Abstract
Although most cases of chronic obstructive pulmonary disease (COPD) occur in smokers, only a fraction of smokers develop the disease. We hypothesized distinct molecular signatures for COPD and emphysema in the peripheral blood mononuclear cells (PBMCs) of current and former smokers. To test this hypothesis, we identified and validated PBMC gene expression profiles in smokers with and without COPD. We generated expression data on 136 subjects from the COPDGene study, using Affymetrix U133 2.0 microarrays (Affymetrix, Santa Clara, CA). Multiple linear regression with adjustment for covariates (gender, age, body mass index, family history, smoking status, and pack-years) was used to identify candidate genes, and ingenuity pathway analysis was used to identify candidate pathways. Candidate genes were validated in 149 subjects according to multiplex quantitative real-time polymerase chain reaction, which included 75 subjects not previously profiled. Pathways that were differentially expressed in subjects with COPD and emphysema included those that play a role in the immune system, inflammatory responses, and sphingolipid (ceramide) metabolism. Twenty-six of the 46 candidate genes (e.g., FOXP1, TCF7, and ASAH1) were validated in the independent cohort. Plasma metabolomics was used to identify a novel glycoceramide (galabiosylceramide) as a biomarker of emphysema, supporting the genomic association between acid ceramidase (ASAH1) and emphysema. COPD is a systemic disease whose gene expression signatures in PBMCs could serve as novel diagnostic or therapeutic targets.
Keywords: airflow obstruction, chronic, microarray analysis, leukocytes, mononuclear
Clinical Relevance
Chronic obstructive pulmonary disease (COPD) and emphysema are lung diseases with systemic manifestations in blood. We identify peripheral blood mononuclear cell gene expression profiles for both COPD and emphysema, and replicate some of the top candidates in subjects from the COPDGene cohort. Pathways associated with COPD and emphysema include cell signaling, transcription factors, and sphingolipid metabolism. These genes and pathways could serve as both diagnostic tests and therapeutic targets.
Chronic obstructive pulmonary disease (COPD) results from a combination of environmental exposures and genetic susceptibility, and is the third leading cause of death in the United States (1). Although cigarette smoking is the major environmental risk factor and is present in greater than 80% of patients with COPD (2), only approximately 20% of smokers develop COPD. Previous studies have shown that there are heritable risk factors (3, 4), but the relationship between genetic risk and the molecular changes associated with COPD remains unknown. Although the traditional definition of COPD is based on spirometry measurements, recent work suggests that distinct COPD subtypes are independent of spirometry, such as those with emphysema predominance (5), chronic bronchitis (6), or frequent exacerbations (7). The molecular mechanisms for these phenotypes remain largely unknown, but several studies using small panels of selected biomarkers suggest that inflammation plays a role in distinguishing phenotypes (8, 9).
Although the primary site of disease in COPD is the lung, increasing evidence indicates that COPD is a systemic disease with such manifestations as inflammation, weight loss, and skeletal muscle dysfunction (10, 11). Several systemic biomarkers (e.g., serum amyloid A and procalcitonin) have been associated with exacerbations of COPD (12). Furthermore, biomarkers in plasma, sputum, and exhaled breath are also associated with fat-free mass in subjects with mild COPD (13). Because of its systemic nature, we hypothesized that genome-wide expression profiling of peripheral blood may be used to identify signatures of COPD phenotypes.
Genome-wide expression studies in subjects with COPD and emphysema have been performed in lung tissue, but typically have involved small sample sizes (n = 16–75) because of the invasiveness of sample collection (14–18). As an alternative, we propose the use of peripheral blood to study gene expression profiles in smokers with and without COPD because of its noninvasive availability and ease of use for biomarker screening. The utility of peripheral blood gene expression profiling for the identification of molecular signatures has been demonstrated in a wide range of diseases and disorders, including cancers and neurologic disease (19). Furthermore, studies of COPD have found overlapping gene expression signatures between blood and lung tissue (20) or alveolar macrophages (21). However, those recent COPD studies of blood involved small sample sizes (n = 24–38) (20, 21) and therefore low statistical power to observe real differences, or they primarily examined subjects with less severe COPD (22). To overcome these limitations, we describe an investigation, which to our knowledge is the largest to use gene expression profiling (n = 211) in blood, with a comprehensive representation of COPD severity.
Materials and Methods
Study Population
This study was reviewed and approved by the Institutional Review Board at National Jewish Health. All subject participants provided written, informed consent and were part of the COPDGene cohort of current and former smokers aged 45–80 years, with a history of smoking at least 10 pack-years, and who self-identified as either non-Hispanic white or African-American, and who had not experienced an acute exacerbation of COPD for at least 30 days (23). Subjects were initially divided into either a hypothesis (n = 136) or a replication/testing (n = 75) group. The hypothesis group was designed to represent a COPD cohort with a broad range of airflow obstruction, and was balanced with respect to demographic and clinical covariates (Table 1). To validate our top findings using an independent platform (as will be discussed), 74 subjects in the hypothesis group were randomly selected as a confirmation group, in addition to the 75 independent replication subjects (see Figure E1 and Table E1 in the online supplement).
TABLE 1.
