located on more central MTs, but this pool is obscured in our analysis by the greater cell depth and larger fraction of motor in the compact form in these regions. Extended KIF17 was also detected above/below nuclei, where the cytoplasmic depth is narrow, and compact KIF17 did not obscure detection of an extended motor.
Table 1.
Summary of FRET efficiencies and percentage of active, extended KIF17 measured under all conditions described
| Expressions | Treatments | EAV | P-value | Active pool | P-value |
| % | % | ||||
| mCh-KIF17-EmGFP | Untreated | 6.8 ± 0.2 | NA | 25.9 ± 1.4 | NA |
| mCh-KIF17-EmGFP | 1 mM AMP-PNP | 2.4 ± 0.9 | <0.001a | 58.4 ± 4.8 | <0.001a |
| mCh-KIF17G754E-EmGFP | Untreated | 2.3 ± 0.6 | <0.001a | 70.1 ± 4.0 | <0.001a |
| mCh-KIF17-EmGFP | 33 µM NZ | 7.0 ± 0.4 | >0.5a | 25.9 ± 3.7 | >0.5a |
| mCh-KIF17-EmGFP | Cold + 33 µM NZ | 9.7 ± 0.5 | <0.001a | 15.2 ± 3.5 | <0.001a |
| mCh-KIF17-EmGFP | 100 nM OA | 7.8 ± 0.4 | <0.05a | 15.9 ± 2.6 | <0.005a |
| mCh-KIF17-EmGFP | 10 µM taxol | 9.3 ± 0.6 | <0.001a | 11.4 ± 2.5 | <0.001a |
| mCh-KIF17-EmGFP | 20 µM BIM-1 | 8.5 ± 0.5 | <0.001a | 14.6 ± 1.7 | <0.001a |
| mCh-KIF17-EmGFP + Myc-EB1 | Untreated | 5.4 ± 0.4 | <0.001a | 40.2 ± 4.3 | <0.001a |
| mCh-KIF17-EmGFP + Myc-EB1 | 33 µM NZ | 6.7 ± 0.5 | <0.005 b | 24.2 ± 3.9 | <0.001b |
| mCh-KIF17-EmGFP + Myc-EB1 | Cold + 33 µM NZ | 8.5 ± 0.6 | <0.001b | 16.3 ± 1.8 | <0.001b |
NA, not applicable.
Value distribution as compared with mCh-KIF17-EmGFP using a two-tailed Student’s t test.
Value distribution as compared with mCh-KIF17-EmGFP + Myc-EB1 using a two-tailed Student’s t test.
To confirm that the KIF17 we detect in an extended conformation represents the population of KIF17 on MTs, we permeabilized living cells in the presence of AMP-PNP, a nonhydrolyzable ATP analogue that recruits kinesins to MTs (Lasek and Brady, 1985). Under these conditions, mCh-KIF17-EmGFP EAV decreased 65%, with a corresponding 2.3-fold increase in KIF17 in an extended form (Fig. S1, D and E; and Table 1), consistent with previous studies (Hammond et al., 2010). Thus, KIF17 in an open conformation by FRET represents the population of motor on MTs.
To verify that different KIF17 conformations detected with FRET reflect motor activity, we next analyzed the constitutively active hinge mutant mCh-KIF17G754E-EmGFP. The FRET efficiency of mCh-KIF17G754E-EmGFP was 66% less than that of mCh-KIF17-EmGFP (EAV = 2.3 ± 0.6%; Fig. 1 D and Table 1). Phasor analysis showed that 70.1 ± 4.0% of KIF17G754E is in an extended conformation, a 2.7-fold increase as compared with KIF17; only 13.8 ± 2.0% of KIF17G754E is detected in a compact conformation (Fig. 1 D and Table 1). Conformationally extended mCh-KIF17G754E-EmGFP localized throughout the cell and accumulated at MT plus ends in cell protrusions (Fig. 1 C, red mask and inset grayscale image), as shown previously for GFP-KIF17G754E (Jaulin and Kreitzer, 2010). Some FRET was detected in these protrusions (Fig. 1 C, green mask) and likely arises from crowding-induced intermolecular interactions rather than motors in a folded state, as suggested previously for kinesin-1 (Cai et al., 2007).
Further evidence that KIF17 conformation reflects its activity comes from data showing that KIF17 motor and tail domains interact and that full-length KIF17 is not efficient for MT-based motility but that removal of the C-terminal tail or mutation of the hinge produces a motor that moves effectively on MTs (Imanishi et al., 2006; Hammond et al., 2010; Jaulin and Kreitzer, 2010). Similarly, recombinant OSM-3, the Caenorhabditis elegans homologue of KIF17, has little to no motility in vitro, but both a hinge mutant and C-terminally truncated OSM-3 move efficiently on MTs (Imanishi et al., 2006; Hammond et al., 2010; Jaulin and Kreitzer, 2010). Here, using purified recombinant protein, we found that the KIF17 tail domain (amino acids 850–1,029) ablates MT gliding and inhibits MT-stimulated ATPase activity in vitro of C-terminally truncated KIF17 (amino acids 1–490, KIF17M-490; Fig. S1 A and Table 2). In these assays, KIF17M-490 had a maximal MT-stimulated ATPase rate (kcat) of 13.6 ± 1.8 s−1, which decreased to 3.9 ± 0.7 s−1 in the presence of the KIF17 tail domain. Interestingly, addition of KIF17 tail also decreases K0.5, MT of KIF17M-490, suggesting that functional inhibition of motor activity is not necessarily caused by displacement of the kinesin from MTs (Dietrich et al., 2008; Acharya et al., 2013). From these and previous studies we conclude the FRET conformation sensor is an accurate reporter of active (extended) and inactive (compact) KIF17 in living cells.
Table 2.