CHARACTERISTICS OF THE MICROARRAY COHORT
| |
GOLD Stage Total (n = 136) |
||||||
|---|---|---|---|---|---|---|---|
| GOLD U (n = 10) | GOLD Stage 0, Control (n = 42) | GOLD Stage 1 (n = 8) | GOLD Stage 2 (n = 34) | GOLD Stage 3 (n = 25) | GOLD Stage 4 (n = 17) | P Value | |
| Sex*, percent female |
80 |
48 |
25 |
50 |
32 |
41 |
0.1321 |
| Age† |
60.6 (7.6) |
60.5 (9.1) |
65.8 (10.5) |
61.8 (9.1) |
68.6 (6.0) |
64.4 (6.0) |
0.0033 |
| Pack-years† |
38.1 (17.9) |
44.5 (28.5) |
51.0 (38.2) |
44.8 (23.2) |
53.8 (22.8) |
57.34 (30.0) |
0.0338 |
| Current smokers*, % |
50 |
32 |
12 |
32 |
12 |
12 |
0.0903 |
| Body mass index† |
29.1 (6.9) |
28.0 (5.0) |
25.9 (2.0) |
30.0 (6.1) |
27.3 (5.4) |
24.8 (6.7) |
0.1564 |
| FEV1 percentage* |
72.0 (5.4) |
97.9 (13.9) |
87.0 (11.5) |
64.0 (8.1) |
40.0 (4.9) |
22.5 (5.8) |
< 0.0001 |
| FEV1/FVC† |
0.75 (0.02) |
0.77 (0.04) |
0.64 (0.05) |
0.59 (0.09) |
0.42 (0.08) |
0.31 (0.06) |
< 0.0001 |
| Emphysema† |
0.5 (0.7) |
1.4(1.5) |
5.6 (6.0) |
6.0 (6.9) |
21.5 (10.9) |
22.1 (9.3) |
< 0.0001 |
| Gas trapping† |
8.0 (8.4) |
8.4 (5.5) |
21.3 (15.6) |
23.0 (16.8) |
51.9 (13.3) |
59.6 (12.4) |
< 0.0001 |
| Six-minute-walk distance† | 1,499.5 (329.8) | 1,656.6 (419.1) | 1,630.3 (315.8) | 1,367.7 (446.6) | 1,146.0 (407.2) | 750.7 (379.7) | < 0.0001 |
Definition of abbreviations: FEV1, forced expiratory volume in 1 second; FVC, forced expiratory volume; GOLD, Global Initiative for Chronic Obstructive Lung Disease; U, unclassified.
For categorical covariates, a GOLD stage–specific percentage is given, along with a P value based on a χ2 test of association. Standard deviations are provided in parentheses.
For continuous covariates, a GOLD stage–specific mean is given, along with a P value based on an overall F-test for equality of means.
Clinical Variables
COPD is defined as post–bronchodilator airflow obstruction with a ratio of forced expiratory volume in 1 second (FEV1) to forced expiratory volume (FVC) of less than 0.70, and is further subdivided into Stages 1–4, based on Global Initiative for Chronic Obstructive Lung Disease (GOLD) guidelines (24). Subjects with FEV1/FVC greater than or equal to 70 and FEV1 percentage of less than 80% post–bronchodilator were considered GOLD unclassified (25). Secondary endpoints included percent emphysema, gas trapping, and 6-minute walking distance (please see the online supplement for further details) (23).
PBMC and RNA Isolation
Peripheral blood was drawn into a BD Vacutainer Cell Preparation Tube (Becton Dickinson, Franklin Lakes, NJ), which was processed within 60 minutes according to the manufacturer’s instructions. Upon centrifugation, lymphocytes and other mononuclear cells appeared in a distinct band and were isolated from the supernatant. RNA isolation was performed using Qiagen RNeasy RNA isolation spin-column kits and protocol, combined with the fully automated Qiagen QIAcube (both from Qiagen, Valencia, CA).
Microarray Experiment and Analysis
The expression of 54,675 transcripts was measured using Affymetrix Human Genome U133 plus 2.0 Gene Array (GEO accession number GSE 42057; Affymetrix, Santa Clara, CA). Quality control was performed, and data were filtered and normalized (see the online supplement). For each probe set, a linear model was fit for the association between gene expression and lung function while controlling for age, sex, body mass index, parental history of COPD, and two smoking variables (smoking status and pack-years). Afterward, the statistical significance of the slope for expression was tested and controlled at a 0.05 false-discovery rate (FDR) (26).
Pathway Analysis
Genes whose expression levels were associated with lung function were further investigated using IPA (Ingenuity Systems, Redwood City, CA). The overrepresentation of significant genes within pathways was assessed using Fisher's exact test, and the FDR was controlled at 0.10.
High-Throughput Quantitative RT-PCR
Based upon preliminary results, 46 genes of interest were selected for validation, using the Applied Biosystems (Foster City, CA) Open Array Platform (see the online supplement). Expression was determined as fold change compared with a panel of five control genes. The Spearman correlation coefficient was calculated for primary clinical endpoints, and P values were controlled at an FDR of 0.05. Combined P value calculations between platforms were performed using the Stouffer combined P value test in both directions (up-regulated or down-regulated) (27).
Results
The demographics of subjects used for microarray analysis are listed by GOLD stage in Table 1. Attempts were made to balance subjects by age, sex, and pack–years for different GOLD stages, and to include a spectrum of emphysema severity. Clinical phenotypes were associated with GOLD stage, pack-years of smoking, and current smoking status.
Microarray Studies of PBMCs
After microarray data normalization and filtering (see the online supplement), separate linear models for the remaining 12,531 transcripts were fit for post-bronchodilator FEV1 percentage and FEV1/FVC across the 136 subjects. The total number of transcripts whose relative abundance was associated with FEV1 percentage, while controlling for demographic and smoking covariates, was 1,090. The total number of transcripts whose relative abundance was associated with FEV1/FVC while controlling for covariates was 1,745. Their intersection involved 993 transcripts (Table E2). For the most statistically significant genes, gene expression explained almost 20% of the variation in FEV1 percentage (Figure E2). With covariates, the entire clinical and gene expression model explained about 25% of the variation in FEV1 percentage.
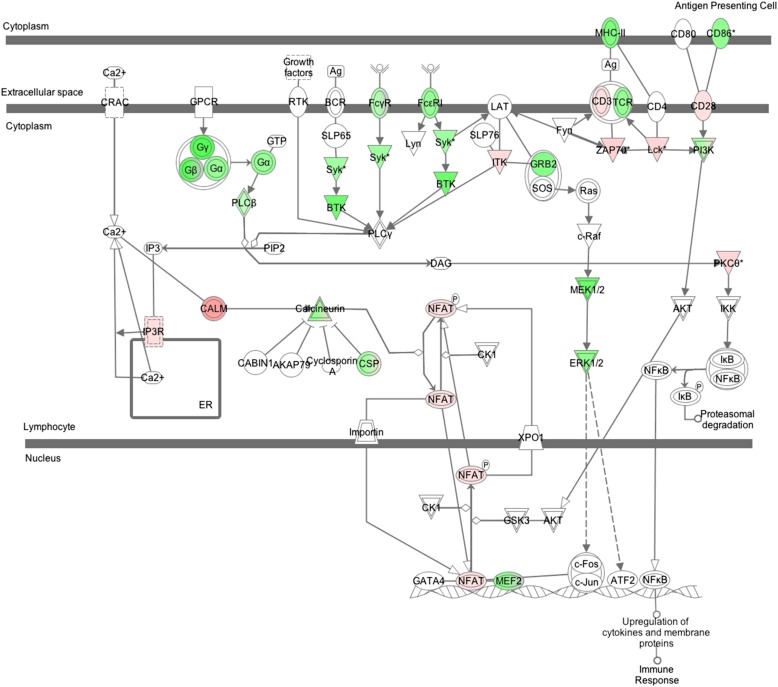
A preliminary investigation revealed that 830 genes of the 993 statistically significant transcripts mapped to 266 canonical pathways (Table E3). Thirty of these had more statistically significant genes represented than would be expected by chance. A large fraction of those (15 out of 30) were related to immunity and inflammatory responses (e.g., Calcium-Induced T Lymphocyte Apoptosis, T Cell Receptor Signaling, Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis). An example in Figure 1 shows the role of the nuclear factor of activated T cells (NFAT) in the regulation of the immune response pathway, where the transcription factor NFAT is up-regulated in subjects with more severe COPD. Metabolic pathways were also overrepresented in our list (e.g., sphingolipid metabolism) (28).
Figure 1.
Role of nuclear factor of activated T-cells (NFAT) in the regulation of the immune response pathway. Genes that are in color are those found to be statistically significant in the analysis of forced expiratory volume in 1 second/forced expiratory volume. Red represents a positive slope with the phenotype, and green represents a negative slope with the phenotype. Each node may represent related molecules, and double lines indicate a family of molecules. *Genes identified more than once in transcripts correlated to phenotype.
Confirmation and Replication of Gene Expression Findings
A quantitative RT-PCR strategy was used for the confirmation and replication of PBMC gene expression in 46 selected genes (Table E4). Candidate genes were selected from preliminary microarray results, based on their overall significance according to P value (10), and their occurrence in significant pathways related to the immune system (11), sphingolipid metabolism and signaling (17), or multiple significant pathways (8). Confirmation involved 74 random subjects profiled in the microarray study (Figure E1). Another 75 independent subjects were not used in the microarray experiments, but were used for the “replication” (or testing) cohort (Table E1).
Twenty-six of the 46 genes profiled showed significant correlation between expression and FEV1/FVC over the complete cohort (FDR-adjusted P < 0.05; see Table E4 in the online data supplement; significant results for FEV1 percentage involve a subset of results for FEV1/FVC, and therefore FEV1/FVC is reported). The magnitude of adjusted P values and the direction of effects (correlation) were similar between the confirmation and replication cohorts. For the 46 genes, the effects of smoking on expression were examined in the larger microarray study, and current smoking status was not predictive of gene expression for any individual probe set on the microarray. However, pack-years were significant for one probe set in the PLCL2 gene (Table E5).
For the final selection of genes, we examined the most significant discovered, based on results from both platforms, namely, the microarray (n = 136) and quantitative RT-PCR (n = 75, i.e., the replication cohort not contained in the original microarray study). Table 2 lists the top 16 candidates for both panels (FDR < 0.05).
TABLE 2.
FALSE DISCOVERY RATE–ADJUSTED P VALUES FOR GENES SELECTED FROM MICROARRAY AND QUANTITATIVE RT-PCR RESULTS
| Gene Identification | Microarray Analysis* |
Quantitative RT-PCR† |
Combined P Value Analysis |
|||
|---|---|---|---|---|---|---|
| FEV1% | FEV1/FVC | FEV1% | FEV1/FVC | FEV1% | FEV1/FVC | |
| ASAH1‡ |
0.0285 |
0.0048 |
0.086 |
0.0109 |
0.0016 |
< 0.0001 |
| BCL2 |
0.0386 |
0.0042 |
0.0762 |
0.2655 |
0.0036 |
0.0017 |
| CASP1‡ |
0.0200 |
0.0054 |
0.0346 |
0.0010 |
0.0008 |
< 0.0001 |
| CEBPD‡ |
0.0298 |
0.0065 |
0.0203 |
0.0065 |
0.0007 |
< 0.0001 |
| FCGR1B‡ |
0.0259 |
0.0164 |
0.0132 |
0.0063 |
0.0006 |
0.0001 |
| FGD2 |
0.0489 |
0.0114 |
0.0008 |
0.0001 |
0.0003 |
< 0.0001 |
| FOXP1‡ |
0.0067 |
0.0010 |
0.0068 |
0.0020 |
0.0014 |
< 0.0001 |
| OFD1 |
0.0032 |
0.0007 |
0.2414 |
0.0417 |
0.0005 |
< 0.0001 |
| PLCB1 |
0.0470 |
0.0142 |
0.1353 |
0.0127 |
0.0069 |
0.0002 |
| RCAN3‡ |
0.0158 |
0.0080 |
0.0190 |
0.0648 |
0.0005 |
0.0005 |
| RHOH |
0.0220 |
0.0035 |
0.2088 |
0.0241 |
0.0047 |
< 0.0001 |
| SULF2 |
0.0173 |
0.0043 |
0.3068 |
0.0310 |
0.0055 |
0.0001 |
| TCF7‡ |
0.0067 |
0.0007 |
0.0002 |
0.0054 |
< 0.0001 |
< 0.0001 |
| TNFRSF1A |
0.0410 |
0.0485 |
0.1198 |
0.0453 |
0.0052 |
0.0026 |
| TNFSF13B‡ |
0.0333 |
0.0105 |
0.0051 |
0.0004 |
0.0005 |
< 0.0001 |
| ZAK | 0.0067 | 0.0007 | 0.0570 | 0.0024 | 0.0003 | < 0.0001 |
Definition of abbreviations: FEV1, forced expiratory volume in 1 second; FVC, forced expiratory volume.
Linear model t test, correcting for covariates in 136 subjects.
Spearman rank correlation test in 75 independent (replication cohort) subjects. Values below 0.01 are in bold, and values between 0.01 and 0.05 are in italics.
Genes with significant fold-change expression correlations in other phenotypes (emphysema, gas trapping, and distance walked).