ATPase properties of KIF17M-490
| Constructs | ATPase activity | Gliding | |||
| kcat | K0.5, MT | Velocity | Motile MT | No. of MT observed | |
| s−1 | µM | µm/s | % | ||
| KIF17 M-490 | 13.6 ± 1.8 | 1.3 ± 0.1 | 0.47 ± 0.02 | 11.6 ± 0.9 | 247 |
| KIF17 M-490 + KIF17-tail | 3.9 ± 0.7 | 0.4 ± 0.3 | ND | ND | 252 |
| KIF17 M-490 + EB1 | 16.6 ± 1.2 | 0.7 ± 0.3 | 0.51 ± 0.03 | 16.6 ± 1.3 | 251 |
ATPase activity of KIF17M-490 in the absence and presence of KIF17-tail or EB1. Data (Fig. S1 A) were fit to a Michaelis–Menton equation to determine kcat and K0.5, MT. Data represent the mean of nine independent experiments at three different MT concentrations ± SEM. kcat refers to the rate of ATP hydrolysis per kinesin dimer. K0.5, MT refers to the concentration of polymerized tubulin to half-maximally stimulate ATPase activity. Errors are ± SEM.
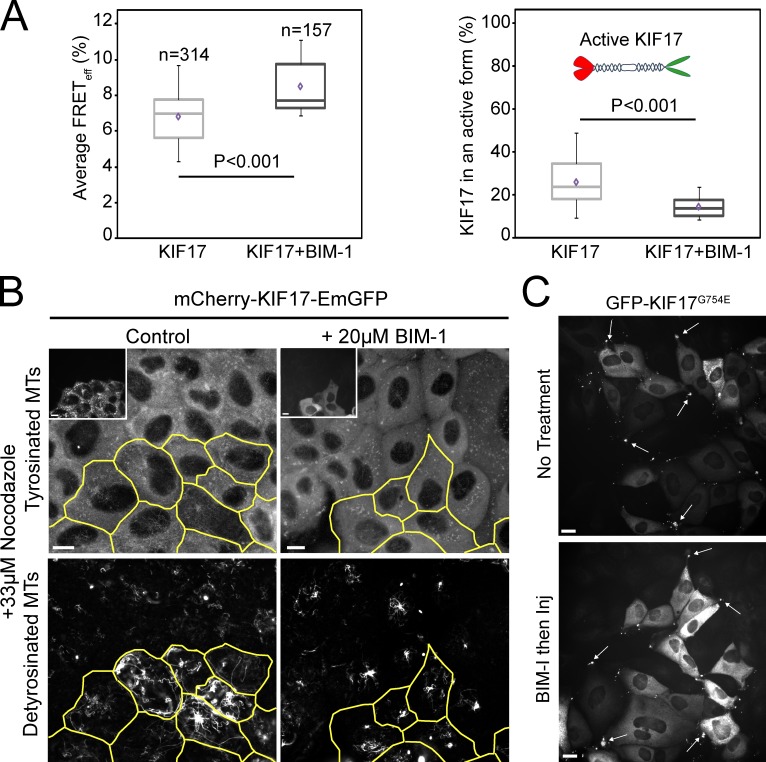
PKC activity is necessary to relieve KIF17 autoinhibition and activate it for MT stabilization
PKC family members have been implicated in regulation of MT dynamics and polarization of several cell types and act in the cortical MT capture and stabilization pathway (Kabir et al., 2001; Fan et al., 2004; Ruiz-Canada et al., 2004; Eng et al., 2006). Because KIF17 may act through this pathway (Jaulin and Kreitzer, 2010), we tested whether PKC affects KIF17 conformation and activity for MT stabilization. We treated MDCK cells expressing mCh-KIF17-EmGFP with the PKC inhibitor BIM-1 (bisindolylmaleimide I; 20 µM) for 1 h (Fig. 2); at this concentration, classical PKCs α and βI and novel PKCs δ and ε are likely inhibited (Martiny-Baron et al., 1993).
Figure 2.
PKC activity is required for KIF17 activation in MDCK cells. (A) Box–whisker plots showing the distribution of FRETeff (left) and the fraction of active KIF17 (right) in control and 20 µM BIM-1–treated cells (1 h). Data were obtained from at least three independent experiments ± SEM. Box–whisker plots show minimum, 25th percentile, median, 75th percentile, maximum, and mean FRET values. (B) Immunostaining of detyrosinated and tyrosinated tubulin in cells expressing mCh-KIF17-EmGFP after cDNA injection (outlined cells and inset) and treatment with BIM-1 as in A. Bars, 10 µm. (C) GFP-KIF17G754E localizes at the plus ends of MTs in cell protrusions (arrows) in control and BIM-1–treated cells as in B. Cells were fixed 2 h after cDNA microinjection. Inj, injection. Bars, 20 µm.
BIM-1 induced a 20% increase over controls in EAV of mCh-KIF17-EmGFP and a 41% decrease in the extended population of KIF17 (Fig. 2, A and B; and Table 1). Furthermore, BIM-1–treated cells expressing mCh-KIF17-EmGFP (or KIF17-EmGFP) failed to generate NZ-resistant, stable MTs (Fig. 2 C). These data implicate PKC in regulating KIF17 conformation and thus its ability to interact with and stabilize MTs in cells. Inhibition of PKC could alternatively prevent generation of stable MTs by affecting downstream components in the MT stabilization pathway (e.g., GSK3-β), as suggested in fibroblasts (Eng et al., 2006); this could inhibit KIF17 activity indirectly by affecting its binding to and unfolding on MTs. However, BIM-1 alone had no effect on levels of posttranslationally modified, stable MTs in MDCK cells (unpublished data). Furthermore, pretreatment of cells with BIM-1 did not affect the localization of the extended mutant GFP-KIF17G754E at MT ends in cell protrusions (Fig. 2 D) or its ability to stabilize MTs (not depicted). Considered together, these data support a model in which active PKC releases KIF17 autoinhibition, promoting binding to and stabilization of MTs. KIF17 contains several predicted PKC phosphorylation sites and thus could be regulated directly by PKC. Alternatively, PKC could affect KIF17 binding to a cargo that would activate the motor in cells (Verhey and Hammond, 2009).