Association of PBMC Gene Expression with Secondary Clinical Phenotypes
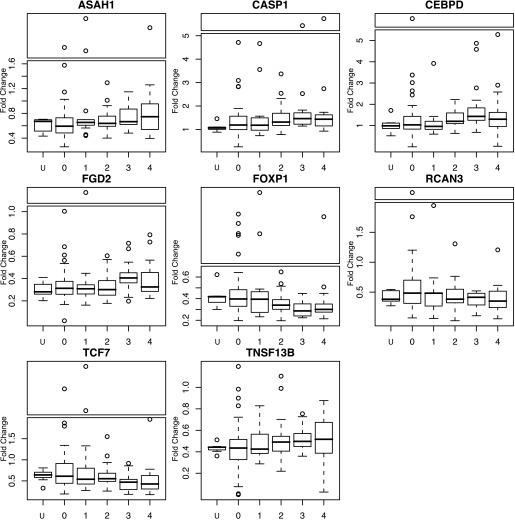
Many of the 16 candidate genes associated with airflow obstruction were also associated with secondary clinical phenotypes (FDR < 0.05). Twelve were correlated with emphysema, 13 were correlated with gas trapping, and eight were correlated with distance walked (Table E6). None of these candidate genes were associated with chronic bronchitis. Correlations were consistent with the microarray results (Table E7) and were based on disease severity, namely, if there was a positive correlation with emphysema and gas trapping, there was a negative correlation with distance walked, FEV1 percentage, and FEV1/FVC, and vice versa. Eight genes produced significant correlations for primary and secondary phenotypes (Figure 2).
Figure 2.
Box plots of expression fold-change are based on quantitative RT-PCR in the complete cohort across Global Initiative for Chronic Obstructive Lung Disease status. For all but TNSF13B, the box plots have been separated into panels, where outliers are displayed at the top. The line segment below each box in the plot is the first quartile, the box segment above this line is the second quartile, the horizontal line within each box represents the median, the box segment above the median is the third quartile, and the line segment above the box is the fourth quartile. Points outside the dashed lines are outliers (>1 box distance from third quartile).
Classification Using Random Forests
To assess the predictive power of the top candidate genes to predict disease status, the supervised learning algorithm “random forests” was used to construct a classifier from the top candidate genes in Table 2. The estimated error rate was approximately 14%, based on the expression of probe sets for these genes, using the microarray data on the original subjects. A similar analysis was performed on the quantitative RT-PCR data using the replication subjects (those not in the microarray analysis), and the estimated error rate was 10%.
Meta-Analysis
Although no other studies this large have investigated PBMC gene expression in COPD, a recent study reported on whole-blood microarray gene expression experiments, using the Sentrix Human WG-6 BeadChips (Illumina, San Diego, CA) microarray platform in 67 test subjects with 69 confirmation subjects (GOLD Stages 1 and 2) (22). A meta-analysis with our results identified a significant overlap of 61 genes (P = 3.1 × 10−10; see the online supplement).
To explore the specificity of our findings in COPD, we examined three other PBMC expression studies in chronic inflammatory diseases unrelated to the lung (Crohn disease and ulcerative colitis) or acute inflammatory disease in the lung (severe acute respiratory syndrome). As with our study, probe sets were filtered if they were not present, and similarity was evident in the number of probe sets filtered in our results compared with each of the three studies (P < 10−3). Comparing retained probe sets, no significant overlap was found with our gene list and the differentially expressed genes between disease and control subjects in any of the three studies (P > 0.20). This indicates that the identified PBMC expression profiles have some specificity to COPD, and are not solely attributable to a general disease state.
Metabolite Profiling
Metabolomics was also used to identify differences in small-molecule profiles among these patients (see the online supplement). Metabolomic investigations identified 19 differentially expressed metabolites between control subjects and those with COPD. A search of the Metlin database resulted in six matches within an acceptable mass tolerance (<5 ppm). One of these, lactosylceramide, also known as galabiosylceramide, was confirmed using high-performance liquid chromatography time of flight (Figure E3). Lactosylceramide was present in all patients with emphysema, a few of the smoking control subjects, and none of the patients with COPD but no emphysema (P = 1.93 × 10−11).
Discussion
To our knowledge, this is one of the largest studies of microarray gene expression profiling for COPD in humans. Unlike most PBMC microarray publications, we were able to confirm many of our most significant findings, using a large independent cohort of subjects. A few of these candidate genes were reported to be associated in smaller previously published studies of PBMC gene expression in similar cohorts, lending weight to the importance of the expression of these genes as biomarkers for COPD. In addition to the large size of both the discovery and testing cohorts, our study contained additional unique features, including a highly phenotyped study population with quantitative computed tomography scans, a balanced study design among GOLD groups, and companion metabolomic investigations.
Most previous studies of gene expression in subjects with COPD have used either lung tissue (15, 17) or bronchial epithelia (16). Because previous studies suggested that COPD is a systemic disease (11, 12, 29, 30), we postulated that the presence of a systemic gene expression profile in subjects with COPD. The choice of studying gene expression in PBMCs versus lung-specific cells such as airway epithelium was primarily determined by the accessibility of suitable samples and the appropriateness of blood draw versus bronchoscopy as a clinical assay. Microarrays of PBMCs have been used to identify gene signatures from diseases as diverse as exertional heat injury (31), sepsis (32), and multiple sclerosis (33). PBMCs have been used to discriminate between subjects with healthy lungs and those with lung diseases such as chronic beryllium disease (34), lung cancer (35), and pulmonary hypertension (36), as well as to distinguish disease activity among patients with severe asthma (37), lung cancer (38), and sickle-cell disease (38). For COPD, the PBMC microarray approach was used previously in small discovery studies of PBMCs from 38 research subjects (21) and 24 subjects undergoing resection for lung tumors (20), as well as whole-blood leukocytes (which contain predominantly neutrophils) in 67 training research subjects (22). These cohorts were limited by small sample size as well as a lack of comprehensive phenotyping (e.g., no emphysema according to high-resolution computed tomography). However, these studies showed promise for using PBMC gene expression as a biomarker COPD. The major limitation of human gene expression studies of COPD, in common with most human studies, is the inability to determine whether differences in gene expression are causative or correlative. However, for one of our candidate genes (ASAH1), we were also able to use a metabolomic approach to identify a related metabolite (lactosylceramide) as a biomarker of COPD and emphysema.
The major analytic strategy to interpret hundreds of statistically significant differences in gene expression involves pathway analysis. Our analysis suggested that subjects with COPD were more likely to exhibit differences in transcriptional control (eukaryotic initiation factors 1 and 4, mechanistic target of rapamycin, G-protein signaling mediated by Tubby, and NFAT; see example in Figure 1), T-cell activation (CD28 signaling in T helper cells, and cytotoxic T lymphocyte–associated protein 4 signaling in cytotoxic T lymphocytes), oxidative stress signaling (NO and reactive oxygen species), and metabolic pathways (sphingolipid metabolism and sphingosine-1–phosphate signaling). The abundance of differentially expressed immune and inflammatory response genes was not unexpected, considering that PBMCs were profiled. Although the candidate pathways were informative, the number of genes in each pathway was prohibitive for replication with a quantitative RT-PCR strategy. Thus, our strategy for the replication study included the selection of 46 genes based on a variety of criteria, including the most highly significant and those represented in pathways of interest. Some of the highlights of the candidate genes that were significant in the validation study will be discussed.
ASAH1 encodes the enzyme N-acylsphingosine amidohydrolase (acid ceramidase), which catalyzes the degradation of ceramide into sphingosine and fatty acid. ASAH1 has been associated with the lysosomal storage disorder Farber disease (39). ASAH1 is highly expressed in the thyroid, lymphocytes, and lung (according to the Gene Atlas) (40). The knockdown of rat alveolar macrophage ASAH1 improves the cigarette smoke–induced inhibition of apoptosis (41). In a recent whole-blood microarray study using 67 training and 65 test subjects, ASAH1 whole-blood expression was the top candidate biomarker for distinguishing subjects with and without airflow obstruction (FEV1/FVC < 0.7 versus FEV1/FVC ≥ 0.7, respectively) (22). In our study, ASAH1 expression in PBMCs was higher in subjects with severe COPD and emphysema in both the training and test cohorts. As further evidence of this pathway’s importance in emphysema and COPD, our metabolomic investigations identified a glycoceramide (galabiosylceramide) as a biomarker of emphysema. Interestingly, an abnormal accumulation of galabiosylceramide occurs in Fabry syndrome (angiokeratoma corporis diffusum), and patients with Fabry syndrome are prone to manifest COPD (42) and emphysema (43). Ceramides are bioactive lipids that mediate cell proliferation, differentiation, apoptosis, adhesion, and migration, and ceramidases such as acid ceramidase (ASAH1) catalyze the hydrolysis of ceramides into sphingosine (44). Thus, acid ceramides play a role in metabolizing ceramides such as galabiosyceramide. We believe that this is the first report identifying galabiosylceramide as a biomarker of COPD. However, additional work will be needed to understand more about the role of the glycoceramides in the pathogenesis of emphysema.
Forkhead box P1 (FOXP1) encodes a transcription factor that is expressed in the lung and is important during lung development (45). FoxP1 protein has been shown to act cooperatively with histone deacetylase 2 to regulate the expression of IL-6, and it modulates the lung epithelial injury response (46). In this study, we found that FOXP1 was also expressed in PBMCs, and that lower expression was associated with both airflow obstruction and emphysema. Low levels of FOXP1 expression were also reported to be associated with COPD in PBMCs from subjects undergoing resection for lung cancer (20). FoxP1 has been shown to play a role in the integrin-dependent modulation of gene expression and macrophage differentiation (47), and thus whether FOXP1 gene expression functions through lymphocytes or is a marker for gene expression in the lung remains unclear.
Toll-like receptor 8 (TLR8) recognizes single-stranded viral RNA, and has been associated with asthma and allergic rhinitis (48). We found that PBMCs expressed significantly lower concentrations of TLR8 in subjects with COPD. The lower expression of TLR8 in patients with COPD could result in increased susceptibility to viral infections (49).
Vanin 2 (VNN2) codes for a pantetheine hydrolase that is expressed in most tissues but demonstrates high expression in leukocytes (50). We observed that subjects with COPD demonstrated lower VNN2 expression in PBMCs. An association between VNN2 expression in whole blood and COPD was also found in the cohort described by Edmiston and colleagues (22). The mechanism by which Vanin 2 might cause COPD is unknown. However, Vanin 2 (formerly called GPI-80) has been shown to play a role in leukocyte trafficking, and is expressed in alveolar macrophages (51).
The major limitation of this study is its cross-sectional basis. This limitation may be assumed to be of minor importance, because the biologic variability of microarray gene expression is very low over short time periods (1 wk), suggesting the stability of the gene expression phenotype (52). However, the stability of PBMC gene expression over longer time periods is unknown, and we do not know whether these gene expression profiles are predictive of declines in lung function or the progression of emphysema. Furthermore, in the parent COPDGene study (23), smokers with normal spirometry were defined as control subjects. Based on these control subjects, we are not able to assess the effects of smoking alone in PBMC expression. Others reported that gene expression is associated with smoking in the oxidative stress response, antiapoptosis and cell-death signaling, and carcinogen metabolism (53–55). However, we found that in our final candidate genes, the correlation of smoking variables with expression was either nonexistent or relatively small compared with the effects of other covariates. Several studies profiled lung tissue in COPD and emphysema subjects. We did not find a significant overlap in the PBMC genes identified in the present work and in those previous studies (data not shown). These results do not suggest a common signature between PBMCs and lung tissue, but the comparisons are also based on small sample sizes (n ≤ 30), which compromises their statistical power. Future work in larger studies or in paired analyses with blood and lung tissue from the same subjects will be able to provide more definitive conclusions. Finally, because of cost and statistical power, the expression profiling was restricted to one racial and ethnic group (non-Hispanic whites). Although this is was a large study with an independent replication cohort, additional gene expression studies in never-smokers and in other ethnicities and races, and different clinical or research studies, would improve the generalizability of our findings.