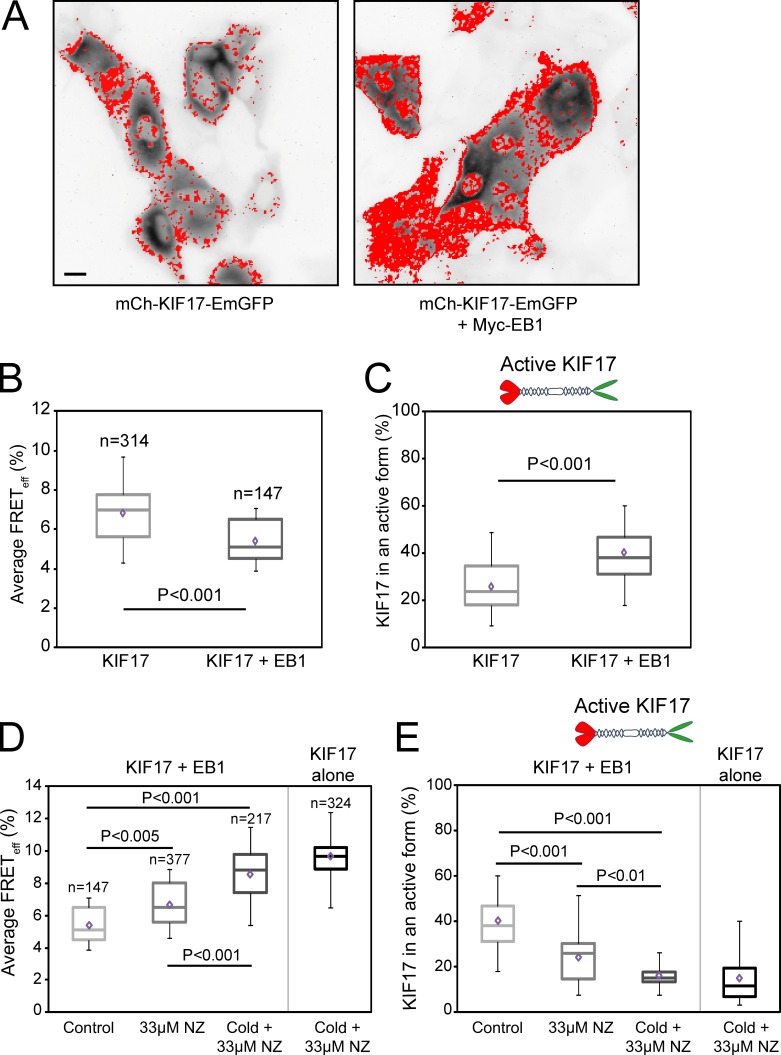
EB1 keeps KIF17 in an active form at MT plus ends in MDCK cells
We showed previously that localization of endogenous KIF17 to MT plus ends in epithelial cells, where it could participate in cortical MT stabilization, is dependent on EB1 but that mutant KIF17G754E accumulates at plus ends and stabilizes MTs independent of EB1 (Jaulin and Kreitzer, 2010). This led us to speculate that EB1 binding to KIF17 at MT ends could either activate the motor for MT stabilization or enhance its accumulation in an active form at MT ends. To test this directly, we coexpressed mCh-KIF17-EmGFP and Myc-EB1 in MDCK cells and analyzed KIF17 conformation with FLIM (Fig. 3). In these experiments, EB1 coexpression resulted in a 20% decrease in EAV as compared with control cells expressing KIF17 alone (Fig. 3 B and Table 1). Phasor analysis showed this change in FRET efficiency corresponded to a 55% increase in the population of extended KIF17 when EB1 was coexpressed (Fig. 3 C and Table 1).
Figure 3.
Coexpression of EB1 increases the pool of extended, active KIF17 in MDCK cells. (A) Fluorescence images of mCh-KIF17-EmGFP expressed alone or with Myc-EB1. Red mask shows the localization of active mCh-KIF17-EmGFP. Bar, 20 µm. (B and D) Box–whisker plots showing the distribution of FRETeff under each experimental condition. (C and E) Box–whisker plots showing the fraction of active KIF17 under each experimental condition. Data were obtained from at least three independent experiments ± SEM. Box–whisker plots show minimum, 25th percentile, median, 75th percentile, maximum, and mean FRET values.
When cells coexpressing EB1 and KIF17 are treated with 33 µM NZ for 45 min to depolymerize dynamic MTs (note that some stable, modified MTs remain even in uninjected cells; Fig. 1 A), the population of extended KIF17 decreases to levels measured in untreated cells expressing KIF17 alone (Fig. 3, D and E; and Table 1). In contrast, when cells are preincubated on ice for 20 min before rewarming in NZ for 45 min, a treatment that results in loss of both stable and dynamic MTs in MDCK cells (Kreitzer et al., 2003), we measure no difference in EAV or the extended population of KIF17 when it is expressed alone or coexpressed with EB1 (Fig. 3, D and E). These data indicate that the EB1-mediated increase in conformationally extended KIF17 is dependent on dynamic MTs and that, in the absence of MTs, EB1 cannot recruit/retain KIF17 on MTs and thus cannot relieve the autoinhibited state.
In ATPase assays in vitro, EB1 did not affect kcat of KIF17M-490 significantly (P > 0.1) but decreased K0.5, MT of KIF17M-490 approximately twofold (P < 0.001; Fig. S1 B and Table 2), similar to what was shown for kinesin-7 (Tea2) in yeast (Browning and Hackney, 2005). In MT-gliding assays, EB1 similarly induced a 70% increase in the number of motile MTs without changing KIF17M-490 velocity significantly. Collectively, these data suggest EB1 may decrease the off rate of KIF17 from MTs and thus retain KIF17 in an extended conformation at dynamic MT ends, promoting MT stabilization.