Despite these limitations, by using COPDGene, we were able to leverage a large, well-phenotyped cohort of subjects with COPD. The candidate genes identified in this study suggest new disease targets and further mechanistic studies that can be designed to identify which candidate genes might be important causes of COPD.
In conclusion, this study is, to our knowledge, the largest gene expression study of smokers with COPD, and demonstrates the potential for PBMC gene expression to serve as a biomarker of COPD. The expression of candidate genes was confirmed in two independent groups with two different assays, and several candidates suggest novel potential targets pathways such as sphingosine-1–phosphate signaling, which were further verified by a plasma metabolomic approach.
Acknowledgments
Acknowledgments
The authors thank Kiel Butterfield for sample handling and RNA isolation.
Footnotes
This study was supported by National Heart, Lung, and Blood Institute grants U01 HL089856 and U01 HL089897 (R.P.B.), National Center for Research Resources/National Institutes of Health grants UL1 RR025780 and S10 RR023703 (N.R.), National Institutes of Health/National Center for Advancing Translational Sciences Colorado Clinical and Translational Science Institute grant UL1 TR000154 (N.R.), and a Butcher Seed Grant.
Author Contributions: T.M.B. and G.J.H. contributed to the analysis and the writing of the manuscript. C.D.C. and M.G.E. assisted with study design and funding. C.S. completed the Institutional Review Board submission and recruited research subjects. R.K. provided the microarrays. D.J.L. performed peripheral blood mononuclear cell RNA isolation and real-time PCR experiments. M.A., R.R., and N.R. performed the metabolomics experiments and data analysis. K.J.K. and R.P.B. were responsible for funding, study design, analysis, and manuscript preparation.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2012-0230OC on April 3, 2013
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Skrepnek GH, Skrepnek SV. Epidemiology, clinical and economic burden, and natural history of chronic obstructive pulmonary disease and asthma. Am J Manag Care. 2004;10:S129–S138. [PubMed] [Google Scholar]
- 2.Eisner MD, Balmes J, Katz PP, Trupin L, Yelin EH, Blanc PD. Lifetime environmental tobacco smoke exposure and the risk of chronic obstructive pulmonary disease. Environ Health. 2005;4:7. doi: 10.1186/1476-069X-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silverman EK, Spira A, Pare PD. Genetics and genomics of chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2009;6:539–542. doi: 10.1513/pats.200904-021DS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silverman EK, Chapman HA, Drazen JM, Weiss ST, Rosner B, Campbell EJ, O’Donnell WJ, Reilly JJ, Ginns L, Mentzer S, et al. Genetic epidemiology of severe, early-onset chronic obstructive pulmonary disease: risk to relatives for airflow obstruction and chronic bronchitis. Am J Respir Crit Care Med. 1998;157:1770–1778. doi: 10.1164/ajrccm.157.6.9706014. [DOI] [PubMed] [Google Scholar]
- 5.Kim WJ, Hoffman E, Reilly J, Hersh C, Demeo D, Washko G, Silverman EK. Association of COPD candidate genes with computed tomography emphysema and airway phenotypes in severe COPD. Eur Respir J. 2011;37:39–43. doi: 10.1183/09031936.00173009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snoeck-Stroband JB, Lapperre TS, Gosman MM, Boezen HM, Timens W, ten Hacken NH, Sont JK, Sterk PJ, Hiemstra PS Groningen Leiden Universities Corticosteroids in Obstructive Lung Disease (GLUCOLD) Study Group. Chronic bronchitis sub-phenotype within COPD: inflammation in sputum and biopsies. Eur Respir J. 2008;31:70–77. doi: 10.1183/09031936.00137006. [DOI] [PubMed] [Google Scholar]
- 7.Hurst JR, Vestbo J, Anzueto A, Locantore N, Mullerova H, Tal-Singer R, Miller B, Lomas DA, Agusti A, Macnee W, et al. Susceptibility to exacerbation in chronic obstructive pulmonary disease. N Engl J Med. 2010;363:1128–1138. doi: 10.1056/NEJMoa0909883. [DOI] [PubMed] [Google Scholar]
- 8.Roy K, Smith J, Kolsum U, Borrill Z, Vestbo J, Singh D. COPD phenotype description using principal components analysis. Respir Res. 2009;10:41. doi: 10.1186/1465-9921-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bon JM, Leader JK, Weissfeld JL, Coxson HO, Zheng B, Branch RA, Kondragunta V, Lee JS, Zhang Y, Choi AMK, et al. The influence of radiographic phenotype and smoking status on peripheral blood biomarker patterns in chronic obstructive pulmonary disease. PLoS ONE. 2009;4:e6865. doi: 10.1371/journal.pone.0006865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agusti AG, Noguera A, Sauleda J, Sala E, Pons J, Busquets X. Systemic effects of chronic obstructive pulmonary disease. Eur Respir J. 2003;21:347–360. doi: 10.1183/09031936.03.00405703. [DOI] [PubMed] [Google Scholar]
- 11.Oudijk EJ, Lammers JW, Koenderman L. Systemic inflammation in chronic obstructive pulmonary disease. Eur Respir J. 2003;46:5s–13s. doi: 10.1183/09031936.03.00004603a. [DOI] [PubMed] [Google Scholar]
- 12.Koutsokera A, Stolz D, Loukides S, Kostikas K. Systemic biomarkers in exacerbations of COPD: the evolving clinical challenge. Chest. 2012;141:396–405. doi: 10.1378/chest.11-0495. [DOI] [PubMed] [Google Scholar]
- 13.Foschino Barbaro MP, Carpagnano GE, Spanevello A, Cagnazzo MG, Barnes PJ. Inflammation, oxidative stress and systemic effects in mild chronic obstructive pulmonary disease. Int J Immunopathol Pharmacol. 2007;20:753–763. doi: 10.1177/039463200702000411. [DOI] [PubMed] [Google Scholar]