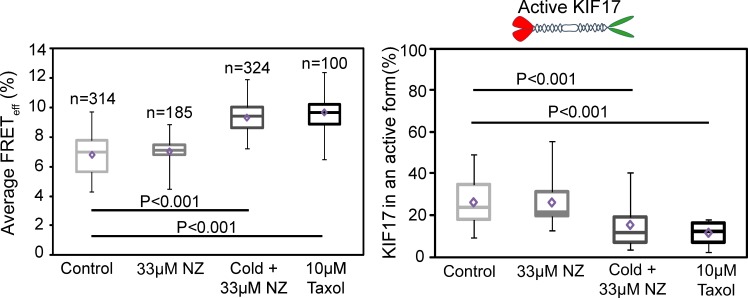
Active KIF17 is on stable MTs in MDCK cells
The constitutively active, extended mutant GFP-KIF17G754E accumulates in large puncta at MT plus ends near the cell cortex in epithelial cells. This localization is observed even in cells treated with NZ to depolymerize dynamic MTs (Jaulin and Kreitzer, 2010), suggesting that active KIF17 is on stable MTs. To determine whether extended KIF17 localizes on a specific subset of MTs, we analyzed mCh-KIF17-EmGFP by FLIM when only dynamic MTs or both dynamic and stable MTs were depolymerized. Depolymerization of dynamic MTs had no effect on EAV or on the fraction of extended conformation mCh-KIF17-EmGFP as compared with untreated controls (Fig. 4, A and B). However, when both dynamic and stable MTs were depolymerized, we measured a 30% increase in EAV accompanied by a 43% decrease in the extended population of KIF17 when compared with control untreated cells (Fig. 4, A and B). To break down stable, modified MTs selectively, we treated cells expressing mCh-KIF17-EmGFP with 100 nM okadaic acid (OA) for 90 min (Fig. S2 A; Gurland and Gundersen, 1993). OA induced a 42% decrease in the extended population of KIF17 to 15.9 ± 2.6% (Fig. S2, B and C), similar to what we observed in cells treated with cold and NZ. Although OA depolymerizes stable, modified MTs preferentially in MDCK cells, we cannot yet exclude the possibility that it affects KIF17 conformation and activity by modifying phosphorylation of the kinesin directly or of another factor that modulates KIF17 activity in our cells.
Figure 4.
Active KIF17 is on stable MTs in MDCK cells. Box–whisker plots showing the distribution of KIF17 FRETeff (left) and the fraction of active KIF17 (right) under each experimental condition. Data were obtained from at least three independent experiments ± SEM. Box–whisker plots show minimum, 25th percentile, median, 75th percentile, maximum, and mean FRET values.
These experiments show that extended KIF17 resides primarily on stable MTs in MDCK cells. As such, we reasoned that stabilizing MTs with taxol (10 µM for 1 h) should lead to an increase in the pool of active, extended mCh-KIF17-EmGFP. Surprisingly, we instead measured a 37% increase in EAV and a corresponding 56% decrease in the extended pool of KIF17 after taxol treatment (Fig. 4, A and B), similar to what was observed in cold and NZ-treated cells. The extended pool of mCh-KIF17-EmGFP was also reduced in cells treated with 50 nM taxol for 1 h, thus the decrease in extended kinesin is not a result of MT bundling induced by high concentrations of the drug (unpublished data). Interestingly, KIF17G754E is lost from MT ends in cell protrusions when cells are treated with taxol, either before or after it was expressed acutely by cDNA injection (Fig. S3 A). Because KIF17G754E remains on MT plus ends in the absence of EB1 (Jaulin and Kreitzer, 2010), it is not likely that loss of EB1 from MTs in taxol (unpublished data) causes the observed changes in KIF17 activity and localization. These results suggest KIF17 uses dynamic MTs to reach MT plus ends where it can participate in MT stabilization and then remain on stable MTs. Consistent with this idea, when living cells are permeabilized in the presence of AMP-PNP, we detect KIF17-EmGFP on both unmodified and posttranslationally modified MTs (Fig. S3 B). Because the majority of extended conformation KIF17 resides on stable MTs and because we measure no change in EAV or extended mCh-KIF17-EmGFP when dynamic MTs are depolymerized selectively, we speculate that the pool of KIF17 being activated on dynamic MTs is small and, thus, below our detection limit with FRET analysis.
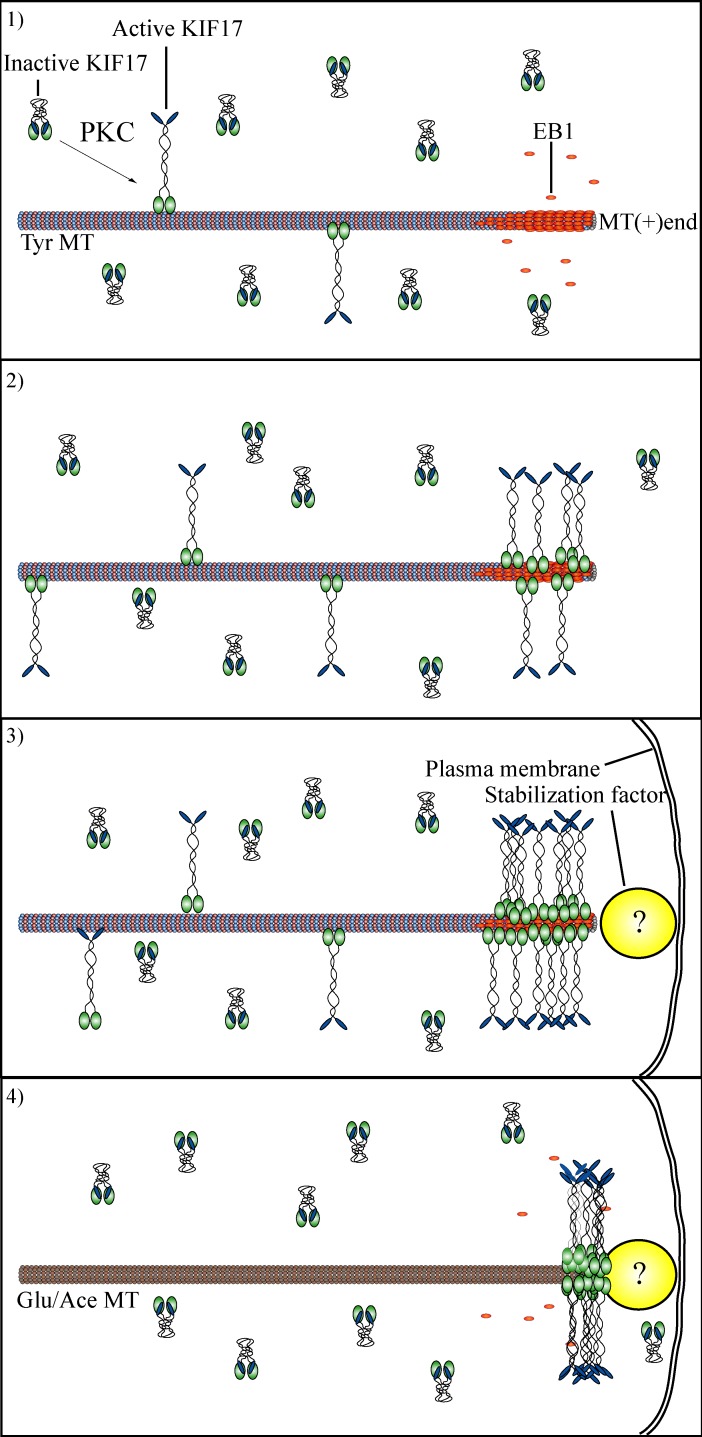
Together, our data support a model (Fig. 5) wherein KIF17 is activated through a pathway involving PKC that induces a conformational change sufficient for binding to MTs. Next, motile KIF17 interacts with EB1 at dynamic MT plus ends, inducing accumulation of activated motor at these sites. Activated KIF17 at MT plus ends could potentially interact directly, or through a cargo, with cortical factors involved in regulating MT stability, forming a “stable” bridge between MTs and the cortex. Alternatively, KIF17 could protect MT ends independent of cargo or cortical factors. Indeed, we find that the KIF17 motor domain is sufficient to protect MTs from depolymerization in vitro (Acharya et al., 2013). Further studies exploring these important questions are underway. The results of this work provide novel mechanistic insight into how KIF17 participates in MT stabilization in epithelial cells. In addition, these studies demonstrate that kinesin biosensors, monitored by FLIM in living cells and analyzed on phasor plots, can be applied widely to study kinesin regulation and to identify physiologically relevant factors that modify kinesin conformation and activity in vivo.
Figure 5.
Proposed model for KIF17 in MT stabilization in epithelial cells. (1) KIF17 is found in the compact, autoinhibited conformation in MDCK cells. After activation, through a pathway involving PKC, KIF17 is activated for binding to MTs. (2) KIF17 binds dynamic MTs and reaches MT plus ends where it interacts with EB1. Interaction with EB1 promotes KIF17 accumulation at MT plus ends. (3) KIF17 is captured at the cortex by an as yet unidentified stabilization factor either directly, via EB1, or via another cargo. Alternatively, KIF17 may stabilize MTs by accumulating at plus ends and inhibiting turnover of tubulin subunits. (4) KIF17 remains at plus ends of stabilized MTs but cannot bind to stable MTs de novo. Ace, acetylated; Tyr, tyrosinated.
Materials and methods
Cell culture, transfection, and microinjection
MDCK cells were cultured in DMEM (4.5 g/liter glucose) with 5% FBS and 20 mM Hepes, pH 7.2. MDCK cells were seeded on sterilized coverslips and used at 80% confluence 2–3 d after plating. Transfections (5 µg DNA) were performed using a nucleofector (Amaxa; Lonza) as recommended. Microinjection was performed essentially as described previously (Jaulin et al., 2007). 5–20 µg/ml cDNAs in HKCl (10 mM Hepes and 140 mM KCl, pH 7.4) were pressure injected using a micromanipulator (MMO-202ND; Narishige). Cells were incubated at 37°C for 60–90 min to allow for expression of cDNAs. NZ and taxol were purchased from Sigma-Aldrich. OA was obtained from EMD Millipore. For FLIM, we analyzed only cells expressing the biosensor at low to medium levels (less than or equal to mean integrated fluorescence intensity/cell of all expressing cells) as there was no significant difference in FRET efficiency or the population of compact and extended KIF17 under these conditions (low expression, EAV = 7.2 ± 0.3% and extended population = 25.4 ± 2.4%; mean/medium expression, EAV = 7.2 ± 0.2% and extended population = 22.2 ± 1.9%). Immunoblots showed that biosensor expression is four to five times above levels of endogenous KIF17.
Immunofluorescence staining
Cells were fixed in −20°C methanol for 1–2 min. Antibodies used were mouse antiacetylated tubulin (611B1; Sigma-Aldrich), rat antityrosinated tubulin (YL1/2), and rabbit antidetyrosinated (Glu) tubulin (SG). Fluorescently conjugated secondary antibodies (aminomethylcoumarin acetate, FITC, Cy3, and Cy5) were obtained from Jackson ImmunoResearch Laboratories, Inc.
Fixed-cell imaging
Fixed-cell images were acquired using E400 or TiE microscopes (Nikon) with 40× (NA 1.0), 60×, or 100× (NA 1.4) Plan Apochromat oil immersion objectives and collected with digital charge-coupled device cameras (6.45-µm pixels, 1 MHz for 14-bit images [ORCA II-ER; Hamamatsu Photonics] or 6.45-µm pixels, 560 MHz for 16-bit images [Neo sCMOS; Andor Technology]). Devices were controlled by either MetaMorph (Molecular Devices) or Elements (Nikon). 14–16-bit images were scaled linearly to subtract background (noncell fluorescence) and highlight features of interest as indicated in the Results and discussion and converted to 8-bit copies for figure assembly. Images shown in Fig. S3 B were also enhanced using the unsharp mask feature in MetaMorph (kernel size = 3, scaling factor = 0.85, and result scale = auto).
Fluorescence lifetime imaging
All FLIM experiments were performed in The Rockefeller University Bio-Imaging Resource Center in New York, NY. To perform FLIM, cells were transferred to recording medium (Hank’s balanced salt solution with 10 mM Hepes, 1% FBS, 4.5 g/liter glucose, and essential and nonessential amino acids) and incubated at 37°C in a stage top incubator (INU series; Tokai Hit Co., Ltd.). The experiments were performed on a Nikon TiE using a 40× Plan Apochromat oil immersion objective (NA 1.3) and a frequency domain lifetime attachment (Lambert Instruments) controlled by the vendor’s LI-FLIM software. EmGFP was excited with ∼4 mW of light from a 488-nm light-emitting diode, modulated at 40 MHz, and emission was collected using a 510–560-nm electromagnetic filter, with an intensified charge-coupled device (10.3-µm pixels; LI2CAM; Lambert Instruments). A reference lifetime of 4.0 ns was determined from a 1-µM solution of fluorescein in saline.