- 14.Bhattacharya S, Srisuma S, Demeo DL, Shapiro SD, Bueno R, Silverman EK, Reilly JJ, Mariani TJ. Molecular biomarkers for quantitative and discrete COPD phenotypes. Am J Respir Cell Mol Biol. 2009;40:359–367. doi: 10.1165/rcmb.2008-0114OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ning W, Li CJ, Kaminski N, Feghali-Bostwick CA, Alber SM, Di YP, Otterbein SL, Song R, Hayashi S, Zhou Z, et al. Comprehensive gene expression profiles reveal pathways related to the pathogenesis of chronic obstructive pulmonary disease. Proc Natl Acad Sci USA. 2004;101:14895–14900. doi: 10.1073/pnas.0401168101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pierrou S, Broberg P, O’Donnell RA, Pawlowski K, Virtala R, Lindqvist E, Richter A, Wilson SJ, Angco G, Möller S, et al. Expression of genes involved in oxidative stress responses in airway epithelial cells of smokers with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2007;175:577–586. doi: 10.1164/rccm.200607-931OC. [DOI] [PubMed] [Google Scholar]
- 17.Spira A, Beane J, Pinto-Plata V, Kadar A, Liu G, Shah V, Celli B, Brody JS. Gene expression profiling of human lung tissue from smokers with severe emphysema. Am J Respir Cell Mol Biol. 2004;31:601–610. doi: 10.1165/rcmb.2004-0273OC. [DOI] [PubMed] [Google Scholar]
- 18.Francis SM, Larsen JE, Pavey SJ, Bowman RV, Hayward NK, Fong KM, Yang IA. Expression profiling identifies genes involved in emphysema severity. Respir Res. 2009;10:81. doi: 10.1186/1465-9921-10-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohr S, Liew CC. The peripheral-blood transcriptome: new insights into disease and risk assessment. Trends Mol Med. 2007;13:422–432. doi: 10.1016/j.molmed.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Bhattacharya S, Tyagi S, Srisuma S, Demeo DL, Shapiro SD, Bueno R, Silverman EK, Reilly JJ, Mariani TJ. Peripheral blood gene expression profiles in COPD subjects. J Clin Bioinform. 2011;1:12. doi: 10.1186/2043-9113-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poliska S, Csanky E, Szanto A, Szatmari I, Mesko B, Szeles L, Dezso B, Scholtz B, Podani J, Kilty I, et al. Chronic obstructive pulmonary disease–specific gene expression signatures of alveolar macrophages as well as peripheral blood monocytes overlap and correlate with lung function. Respiration. 2011;81:499–510. doi: 10.1159/000324297. [DOI] [PubMed] [Google Scholar]
- 22.Edmiston JS, Archer KJ, Scian MJ, Joyce AR, Zedler BK, Murrelle EL. Gene expression profiling of peripheral blood leukocytes identifies potential novel biomarkers of chronic obstructive pulmonary disease in current and former smokers. Biomarkers. 2010;15:715–730. doi: 10.3109/1354750X.2010.512091. [DOI] [PubMed] [Google Scholar]
- 23.Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, Curran-Everett D, Silverman EK, Crapo JD. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7:32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fabbri LM, Hurd SS. Global strategy for the diagnosis, management and prevention of COPD: 2003 update. Eur Respir J. 2003;22:1–2. doi: 10.1183/09031936.03.00063703. [DOI] [PubMed] [Google Scholar]
- 25.Washko GR, Hunninghake GM, Fernandez IE, Nishino M, Okajima Y, Yamashiro T, Ross JC, Estépar RS, Lynch DA, Brehm JM, et al. Lung volumes and emphysema in smokers with interstitial lung abnormalities. N Engl J Med. 2011;364:897–906. doi: 10.1056/NEJMoa1007285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- 27.Stouffer SA, Suchman EA, DeVinney LC, Star SA, Williams RM., Jr . Princeton: Princeton University Press; 1949. The American soldier, volume 1: adjustment during army life. [Google Scholar]
- 28.Petrache I, Petrusca DN, Bowler RP, Kamocki K. Involvement of ceramide in cell death responses in the pulmonary circulation. Proc Am Thorac Soc. 2011;8:492–496. doi: 10.1513/pats.201104-034MW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wouters EF, Creutzberg EC, Schols AM. Systemic effects in COPD. Chest. 2002;121:127S–130S. doi: 10.1378/chest.121.5_suppl.127s. [DOI] [PubMed] [Google Scholar]
- 30.Aldonyte R, Eriksson S, Piitulainen E, Wallmark A, Janciauskiene S. Analysis of systemic biomarkers in COPD patients. COPD. 2004;1:155–164. doi: 10.1081/copd-120030828. [DOI] [PubMed] [Google Scholar]
- 31.Sonna LA, Wenger CB, Flinn S, Sheldon HK, Sawka MN, Lilly CM. Exertional heat injury and gene expression changes: a DNA microarray analysis study. J Appl Physiol. 2004;96:1943–1953. doi: 10.1152/japplphysiol.00886.2003. [DOI] [PubMed] [Google Scholar]
- 32.Tang BM, McLean AS, Dawes IW, Huang SJ, Lin RC. Gene-expression profiling of peripheral blood mononuclear cells in sepsis. Crit Care Med. 2009;37:882–888. doi: 10.1097/CCM.0b013e31819b52fd. [DOI] [PubMed] [Google Scholar]
- 33.Iglesias AH, Camelo S, Hwang D, Villanueva R, Stephanopoulos G, Dangond F. Microarray detection of E2F pathway activation and other targets in multiple sclerosis peripheral blood mononuclear cells. J Neuroimmunol. 2004;150:163–177. doi: 10.1016/j.jneuroim.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 34.Hong-Geller E, Pardington PE, Cary RB, Sauer NN, Gupta G. Chemokine regulation in response to beryllium exposure in human peripheral blood mononuclear and dendritic cells. Toxicology. 2006;218:216–228. doi: 10.1016/j.tox.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 35.Showe MK, Vachani A, Kossenkov AV, Yousef M, Nichols C, Nikonova EV, Chang C, Kucharczuk J, Tran B, Wakeam E, et al. Gene expression profiles in peripheral blood mononuclear cells can distinguish patients with non–small cell lung cancer from patients with nonmalignant lung disease. Cancer Res. 2009;69:9202–9210. doi: 10.1158/0008-5472.CAN-09-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bull TM, Coldren CD, Moore M, Sotto-Santiago SM, Pham DV, Nana-Sinkam SP, Voelkel NF, Geraci MW. Gene microarray analysis of peripheral blood cells in pulmonary arterial hypertension. Am J Respir Crit Care Med. 2004;170:911–919. doi: 10.1164/rccm.200312-1686OC. [DOI] [PubMed] [Google Scholar]