Expression constructs
KIF17 and EB1 were amplified by PCR from human A549 or Caco2 cells and cloned into the p201DONR (Gateway; Invitrogen) vector (Jaulin and Kreitzer, 2010). We engineered the FRET expression vector by modifying pDEST-monomeric mCh-C1a (Invitrogen) to include EmGFP (monomeric EmGFP) at the C terminus. DNA encoding KIF17 sequences were introduced by recombination with LR Clonase as recommended (Invitrogen), resulting in constructs with mCh (acceptor) and EmGFP (donor) fused to the N and C termini of kinesin, respectively, with a 10–amino acid linker between the tags and the kinesin. A previous study in COS cells suggested that large epitope tags connected to kinesins via short linkers to the N (4 amino acids) or C (10 amino acids) termini of KIF17 could interfere with conformational regulation of the protein (Hammond et al., 2010). Therefore, we also tested constructs with long linkers in our experiments. For all of these (≤30 amino acids), we saw no loss of MT association or significant difference in KIF17 activity by FLIM.
Constructs and protein purification
Recombinant, functional mCh-KIF17-EmGFP (or any version of full-length KIF17) could not be purified in sufficient quantities in any host system tested. Therefore, we instead generated a C-terminally truncated protein encoding KIF17M-490 (amino acids 1–490) containing the motor, neck, and first coiled-coil domains of KIF17 for analysis in vitro. The in vitro activity of KIF17M-490 was identical to a version of the protein lacking the first coiled coil (KIF17M-370). KIF17M-490 was amplified by PCR from full-length human KIF17 described in Jaulin and Kreitzer (2010) and cloned into p201DONR (Invitrogen) as described for biosensor constructs. The forward primer used to generate KIF17M-490 was 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGCCTCCGAGGCGGTGAA-3′; the reverse primer was 5′-GGGGACCACTTGTACAAGAAAGCTGGGTCCTAAAAAGCAGGCGGGTACTCAGC-3′. All N-terminal GST fusions were generated by recombination of with LR Clonase into pDEST27 (Invitrogen). GST-KIF17 tail domain (tail 849–1,029 amino acids) and full-length GST-EB1 constructs in pDEST27 were described previously (Jaulin and Kreitzer, 2010). GST fusions were transformed into Escherichia coli BL21-DE3 cells, and protein expression was induced by addition of 1 mM IPTG for 4 h before harvesting cells. GST fusions were immobilized on glutathione–Sepharose 4B (GE Healthcare) for affinity purification. For motor domain constructs, additional purification on anion exchange chromatography (for motor domain; Q Sepharose Fast Flow; Sigma-Aldrich) was required to remove contaminating bands. Purified proteins were dialyzed and stored in 80 mM K-Pipes, 100 mM KCl, 2 mM MgCl2, and 1 mM EGTA, pH 6.9, with 5% glycerol and 0.5 mM DTT. Purified motor domain was stored at >1.5 mg/ml with 50 µM ATP. Protein concentrations were determined by Bradford assay and are reported as monomer concentration if not otherwise stated.
Tubulin purification.
Tubulin was purified from bovine brain by two cycles of temperature-dependent assembly and disassembly followed by chromatography over DEAE-Sephadex (Sigma-Aldrich) as previously described (Borisy et al., 1975). In brief, tubulin was polymerized from brain homogenates in PEM (80 mM Pipes-K, pH 6.8, 1 mM EGTA, and 1 mM MgCl2) supplemented with 0.1 mM GTP and 10% glycerol at 37°C, and assembled MTs were pelleted by centrifugation. The supernatant was removed, and MTs were dissociated in cold PEM containing 1 mM GTP and centrifuged at 4°C to remove aggregated protein. MT-associated proteins were removed by passing cycled tubulin over DEAE-Sephadex. Tubulin purity was analyzed by SDS-PAGE and Coomassie blue staining. After two cycles, tubulin in PEM-GTP1mM was snap frozen in single-use aliquots and stored at −80°C. Before use, an additional cycle of cold disassembly and centrifugation at 4°C was performed to remove protein aggregates. For in vitro assays, 25 µM DEAE tubulin was incubated on ice for 5 min in BRB80 buffer with 1 mM DTT and 1 mM GMPCPP (guanosine-5′-[(α,β)-methyleno]triphosphate; Jena Bioscience) and clarified by centrifugation at 90,000 rpm in a rotor (TLA100.4; Beckman Coulter) for 5 min at 4°C. Clarified tubulin was polymerized at 37°C for 60 min and centrifuged at 90,000 rpm for 5 min at 37°C. The MT pellet was resuspended in prewarmed BRB80 buffer with 25 µM taxol and 1 mM DTT (0.8 volume of starting polymerization volume).
MT-stimulated ATPase activity.
MT-stimulated ATPase activity was determined by measuring phosphate release at 25°C with the PiColorLock reagent (Innova Biosciences) in a modified malachite green assay. 50–150 nM KIF17M-490 alone or with KIF17-tail (equimolar with KIF17-M) and EB1 (2× molar ratio) was incubated with MTs in BRB80, pH 6.9, containing 75 mM KCl and 1 mM DTT. Phosphate release was plotted against MT concentration, and data were fit to a Michealis–Menton equation using Prism (GraphPad Software) to determine kcat and K0.5, MT. No ATP hydrolysis was detected when MTs, EB1, or KIF17-tail were assayed in the absence of KIF17M-490.
The phasor transformation
Globals software (Laboratory for Fluorescence Dynamics, University of Illinois at Urbana-Champaign, Urbana, IL) was used to analyze fluorescence data. Box–whisker plots show minimum, 25th percentile, median, 75th percentile, maximum, and mean FRET values. Statistical significance was determined using a two-tailed Student’s t test. Coordinates s and g in the phasor plot correspond to intensity decay, I(t), and are assigned based on the following expressions as described in Caiolfa et al. (2007):
ω represents the angular frequency of light modulation. i and j are identifiers of individual pixels in the image. When measured in frequency domain
mi,j and ϕi,j represent modulation (m) and phase (ϕ) of the emission relative to excitation. For a single exponential decay, I(t) = Ae − t/τ, phasor coordinates are
For multiexponential components in a pixel, i,j, phasor coordinates are
fk is the intensity-weighted fractional contribution of the component with lifetime τk. For a single exponential decay in which
all components are included on the phasor plot within the universal circle (center: [0.5, 0]; radius: [0.5]). Phasors corresponding to short lifetimes fall closest to coordinate (1, 0). Phasors corresponding to long lifetimes fall closest to coordinate (0, 0). FRET trajectories on the phasor plot are then calculated, after determining those of unquenched donors and autofluorescence with the equation
The phasor corresponding to quenched EmGFP is calculated according to the last equation. The curved trajectory on the phasor plot represents realizations of all possible phasors quenched with different efficiencies.
Circles on the phasor plot include the majority (>95%) of donor phasors. The radius (extending to ∼7% FRET efficiency here) determines the confidence limit for FRET analysis (Caiolfa et al., 2007; Barreiro et al., 2008). Two phasor subsets are shown in the plots, and corresponding pixels are mapped in the FLIM images: pixels highlighted by the red mask have FRET efficiency <7% and represent KIF17 in the extended or active form, and pixels highlighted by the green mask have FRET efficiency between 7–21% and represent KIF17 in the compact, inactive conformation. FRET efficiency is obtained by localizing the positions of phasor clusters in the phasor plot. The position of the measured point on the trajectory gives the FRET efficiency. Because the fluorescence decay in each pixel of the image gives a point in the phasor plot, the location of the measured point is the intensity-weighted mean of FRET, non-FRET, and autofluorescent components. Thus, for each pixel in which the contribution of the non-FRET component is high (i.e., autofluorescence), points on the phasor plot will approach the green line (representing 0% FRET efficiency). Conversely, in pixels in which the contribution of the FRET component is high, points on the phasor plot approach the red line (representing 50% FRET efficiency) following the black trajectory. Because expressed mCh-KIF17-EmGFP is primarily soluble, we expect a FRET contribution in each pixel and thus each point on the phasor plot. As such, active and inactive KIF17 populations do not segregate into distinct peaks. In most cases, some points on the phasor spread outside the circle of confidence and are not used to determine fractions of active and inactive KIF17. Thus, active plus inactive KIF17 populations do not generally add up to 100%.
Online supplemental material
Fig. S1 shows characterization of KIF17 for FLIM. Fig. S2 shows that selective depolymerization of stable MTs with OA results in loss of active KIF17. Fig. S3 shows that KIF17 localizes on dynamic and stable MTs in MDCK cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201305023/DC1.
Supplementary Material
Acknowledgments
We thank E. Rodriguez-Boulan for helpful comments on the manuscript and the Rockefeller University Bio-Imaging Resource Center for use of the FLIM instrument.
This work was supported by grants from the National Institutes of Health (R01 GM087575) and the Irma T. Hirschl Trust to G. Kreitzer.
Footnotes
Abbreviations used in this paper:
- EmGFP
- Emerald GFP
- FLIM
- fluorescence lifetime imaging microscopy
- FRET
- Förster resonance energy transfer
- mCh
- mCherry
- MT
- microtubule
- NZ
- nocodazole
- OA
- okadaic acid
References
- Acharya B.R., Espenel C., Kreitzer G. 2013. Direct regulation of microtubule dynamics by KIF17 motor and tail domains. J. Biol. Chem. 10.1074/jbc.M113.494989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro O., Zamai M., Yáñez-Mó M., Tejera E., López-Romero P., Monk P.N., Gratton E., Caiolfa V.R., Sánchez-Madrid F. 2008. Endothelial adhesion receptors are recruited to adherent leukocytes by inclusion in preformed tetraspanin nanoplatforms. J. Cell Biol. 183:527–542 10.1083/jcb.200805076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiaens P.I., Squire A. 1999. Fluorescence lifetime imaging microscopy: spatial resolution of biochemical processes in the cell. Trends Cell Biol. 9:48–52 10.1016/S0962-8924(98)01410-X [DOI] [PubMed] [Google Scholar]
- Borisy G.G., Marcum J.M., Olmsted J.B., Murphy D.B., Johnson K.A. 1975. Purification of tubulin and associated high molecular weight proteins from porcine brain and characterization of microtubule assembly in vitro. Ann. NY Acad. Sci. 253:107–132 10.1111/j.1749-6632.1975.tb19196.x [DOI] [PubMed] [Google Scholar]
- Browning H., Hackney D.D. 2005. The EB1 homolog Mal3 stimulates the ATPase of the kinesin Tea2 by recruiting it to the microtubule. J. Biol. Chem. 280:12299–12304 10.1074/jbc.M413620200 [DOI] [PubMed] [Google Scholar]
- Cai D., Hoppe A.D., Swanson J.A., Verhey K.J. 2007. Kinesin-1 structural organization and conformational changes revealed by FRET stoichiometry in live cells. J. Cell Biol. 176:51–63 10.1083/jcb.200605097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiolfa V.R., Zamai M., Malengo G., Andolfo A., Madsen C.D., Sutin J., Digman M.A., Gratton E., Blasi F., Sidenius N. 2007. Monomer dimer dynamics and distribution of GPI-anchored uPAR are determined by cell surface protein assemblies. J. Cell Biol. 179:1067–1082 10.1083/jcb.200702151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton A.H., Hanley Q.S., Verveer P.J. 2004. Graphical representation and multicomponent analysis of single-frequency fluorescence lifetime imaging microscopy data. J. Microsc. 213:1–5 10.1111/j.1365-2818.2004.01265.x [DOI] [PubMed] [Google Scholar]
- Coy D.L., Hancock W.O., Wagenbach M., Howard J. 1999. Kinesin’s tail domain is an inhibitory regulator of the motor domain. Nat. Cell Biol. 1:288–292 10.1038/13001 [DOI] [PubMed] [Google Scholar]