- 37.Subrata LS, Bizzintino J, Mamessier E, Bosco A, McKenna KL, Wikström ME, Goldblatt J, Sly PD, Hales BJ, Thomas WR, et al. Interactions between innate antiviral and atopic immunoinflammatory pathways precipitate and sustain asthma exacerbations in children. J Immunol. 2009;183:2793–2800. doi: 10.4049/jimmunol.0900695. [DOI] [PubMed] [Google Scholar]
- 38.Raghavachari N, Xu X, Munson PJ, Gladwin MT. Characterization of whole blood gene expression profiles as a sequel to globin mRNA reduction in patients with sickle cell disease. PLoS ONE. 2009;4:e6484. doi: 10.1371/journal.pone.0006484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koch J, Gartner S, Li CM, Quintern LE, Bernardo K, Levran O, Schnabel D, Desnick RJ, Schuchman EH, Sandhoff K. Molecular cloning and characterization of a full-length complementary DNA encoding human acid ceramidase: identification of the first molecular lesion causing Farber disease. J Biol Chem. 1996;271:33110–33115. doi: 10.1074/jbc.271.51.33110. [DOI] [PubMed] [Google Scholar]
- 40.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petrusca DN, Gu Y, Adamowicz JJ, Rush NI, Hubbard WC, Smith PA, Berdyshev EV, Birokov KG, Lee CH, Tuder RM, et al. Sphingolipid-mediated inhibition of apoptotic cell clearance by alveolar macrophages. J Biol Chem. 2010;285:40322–40332. doi: 10.1074/jbc.M110.137604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown LK, Miller A, Bhuptani A, Sloane MF, Zimmerman MI, Schilero G, Eng CM, Desnick RJ. Pulmonary involvement in Fabry disease. Am J Respir Crit Care Med. 1997;155:1004–1010. doi: 10.1164/ajrccm.155.3.9116979. [DOI] [PubMed] [Google Scholar]
- 43.Kariman K, Singletary WV, Jr, Sieker HO. Pulmonary involvement in Fabry’s disease. Am J Med. 1978;64:911–912. doi: 10.1016/0002-9343(78)90542-9. [DOI] [PubMed] [Google Scholar]
- 44.Mao C, Obeid LM. Ceramidases: regulators of cellular responses mediated by ceramide, sphingosine, and sphingosine-1–phosphate. Biochim Biophys Acta. 2008;1781:424–434. doi: 10.1016/j.bbalip.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shu W, Lu MM, Zhang Y, Tucker PW, Zhou D, Morrisey EE. FOXP2 and FOXP1 cooperatively regulate lung and esophagus development. Development. 2007;134:1991–2000. doi: 10.1242/dev.02846. [DOI] [PubMed] [Google Scholar]
- 46.Chokas AL, Trivedi CM, Lu MM, Tucker PW, Li S, Epstein JA, Morrisey EE. FOXP1/2/4–NuRD interactions regulate gene expression and epithelial injury response in the lung via regulation of interleukin-6. J Biol Chem. 2010;285:13304–13313. doi: 10.1074/jbc.M109.088468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi C, Zhang X, Chen Z, Sulaiman K, Feinberg MW, Ballantyne CM, et al. Integrin engagement regulates monocyte differentiation through the Forkhead transcription factor FOXP1. J Clin Invest. 2004;114:408–418. doi: 10.1172/JCI21100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moller-Larsen S, Nyegaard M, Haagerup A, Vestbo J, Kruse TA, Borglum AD. Association analysis identifies TLR7 and TLR8 as novel risk genes in asthma and related disorders. Thorax. 2008;63:1064–1069. doi: 10.1136/thx.2007.094128. [DOI] [PubMed] [Google Scholar]
- 49.Varkey JB, Varkey B. Viral infections in patients with chronic obstructive pulmonary disease. Curr Opin Pulm Med. 2008;14:89–94. doi: 10.1097/MCP.0b013e3282f4a99f. [DOI] [PubMed] [Google Scholar]
- 50.Kaskow BJ, Michael Proffit J, Blangero J, Moses EK, Abraham LJ. Diverse biological activities of the vascular non-inflammatory molecules: the Vanin pantetheinases. Biochem Biophys Res Commun. 2012;417:653–658. doi: 10.1016/j.bbrc.2011.11.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koike S, Takeda Y, Hozumi Y, Okazaki S, Aoyagi M, Sendo F. Immunohistochemical localization in human tissues of GPI-80, a novel glycosylphosphatidyl inositol–anchored protein that may regulate neutrophil extravasation. Cell Tissue Res. 2002;307:91–99. doi: 10.1007/s00441-001-0481-z. [DOI] [PubMed] [Google Scholar]
- 52.Bryant PA, Smyth GK, Robins-Browne R, Curtis N. Technical variability is greater than biological variability in a microarray experiment but both are outweighed by changes induced by stimulation. PLoS ONE. 2011;6:e19556. doi: 10.1371/journal.pone.0019556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Charlesworth JC, Curran JE, Johnson MP, Goring HH, Dyer TD, Diego VP, Kent JW, Jr, Mahaney MC, Almasy L, MacCluer JW, et al. Transcriptomic epidemiology of smoking: the effect of smoking on gene expression in lymphocytes. BMC Med Genomics. 2010;3:29. doi: 10.1186/1755-8794-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Leeuwen DM, van Agen E, Gottschalk RW, Vlietinck R, Gielen M, van Herwijnen MH, et al. Cigarette smoke–induced differential gene expression in blood cells from monozygotic twin pairs. Carcinogenesis. 2007;28:691–697. doi: 10.1093/carcin/bgl199. [DOI] [PubMed] [Google Scholar]
- 55.van Leeuwen DM, Gottschalk RW, van Herwijnen MH, Moonen EJ, Kleinjans JC, van Delft JH. Differential gene expression in human peripheral blood mononuclear cells induced by cigarette smoke and its constituents. Toxicol Sci. 2005;86:200–210. doi: 10.1093/toxsci/kfi168. [DOI] [PubMed] [Google Scholar]