- Dietrich K.A., Sindelar C.V., Brewer P.D., Downing K.H., Cremo C.R., Rice S.E. 2008. The kinesin-1 motor protein is regulated by a direct interaction of its head and tail. Proc. Natl. Acad. Sci. USA. 105:8938–8943 10.1073/pnas.0803575105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digman M.A., Caiolfa V.R., Zamai M., Gratton E. 2008. The phasor approach to fluorescence lifetime imaging analysis. Biophys. J. 94:L14–L16 10.1529/biophysj.107.120154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng C.H., Huckaba T.M., Gundersen G.G. 2006. The formin mDia regulates GSK3beta through novel PKCs to promote microtubule stabilization but not MTOC reorientation in migrating fibroblasts. Mol. Biol. Cell. 17:5004–5016 10.1091/mbc.E05-10-0914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S., Hurd T.W., Liu C.J., Straight S.W., Weimbs T., Hurd E.A., Domino S.E., Margolis B. 2004. Polarity proteins control ciliogenesis via kinesin motor interactions. Curr. Biol. 14:1451–1461 10.1016/j.cub.2004.08.025 [DOI] [PubMed] [Google Scholar]
- Gierke S., Wittmann T. 2012. EB1-recruited microtubule +TIP complexes coordinate protrusion dynamics during 3D epithelial remodeling. Curr. Biol. 22:753–762 10.1016/j.cub.2012.02.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen G.G. 2002. Evolutionary conservation of microtubule-capture mechanisms. Nat. Rev. Mol. Cell Biol. 3:296–304 10.1038/nrm777 [DOI] [PubMed] [Google Scholar]
- Gurland G., Gundersen G.G. 1993. Protein phosphatase inhibitors induce the selective breakdown of stable microtubules in fibroblasts and epithelial cells. Proc. Natl. Acad. Sci. USA. 90:8827–8831 10.1073/pnas.90.19.8827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney D.D., Stock M.F. 2000. Kinesin’s IAK tail domain inhibits initial microtubule-stimulated ADP release. Nat. Cell Biol. 2:257–260 10.1038/35010525 [DOI] [PubMed] [Google Scholar]
- Hammond J.W., Cai D., Blasius T.L., Li Z., Jiang Y., Jih G.T., Meyhofer E., Verhey K.J. 2009. Mammalian Kinesin-3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. PLoS Biol. 7:e72 10.1371/journal.pbio.1000072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond J.W., Blasius T.L., Soppina V., Cai D., Verhey K.J. 2010. Autoinhibition of the kinesin-2 motor KIF17 via dual intramolecular mechanisms. J. Cell Biol. 189:1013–1025 10.1083/jcb.201001057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanishi M., Endres N.F., Gennerich A., Vale R.D. 2006. Autoinhibition regulates the motility of the C. elegans intraflagellar transport motor OSM-3. J. Cell Biol. 174:931–937 10.1083/jcb.200605179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaulin F., Kreitzer G. 2010. KIF17 stabilizes microtubules and contributes to epithelial morphogenesis by acting at MT plus ends with EB1 and APC. J. Cell Biol. 190:443–460 10.1083/jcb.201006044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaulin F., Xue X., Rodriguez-Boulan E., Kreitzer G. 2007. Polarization-dependent selective transport to the apical membrane by KIF5B in MDCK cells. Dev. Cell. 13:511–522 10.1016/j.devcel.2007.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabir N., Schaefer A.W., Nakhost A., Sossin W.S., Forscher P. 2001. Protein kinase C activation promotes microtubule advance in neuronal growth cones by increasing average microtubule growth lifetimes. J. Cell Biol. 152:1033–1044 10.1083/jcb.152.5.1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitzer G., Schmoranzer J., Low S.H., Li X., Gan Y., Weimbs T., Simon S.M., Rodriguez-Boulan E. 2003. Three-dimensional analysis of post-Golgi carrier exocytosis in epithelial cells. Nat. Cell Biol. 5:126–136 10.1038/ncb917 [DOI] [PubMed] [Google Scholar]
- Lasek R.J., Brady S.T. 1985. Attachment of transported vesicles to microtubules in axoplasm is facilitated by AMP-PNP. Nature. 316:645–647 10.1038/316645a0 [DOI] [PubMed] [Google Scholar]
- Martiny-Baron G., Kazanietz M.G., Mischak H., Blumberg P.M., Kochs G., Hug H., Marmé D., Schächtele C. 1993. Selective inhibition of protein kinase C isozymes by the indolocarbazole Gö 6976. J. Biol. Chem. 268:9194–9197 [PubMed] [Google Scholar]
- Padilla-Parra S., Tramier M. 2012. FRET microscopy in the living cell: different approaches, strengths and weaknesses. Bioessays. 34:369–376 10.1002/bies.201100086 [DOI] [PubMed] [Google Scholar]
- Piston D.W., Kremers G.J. 2007. Fluorescent protein FRET: the good, the bad and the ugly. Trends Biochem. Sci. 32:407–414 10.1016/j.tibs.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Redford G.I., Clegg R.M. 2005. Polar plot representation for frequency-domain analysis of fluorescence lifetimes. J. Fluoresc. 15:805–815 10.1007/s10895-005-2990-8 [DOI] [PubMed] [Google Scholar]
- Ruiz-Canada C., Ashley J., Moeckel-Cole S., Drier E., Yin J., Budnik V. 2004. New synaptic bouton formation is disrupted by misregulation of microtubule stability in aPKC mutants. Neuron. 42:567–580 10.1016/S0896-6273(04)00255-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliwa M., Woehlke G. 2003. Molecular motors. Nature. 422:759–765 10.1038/nature01601 [DOI] [PubMed] [Google Scholar]
- Vale R.D. 2003. The molecular motor toolbox for intracellular transport. Cell. 112:467–480 10.1016/S0092-8674(03)00111-9 [DOI] [PubMed] [Google Scholar]
- Verhey K.J., Hammond J.W. 2009. Traffic control: regulation of kinesin motors. Nat. Rev. Mol. Cell Biol. 10:765–777 10.1038/nrm2782 [DOI] [PubMed] [Google Scholar]
- Wallrabe H., Periasamy A. 2005. Imaging protein molecules using FRET and FLIM microscopy. Curr. Opin. Biotechnol. 16:19–27 10.1016/j.copbio.2004.12.002 [DOI] [PubMed] [Google Scholar]
- Wu X., Xiang X., Hammer J.A., III 2006. Motor proteins at the microtubule plus-end. Trends Cell Biol. 16:135–143 10.1016/j.tcb.2006.01.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.